Abstract
The phyllosphere, which is defined as the parts of terrestrial plants above the ground, is a large habitat for different microorganisms that show a high extent of adaption to their environment. A number of hypotheses were generated by culture-independent functional genomics studies to explain the competitiveness of specialized bacteria in the phyllosphere. In contrast, in situ data at the metabolome level as a function of bacterial colonization are lacking. Here, we aimed to obtain new insights into the metabolic interplay between host and epiphytes upon colonization of Arabidopsis thaliana leaves in a controlled laboratory setting using environmental metabolomics approaches. Quantitative nuclear magnetic resonance (NMR) and imaging high-resolution mass spectrometry (IMS) methods were used to identify Arabidopsis leaf surface compounds and their possible involvement in the epiphytic lifestyle by relative changes in compound pools. The dominant carbohydrates on the leaf surfaces were sucrose, fructose and glucose. These sugars were significantly and specifically altered after epiphytic leaf colonization by the organoheterotroph Sphingomonas melonis or the phytopathogen Pseudomonas syringae pv. tomato, but only to a minor extent by the methylotroph Methylobacterium extorquens. In addition to carbohydrates, IMS revealed surprising alterations in arginine metabolism and phytoalexin biosynthesis that were dependent on the presence of bacteria, which might reflect the consequences of bacterial activity and the recognition of not only pathogens but also commensals by the plant. These results highlight the power of environmental metabolomics to aid in elucidating the molecular basis underlying plant–epiphyte interactions in situ.
Introduction
The vast extent of leaf surfaces corresponds to the equivalent of more than 10 times of the Earth's vegetated surface area (Lambin and Meyfroidt, 2011) and represents a habitat for distinct microbial communities, a large part of which are Proteobacteria (for example, species of Rhizobiales and Sphingomonadales), Actinobacteria and Bacteroidetes (Vorholt, 2012). Major community selection factors are substrate and water availability, which drive the co-existence of bacteria that have complementary niches (Wilson and Lindow, 1994; Lindow and Brandl, 2003). The primary source of nutrients on the phylloplane are plant photo-assimilates that diffuse through the hydrophobic cuticle lining the leaf epidermal cell layer (van der Wal and Leveau, 2011). In addition, epiphytes counteract physicochemical stressors such as UV, salt and temperature, among others, which are sensed by bacterial stress response systems (Gourion et al., 2008; Kaczmarczyk et al., 2014). These regulatory components and assimilatory enzymes attributable to the oligotrophy of the phyllosphere have been revealed to be epiphytic fitness factors by several culture-independent functional genomics studies (Delmotte et al., 2009; Atamna-Ismaeel et al., 2012; Goudeau et al., 2013; Knief, 2014). Bacteria lacking such genetic prerequisites are quickly outcompeted in this highly variable and mostly short-lived habitat (Vorholt, 2012), which is exemplified by the fact that the epiphytic community composition shows a certain degree of consistency among individuals of the same plant species under field conditions (Knief et al., 2010b).
Earlier studies characterized potential leaf surface substrates with an emphasis on phytopathological relevance in maize, leek, bean, tobacco and poplar under greenhouse or field conditions. A number of carbohydrates, lipids and amino acids were detected in varying quantities using mostly gas or liquid chromatography and enzymatic assays (Tukey, 1970; Fiala et al., 1990; Mercier and Lindow, 2000). In addition, plant epidermal cell type-specific metabolic profiling as well as volatile organic compounds emitted through plant stomata have been measured using mass spectrometry (Steeghs et al., 2004; Ebert et al., 2010), the most prominent being methanol produced as a byproduct of pectin biosynthesis (Folkers et al., 2008). Other studies focused on the depletion of carbohydrates or volatile organic compounds due to bacterial colonization alone or in combination with in planta microscopy of fluorescent reporter strains to show the activity of regulatory genetic elements associated with the assimilation of fructose or one-carbon compounds for example (Joyner and Lindow, 2000; Leveau and Lindow, 2001; Abanda-Nkpwatt et al., 2006; Kawaguchi et al., 2011). The resulting spatial patterns were used to corroborate the hypothesis that leaf surface compounds are heterogeneously distributed depending on the degree of cuticle wetting and micro-topography and that a significant portion may therefore not be available to phyllosphere microorganisms. Consequently, individual leaves are unequally colonized, with most bacteria residing in epidermal grooves and near stomata or trichomes (Monier and Lindow, 2003; Remus-Emsermann et al., 2012, 2014).
