Abstract
The biological diversity and composition of microorganisms influences both human health outcomes and ecological processes; therefore, understanding the factors that influence microbial biodiversity is key to creating healthy, functional landscapes in which to live. In general, biological diversity is predicted to be limited by habitat size, which for green areas is often reduced in cities, and by chronic disturbance (stress). These hypotheses have not previously been tested in microbial systems in direct comparison to macroorganisms. Here we analyzed bacterial, fungal and ant communities in small road medians (average area 0.0008 km2) and larger parks (average area 0.64 km2) across Manhattan (NYC). Bacterial species richness was not significantly different between medians and parks, but community composition was significantly distinct. In contrast, ant communities differed both in composition and richness with fewer ant species in medians than parks. Fungi showed no significant variation in composition or richness but had few shared taxa between habitats or sites. The diversity and composition of microbes appears less sensitive to habitat patchiness or urban stress than those of macroorganisms. Microbes and their associated ecosystem services and functions may be more resilient to the negative effects of urbanization than has been previously appreciated.
Introduction
Urban habitats are among the most rapidly expanding biomes on Earth and are the one in which most people live (Grimm et al., 2008). But cities are not uniform. Each city contains a mosaic of green habitat patches of varying sizes that are subject to differing degrees of chronic environmental stress, where stressors are factors such as increased heat, pollution, disturbance and habitat fragmentation (Niemelä et al., 2011) that alter the rates of biochemical reactions or exert physical force on organisms (Menge and Sutherland, 1987). This stress is predicted to reduce diversity beyond the predicted reduction owing to small habitat patch size, while intermediate disturbances (for example, suburban development) may lead to local diversity peaks. Plant and animal communities often show these expected patterns of intermediate optima, while minimal diversity is seen in the most urbanized sites (for example, Blair, 1996; Blair and Launer, 1997; Cam et al., 2000; Marzluff, 2005; McKinney, 2008; Minor and Urban, 2010; Saito and Koike, 2013). Response to urbanization varies within a city depending on site characteristics (Sushinsky et al., 2013) as well as on the species assemblage and the spatial scale investigated (Concepción et al., 2015). It also varies between cities, with the amount of species loss best explained best by land cover and city's age (Aronson et al., 2014).
It is worth noting, however, that the very most urbanized sites are often not studied. Little is known about the walls of buildings, for instance, although they occupy a large surface area. Or, if our focus is confined to green spaces, the most common green spaces in growing cities are the ones embedded in cement, such as street medians or tree pits, and these have been little considered. Although these small patches are unlikely to provide habitat to many charismatic vertebrates, their ability to sustain the smaller organisms in cities, including not only insects but also microbes, is unclear.
Microbes in soils are central not only to global biogeochemical cycles (Falkowski et al., 2008), but also to local nutrient cycles (Wall, 2004), such that the composition of microbes in urban soils is likely to govern the fate of nutrients much as it does elsewhere. Globally, soil microbes are also a source of discovery in medicine and biotechnology (Bull, 2004; Challis, 2008), and conservation of microbial biodiversity is increasingly discussed (Colwell, 1997; Cockell and Jones, 2009; Sutherland et al., 2010; Bodelier, 2011). Furthermore, a growing literature suggests that the failure of children to be exposed to a diversity of microbes threatens the health of those children, particularly through its relation to allergic and autoimmune diseases (for example, Rook, 2009; Hanski et al., 2012). The large literature linking microbial diversity and human health tends to assume that the diversity of microbes to which children are exposed in urban environments is less than that in rural environments (for example, Hanski et al., 2012). However, although urbanization can be a major threat to conservation efforts of macroorganisms (Grimm et al., 2008), our understanding of the response of microorganisms to urbanization is poor.
We might expect the loss of diversity with urbanization for microbes, including archaea, bacteria and fungi, if microscopic taxa respond similarly to urbanization and habitat fragmentation as macroscopic organisms. Species–area relationships, with fewer species in smaller sites, have been observed for microbes (Green et al., 2004; Horner-Devine et al., 2004; Bell et al., 2005) in habitats such as tree holes owing to the greater potential for colonization and quantity and diversity of available resources and habitats. At least for macroorganisms, urban, habitat area has a positive influence on biodiversity (Basham et al., 2010; Shanahan et al., 2011; Beninde et al., 2015). If urban habitat patches act analogously to tree holes, we expect to find fewer microbial species in smaller habitats in cities. In addition, stress associated with urban development and pollution can disturb soil, altering its properties and leading to degradation (Parker, 2010), and this stress may be greatest in small patches. For instance, small patches of habitat in New York City, those associated with street medians, have been found to be hotter, drier, have less canopy and are surrounded by more impervious surface relative to nearby parks (Youngsteadt et al., 2015). These stresses may reduce microbial diversity in addition to the direct influence of area.
On the other hand, when large habitats in cities are compared with habitats outside of cites, the total diversity of urban microbes appears high. Ramirez et al. (2014) recently found similar diversity of bacteria in a global data set as what they found in the soil of Central Park. Barberán et al. (2015) also found that the microbes present in dust in cities were no less diverse than those in samples from outside of cities. However, compared with most green patches within cities, Central Park is very large (3.41 km2 compared with the average park size of 0.004 km2) as is the scale of analysis in Barberán et al. (2015). It remains unknown whether the diversity of microbes that persists in smaller and more isolated patches in cities is comparable to the diversity encountered across whole cities or in the largest parks.
