Abstract
Microbiome studies have demonstrated the high inter-individual diversity of the gut microbiota. However, how the initial composition of the microbiome affects the impact of antibiotics on microbial communities is relatively unexplored. To specifically address this question, we administered a second-generation cephalosporin, cefprozil, to healthy volunteers. Stool samples gathered before antibiotic exposure, at the end of the treatment and 3 months later were analysed using shotgun metagenomic sequencing. On average, 15 billion nucleotides were sequenced for each sample. We show that standard antibiotic treatment can alter the gut microbiome in a specific, reproducible and predictable manner. The most consistent effect of the antibiotic was the increase of Lachnoclostridium bolteae in 16 out of the 18 cefprozil-exposed participants. Strikingly, we identified a subgroup of participants who were enriched in the opportunistic pathogen Enterobacter cloacae after exposure to the antibiotic, an effect linked to lower initial microbiome diversity and to a Bacteroides enterotype. Although the resistance gene content of participants' microbiomes was altered by the antibiotic, the impact of cefprozil remained specific to individual participants. Resistance genes that were not detectable prior to treatment were observed after a 7-day course of antibiotic administration. Specifically, point mutations in beta-lactamase blaCfxA-6 were enriched after antibiotic treatment in several participants. This suggests that monitoring the initial composition of the microbiome before treatment could assist in the prevention of some of the adverse effects associated with antibiotics or other treatments.
Introduction
The human digestive tract is a complex ecosystem in which communities of microorganisms interact with each other and with their host. Unbalancing this environment modifies these endemic microbial communities and can promote the emergence of opportunistic infections such as Clostridium difficile (Chang et al., 2008; Manges et al., 2010), affect weight and obesity (Turnbaugh et al., 2009) and even influence mental health (Hsiao et al., 2013). The administration of antibiotics can perturb this microflora temporarily and in certain cases permanently (Dethlefsen et al., 2008; Dethlefsen and Relman, 2011; Hernández et al., 2013; Pérez-Cobas et al., 2013). Bacterial resistance to antibiotics is a current public health concern as it can transform diseases that were easily treatable 30 years ago into deadly threats (WHO, 2014). Indeed, microorganisms themselves can produce antibiotics (Donia et al., 2014) with resistance to these natural molecules being a selective factor aiding the survival of these strains in this environment (Forsberg et al., 2012, 2014). As most clinically relevant antibiotics are derived from natural molecules, bacterial gene evolution potentially allows the development of resistance to current drugs. Mobility of resistance genes from commensal species to pathogens through conjugation, transduction and transformation has, as a consequence, rendered part of our therapeutic arsenal inefficient and in some cases obsolete. Our responsibility for enhancing resistance is demonstrated by the clinical and agricultural use of antibiotics throughout the world, which increases the incidence of resistance genes in the gut microbiota (Ghosh et al., 2013; Forslund et al., 2013, 2014).
Several studies have examined the impact of antibiotics on the microbiota of individuals (Pallav et al., 2014; Dethlefsen and Relman, 2011; Pérez-Cobas et al., 2013) using 16S rRNA gene sequencing alone or combined with metatranscriptomics (Maurice et al., 2013). As expected, the composition of microbial communities found in the gut and the regulation of bacterial genes were shown to be affected by antibiotics. However, the high inter-individual heterogeneity of the human gut microbiome and its quick response to changes in lifestyle and diet affect the impact of these compounds and of diseases on gut microbiota (David et al., 2014). In this study, we exposed 18 healthy volunteers (E) selected using stringent inclusion and exclusion criteria, to a therapeutic dosage of the antibiotic cefprozil, a commonly used second-generation cephalosporin clinically recognized for having minimal side effects (Lode et al., 1992; Wise, 1994; Edlund and Nord, 2000). Using the same selective criteria, we enrolled six controls (C) who received no antibiotic. Stool samples were collected before treatment (E0 and C0: initial state), at the end of the treatment (E7 and C7: peak effects) and 3 months after the end of the treatment (E90 and C90: long-term effects). Metagenomic DNA was extracted from the stools and sequenced at an average of 15 Gb per sample (Supplementary Table S1). Shotgun metagenomic sequencing allowed the assembly of an average of 124 million nucleotides per stool sample, which enabled monitoring of metagenome composition down to the species rank, including the identification of resistance genes and mobile elements. In summary, the results from this study indicate that, while cefprozil altered the microbiome of healthy individuals in a reproducible manner, it also permitted the emergence of potentially pathogenic Enterobacteriaceae and of rare resistance genes that were not detected prior to the treatment.
Materials and methods
Sample collection
The clinical protocol was approved by the ethical committee of the CHU de Québec–Université Laval. Informed written consent was obtained from participants. Participants exposed to the antibiotic and controls were selected following the same stringent criteria. Healthy volunteers recruited in the Quebec City region (Canada) were aged between 21 and 35 years and had normal intestinal transit. Characteristics of the participants are described in Supplementary Table S2. Subjects with any of the exclusion criteria below were not eligible for entry into the present study:
Working in a health-care facility or living with someone working in a health-care facility.
Working on a farm or household contact in the last 2 weeks.
Slaughterhouse worker or household contact.
Animal care worker or household contact.
Vegetarians.
Smokers.
Chronic alcohol consumption (more than one 1.5-ounce servings of 80 proof distilled spirits, five 12-ounce servings of beer or five 5-ounce servings of wine per day).
Antibiotic therapy or history of hospitalization (>24 h) in the past 12 months prior to the study.
Living with someone or an animal that has been on antibiotic therapy in the last month.
Any gastrointestinal or underlying pathology.
Any chronic illness.
Any infection requiring chemo/antibiotic therapy.
Diarrhoeal disease (World Health Organization definition) in the last 3 months prior to the study.
Gastro-intestinal-related medication (prescription antibiotics).
Immunomodulating medication such as antitumour necrosis factor or steroids.
Allergy to β-lactams.
Pregnant or lactating women.
Taking alimentary supplements.
Body mass index abnormal defined as <18.5 or >30 kg m2.
Eighteen participants were treated twice a day with an oral dose of 500 mg cefprozil, a second-generation cephalosporin, for 7 days. The antibiotic was self-administered. Six controls who did not receive antibiotics were also enrolled. Fresh stool specimens were self-collected by participants at three time points: before the antibiotic treatment (0), at the end of the treatment (7), and 90 days after the end of the treatment (90). The stool collection procedure was explained to the participants at the time of enrolment. Participants were also given an illustrated document detailing the procedure. Samples were collected following human microbiome project protocols (Aagaard et al., 2013) using stool specimen collection kits comprising: one Ziploc bag (Johnson and Son, Brantford, ON, Canada) and one kraft paper bag for transport, one GasPak EZ Anaerobe sachet (Becton, Dickinson and Company, Sparks, MD, USA), one M40 Transystem 408C transport swab (Copan Italia S.P.A., Brescia, Italy), one Commode Specimen Collection System (Biomedical Polymers, Inc., Gardner, MA, USA). All samples were brought to the laboratory within 2 h of collection and placed immediately in an anaerobic chamber for processing. Stool aspect was classified according to the Bristol scale (Lewis and Heaton, 1997). Stools were aliquoted and stored at −80 °C.