The experiments described above significantly contributed to our understanding of bacterial physiology in the phyllosphere and helped to form new hypotheses regarding the distinct biological processes observed in this habitat. Metagenome and metaproteome information is a prerequisite for a systems-level description of epiphyte in situ physiology, but is per se not sufficient to adequately explain the observable microbial phenotypes described above. Bacterial bioreporters allow one or a few metabolites or physicochemical parameters to be detected, which must be experimentally validated for their specificity and sensitivity, and require the presence of the bacteria at the area of interest. Consequently, direct evidence of potential substrate assimilation by epiphytes is limited to a small number of bacteria and compounds. Moreover, it has not been shown which secondary plant metabolites reach the leaf cuticle or accumulate there in response to bacterial colonization, which is in contrast to the detection of signaling compounds inside plants as a consequence of bacterial presence (Ryu et al., 2004; Effantin et al., 2011; Pieterse et al., 2014).
In this study, we aimed to apply an environmental metabolomics approach to unravel the consequences of bacterial colonization on the phylloplane at the metabolite level, that is, the metabolic footprint (see Supplementary Figure S1). A reductionist laboratory setting for controlled Arabidopsis thaliana cultivation was used to inoculate plants with well-characterized epiphytic model strains for subsequent colonization, resulting in different metabolic modes and various types of plant–microbe interactions. We included the methylotroph Methylobacterium extorquens PA1 (Knief et al., 2010a) and the organoheterotroph Sphingomonas melonis Fr1 (Innerebner et al., 2011), which are both plant commensals, members of the Alphaproteobacteria and representatives of species ubiquitously and abundantly found in phyllosphere microbiota according to environmental studies (Vorholt, 2012). Both strains have been shown to colonize endophytic sites but at more than ten-fold lower cell counts, which may suggest that they thrive on substrates and/or at physicochemical conditions found on the phylloplane (Sy et al., 2005; Innerebner et al., 2011). Pseudomonas spp. are also regularly found in the phyllosphere of Arabidopsis, although their numbers appear to be variable (Delmotte et al., 2009; Bodenhausen et al., 2013; Reisberg et al., 2013). Pseudomonas syringae pv. tomato DC3000 is the best-studied bacterial phytopathogen model (Cuppels, 1986; Mansfield et al., 2012) and has been intensively investigated for its effector repertoire allowing it to manipulate the plant host and overcome plant defenses once the bacterium reaches the apoplast (Lindeberg et al., 2012; Wessling et al., 2014). Another reason to include the strain in this study was because insufficient in situ physiological data on its initial epiphytic growth phase currently exist. This is in contrast to known metabolic adaptions to leaf interior sites in subsequent infection steps, which have been studied by others (Beattie and Lindow, 1999; Rico and Preston, 2008; Rico et al., 2011). Our analytical rationale was to achieve ample coverage, sensitivity and robustness required to monitor a phylloplane-specific plant metabolome (that is, exometabolome). For this purpose, we combined quantitative nuclear magnetic resonance (NMR) for absolute quantification in leaf surface washes and imaging high-resolution mass spectrometry (IMS) as direct (that is, in situ) phylloplane compound measurements with spatial information (Grivet et al., 2003; Phelan et al., 2012). The present study showed striking differences in phylloplane carbohydrate levels and plant secondary metabolism in dependence on bacterial colonization, which provide valuable information on the bacterial lifestyle in the phyllosphere.