Here we consider the influence of habitat area and stress on soil microbes and one group of insects, ants, in Manhattan. In addition to having a number of relatively large parks, New York City, and Manhattan in particular, has many smaller patches of green, including not only gardens and abandoned lots but also the smallest common type of green patch, street medians. In doing so, we addressed the following questions: (i) What is the relative microbial diversity in the soil of small, high-stress urban habitats (street medians) compared with larger, low-stress urban habitats (city parks)? Based on studies of animals and plants in urban ecosystems, we expected that species richness would be lower in street medians than in city parks. Additionally, we expected the composition of microbial communities to be distinct in the two habitat types because previous studies have shown that persisting in high-stress habitats and/or maintaining viable populations in small habitat patches can be strongly influenced by species-specific traits (Schleicher et al., 2011; Slade et al., 2013; Concepción et al., 2015).
Next we asked (ii) How do the patterns of bacterial, fungal and ant diversity compare? Fungi are intermediate in size (typical eukaryotic cells are between 1–2 orders of magnitude larger than bacteria and archaea) and perhaps have intermediate dispersal ability relative to ants and bacteria. Both ants and fungi are common and abundant in ground communities in urban ecosystems, and previous work has shown that these groups can be sensitive to urbanization and habitat patch size (Newbound et al., 2010; Savage et al., 2015). To our knowledge, this is the first analysis of intra-city diversity patterns for microbes with direct comparison to macroorganisms.
Materials and methods
Sampling: bacteria, archaea, and fungi
We collected soil samples between 30 May and 6 June 2013 from medians (n=21) and parks (n=25) in Manhattan (NYC). The medians were located along Broadway (n=7) and the corridor along 12th Avenue, 11th Avenue and West Street (n=14, Supplementary Figure S1). Parks were chosen based on their proximity to street medians and included parklands (with sports fields and playgrounds) and urban forests (Supplementary Figure S1). We sampled two sites for each park except Central Park (3 sites), Battery Park (3 sites) and Washington Market Park (1 site). A single site was sampled for each median.
At each site, we used an open end soil probe (AMS, American Falls, ID, USA) to collect a total of five soil cores (2.5 cm wide × 10 cm deep, with one collected every 4 m along a 20-m transect). We immediately pooled the soil cores from each site by combining them in a sterile Whirlpak bag (Forestry Suppliers, Jackson, MS, USA). The soil corer was cleaned with 70% ethanol between samples to avoid cross-contamination. Soils were frozen at −20 °C on the same day of collection. Across all sites, we sampled equal areas to control sampling effort. We treated individual sites within a single park as separate sites; they had equal area and were a minimum of 65 m apart.
Soil analyses
To homogenize soil samples and remove non-soil fragments, soils were passed through a sterile, 2.0-mm sieve prior to processing. Total carbon and nitrogen were quantified by combustion using an Elementar Vario Macro CNS analyzer (Elementar, Hanau, Germany) and macronutrients and micronutrients were measured using a Mehlich 1 extraction (Mehlich, 1953) followed by inductively coupled plasma spectrometry (Varian Vista MPX Radial ICP-OES, Varian Inc., Palo Alto, CA, USA). All soil nutrient analyses were completed at the Auburn University Soil Testing Laboratory (Auburn, AL, USA).
Microbial DNA was isolated using the Power Soil Extraction Kit (MoBio, Carlsbad, CA, USA), and three DNA extractions were pooled for each sample prior to sequencing. Bacterial DNA was sequenced for all sites using the16S rRNA gene by using the 515-F and 806-R primer pair (Fierer et al., 2012). Fungal DNA was sequenced for 26 sites (13 parks and 13 medians; Supplementary Figure S1) using the internal transcribed spacer (ITS1) region of rRNA, which was amplified using the ITS1-F and ITS2 primer pair (McGuire et al., 2013). Quantification of PCR products was accomplished using the PicoGreen dsDNA assay (ThermoFisher Scientific, Waltham, MA, USA), and equimolar concentrations of amplicons were sequenced using the Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA) at the University of Colorado Next Generation Sequencing Facility. Raw sequence data were processed using a custom Python script (https://github.com/leffj/helper-code-for-uparse) and processed with the UPARSE pipeline (Edgar, 2013). Samples were rarified to 10 000 sequences per sample for fungal analyses and 14 300 sequences per sample for bacterial analyses. Operational taxonomic units were delineated using a 97% similarity threshold, and taxonomy was determined using the RDP classifier for bacteria (Wang et al., 2007) and the UNITE database for fungi (Abarenkov et al., 2010).
Sampling: ants
We collected ants across medians and parks in the Upper West Side of Manhattan (NYC) across 2 consecutive years (14–24 June 2011 and 20 August–7 September 2012). In sum, the samples included 21 street medians along Broadway and 26 parks (Supplementary Figure S1). As above, we sampled equal areas of all sites. As described in more detail in Savage et al. (2015), ants were sampled by extraction from leaf-litter, such that an ant sample is a measure of the composition of ants in a standardized amount of litter on the forest floor. In each site, we collected ants using both hand collection and Winkler sifting. Because we were focused on comparing street medians to all parks, we used a single park category in our analyses rather than splitting the sites into forests and parklands, as Savage et al. (2015) did.