DNA extraction
Genomic DNA was extracted from 500 mg aliquots of each of the 72 frozen stool specimens using the MO BIO PowerMax Soil DNA Extraction Kit (MO BIO Laboratories, Carlsbad, CA, USA) following a modified human microbiome project procedure (Aagaard et al., 2013). Briefly, each stool sample (500 mg) was homogenized by vortexing in 15 ml PowerBead Solution (MO BIO cell lysis buffer supplied with the DNA Extraction Kit) and centrifuged at 500 r.c.f. for 5 min at room temperature. Supernatant was transferred to collection tubes, incubated for 10 min at 65 °C and then at 95 °C for an additional 10 min. The supernatant was transferred to PowerMax bead tubes containing Garnet beads (supplied with the DNA Extraction Kit) for mechanical cell lysis and DNA extraction according to the directions of PowerMax Soil DNA Isolation Kit. All DNAs were eluted using PCR grade water and stored at 4 °C. The quantity and quality of isolated DNA was measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), agarose gel electrophoresis and a QuantiFluor dsDNA System (Promega, Madison, WI, USA).
Sequencing
Metagenomic DNA was quantified using the QuantiFluor dye (Promega). Sequencing libraries were prepared using the Nextera Sample Preparation Kit (Illumina, San Diego, CA, USA) with 50 ng of purified DNA. Dual indices were added during library preparation. Libraries were validated using Agilent Bioanalyzer 2100 high sensitivity DNA chips (Agilent Technologies, Santa Clara, CA, USA) and were quantified again with QuantiFluor. Libraries were diluted to a 2 nm concentration prior to pooling. Two libraries were pooled per HiSeq lane. Sequencing was performed on the HiSeq 1000 sequencer (Illumina) using v3 chemistry and paired-end 101 bp reads. A total of 72 metagenomes were sequenced with an average of 15 Gb per sample (Supplementary Table 1). Metagenomic reads and assemblies have been deposited in the European Nucleotide Archive database in project PRJEB8094.
Assembly, profiling and data mining
Metagenomes were assembled and profiled for taxonomy and resistance genes using the Ray Meta 2.0 assembler (Boisvert et al., 2010, 2012). The Ray Meta software first constructs a de Bruijn graph based on the sequenced reads using a defined k-mer length, in this case 31 nucleotides. Contigs are assembled by finding the best paths in the graph. Subsequently, the k-mers in the graph are coloured based on their occurrence in a reference database comprising a selection of the genomes available at National Center for Biotechnology Information (NCBI; last updated in September 2014, see Supplementary Table S3). Using the NCBI taxonomy, the abundance of k-mers is processed to determine taxa frequencies at relevant taxonomical ranks. Contig identification is based on de Bruijn graph-based contig assignation (colouring) performed by Ray Meta. In brief, Ray Meta identifies the organisms that share k-mers with each contig in the reference genome database (Supplementary Table S3). Contigs longer than 2.5 kb are identified to taxa using a last-common ancestor algorithm that determines the taxonomic rank by calculating the sum of child nodes and selecting the taxa for which the sum comprises at least 90% of k-mers (https://github.com/plpla/laughing-nemesis).
Genes were found on contigs using Prodigal 2.6 with the metagenomic option (Hyatt et al., 2010). Putative genes were aligned to the MERGEM database using FASTA36 to identify putative resistance genes and mobile elements (Pearson, 2000). Genes were annotated as resistance genes if they had an amino-acid identity >40% with a type gene found in MERGEM. This version of MERGEM included 1760 type genes. The abundance of a given resistance gene or mobile element in a sample was computed by summing the mode of k-mer depths for all the contigs harbouring the gene. This provides an estimation of the abundance of specific genes. This approach is less sensitive than read-based approaches but allows for more precise gene identification. The reference sequences used for resistance genes and mobile elements profiling are available in the MERGEM database (http://www.mergem.genome.ulaval.ca). Read alignment was visualized using the Integrative Genomic Viewer (Thorvaldsdóttir et al., 2013).
Statistical analysis was carried out in R 3.0.2. Taxon frequencies were processed using the Hellinger transformation (Legendre and De Cáceres, 2013) before fitting them to a mixed linear-effect model (NLME 3.1). Analysis of variance P-values were corrected for false discovery rate using the Benjamini–Hochberg–Yekutieli procedure (Benjamini and Yekutieli, 2001). Microbial diversity was computed using the VEGAN package (Dixon, 2003). Plots to visualize data distribution and density were created using the Beanplot package (Kampstra, 2008). Multiple factor analysis (MFA) was performed using FactoMineR 1.25 (Lê et al., 2008). MFA is a method conceptually similar to principal component analysis or multiple correspondence analysis. The main advantage of MFA is that it allows the integration of structured groups of variables. The current analysis included 6070 quantitative variables, which belonged to 11 variable groups. Therefore, calculation of the multidimensional distance between the samples was based on the profiling results of the class, family, genus, species, subspecies and species group taxonomical ranks, the abundance of resistance genes or mobile elements, diversity indices, sequencing statistics and Bristol stool category. The influence of each variable group on the principal components can also be estimated by the MFA. Bioinformatics and statistical analysis are detailed in Supplementary Information.
Results
Cefprozil intake affects the microbiota
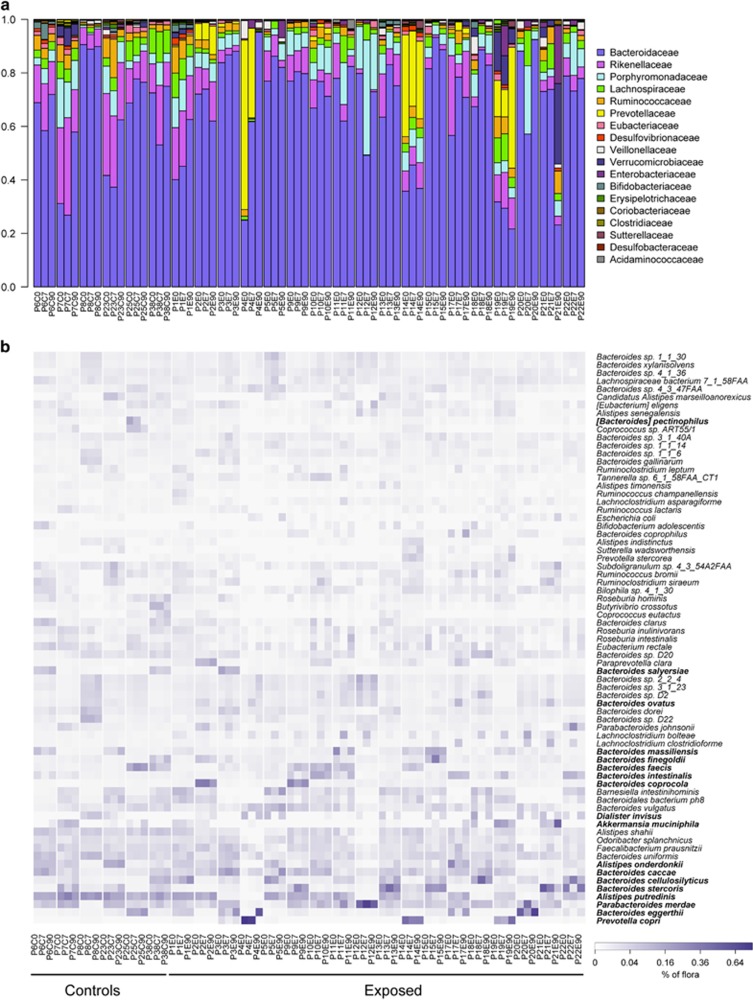
Eighteen healthy participants received the antibiotic cefprozil for 7 days. Six control participants, unexposed to antibiotics, were also enrolled to evaluate variability. Stool samples were obtained before (C0 and E0) and after the treatment (C7 and E7) and 90 days after the end of the antibiotic course (C90 and E90). These samples were then submitted to deep shotgun sequencing. The Ray Meta assembler (Boisvert et al., 2012) was used to assemble and profile the taxonomic composition, gene function and resistance gene content of sequenced metagenomes. In most samples, microbiome composition at the family rank shows an overall dominance of Bacteroidaceae (Figure 1a). Still, diversity at the species rank highlights differences not observed at the family level. Each participant was found to harbour a specific subset of Bacteroides species (Figure 1b). As shown by Franzosa et al. (2015), the composition of the gut microbiome can serve to uniquely identify individuals. Our findings indicated that inter-individual variability at the species level was greater than the effect of the antibiotic for most participants (Supplementary Figure S1). The dominant species detected in stools were typically associated with 1 of the 18 species belonging to the Akkermansia, Alistipes, Bacteroides, Dialister, Parabacteroides or Prevotella genera (Figure 1b), with Bacteroides being the dominant genus for 40 out of the 72 samples. Although changes in dominant species were observed between time points, we could not link such changes to antibiotic exposure, an observation suggesting that cefprozil has a limited impact on the most abundant gut microbes, as previously observed using culture (Lode et al., 1992).