Materials and methods
Plant cultivations and inoculation with epiphytes
A. thaliana ecotype Col-0 was gnotobiotically grown in microboxes (Combiness, Nazareth, Belgium) as described elsewhere (Innerebner et al., 2011). In brief, plant seeds were surface-sterilized with 70% ethanol and 7% NaOCl and placed onto plant agar (Duchefa, Haarlem, the Netherlands). Sown seeds were inoculated with 5μl bacterial suspension corresponding to 1.25 × 106 colony forming units (CFU) in 10 mM MgCl2 for plants to be colonized with M. extorquens PA1 (Knief et al., 2010a) or S. melonis Fr1 (Innerebner et al., 2011). All bacterial cultures were grown in liquid minimal medium published by Kiefer et al. (2009) and Peyraud et al. (2009), supplemented with 27 mM D-glucose ('microbiology grade', Sigma-Aldrich, Buchs, Switzerland) and 125 mM methanol (LC-MS grade, Fisher Scientific, Loughborough, Leicestershire, UK) at pH 6.5 in 100 ml baffled shake flasks, inoculated directly from cryo-stocks. Germination was allowed in growth chambers (Percival, Perry, IA, USA) under long day conditions (16 h photoperiod at 25 °C day-temperature and 22 °C night-temperature) for 1 week before switching to a short day (9 h photoperiod) regime until harvest. Pseudomonas syringae pv. tomato DC3000 (Cuppels, 1986) infection was performed by nebulizer mediated spraying of a 105 CFU/ml infection inoculum in 10 mM MgCl2 on three-week-old plants.
Leaf washes
After 4 weeks, plants were removed from microboxes by detaching the roots with a scalpel, and abaxial leaf surfaces were washed extensively with approximately 1 ml 5% (v/v) methanol in ultra-pure water for seven to eight plants per sample using a mechanical pipette (which corresponds to up to 100 leaves). The collected leaf wash was centrifuged at 8000 g for 5 min, and the supernatant was frozen immediately in liquid nitrogen and stored at −80 °C. Bacterial colonization densities were determined by epiphyte harvest of the plant material used for leaf washings, involving rigorous shaking in phosphate-buffered Silwet L-77 (GE Bayer Silicones, Leverkusen, Germany) solution and dilution plating on agar containing the minimal medium described above following published protocols (Innerebner et al., 2011). The plant material used for surface washing and epiphyte harvesting was removed from the harvest buffer and dried under vacuum ('SpeedVac', Savant Instruments, Farmingdale, NY, USA) overnight followed by continued drying at 80 °C for 2 days to determine its dry weight.
NMR identification and quantification of phylloplane compounds
1H-MMR spectra were recorded on a Bruker Avance III 800, 13 MHz instrument (Bruker, Bremen, Germany), equipped with a 5 mm cryoprobe QPCI (1H, 13C, 31P and 15N). Phylloplane washes (120 μl) were mixed with 40 μl of a 1 mM 2-(trimethylsilyl)propionic-2,2,3,3-d4 acid (TSP-d4) solution in D2O as an internal intensity and chemical shift standard without further sample pretreatment for metabolite quantification. Measurements were performed at 286 K with 64 scans per spectrum and a flip angle of 30° (d1: 15 s, TD: 128k and presaturation of the 1H2O signal - O1P: 4700 p.p.m.). Basic analysis of the acquired spectra and peak integrations were performed using the Bruker 'TopSpin' software suit. Unpaired t-tests were performed to check for statistical significance among different treatments, after normal distribution of the measured metabolite quantities was verified by QQ-plotting. Where ratios of average metabolite pool values from independent sets of samples were calculated, standard deviations were derived using error propagation (that is, due to the destructive nature of the leaf wash extraction procedure reported here).
IMS of phylloplane compounds
Direct, spatially resolved mass spectrometric detection of phylloplane compounds was achieved by applying IMS to gnotobiotically inoculated A. thaliana plant leaves. IMS was performed using matrix-assisted laser desorption ionization-mediated Fourier-transform mass spectrometry (MALDI-FTMS) in tissue-imaging mode operation. This was performed by coating leaf surfaces immobilized to stainless steel target plates with a conductive, double-sided adhesive tape (Plano GmbH, Wetzlar, Germany), with 'universal MALDI matrix' (Sigma-Aldrich) dissolved in methanol with approximately 5–10 mg per microscope slide-sized plate using a robotic sprayer ('SunCollect', Sunchrome, Friedrichsdorf, Germany). Such plant leaf preparations were measured with a MALDI 'LTQ-Orbitrap' (Thermo Fisher Scientific GmbH, Bremen, Germany) instrument in positive ionization mode in a mass range between 100 and 1000 m/z with a resolution of 100 000 at 400 m/z and 2 microscans per step. The spatial resolution was set to 100 μm, and the UV-laser energy was 30 μJ. Basic raw MALDI-IMS data set processing and visualizations were performed with 'Spectviewer' (imzml.org, Schramm et al. (2012)).