Statistical analyses
All statistical analyses were conducted using R (version 3.1.1; www.r-project.org). Soil chemistry differences between habitat types were tested using Mann–Whitney U-test. Richness of each sample was calculated using the package BiodiversityR (Roeland Kindt R) and compared between habitat types using a Mann–Whitney U-test. We calculated sample-based species accumulation curves for each habitat type using 100 permutations in the specaccum function in vegan (Jari Oksanen R). We tested for differences between the accumulation curves using a two-way Kolmogorov–Smirnov test. We ran a linear regression to test for a relationship between richness and habitat area.
We conducted non-metric multidimensional scaling ordinations with the metaMDS function in vegan. We then tested whether community-level compositional variation was related to our environmental predictors by fitting the ordination scores to habitat type and size with the envfit function in vegan. Phyla- and class-level abundance was analyzed using analysis of variance with post-hoc Tukey's honest significant difference test. The abundance of potentially pathogenic bacteria was calculated following Kembel et al. (2012) and analyzed using a t-test. Indicator family analysis was conducted with 9999 permutations using the multipatt function in indicspecies (Miquel De Caceres R). To test for the relationship between physical distances and community dissimilarity in microbial samples, we ran a Mantel Test with 9999 permutations using R package ade4 (Aurélie Siberchicot R). We compared linear distances between sampling points (measured in Google Earth) with Bray–Curtis dissimilarity table produced using the vegdist function in vegan.
Nucleotide sequence accession numbers
The 16S rRNA and ITS nucleotide sequences generated in this study can be downloaded from the European Nucleotide Archive under study accession number PRJEB9616.
Results
Median soils had significantly more copper (P=0.0016), less nitrogen (P=0.0066) and more zinc (P=0.0029) than parks. They were also significantly more basic (P=0.0116), and altogether, the medians were smaller than parks (P<0.0001). Park and median soils did not differ, however, in lead, carbon, calcium, potassium, magnesium, phosphorous, aluminum, boron, barium, iron, manganese, sodium or nickel (Table 1).
Table 1. Size and some soil characteristics vary between parks and medians.
| Variable |
Median (n=19) |
Park (n=22) |
||
|---|---|---|---|---|
| Mean | 95% CI | Mean | 95% CI | |
| Size (m2)* | 819.38 | 422.9–1215.9 | 567 980.48 | 130 871.6–1 005 089.4 |
| Aluminum | 322.42 | 287.6–357.2 | 419.59 | 334.5–504.7 |
| Barium | 14.83 | 13.3–16.4 | 16.31 | 15–17.7 |
| Boron | 2.5 | 1.9–3.1 | 2.01 | 1.4–2.6 |
| Calcium | 3979.32 | 3295.1–4663.5 | 3875.27 | 2960.0–4790.5 |
| Carbon | 6.07 | 5.4–6.8 | 6.80 | 5.6–8.0 |
| Copper* | 15.98 | 12.0–17.9 | 8.05 | 4.9–11.2 |
| Iron | 61.04 | 52.3–69.8 | 59.49 | 37.1–81.9 |
| Lead | 32.21 | 6.5–57.9 | 21.64 | 11.9–31.3 |
| Magnesium | 591.89 | 502.7–681.1 | 489.91 | 381.8–598.0 |
| Manganese | 71.89 | 62.2–81.6 | 76.95 | 66.0–87.9 |
| Nickel | 1.81 | 1.4–2.3 | 2.02 | 1.4–2.6 |
| Nitrogen* | 0.33 | 0.3–0.4 | 0.45 | 0.4–0.5 |
| pH* | 7.7 | 7.5–8 | 6.95 | 6.5–7.4 |
| Phosphorous | 100.73 | 71.8–129.7 | 159.14 | 109.4–208.9 |
| Potassium | 165 | 146.5–183.5 | 223.59 | 183.6–263.6 |
| Sodium | 429.21 | 318.0–540.4 | 333.23 | 231.3–435.1 |
| Zinc* | 82.68 | 57.6–107.7 | 64.59 | 32.2–97.0 |
Abbreviation: CI, confidence interval. *P<0.05.
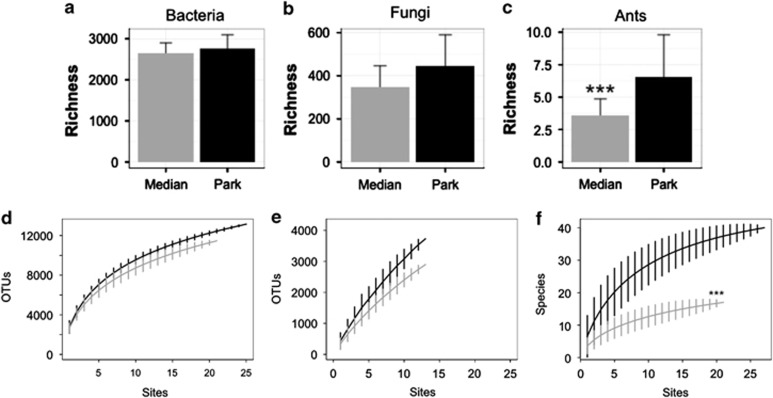
Richness of bacteria did not differ between parks and medians (P=0.117; Figure 1a), and there was no difference between park and median species accumulation curves (P=0.076; Figure 1d). Archaea also do not show a difference in richness (P=0.5866), although species accumulated more slowly across medians (P=0.0023; Supplementary Figure S2). (Archaea represent a small portion (0.5%) of the phylotypes observed likely owing to the sequencing protocol so henceforth we will report only 16S rRNA results for bacteria.)
Figure 1.