Figure 1.
Composition of bacterial communities in all participants before exposure, 7 days later and 90 days after exposure to cefprozil. (a) Bar plot representing the relative abundance of bacterial families in controls and exposed participants. (b) Heat map representing the relative abundance of bacterial species that constituted >1% of the community in at least one participant. Species are grouped based on hierarchical clustering. Species in bold are the most abundant in at least one microbiome. P precedes the identification number of the participant, E indicates exposed and C indicates control.
A common pattern of response to the antibiotic was observed in the exposed participants. Indeed, six bacterial families were significantly decreased after exposure to antibiotics: Bifidobacteriaceae, Coriobacteriaceae, Eubacteriaceae, Oxalobacteraceae, Pasteurellaceae, and Veillonellaceae (Supplementary Figure S2, Supplementary Table S4). No bacterial family uniformly increased after antibiotic treatment, but the genera Flavonifractor, Lachnoclostridium and Parabacteroides increased significantly (Supplementary Figure S2). Strikingly, Lachnoclostridium bolteae increased in 16 out of the 18 participants exposed to cefprozil, an average rise of 20-fold, and returned to its initial (E0) level at E90 (Supplementary Figure S2). Similarly, abundance of taxa such as Dialister invisus, Eubacterium rectale and Veillonellaceae were drastically reduced at E7 but returned to initial E0 levels at the E90 sampling time point. In contrast, the abundance of these three taxa in all six control individuals remained stable throughout the experiment. The size of the assembled metagenomes decreased from an average of 118 million nucleotides at E0 to 97 million at E7, a 17.3% drop (P=0.01). This is further supported by the N50 of E7 samples, which was 30% higher than that of the E0 assemblies (P=0.09). However, the Simpson diversity of the microbiomes was only slightly reduced by 7.6% on average (P=0.42).
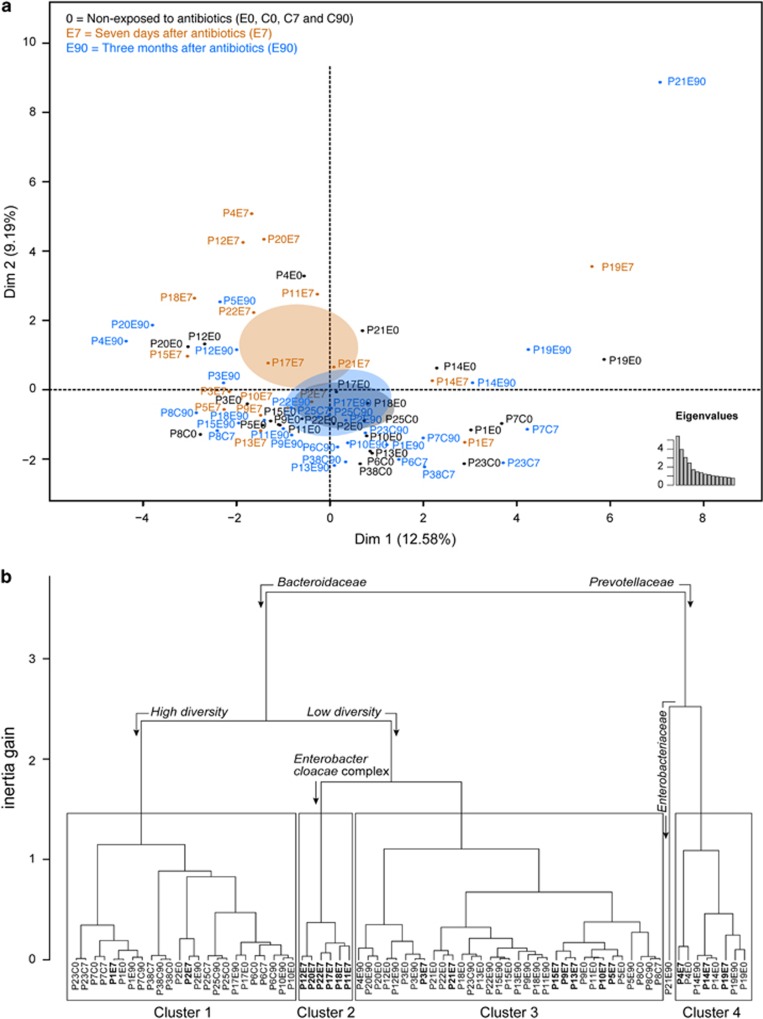
In order to understand the interaction between antibiotic intake and the microbiome, we used MFA to integrate variable groups in a principal component analysis-like procedure (Lê et al., 2008). This analysis included 11 groups of variables, including taxonomic composition at the level of four ranks, sequencing statistics and abundance of resistance genes and mobile elements. The principal component map of the MFA revealed that cefprozil had a notable effect on the microbiome of exposed participants (Figure 2a). This effect was discernible on the second axis of the MFA, whereby the ellipse representing E7 had only a slight overlap with the ellipse of non-exposed samples (E0, C0, C7 and C90). This is due to changes in bacterial composition, resistance genes and mobile elements (Supplementary Figures S3 and S4). The ellipses of controls at C0, C7 and C90 had a strong overlap (Supplementary Figure S5), indicating a lower temporal variability in controls compared with exposed participants. Hierarchical clustering based on MFA results separated samples between Bacteroidaceae (Clusters 1, 2 and 3) and Prevotellaceae (Cluster 4) dominant microbiomes (Figure 2b). The Bacteroidaceae-rich samples were further separated between high (Cluster 1) and low (Clusters 2 and 3) taxonomical diversity samples, in a way similar to previously described enterotypes (Arumugam et al., 2011; Figure 2b). Aside from individuals P4 and P21, all individuals returned to their E0 cluster at the E90 sampling point. By contrast, the C7 stool samples from all controls remained similar to C0, with only one (P23) of six controls shifting from Cluster 1 to Cluster 3 after 90 days. It should be noted that stool samples from controls (C0, C7 and C90) are dominant in Cluster 1 (14 samples out of 18), while only 4 out of 18 E0 participants were in this cluster. By contrast, only 4 control specimens were in cluster 3, compared with 11 E0 samples. In addition, none of the control specimens were of the Prevotellaceae enterotype associated with our Cluster 4. This lower representation of controls outside of Cluster 1 could explain the separation between the ellipses of controls and exposed participants at E0 (Supplementary Figure S5).