For spatially resolved mass spectrometric fragmentation of phylloplane analytes, leaf surfaces were analyzed analogously to the method described above; excised Arabidopsis leaves were immobilized on a standard microscope slide with double-sided adhesive tape. The analysis was performed at ambient conditions using a 'Flowprobe' in situ micro-extraction system (Prosolia, Indianapolis, IN, USA), which was hyphenated via an electrospray ionization source to a Thermo Scientific 'QExactive' FTMS in positive and negative mode. The spray voltage was set to 3 kV with a capillary temperature of 150 °C. The solvent used was 5% methanol with a flow-rate of 10 μl per minute. Leaf surfaces were measured in adjacent, one-mm-wide lanes with a probe-speed of 50 μm per second. Chemical entities were monitored in a mass range between 100 and 1000 m/z at a resolution of 70 000 at 400 m/z with data-dependent fragmentation of 10 precursors per MS1 scan at a resolution of 17 500 at 400 m/z. All raw data including extracted peak lists and fragment spectra were made publicly available under the following link: ftp://MSV000078981@massive.ucsd.edu (password: a).
MS-derived metabolome data analysis
Downstream data processing and analysis, Flowprobe-FTMS data, mass slice image visualization and summed spectra extraction from regions of interest were performed with custom-built metabolome analysis pipelines based on the open-source MS data analysis framework 'emzed' (version 1.3, http://emzed.ethz.ch, Kiefer et al. (2013)). Raw data processing included post-acquisition m/z shift correction, basic isotope clustering, and satellite and background peak removal. For metabolite identification via accurate molecular mass determination (⩽8 p.p.m. mass error), a subset of different public databases (that is, AraCyc 11.5, Kegg (Kyoto Encyclopedia of Genes and Genomes), LipidMaps (LIPID Metabolites and Pathways Strategy) and Knapsack) was compiled to provide a probabilistic phylloplane compound solution space. Fragment spectra were matched and/or predicted using the mass bank online applications (http://www.massbank.jp, Horai et al. (2010)). As an untargeted analysis, pairwise Euclidean and Bray-Curtis distances between mass spectrometric imaging data sets of individual plant treatments were calculated using whole leaves as the region of interest. For increased information value, mass features were filtered globally (that is, over all samples) for a minimum level of reproducibility (for example, detection in all replicates of at least one treatment). All intensity values were total ion current- and log-normalized.
Whole leaf epifluorescence microscopy
Epifluorescence microscopy of whole A. thaliana leaves colonized with reporter strain variants having chromosomally integrated fluorophore genes of the epiphyte model strains listed above was performed on a Leica DMI6000B (Leica Microsystems GmbH, Wetzlar, Germany) inverted microscope equipped with a robotic stage. S. melonis Fr1 mCherry (Remus-Emsermann and Vorholt, unpublished) was analyzed with a Cy3 filter and P. syringae pv. tomato DC3000 YFP (Remus-Emsermann and Vorholt, unpublished) and M. extorquens PA1 5xGFP (wild-type PA1 background transformed with pBRI-gfp5 published in Choi et al. (2006)), were visualized with a broad GFP filter. The MALDI slide preparations for phylloplane IMS described above was used for microscopic imaging prior to spray coating with MALDI matrix and mass spectrometric measurements. Epifluorescent image overlays with MALDI-MS-based mass slice images were performed using 'GNU image manipulation program' (www.gimp.org).