Species richness responses differ across taxonomic groups. Richness in medians (gray) and parks (black) for bacteria (a), fungi (b) and ants (c). ***P<0.001. Species accumulation curves for medians (gray) and parks (black) for bacteria and archaea (d), fungi (e) and ants (f).
Fungi trended towards decreased richness in medians (P=0.08; Figure 1b) but did not show significant difference in the accumulation curve functions (P=0.5882; Figure 1e). To test whether the fungal species missing from medians were primarily plant associated, we explored the diversity patterns of just arbuscular mycorrhizal fungi (phylum Glomeromycota). These fungi show no differences in richness (P=0.7968) or species accumulation (P=0.8143; Supplementary Figure S3), such that their absence does not account for the trend toward reduced fungal richness reduction in medians.
The microbial results contrast with those for ant communities, which were significantly less rich in medians than parks (P=0.0003; Figure 1c) and showed significant reduction in diversity in the median species accumulation curve (P<0.0001; Figure 1f).
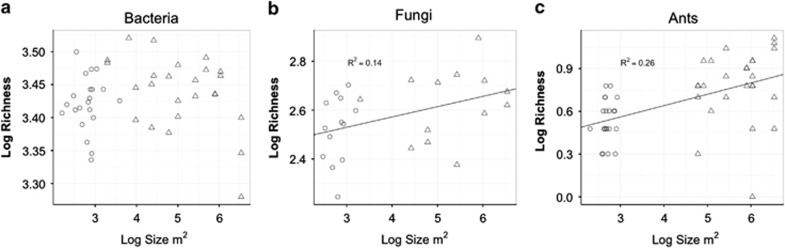
Across all green spaces sampled, bacterial communities did not exhibit a relationship between size of habitat and species richness (P=0.8043; Figure 2a). In contrast, fungal richness increased with habitat area (P=0.0362, R2=0.1357; Figure 2b). Habitat area explained the most variation in species richness for ant communities (P=0.0001, R2=0.2597; Figure 2c). The observed relationships between habitat area and species richness did not hold when considered solely within medians or within parks for fungi (P=0.34 and P=0.43, respectively) or ants (P=0.48 and P=0.48, respectively) such that these relationships appear driven by differences between parks and medians.
Figure 2.
Species richness is a function of habitat size (m2) for fungi (b) and ants (c) but not for bacteria (a). Median sites are indicated with circles; park sites are indicated with triangles.
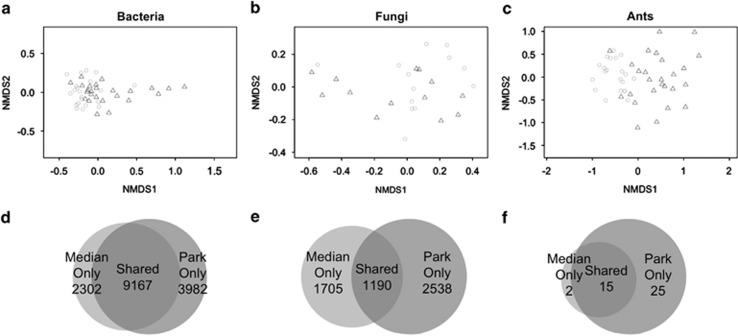
In contrast to the large differences in richness response between ants and bacteria, these groups had similar patterns of species composition response. Specifically, bacterial composition was significantly different in medians and parks (2D stress=0.149, P=0.004, R2=0.163; Figure 3a) as it was in ants (2D stress=0.180, P=0.001, R2=0.38; Figure 3c). Similarly, size was significantly correlated with composition in both bacteria (P=0.001, R2=0.367) and ants (P=0.044, R2=0.13). However, fungal species composition did not differ significantly between parks and medians (2D stress=0.164, P=0.187; Figure 3b) and did not correlate with size (P=0.144).
Figure 3.
Composition differs between medians and parks for some taxa. Non-metric multidimensional scaling ordination plots for bacteria (a), fungi (b) and ants (c). Circles are median plots, and triangles are parks. Species/operational taxonomical unit overlap is unique for bacteria and archaea (d), fungi (e) and ants (f).
Each taxonomic group demonstrated different patterns of species overlap between medians and parks. For bacteria, the majority of operational taxonomical units were shared between medians and parks, but each habitat type still had a significant fraction (>20%) of unique taxa (Figure 3d). For fungi, less than half of operational taxonomical units were shared between medians and parks, with each habitat type supporting mostly unique communities (Figure 3e). For ants, the median communities were almost entirely a nested subset of the taxa found in the parks (Figure 3f).
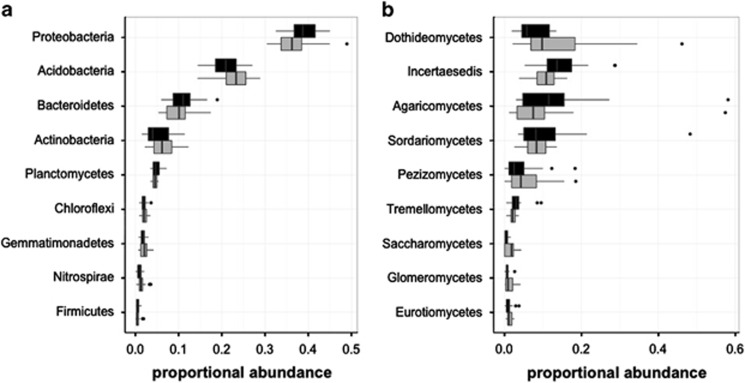
There were few patterns of differences between microbial composition in parks and medians at higher taxonomic levels. We found no differences between the abundance of any of the nine most common bacterial phyla (Figure 4a). Of the 252 named bacterial families, 9 were significantly associated with either medians or parks. Three families from the phylum Actinobacteria were associated with medians: Promicromonosporaceae (P=0.0062), Rubrobacteraceae (P=0.0252), and AKIW874 (P=0.0194). However, in medians the summed abundance of these taxa represent, on average, only 1.76 × 10−4±2.09 × 10−4%. Six families were associated with parks: Aeromonadaceae (P=0.0004), JTB38 (P=0.045), Gallionellaceae (P=0.0042), Spirochaetaceae (P=0.0069), Ktedonobacteraceae (P=0.0229), and Ruminococcaceae (P=0.0348). The average sum abundance of these families in parks is only 0.0007±0.0005%. In addition, the abundance of potentially pathogenic bacteria was not significantly different between parks and medians (P=0.8413; Supplementary Figure S4).