Figure 2.
MFA reveals the effect of cefprozil on the microbiome of healthy individuals. MFA was performed using 11 variable groups representing data of taxonomic profiling, abundance of resistance genes or mobile elements, diversity indices, sequencing depth and assembly statistics. (a) The factorial map presents the impact of exposure to antibiotics on the microbial flora of participants based on the MFA. Black identifiers and ellipses indicate samples non-exposed to the antibiotic (E0, C0, C7 and C90), tan samples taken at the end of the antibiotic treatment (E7) and blue samples taken 90 days after the end of the treatment (E90). The ellipses represent the barycentre of the sample groups with 95% confidence. Eigenvalues show the significance of dimensions 1 and 2. (b) Hierarchical clustering based on results from the MFA. Four clusters are identified, along with a singleton sample. The quantitative variables driving the separation of samples in the clustering are indicated by bent arrows over the dendrogram. Association of microbiome characteristics with clusters was undertaken using an F-test in a one-way analysis of variance. Samples at E7 are in bold to emphasis the clustering of exposed participants.
The taxonomical diversity of Cluster 1 was correlated to a higher abundance of Rikenellaceae and of members of the class Clostridia, notably Eubacteriaceae and Ruminococcaceae, compared with the other clusters (Supplementary Figure S5 and S6). Cluster 2 was associated with a higher abundance of Enterobacter cloacae complex bacteria, L. bolteae and C. clostridioforme. Interestingly, this cluster comprised E7 samples from six participants exposed to cefprozil, indicating a reproducible effect for the antibiotic treatment specific to these six participants. Stools of all six participants returned to their cluster of origin at E90. We observed strong alterations in the microbiome of one (P4) of the three participants dominated by Prevotellaceae (Cluster 4), which saw its Bacteroidaceae level increase from 25 to 95% at E7 (Figure 1a), although the antibiotic had limited impact on the microbiome composition of the two other participants (P14 and P19). Sample P21E90 behaved as an outlier in showing important alterations from its initial composition. Compared with E0, the microbiome of P21E90 had a fourfold increase in Enterobacteriaceae and a threefold increase in Verrucomicrobiaceae relative abundance (Figure 1a). Interestingly, this participant started taking ferrous sulphate as a treatment for anaemia during the antibiotic course. It has been reported that ferrous sulphate can alter the microbiome in rats, increasing Enterobacteriaceae (Dostal et al., 2012).
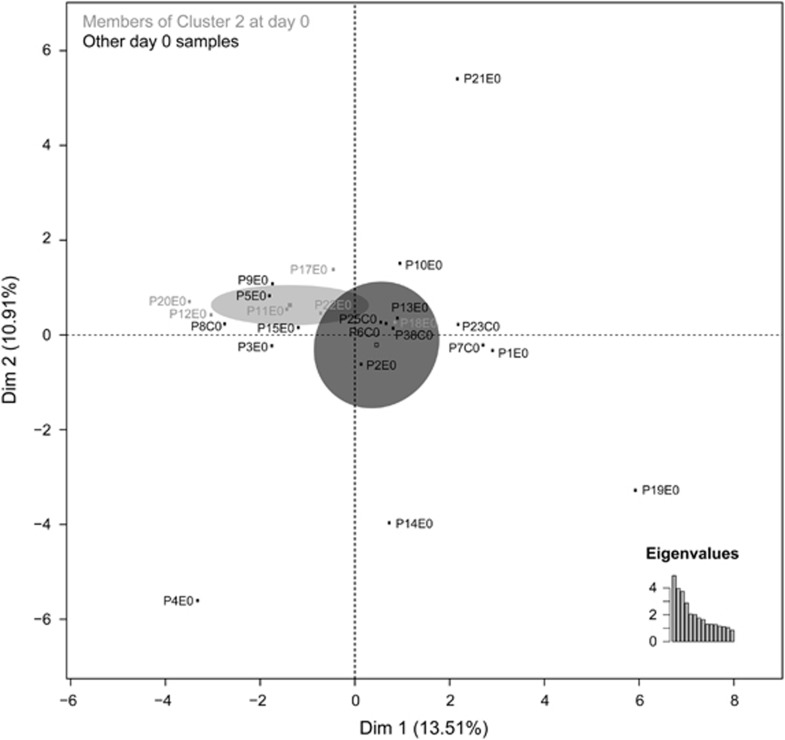
An additional MFA analysis that focussed specifically on samples recovered prior to antibiotic exposure (E0 and C0) revealed that the six samples from Cluster 2 (Figure 2b) that were enriched in E. cloacae after cefprozil exposure also tightly grouped together at baseline (Figure 3). When compared with the other E0 microbiomes using a linear model-based statistical analysis (Supplementary Table S1), these E0 samples were associated with an average species diversity 29% lower than that of all other clusters combined (P=0.04). They were also associated with reduced levels of lower abundance taxa (that is, <0.1%) such as the classes Bacilli, Methanobacteria and Mollicutes, the genera Streptomyces and Xanthomonas and species Dorea formicigenerans, Gemmata obscuriglobus, Marvinbryantia formatexigens and Prevotella marshii (Supplementary Figures S7 and S8). Levels of the E. cloacae complex at E0 in these Cluster 2 members did not differ from those detected in the other E0 samples.
Figure 3.
Participants enriched in E. cloacae complex after antibiotic exposure were clustered together before exposure. MFA was performed on day 0 samples only. Clustered participants enriched in the E. cloacae complex at E7 (Cluster 2) are represented in grey with the remainder of the samples presented in black.
Genes altered by exposure to antibiotic
Gene ontology terms that significantly increased after exposure to antibiotics had an initial relative abundance higher than ontology terms that were decreased after drug intake. On average, the k-mer abundances of significantly increased gene ontology terms were 6.9-fold higher than significantly decreased biological processes terms (15.4-fold for molecular functions and 2.5-fold for cellular components; Supplementary Figure S9 and Supplementary Table S6). Transposition was the most abundant of the significantly increased biological process terms (93th percentile). The beta-lactam antibiotic catabolic process category was also significantly increased at E7 compared with E0 (74th percentile). Annotation of the assembled contigs allowed the detection of a total of 10.2 million putative genes, including 24 686 putative insertion sequences (IS) and 43 353 putative resistance genes. We found 2101 resistance genes located on the same contig as an IS. The sequences for 1439 putative beta-lactamases were retrieved from the assembled metagenomes.