Results and discussion
Phylloplane carbohydrates detected by NMR
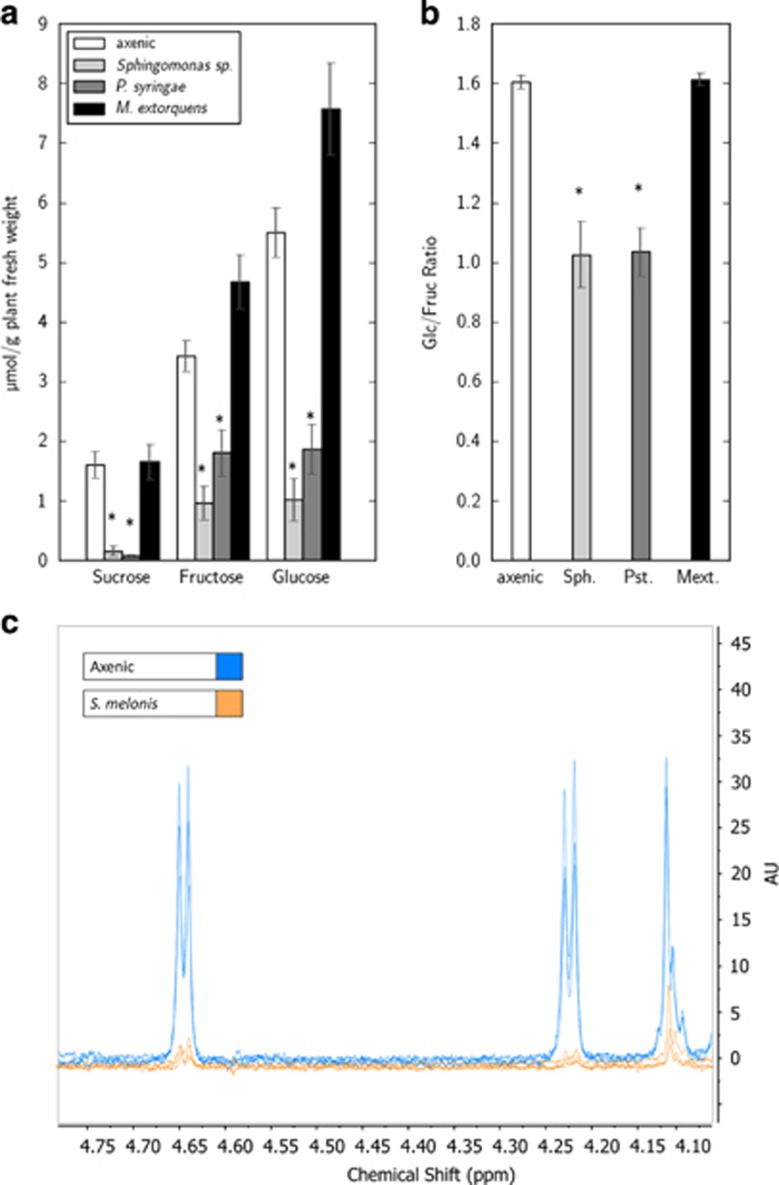
To detect the dominant phylloplane compounds as a function of bacterial colonization, leaf washes were made from axenic plants and plants inoculated with the model strains M. extorquens PA1, S. melonis Fr1 and P. syringae pv. tomato DC3000. 1H-NMR was used as a quantitative method to identify and quantify major compounds from the phylloplane without sample-pretreatment other than the removal of solid particles by centrifugation and the addition of an internal standard. Because we expected the sugar yields in our leaf washes to be low, we optimized our analytical strategy for improved sensitivity. We found that pooling approximately seven plants per sample gave peaks in the carbohydrate region with a signal-to-noise ratio amenable to integration (Figure 1c). The dominant sugars were found to be glucose, fructose and sucrose under all conditions tested. Other high-intensity peaks could be attributed to volatile organic compounds, namely acetate, ethanol and a volatile ketone (putatively acetone). To further improve the sensitivity of our method, the samples were concentrated by evaporation and measured with 1H-NMR with a larger number of scans using a micro-cryoprobe. In that way, additional compounds such as amino acids, oligosaccharides and inositol could be detected, for which the high signal density in the carbohydrate region masked peaks or precluded their integration. Quantification of the carbohydrate peaks showed that the dominant phylloplane sugars were present in average concentrations of 3–8 μmol per gram plant fresh weight for the monosaccharides glucose and fructose and 1–2 μmol per gram plant fresh weight for sucrose (Figure 1a), which corresponds to 40–100 nmol per cm2 leaf area and 14–28 nmol per cm2 leaf area, respectively. These values obtained from our model system are an order of magnitude higher than previously reported sugar quantities on surfaces of other plant species (Fiala et al., 1990; Mercier and Lindow, 2000), which can likely be explained by the thin cuticle of A. thaliana compared with that of the other plants (Schreiber and Schönherr, 2009). When comparing glucose, fructose and sucrose concentrations across the different bacterial inoculation treatments, a decrease in samples of epiphytes with glycolytic capacity (that is, P. syringae and S. melonis) becomes apparent. The most striking decrease in concentration was observed with sucrose with a difference of an order of magnitude between both P. syringae or S. melonis and samples without such bacterial glycolytic capacity (that is, M. extorquens or axenic plants), although the quantities never decreased below the limit of detection. In samples from leaves colonized by M. extorquens, which lacks the ability to metabolize these monosaccharides and disaccharides (Knief et al., 2010a), sugar depletion was not observed (Figure 1a). On the contrary, the sugar levels appeared to be generally elevated, but only with marginal significance (that is, t-test, 0.05⩽P-value⩽0.06). Notably, the ratio of the individual monosaccharides changed as a function of bacterial colonization. While glucose and fructose were in the same range in the samples obtained from plants colonized by glycolytic epiphytes, the levels of glucose were 1.5-fold above fructose levels in axenic controls and M. extorquens leaf wash samples (Figure 1b), which suggests bacterial co-consumption of monosaccharides with a preference for