Figure 4.
Park and median composition are similar among the most common higher order taxa. Boxplots of the relative abundances of the most dominant bacterial phyla (a) and fungal classes (b) show no significant differences in relative abundance between parks (black) and medians (gray).
Similarly, there were no significant differences between any of the 10 most common fungal classes (Figure 4b). Only one fungal family was significantly indicative of medians—Liptomycetaceae (P=0.0091)—but its average abundance in medians was only 0.0144±0.016%. Only two families were associated with parks—Hyaloscyphaceae (P=0.0005) and Annulatascaceae (P=0.0357)—but together these were, on average, only 0.0087±0.0072% of the fungi found in parks.
To assess whether distance between sites was a potential confounding variable in our analysis of compositional variation, we conducted Mantel tests comparing physical distance matrices with Bray–Curtis dissimilarity matrices. The test was not significant for bacteria (P=0.3473) or fungi (P=0.13)—indicating that microbial samples from sites closer together were not necessarily more compositionally similar. Similarly, Savage et al. (2015) found no effect of distance on ant community composition across Manhattan when controlling for habitat type.
Discussion
Soils harbor vast repertoires of microbial diversity, which are critical for ecosystem functions, drug discovery and perhaps even human health. Urban stress can limit diversity of aboveground macroorganisms, and we tested whether this pattern also holds for microbes. We predicted that microbial diversity would be limited in urban medians, which, compared with larger urban parks, are small alkaline, nitrogen poor, isolated, hot, dry, exposed, high in copper and surrounded by cement and other impervious surfaces (this study; Youngsteadt et al., 2015). Instead, bacterial diversity in these small, high-stress median sites was as high as that of the parks and was not associated with habitat area. In contrast, the diversity of ants was lower in medians than in parks and positively correlated with habitat area. Fungi showed an intermediate pattern. Ant and bacterial (but not fungal) community composition differed between parks and medians.
As has already been shown for ants in Manhattan (Savage et al., 2015), as well as for plants, vertebrates and insects elsewhere (Blair, 1996; Blair and Launer, 1997; Cam et al., 2000; Minor and Urban, 2010; Saito and Koike, 2013; Concepción et al., 2015), we re-acknowledge that many macroorganisms decline in diversity in cities relative to rural areas and that some species are absent in cities, resulting in changes in composition. Recent work (Beninde et al., 2015) has surveyed the causes of intra-urban biodiversity variation for a range of macroorganisms and found that, to a great extent, shifts in composition and declines in diversity in cities are due to the effects of habitat area and connectivity, with the most affected green spaces in cities being both small and disconnected.
Notably, we found that habitat area was related only to fungal and ant richness, not bacterial richness. Habitat size is known to affect microbial richness elsewhere. For example, taxa–area relationships predict fungal richness in undeveloped arid soil (Green et al., 2004) and bacterial richness in a salt marsh (Horner-Devine et al., 2004), but in both cases the slope of the species–area relationship was low relative to relationships for macroorganisms. We did not consider connectivity, though we note that the medians considered are surrounded by pavement and hence relatively disconnected for most organisms. However, we found that physical distance between sites—a proxy for dispersal limitation and thus potentially related to connectivity—was not predictive of community similarity for the microbes or ants. Future work on urban microbes might consider additional site characteristics assessed in Beninde et al. (2015), including management history and biotic and abiotic factors.
In this study, we show that overall fungi showed some tendency toward the loss of biodiversity with decreases in habitat size and increases in stress while bacteria did not. Similar results have been found in Central Park where microbial eukaryotes but not bacteria and archaea were a subset of the global population (Ramirez et al., 2014). Additionally, on treetop ‘islands' macro- and meso-fauna diversity tended to decrease with reductions in habitat size, whereas microbial diversity did not (Wardle et al., 2003). Our results extend these patterns to one of the most extreme terrestrial habitats on Earth in which plants are still the primary producers. Whether these patterns hold in other mega-cities or smaller, more suburban areas of human development remains to be seen.
The compositional changes we observed in the fungal community are not consistent with a nested subset pattern wherein a subset of the species found in parks would be found in medians. We might also have expected a decline in arbuscular mychorrhizal fungi, which are tightly associated with host plants, in response to the decline in plant diversity in medians. However, the arbuscular mycorrhizal fungi did not decline in diversity in medians. Instead, the majority of arbuscular mycorrhizal fungal phylotypes found in medians were not also found in parks. One of the only other studies evaluating fungi in NYC also found that arbuscular mycorrhizal fungi were distinct in different green spaces (McGuire et al., 2013), suggesting that this functional guild of fungi is strongly structured by microsite conditions. The only family of fungi found to be indicative of medians was a member of the class Saccharomycetes and the families indicative of parks were a Leotimycetes and a Sordariomycetes. Both of those families are associated with growth on dead wood.