Using the MERGEM database, we assigned these 43 353 putative resistance genes to 326 type genes classified by current resistance gene taxonomy (Liu and Pop, 2009; McArthur et al., 2013). Only 21 of these 326 resistance genes types were found in all metagenomes while 47 of 326 were found in a single sample (Supplementary Table S7). Five resistance gene types were significantly increased at E7 compared with E0, notably arr2, blaCepA and mef(G) (Supplementary Table S4). The β-lactamase blaCepA was found in contigs that could be associated with the order Bacteroidales. It had several sequence variants and was present in 66 of the 72 samples. Increased abundances of arr2 and mef(G) could be associated with L. bolteae. The arr2-related genes were detected in six E7 samples in contigs assigned to L. bolteae. They were 52% identical to the arr2-type sequence of MERGEM. This resistance gene functions as Rifampin ADP-ribosyl transferase, an enzyme that is widely distributed in the genomes of bacteria (Baysarowich et al., 2008). Genes sharing 40–50% identity with mef(G) were detected in 3 E0 samples and in 13 E7 samples. One subgroup of mef(G) orthologs was similar to a major facilitator transporter found in a Bacteroides context and a second subgroup was related to a macrolide efflux pump in a Lachnoclostridium context.
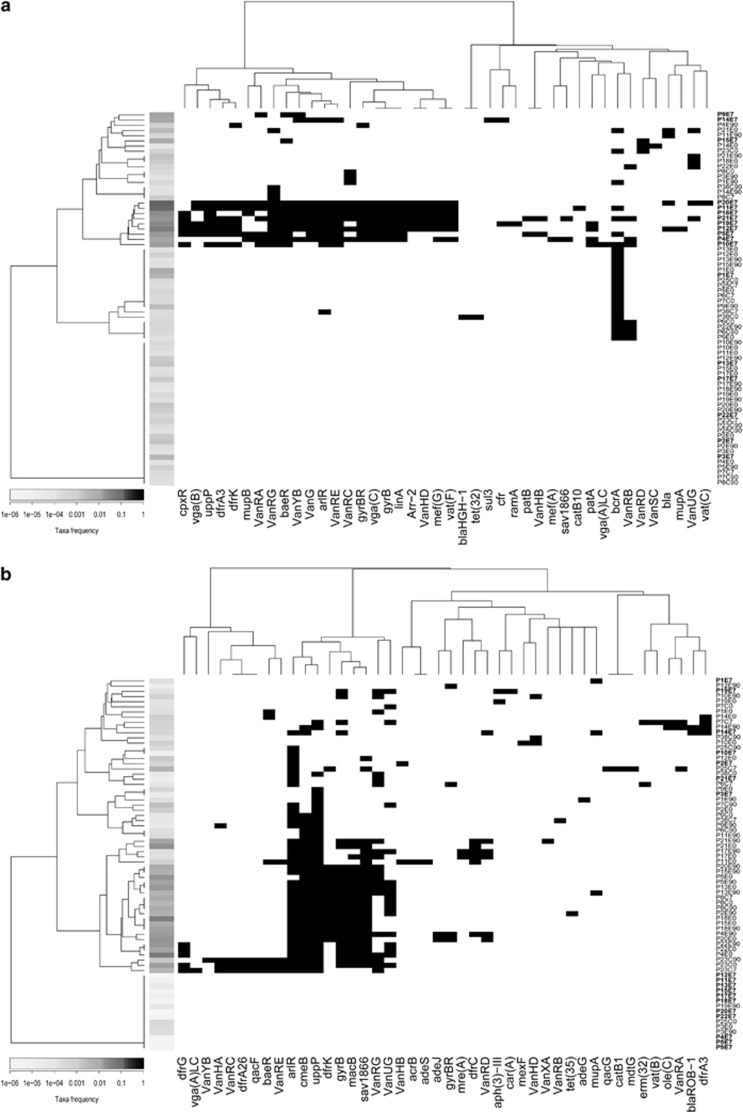
In order to understand the relationship between antibiotic resistance genes and the modulation of taxa, we evaluated the occurrence of resistance genes in taxa that were affected by antibiotic treatment. We annotated a total of 42 resistance gene types in contigs assigned to L. bolteae, which is favoured by the antibiotic (Figure 4a). Hierarchical clustering of gene distribution shows a cluster of E7 samples that contained up to 26 different resistance gene types in a L. bolteae context. In contrast, E7 samples contained few occurrences of resistance genes associated with contigs related to Veillonellaceae, a family that is strongly reduced by antibiotic treatment (Figure 4b). Most E7 samples had more than 10-fold reductions of Veillonellaceae compared with corresponding E0 samples. However, Veillonellaceae in P14E7 was only reduced twofold. This participant was the only one positive for the presence of blaROB-1 in a Veillonellaceae context.
Figure 4.
Association of resistance genes with genomic context for (a) L. bolteae and (b) Veillonellaceae. The presence of a resistance gene (black rectangle) was considered positive when it was annotated on a contig originating from the taxon of interest. Ordering of the resistance genes and samples was based on the hierarchical clustering of binary distances. Abundance of taxa in samples is represented by a scale of grey on the left side of the plot. The gene identified bla is an uncharacterized putative beta-lactamase previously observed in C. clostridioforme (KC188672.1).
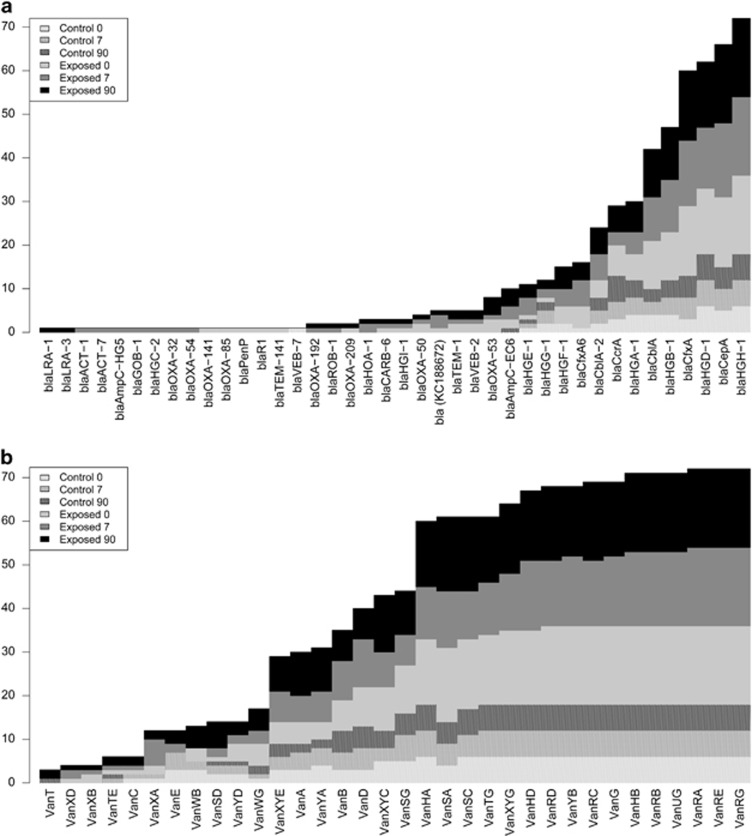
Analysis of individual participants revealed striking modulation of beta-lactamase gene abundances following antibiotic intake. For example, we monitored a 90-fold increase in abundance of blaTEM-1 at E7 for participant P4 (Supplementary Figure S10). Interestingly, this blaTEM-1 was located between IS26 mobile elements as previously detected on an Escherichia coli plasmid associated with multidrug resistance (Wang et al., 2014). We also observed the postantibiotic emergence (Figure 5a) of blaOXA-32 in P11E7, blaOXA-54 in P9E7, blaOXA-192 in P1E7, blaROB-1 in participant P14E7 (in Veillonella), blaHOA-1 in participants P15E7 (in Akkermansia) and P4E7, blaHGC-2 in P1E7, blaGOB-1 in P2E7, blaACT-1 in P11E7 (in Enterobacter), blaACT-7 in P18E7 (in Enterobacter), blaAmpC-HG5 in P19E7 (in E. coli), as well as a conserved uncharacterized putative beta-lactamase in P12E7, P20E7 and P21E7, which was previously observed in C. clostridioforme (KC188672.1). With the exception of this last gene in P21E0, none of these beta-lactamase-coding genes were detected in the corresponding E0 samples. In contrast to beta-lactamases, resistance genes associated with vancomycin resistance were commonly detected in the samples and were not associated with E7 (Figure 5b).