glucose. However, this phenomenon could also be attributed to an indirect, plant-mediated effect. The latter could either be a cellular regulatory response of the plant host to glycolytic bacterial overgrowth (for example, regulation of its starch content) or a compound-specific alteration in diffusion across the cuticle because of bacterial surfactants (Burch et al., 2011), if one assumes that the main source of monosaccharides on the leaf surface stems from sucrose spilt by plant cell wall-bound invertase or sucrose synthase (Sonnewald et al., 1991). Similarly, the small increase in monosaccharides on M. extorquens-colonized plant surfaces compared with axenic controls might be a consequence of physicochemical surface modification by the non-glycolytic bacterial colonization (for example, extracellular cellulose production), which is a wide-spread evolutionary adaption of natural phyllosphere microbiota. M. extorquens-colonized plants may therefore serve as a better control for sugar pools in a system without bacterial glycolytic activity than axenic plants. All these findings were confirmed in a second independent experiment, except for the decrease of the monosaccharides in P. syringae-infected samples compared with axenic controls (Supplementary Figure S2).
Figure 1.
Substrate quantities as determined by NMR in dependence on bacterial colonizations. Absolute quantities are shown in panel (a). Monosaccharide pool ratios given in panel (b) show a clear alteration by leaf colonization of potentially glycolytic strains (calculated as the average of ratios of all samples of a treatment individually). Abbreviations used here and typical colonization density reached at the time of experiments (CFU per gram plant fresh weight): Sph.: S. melonis Fr1 (2 × 108), Pst.: P. syringae pv. tomato DC3000 (5 × 108), Mext.: M. extorquens PA1 (8 × 107). Bars represent the mean of at least four replicates each with standard errors of the mean shown as error bars. Asterisks signify t-test-derived P-values below 0.05. Panel (c) shows exemplary 1H-NMR spectra excerpts from three axenic (blue) and S. melonis (yellow) phylloplane wash extracts, with clearly visible anomeric proton peaks of glucose (doublet at 4.62 p.p.m.) and sucrose (doublet at 4.22 p.p.m.). The rightmost quadruplet belongs to fructose.
The data described above were generated from plants that were sampled during the daytime. To investigate whether diurnal phylloplane nutrient fluctuations were influenced by bacterial colonization, we performed the leaf wash harvest experiment during the nighttime. When comparing phylloplane sucrose pools on axenic plants at night with those at the end of the day, larger amounts were detected at the end of the day compared with night (Supplementary Figure S3), which has also been postulated by others but without taking bacterial colonization into account (Fiala et al., 1990). Interestingly, the accumulation of sugars during the day appears to be diminished when the leaf surface is colonized by glycolytic epiphytes (Supplementary Figure S3, right plot). Because intracellular sugar concentrations are lower at night (Gibon et al., 2006), these results may indicate diffusion from the leaf surface back into interior compartments of the leaf, which might be diminished by the assimilation activity of glycolytic epiphytes. In view of a diurnal change of substrate availability, regulation of the bacterial metabolism might occur which could be influenced by both plant signals and bacterial light sensors.