In contrast to the fungi, bacterial richness did not differ between parks and medians nor did it correlate with habitat size; however, bacterial composition did differ between parks and medians, albeit subtly. The pattern in richness is interesting in terms of the basic biology of bacteria, because it suggests that there is both very effective dispersal and sufficient environmental heterogeneity—even at these small scales—to maintain relatively high diversity. The imperfect overlap between parks and medians in composition may be due to differences in the microhabitat of these sites and the novel microhabitats and resources present in medians, including those associated with pollution, such that a significant number of bacterial taxa (>20%) were only present in medians. Interestingly, while Ramirez et al. (2014) documented an elevated abundance of pathogens in Central Park relative to a global data set, we did not find a concomitant difference in abundance between parks and medians. Medians (and, in our data set, parks) are unlike indoor environments, which are dominated by human-associated microbes (Lax et al., 2014). This is true even though the medians are, in many cases, just feet above the subway system in which human-associated microbes are most abundant (Afshinnekoo et al., 2015).
For ants, differences in composition appear associated with differences in function (for example, how fast discarded food is removed; Youngsteadt et al., 2015). The same might be true for microbes; a key next step is to directly assay microbial function in these habitats as a function of composition and diversity. Experimental loss of diversity and simplification of soil communities has recently been shown to lead to changes in decomposition, nutrient retention and nutrient cycling as well as affecting aboveground plant communities in grassland microcosms (Waag et al., 2014). Assaying function is particularly important in these urban soils, because urbanization has been found to lead to enhanced C cycling rates, altered regional C budgets and heightened soil respiration in lawns relative to adjacent native and agricultural systems (Kaye et al., 2005), but it is unknown whether these changes were due to changes in bacterial composition, fungal composition or simply the physiology of key species. Additionally, green roofs have been found to harbor many fungi that are associated with organic contaminants, but it is unknown whether these taxa are present because they are resistant to pollutants or because they are actively involved in their degradation (McGuire et al., 2013). We found some of these contaminant-associated taxa in our samples but only at very low abundance and with no relationship to habitat type.
Beyond the potential for differences in function, our results also have implications both for conservation and for human health. The finding that increased urban stress does not reduce bacterial or fungal diversity relative to parks implies that cities may have very different consequences with regard to microbial conservation relative to the conservation of animals and plants. In fact, the presence of unique taxa found only in medians highlights the possibility that extreme habitats, even if they are man-made, can have unique conservation value. Nevertheless, these extreme sites could not serve as replacements if we were to lose less urbanized spaces. Conservation of the diversity and function of soil microbial communities will not only need to involve but also extend beyond urban sites.
It has been shown that the psychological benefits that humans gain from encountering urban greenspaces increase with the species richness of these greenspaces (Fuller et al., 2007). The medians in New York (and we suspect similar patches of habitat in other megacities) are sufficiently extreme to reduce diversity, even of disturbance-tolerant taxa such as ants. Thus the public, who encounter nature primarily in such patches (Dunn et al., 2006), are left with a narrow set of visually observable species with which to engage. Surprisingly, even this narrow set is relatively poorly studied—for example, one of the most common ant species in the medians does not have a species name and one has only been studied three times in North America (Savage et al., 2015).
We know of no evidence that the diversity of microbes has psychological benefits (though one might imagine such benefits via the influence of microbes on volatile chemicals and, more generally, the odor of soils and plants). However, in the context of human health, it has been suggested that a reduced exposure to a diversity of microbes predisposes children in modern, developed environments to autoimmune disorders (for example, Riedler et al., 2001; Hanski et al., 2012). Our results suggest that if such an effect does exist it is likely not simply due to people in developed areas having lower diversity of soil microbes in their environment. Much of the diversity of the bacteria found in the world is present in Central Park (Ramirez et al., 2014) and the diversity of bacteria in the medians and in Central Park is similar (this study). Instead, it may be differences in exposure to environmental microbes or the identity of the environmental microbes present between rural and developed areas that contributes to altered immune development.
Our results indicate that how we think about a global theory of diversity needs to be adjusted when we include microbes. The scale of both movement and habitat are critical elements affecting which individuals are able to reach and thrive in stressful environments, such as those commonly found in cities. As stressful habitats become more common owing to human development and global climate change (Grimm et al., 2008; Niemelä et al., 2011; Elmqvist et al., 2013), a cohesive theory about how diversity changes in these environments will be crucial. Birds, mammals and even insects may respond strongly to the stresses of urban life, but smaller organisms may not be as responsive. This difference may, in part, be one of scale. Relative to an individual bacterial cell, a street median is still enormous, virtually a continent of heterogeneity. Therefore, forecasts of impact on microbial responses to environmental change may need to consider different spatial grains and extremes than those typically evaluated in the context of macroorganisms. This task may prove challenging. Yet, because microbial taxa have an outsized role in major ecosystem services, it is also crucial to include them if we are to fully understand global change.