Figure 5.
Number of samples positive for (a) beta-lactamases and (b) vancomycin resistance genes for antibiotic-exposed participants and controls at three time points. A stacked bar graph shows how many samples of each six categories were positive for resistance genes. The vertical axis represents the total number of reads positive for a resistance gene. Time points are coded as indicated in the upper left corner of the figure.
A unique observation from our metagenomic data was that exposure to the antibiotic selected for specific alleles of resistance genes as exemplified by the blaCfxA-6 variants detected in P3, P4, P14 and P19. The sequences of this gene in P3E0 and P4E0 had amino-acid identity >99%. The Y259 allele observed at E0 disappeared at E7 while the D259 allele appeared after antibiotic exposure (P3E7 and P4E7; Supplementary Table S8, Supplementary Figure S11). In both participants, the blaCfxA-6 gene was located near an IS942 mobile element. Interestingly, while the total abundance of all blaCfxA-6 alleles was reduced sixfold at P3E7 compared with P3E0, the D259 blaCfxA-6 could only be detected in P3E7, where it was the dominant blaCfxA-6 allele. In P3E0 and P4E0, the D259 allele was at very low abundance (1 and 9 reads, respectively), whereas it became dominant in P3E7 and P4E7 (29 and 1509 reads), suggesting that it is not a de novo mutation but a selection of preexisting strains by the antibiotic treatment.
Discussion
We exposed 18 carefully selected young healthy volunteers to a 7-day course of cefprozil, an antibiotic known to cause minimal disturbance of the microbiota (Lode et al., 1992; Wise, 1994; Edlund and Nord, 2000). Our main objective was to assess the effect of the antibiotic on the microbiome and on resistance genes. Studies by others on the effects of antibiotics on the microbiota were conducted on eight or fewer healthy volunteers and used only 16S-based metagenomics. They demonstrated that microbiota recovery following antibiotic treatment can be incomplete and that individual floras react differently to the same treatment (Dethlefsen et al., 2008; Pallav et al., 2014). In order to reduce variability of participants for both controls and exposed volunteers, we applied nineteen strict exclusion criteria at the enrolment stage. Exclusion criteria were devised to exclude antibiotic intake in the previous 12 months as well as to limit exposure to microbiota potentially selected by antibiotics (occupational and household contacts). Despite careful volunteer selection, microbiome composition measured by shotgun sequencing was different between participants, with controls more homogeneous than exposed participants. Interestingly, taxa observed in the participants allowed clustering of their microbiomes into groups similar to previously described enterotypes (Arumugam et al., 2011). Our use of deep shotgun sequencing allowed assembly of microbiomes and measurement of changes in gene content. Metagenomic sequences showed that the inter-individual variability of resistance genes was even higher than differences in flora, with most resistance genes observed in a limited number of participants, in some cases only after antibiotic treatment. These results show that detailed exclusion criteria are not sufficient to generate cohorts with homogenous microbiomes. Increasing the number of participants and prescreening the microbiome of candidates could be approaches to reduce inter-individual variability in future studies.
Even though we observed a high variability between individuals, we detected that the antibiotic had a reproducible effect on microbiomes. Our results suggest that exposure to the antibiotic reduced richness as it caused a significant decrease in the level of six bacterial families (40 species; Supplementary Table S4). By contrast, only three genera systematically increased during antibiotic treatment (11 species). The total size of the assembled metagenomes was reduced at E7 compared with E0 and the N50 of assemblies increased. Microbiomes with lower richness require smaller sequencing efforts compared with richer microbiomes, yielding larger assemblies with longer contigs for the same number of reads (Rodriguez-R and Konstantinidis, 2014). On the other hand, the Simpson diversity was only marginally reduced by treatment. It is possible that the diversity index did not reflect the reduction of bacterial richness because the reduced taxa were of low abundance. Diversity indices have been criticized for not taking into account lower abundance taxa (Li et al., 2012; Haegeman et al., 2013). In our study, the dominant taxa were not perceptibly affected by the antibiotic and the drug mostly affected lower abundance taxa. This resulted in a reduced abundance of k-mers associated with lower percentile gene ontology categories, for example, sporulation.
The most consistent effect of the antibiotic was the increase of L. bolteae in 16 out of the 18 cefprozil-exposed participants. In the current study, we were able to associate several resistance genes with contigs related to L. bolteae. In general, samples with the highest observed abundance of L. bolteae had more resistance genes related to this species than samples with lower abundance. However, samples presenting L. bolteae abundance <1% are more difficult to assemble, and this could explain the non-detection of resistance genes in these samples. The very nature of mobile elements can lead to incorrect contig identification as it is difficult to ascertain the host of such mobile genes. Nonetheless, L. bolteae is known to be resistant to the second-generation cephalosporin cefoxitin (Song et al., 2003). This species has previously been associated with dysbiotic states, such as autism (Finegold et al., 2002; Song et al., 2004; Yutin and Galperin, 2013), acute rejection of liver transplant (Ren et al., 2014) and with low microbiome diversity, a property associated with obesity and with a marked inflammatory phenotype (Le Chatelier et al., 2013). Although the exact role of L. bolteae in these diverse conditions remains unknown, it may be linked to host or microbial imbalance, making it a potential marker for dysbiosis, a state also induced by antibiotics.