IMS reveals the heterogeneous distribution of phylloplane compounds
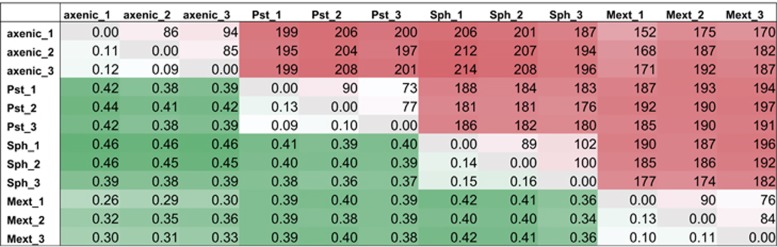
The NMR-based analytical workflow described above—with all of its analytical power—does not capture biological variation, which is smaller in leaf area than the sample size of several pooled plants. Because epiphyte in situ physiology can involve habitat heterogeneity at or above the micrometer scale (Lindow and Leveau, 2002), we chose to complement our environmental metabolomics approach with phylloplane IMS to monitor the spatial distribution of plant leaf surface compounds. Using MALDI-Orbitrap MS, we recorded IMS data sets from matrix-coated leaves at the small molecule mass range (100–1000 m/z) with a 100 μm raster size. Although the scan density did not completely resolve small anatomic sub-structures such as stomata or small microbial aggregates, it was sufficient to probe the important topographical regions of a whole leaf, such as vessels or pathogen-elicited lesions. Mass slice image generation confirmed that MALDI-based phylloplane IMS allowed us to resolve the spatial distribution of small molecules with high ion counts for abundant, well-ionizable plant metabolites (see below). After various filtering steps described in the Material and methods section, an average of approximately 1000 mass features specifically found on the leaf surface area were detected for each colonization treatment, about half of which were found either in all conditions tested here or shared among axenic plants and after M. extorquens colonization. Pairwise distance analyses on these data sets using multiple metrics showed that, first, the A. thaliana epiphyte colonization treatment applied here resulted in a reproducible perturbation that could be captured by phylloplane MALDI-IMS, and, second, that Methylobacterium colonization was more similar to axenic conditions than S. melonis colonization and infection with P. syringae pv. tomato (Figure 2). This, together with a principal component analysis of the same data set shown in Supplementary Figure S4, corroborated the hypothesis that S. melonis occupies a phylloplane niche that has a larger overlap with the one of P. syringae pv. tomato compared with that of Methylobacteria for instance (Innerebner et al., 2011), and therefore may be in competition with the phytopathogen for growth-promoting substances. As mentioned above, the universally present mass features comprise a large part of the entire filtered data set. Compared with that, potential compounds that were specifically found in only one treatment were less common, and very rarely, a specific feature disappeared after a given treatment but was detectable in all others (Supplementary Figure S5). This pattern was also valid if only features for which a putative identification could be obtained with exact monoisotopic mass matching were compared. The latter subset comprised almost 20% of the filtered feature list.
Figure 2.
Pairwise distance matrix of individual plant colonization treatments in replicates derived from MALDI-FTMS phylloplane imaging data sets. The abbreviations used to label rows and columns are the same as in Figure 1. Color shading mark small to large distances, respectively, which means dark-shaded comparisons of colonization treatments deviate more from each other in terms of phylloplane exometabolome composition than light-shaded comparisons. Red-colored fields in the upper right half of the distance matrix are pairwise Euclidian distances. For the green-colored fields in the lower left half, a Bray-Curtis metric was used. Permanova testing confirmed, that the individual treatments are significantly different from each other, with both Euclidian and Bray-Curtis metrics (i.e., P-value<0.001, using 1000 permutations).