Acknowledgments
We thank Britne Hackett, Benoit Guenard, Shelby Anderson, Andrew Ernst and Ryanna Henderson for field assistance. Noah Fierer and Jon Leff carried out the sequencing and bioinformatics. Lawrence David provided helpful commentary on the manuscript, and Greg Ames provided useful advice on the Kolmogorov–Smirnov test. The manuscript was greatly improved through the comments of two anonymous reviewers. Thanks also to the New York Urban Field Station. This study was funded by an NSF RAPID (1318655) to SDF and RRD. It was also supported by Cooperative Agreement No. G11AC20471 and G13AC00405 from the United States Geological Survey to RRD and SDF. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE 1106401 to ATR. Its contents are solely the responsibility of the authors and do not necessarily represent the views of the Department of the Interior Southeast Climate Science Center or the USGS. This manuscript is submitted for publication with the understanding that the United States Government is authorized to reproduce and distribute reprints for Governmental purposes.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abarenkov K, Nilsson R, Larsson K, Alexander I, Eberhardt U, Erland S. (2010). The UNITE database for molecular identification of fungi—recent updates and future perspectives. New Phytol 186: 281–285. [DOI] [PubMed] [Google Scholar]
- Afshinnekoo E, Meydan C, Chowdhury S, Jaroudi D, Boyer C, Bernstein N et al. (2015). Geospatial resolution of human and bacterial diversity with city-scale metagenomics. CELS 1: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson MFJ, La Sorte FA, Nilon CH, Katti M, Goddard MA, Lepczyk CA et al. (2014). A global analysis of the impacts of urbanization on bird and plant diversity reveals key anthropogenic drivers. Proc R Soc B 281: 20133330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberán A, Ladau J, Leff JW, Pollard KS, Menninger H, Dunn RR, Fierer N. (2015). Continental-scale distributions of dust-associated bacteria and fungi. PNAS 112: 5756–5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basham R, Law B, Banks P. (2010). Microbats in a 'leafy' urban landscape: are they persisting, and what factors influence their presence? Austral Ecol 36: 633–678. [Google Scholar]
- Bell T, Ager D, Song J-I, Newman JA, Thompson IP, Lilley AK et al. (2005). Larger islands house more bacterial taxa. Science 308: 1884–1884. [DOI] [PubMed] [Google Scholar]
- Beninde J, Veith M, Hochkirch. (2015). Biodiversity in cities needs space: a meta-analysis of factors determining intra-urban biodiversity variation. Ecol Lett 18: 581–592. [DOI] [PubMed] [Google Scholar]
- Blair RB. (1996). Land use and avian species diversity along an urban gradient. Ecol Appl 6: 506–519. [Google Scholar]
- Blair RB, Launer AE. (1997). Butterfly diversity and human land use: species assemblages along an urban gradient. Biol Conserv 80: 113–125. [Google Scholar]
- Bodelier PLE. (2011). Toward understanding, managing, and protecting microbial ecosystems. Front Microbiol 2: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull AT. (2004) Microbial Diversity and Bioprospecting. American Society for Microbiology Press: : Washington, DC, USA. [Google Scholar]
- Cam E, Nichols J, Sauer J, Hines J, Flather C. (2000). Relative species richness and community completeness: birds and urbanization in the Mid-Atlantic states. Ecol Appl 10: 1196–1210. [Google Scholar]
- Challis GL. (2008). Mining microbial genomes for new natural products and biosynthetic pathways. Microbiology 154: 1555–1569. [DOI] [PubMed] [Google Scholar]
- Cockell CS, Jones HL. (2009). Advancing the case for microbial conservation. Oryx 43: 520–526. [Google Scholar]
- Colwell RR. (1997). Microbial diversity: the importance of exploration and conservation. J Ind Microbiol Biotechnol 18: 302–307. [DOI] [PubMed] [Google Scholar]
- Concepción ED, Moretti M, Altermatt F, Nobis MP, Obrist MK. (2015). Impacts of urbanisation on biodiversity: the role of species mobility, degree of specialisation and spatial scale. Oikos e-pub ahead of print 4 March 2015 doi:10.1111/oik.02166.
- Dunn RR, Gavin M, Sanchez M, Solomon J. (2006). The pigeon paradox: dependence of global conservation on urban nature. Conserv Biol 20: 1814–1816. [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10: 996–998. [DOI] [PubMed] [Google Scholar]
- Elmqvist T, Fragkias M, Goodness J, Güneralp B, Marcotullio PJ, McDonald RI et al. (2013) Urbanization, Biodiversity, and Ecosystem Services: Challenges and Opportunities. Springer: Netherlands. [Google Scholar]
- Falkowski PG, Fenchel T, Delong EF. (2008). The microbial engines that drive earth's biogeochemical cycles. Science 320: 1034–1039. [DOI] [PubMed] [Google Scholar]
- Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R. (2012). Comparative metagenomic, phylogenetic, and physiological analyses of soil microbial communities across nitrogen gradients. ISME J 6: 1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller RA, Irvine KN, Devine-Wright P, Warren PH, Gaston KJ. (2007). Psycholgoical benefits of greensapce increase with biodiversity. Biol Lett 3: 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JL, Holmes AJ, Westoby M, Oliver I, Briscoe D, Dangerfield M et al. (2004). Spatial scaling of microbial eukaryote diversity. Nature 432: 747–750. [DOI] [PubMed] [Google Scholar]
- Grimm NB, Faeth SH, Golubiewski NE, Redman CL, Wu J, Bai X et al. (2008). Global change and the ecology of cities. Science 319: 756–760. [DOI] [PubMed] [Google Scholar]
- Hanski I, von Hertzen L, Fyhrquist N, Koskinen K, Torppa K, Laatikainen T et al. (2012). Environmental biodiversity, human microbiota, and allergy are interrelated. PNAS 109: 8334–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner-Devine MC, Lage M, Hughes JB, Bohannan B. (2004). A taxa-area relationship for bacteria. Nature 432: 750–753. [DOI] [PubMed] [Google Scholar]
- Kaye J, McCUlley RL, Burke IC. (2005). Carbon fluxes, nitrogen cycling, and soil microbial communities in adjacent urban, native, and agricultural ecosystems. Global Change Biol 11: 575–587. [Google Scholar]
- Kembel SW, Jones E, Kline J, Northcutt D, Stenson J, Womack AM et al. (2012). Architectural design influences the diversity and structure of the built environment microbiome. ISME J 8: 1469–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM et al. (2014). Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345: 1048–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff JM. (2005). Island biogeography for an urbanizing world: how extinction and colonization may determine biological diversity in human dominated landscapes. Urban Ecosyst 8: 157–177. [Google Scholar]
- McGuire KL, Payne SG, Palmer MI, Gillikin CM, Keefe D, Kim SJ et al. (2013). Digging the New York City skyline: soil fungal communities in green roofs and city parks. PLoS One 8: e58020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney ML. (2008). Effects of urbanization on species richness: a review of plants and animals. Urban Ecosyst 11: 161–176. [Google Scholar]
- Mehlich A (1953). Determination of P, Ca, Mg, K, Na, and NH4. North Carolina Soil Test Division (Mimeo 1953)..