In addition to the response observed in all treated participants, we detected changes specific to a subset of volunteers. Six of the 18 exposed participants had their level of E. cloacae complex bacteria increase from an average of 0.0001 to 0.1% after a 7-day course of cefprozil. Five of these six participants were initially associated with the low diversity, Bacteroides-rich Cluster 3 (Figure 2b), which is related to the proposed Bacteroides enterotype (Arumugam et al., 2011). We find the association between E. cloacae complex blooming and lower initial microbiota diversity reminiscent of C. difficile infection (Chang et al., 2008). Moreover, blooms of Enterobacteriaceae have previously been reported after treatment with antibiotics (Buffie et al., 2012). Expansion of Enterobacter after antibiotic treatment is not unexpected, as Enterobacteriaceae such as E. cloacae are known carriers of chromosomal beta-lactamases that confer resistance to cefprozil (Kayser, 1994; Wise, 1994). In two participants, we observed an increase of blaACT abundance in a contig closely related to an Enterobacter chromosome at the E7 time point. Genes related to tigecycline resistance (ramA, acrA, acrB) were also observed in E. cloacae (Veleba et al., 2013). Lineages of E. cloacae that are considered to have epidemic potential harbour multiple beta-lactamases (Izdebski et al., 2014). However, we cannot exclude that blooms of the E. cloacae are not only triggered by antibiotic resistance and that other fitness factors may be involved.
In all six participants, levels of the E. cloacae complex returned to initial levels at day 90, indicating that the antibiotic had a transient effect on this phenomenon. In a similar way, significantly modulated taxa returned to their initial abundance level at E90. The variability of the microbiome after 90 days was comparable to that of the controls (Figure 2). Only two out of 18 participants exposed to the antibiotic had a strikingly different microbiome composition 3 months after the antibiotic treatment than before exposure to cefprozil. We suspect that these two participants could have been affected by external factors. For example, P21 took iron supplements during the antibiotic treatment. We also hypothesize that the shift from a Prevotella-dominated microbiota to high levels of Bacteroides in P4 could be linked to factors other than cefprozil. Modification in diet has been shown to induce such changes in this enterotype (Wu et al., 2011). However, the limited number of Prevotella-dominant participants enrolled in our study and the absence of data on their diet make further interpretation difficult.
We observed individual-specific changes in resistance gene abundances, confirming that potentially important resistance genes, which may be below detection levels, are present in low abundance in the microbiome and can be selected during in vivo exposure to antibiotics. For example, beta-lactamases such as blaOXA, blaACT, blaAmpC and blaTEM were drastically increased at E7, whereas they could not be assembled at E0 (Figure 5a). These genes were often in proximity to mobile elements. Selection of beta-lactamase variants was also observed after antibiotic treatment, for example, the emergence of the same non-synonymous mutations in blaCfxA-6 was detected in two participants, a mutation that was found in very low abundance at E0. Most of these resistance genes were not detected 90 days later. Still, it remains difficult to discern with metagenomics whether resistance genes drive taxa abundance or whether modulation of specific bacteria affects resistance genes related or not to the antibiotic. For example, mutation of blaCfxA-6 could represent gene selection while genes found in L. bolteae could relate to species enrichment. Nonetheless, initially undetectable antibiotic resistance genes were selected toward higher abundance after antibiotic exposure and returned to lower levels following antibiotic withdrawal.
Overall, the impact of the antibiotic could be summarized in three categories. First, cefprozil had a reproducible effect on the microbiomes in almost all participants, leading to the increase of a limited number of specific genera and to the decrease of several bacterial families. The second effect was observed in a subset of the exposed participants, who had a bloom of a specific group of bacteria after antibiotic treatment. This increase in E. cloacae complex bacteria was observed in participants who initially had a Bacteroides enterotype with lower microbial diversity. The third effect consisted in the appearance of resistance genes at E7, in some cases revealing specific alleles, which were undetectable before the treatment. Increased abundance of resistance genes can either be modulated by gene selection or by strain enrichment. In both cases, they are the result of antibiotic treatment, a situation that could favour their transfer to pathogenic species (Beaber et al., 2004; Ubeda et al., 2005; Prudhomme et al., 2006). Situations similar to blooms of E. cloacae could have clinical importance because Enterobacteriaceae are known for their propensity to transfer genes, especially in the presence of gut inflammation (Stecher et al., 2012). For this reason, knowledge of how antibiotics or other molecules, for example iron, can affect gut microbiota would result in better treatment of at-risk patients. In conclusion, knowing how the initial state of the microbiota determines the impact of antibiotics could allow personalized treatment in order to minimize adverse effects. Moreover, we believe that the analysis of the microbiota, microbiome and resistome will become an essential part of the industrial discovery and evaluation of new antibiotics and most likely other drugs. This could guide the development of better therapeutic interventions, minimizing the impact on the microbiota and even taking advantage of the microbiome.
Acknowledgments
This study was financed by the Québec Consortium for Drug Discovery (CQDM) and by the Canada Research Chair Program (JC and MO). FR, AAO, MD, NI and BD were supported by Mitacs through the Mitacs-Accelerate program. Computations were performed on the Guillimin supercomputer at McGill University and the Colosse supercomputer at Université Laval, under the auspices of Calcul Québec and Compute Canada. The operations of Guillimin and Colosse are funded by the Canada Foundation for Innovation (CFI), the National Science and Engineering Research Council (NSERC), NanoQuébec and the Fonds Québécois de Recherche sur la Nature et les Technologies (FQRNT).
Author contributions
FR contributed to experimental design, prepared sequencing libraries, sequenced samples, performed bioinformatics and data mining, analysed results and wrote the manuscript with MB. FR, AAO, NI and HG prepared sequencing libraries. EB managed stool samples. AAO and RG extracted nucleic acids. MD created and curated reference databases. MD, PLP, DKB and PHR performed bioinformatics analyses. IC and ST were in charge of the clinical study. BD, PL, JF, JLS and MCD offered technical and scientific support for the study. ST, MB, AH, PHR, MO, MGB and JC designed the study and supervised the project. All authors critically reviewed the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Aagaard K, Petrosino J, Keitel W, Watson M, Katancik J, Garcia N et al. (2013). The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB J 27: 1012–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR et al. (2011). Enterotypes of the human gut microbiome. Nature 473: 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysarowich J, Koteva K, Hughes DW, Ejim L, Griffiths E, Zhang K et al. (2008). Rifamycin antibiotic resistance by ADP-ribosylation: structure and diversity of Arr. Proc Natl Acad Sci USA 105: 4886–4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaber JW, Hochhut B, Waldor MK. (2004). SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427: 72–74. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D. (2001). The control of the false discovery rate in multiple testing under dependency. Ann Stat 29: 1165–1188. [Google Scholar]