- Menge BA, Sutherland JP. (1987). Community regulation: variation in disturbance, competition, and predation in relation to environmental-stress and recruitment. Am Nat 130: 730–757. [Google Scholar]
- Minor E, Urban D. (2010). Forest bird communities across a gradient of urban development. Urban Ecosyst 13: 51–71. [Google Scholar]
- Newbound M, Mccarthy MA, Lebel T. (2010). Fungi and the urban environment: a review. Landscape Urban Plann 96: 238–145. [Google Scholar]
- Niemelä J, Breuste J, Guntenspergen G, McIntyre N, Elmqvist T. (2011) Urban Ecology: Patterns, Processes, and Applications. Oxford University Press: : New York, USA. [Google Scholar]
- Parker SS. (2010). Buried treasure: soil biodiversity and conservation. Biodivers Conserv 19: 3473–3756. [Google Scholar]
- Ramirez KS, Leff JW, Barberan A, Bates ST, Betley J, Crowther TW et al. (2014). Biogeographic patterns in below-ground diversity in New York City's Central Park are similar to those observed globally. P Roy Soc B Biol Sci 281: 20141988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedler J, Braun-Fahrlander C, Eder W, Schreuer M, Waser M, Maisch S et al. (2001). Exposure to farming in early life and development of asthma and allergy: a cross-sectional survey. Lancet 358: 1129–1133. [DOI] [PubMed] [Google Scholar]
- Rook GAW. (2009). Review series on helminths, immune modulation and the hygiene hypothesis: the broader implications of the hygeine hypothesis. Immunology 126: 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito MA, Koike F. (2013). Distribution of wild mammal assemblages along an urban-rural-forest gradient in warm-temperate East Asia. PLoS One 8: e65464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage AM, Hackett B, Guénard B, Youngsteadt EK, Dunn RR. (2015). Fine-scale heterogeneity across Manhattan's urban habitat mosaic is associated with variation in ant composition and richness. Insect Conserv Divers 8: 216–228. [Google Scholar]
- Schleicher A, Biedermann R, Kleyer M. (2011). Dispersal traits determine plant response to habitat connectivity in an urban landscape. Landscape Ecol 26: 529–540. [Google Scholar]
- Shanahan D, Miller C, Possingham HP, Fuller RA. (2011). The influence of patch area and connectivity on avian communities in urban revegetation. Biol Conserv 144: 722–729. [Google Scholar]
- Slade EM, Merckx T, Riutta T, Bebber DP, Redhead D, Riordan P et al. (2013). Life-history traits and landscape characteristics predict macro-moth responses to forest fragmentation. Ecology 94: 1519–1530. [DOI] [PubMed] [Google Scholar]
- Sushinsky JR, Rhodes JR, Possingham HP, Gill TK, Fuller RA. (2013). How should we grow cities to minimize their biodiversity impact. Glob Change Biol 19: 401–410. [DOI] [PubMed] [Google Scholar]
- Sutherland WJ, Clout M, Cote IM, Daszak P, Depledge MH, Fellman L et al. (2010). A horizon scan of global conservation issues for 2010. Trends Ecol Evol 25: 1–7. [DOI] [PubMed] [Google Scholar]
- Waag C, Bender SF, Widmer F, van der Heijden MGA. (2014). Soil biodiversity and soil community composition determine ecosystem multifunctionality. PNAS 111: 5266–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall D. (2004). Sustaining biodiversity and ecoystems services in soils and sediments. SCOPE Series. Island Press: Washington, DC, USA. [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. (2007). Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardle DA, Yeates GW, Barker GM, Bellingham PJ, Bonner KI, Williamson WM. (2003). Island biology and ecosystem functioning in epiphytic soil communities. Science 301: 1717–1720. [DOI] [PubMed] [Google Scholar]
- Youngsteadt EK, Henderson RC, Savage AM, Ernst AF, Dunn RR, Frank SD. (2015). Habitat and species identity, not diversity, predict the extent of refuse consumption by urban arthropods. Glob Change Biol 21: 1103–1115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.