- Boisvert S, Laviolette F, Corbeil J. (2010). Ray: Simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol 17: 1519–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert S, Raymond F, Godzaridis E, Laviolette F, Corbeil J. (2012). Ray Meta: scalable de novo metagenome assembly and profiling. Genome Biol 13: R122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A et al. (2012). Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun 80: 62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM et al. (2008). Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis 197: 435–438. [DOI] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500: 541–546. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505: 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, Relman DA. (2008). The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16 S rRNA sequencing. PLoS Biol 6: e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Relman DA. (2011). Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA 108: 4554–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon P. (2003). VEGAN, a package of R functions for community ecology. J Veg Sci 14: 927–930. [Google Scholar]
- Donia MS, Cimermancic P, Schulze CJ, Brown LCW, Martin J, Mitreva M et al. (2014). A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158: 1402–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostal A, Chassard C, Hilty FM, Zimmermann MB, Jaeggi T, Rossi S et al. (2012). Iron depletion and repletion with ferrous sulfate or electrolytic iron modifies the composition and metabolic activity of the gut microbiota in rats. J Nutr 142: 271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund C, Nord C. (2000). Effect on the human normal microflora of oral antibiotics for treatment of urinary tract infections. J Antimicrob Chemother 46: 41–48. [PubMed] [Google Scholar]
- Finegold SM, Molitoris D, Song Y, Liu C, Vaisanen M-L, Bolte E et al. (2002). Gastrointestinal microflora studies in late-onset autism. Clin Infect Dis 35: S6–S16. [DOI] [PubMed] [Google Scholar]
- Forsberg KJ, Patel S, Gibson MK, Lauber CL, Knight R, Fierer N et al. (2014). Bacterial phylogeny structures soil resistomes across habitats. Nature 509: 612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MOA, Dantas G. (2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337: 1107–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K, Sunagawa S, Coelho LP, Bork P. (2014). Metagenomic insights into the human gut resistome and the forces that shape it. Bioessays 36: 316–329. [DOI] [PubMed] [Google Scholar]
- Forslund K, Sunagawa S, Kultima JR, Mende DR, Arumugam M, Typas A et al. (2013). Country-specific antibiotic use practices impact the human gut resistome. Genome Res 23: 1163–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Huang K, Meadow JF, Gevers D, Lemon KP, Bohannan BJM et al. (2015). Identifying personal microbiomes using metagenomic codes. Proc Natl Acad Sci 112: E2930–E2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh TS, Gupta S, Sen, Nair GB, Mande SS. (2013). In silico analysis of antibiotic resistance genes in the gut microflora of individuals from diverse geographies and age-groups. PLoS One 8: e83823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegeman B, Hamelin J, Moriarty J, Neal P, Dushoff J, Weitz JS. (2013). Robust estimation of microbial diversity in theory and in practice. ISME J 7: 1092–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández E, Bargiela R, Diez MS, Friedrichs A, Pérez-Cobas AE, Gosalbes MJ et al. (2013). Functional consequences of microbial shifts in the human gastrointestinal tract linked to antibiotic treatment and obesity. Gut Microbes 4: 306–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T et al. (2013). Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155: 1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izdebski R, Baraniak A, Herda M, Fiett J, Bonten MJM, Carmeli Y et al. (2014). MLST reveals potentially high-risk international clones of Enterobacter cloacae. J Antimicrob Chemother 70: 48–56. [DOI] [PubMed] [Google Scholar]
- Kampstra P. (2008). Beanplot : a boxplot alternative for visual comparison of distributions. J Stat Softw 28: 1–9. [Google Scholar]
- Kayser FH. (1994). In vitro activity of cefpodoxime in comparison with other oral beta-lactam antibiotics. Infection 22: 370–375. [DOI] [PubMed] [Google Scholar]
- Lê S, Josse J, Husson F. (2008). FactoMineR : An R package for multivariate analysis. J Stat Softw 25: 1–18. [Google Scholar]
- Legendre P, De Cáceres M. (2013). Beta diversity as the variance of community data: dissimilarity coefficients and partitioning. Ecol Lett 16: 951–963. [DOI] [PubMed] [Google Scholar]
- Lewis SJ, Heaton KW. (1997). Stool form scale as a useful guide to intestinal transit time. Scand J Gastroenterol 32: 920–924. [DOI] [PubMed] [Google Scholar]
- Li K, Bihan M, Yooseph S, Methé BA. (2012). Analyses of the microbial diversity across the human microbiome. PLoS One 7: e32118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Pop M. (2009). ARDB—Antibiotic Resistance Genes Database. Nucleic Acids Res 37: D443–D447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lode H, Müller C, Borner K, Nord CE, Koeppe P. (1992). Multiple-dose pharmacokinetics of cefprozil and its impact on intestinal flora of volunteers. Antimicrob Agents Chemother 36: 144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manges AR, Labbe A, Loo VG, Atherton JK, Behr MA, Masson L et al. (2010). Comparative metagenomic study of alterations to the intestinal microbiota and risk of nosocomial Clostridum difficile-associated disease. J Infect Dis 202: 1877–1884. [DOI] [PubMed] [Google Scholar]
- Maurice CF, Haiser HJ, Turnbaugh PJ. (2013). Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152: 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ et al. (2013). The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57: 3348–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallav K, Dowd SE, Villafuerte J, Yang X, Kabbani T, Hansen J et al. (2014). Effects of polysaccharopeptide from Trametes versicolor and amoxicillin on the gut microbiome of healthy volunteers: a randomized clinical trial. Gut Microbes 5: 458–467. [DOI] [PubMed] [Google Scholar]
- Pearson WR. (2000). Flexible sequence similarity searching with the FASTA3 program package. Methods Mol Biol 132: 185–219. [DOI] [PubMed] [Google Scholar]
- Pérez-Cobas AE, Gosalbes MJ, Friedrichs A, Knecht H, Artacho A, Eismann K et al. (2013). Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut 62: 1591–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys J-P. (2006). Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313: 89–92. [DOI] [PubMed] [Google Scholar]
- Ren Z, Jiang J, Lu H, Chen X, He Y, Zhang H et al. (2014). Intestinal microbial variation may predict early acute rejection after liver transplantation in rats. Transplantation 98: 844–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-R LM, Konstantinidis KT. (2014). Estimating coverage in metagenomic data sets and why it matters. ISME J 8: 2349–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Liu C, Finegold SM. (2004). Real-time PCR quantitation of clostridia in feces of autistic children. Appl Environ Microbiol 70: 6459–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Liu C, Molitoris DR, Tomzynski TJ, Lawson PA, Collins MD et al. (2003). Clostridium bolteae sp. nov., isolated from human sources. Syst Appl Microbiol 26: 84–89. [DOI] [PubMed] [Google Scholar]
- Stecher B, Denzler R, Maier L, Bernet F, Sanders MJ, Pickard DJ et al. (2012). Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc Natl Acad Sci USA 109: 1269–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE et al. (2009). A core gut microbiome in obese and lean twins. Nature 457: 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C, Maiques E, Knecht E, Lasa I, Novick RP, Penadés JR. (2005). Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol Microbiol 56: 836–844. [DOI] [PubMed] [Google Scholar]
- Veleba M, De Majumdar S, Hornsey M, Woodford N, Schneiders T. (2013). Genetic characterization of tigecycline resistance in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes. J Antimicrob Chemother 68: 1011–1018. [DOI] [PubMed] [Google Scholar]
- Wang J, Stephan R, Power K, Yan Q, Hächler H, Fanning S. (2014). Nucleotide sequences of 705 16 transmissible plasmids identified in nine multidrug-resistant Escherichia coli isolates expressing an ESBL phenotype isolated from food-producing animals and healthy humans. J Antimicrob Chemother 69: 2658–2668. [DOI] [PubMed] [Google Scholar]
- WHO. (2014) Antimicrobial Resistance: Global Report on Surveillance 2014. World Health Organization: Geneva, Switzerland, http://www.who.int/mediacentre/news/releases/2014/amr-report/en/ (accessed 3 July 2014). [Google Scholar]
- Wise R. (1994). Comparative microbiological activity and pharmacokinetics of cefprozil. Eur J Clin Microbiol Infect Dis 13: 839–845. [DOI] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y-Y, Keilbaugh SA et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334: 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Galperin MY. (2013). A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol 15: 2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.