In addition to the analytic workflow described above, we undertook additional efforts to improve the identification of known or unknown candidate metabolites. We established in situ micro-scale solvent extraction hyphenated to mass spectrometry (Flowprobe-Orbitrap MS2), which can be used to probe the same phyllosphere model system. In that manner, all the candidate metabolites further discussed below have also been confirmed by fragment spectral matching using public databases or were at least putatively predicted by substructure analysis if no standard fragment spectra had been published (Supplementary Figure S8).
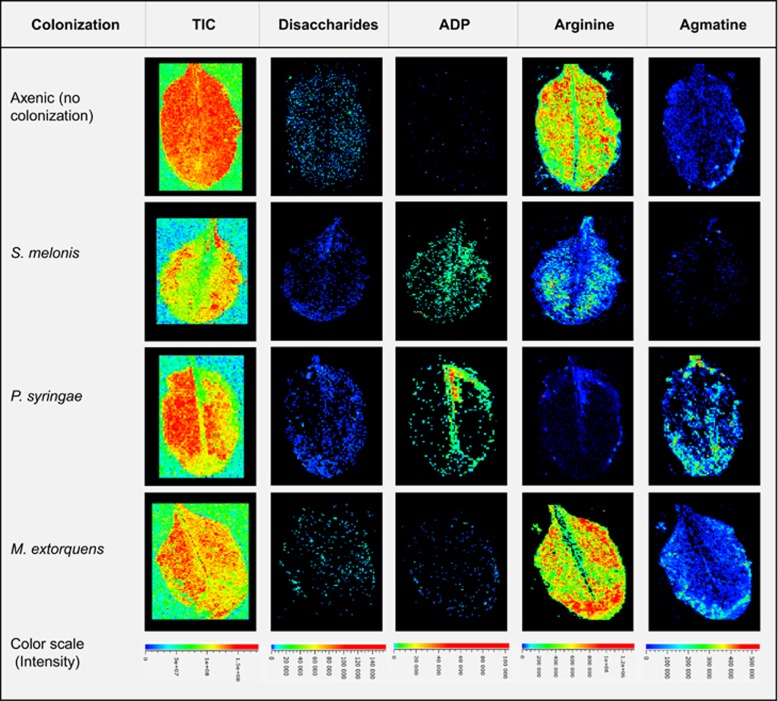
As described above, heterogeneous distribution of various phylloplane compounds could be shown by mass slice image generation, which indicated treatment-specific ion count differences and spatial characteristics. Exemplary images are shown in Figure 3 for disaccharides (based on the NMR results, this should be mainly sucrose), adenosine diphosphate (ADP), arginine and agmatine. The mass slice image for disaccharides shows a patchy distribution across the leaf surface with localized high-intensity regions. Regions with high relative intensity, could also be found along leaf vessels or edges on leaf surfaces of all treatments. On the other hand, S. melonis- and P. syringae-colonized leaf samples show a more equally distributed disaccharides signal albeit at lower relative intensities indicative of sugar consumption by these epiphytes (see above).
Figure 3.
Examples of phylloplane mass slice images (MALDI-IMS) plus the total ion current image (TIC) in the left-hand image series, showing heterogeneous distributions of potential substrates and bacterial metabolites, with ion counts encoded by false colors. The color scales shown at the bottom have been unified for each metabolite image series (i.e., columns), but not for the individual treatments (i.e., rows).
Nucleotide signals on colonized leaves are co-localized with regions of high bacterial density
In addition to primary plant metabolites such as sugars, IMS showed clear nucleotide signals (for example, adenosine mono- and poly-phosphates) on leaf surfaces. Notably, however, these signals were only detected on colonized leaves, but not in axenic controls. To examine whether the signals are caused by bacteria, we imaged entire leaves colonized with fluorescent reporter strain variants of all three strains used here, with both epifluorescence microscopy to locate regions of main bacterial colonization and with MALDI-based IMS as described above. The epifluorescence image was then overlaid with the mass slice image for ADP to check for co-localization. Indeed, co-localizing macro-colony-shaped fluorescence signals and ADP-positive MALDI-MS scans could be observed, as shown in Supplementary Figure S6, indicating that nucleotide signals could be used as a proxy for principle epiphyte cell mass localization. For instance, the ADP signal of P. syringae does not extend into lesioned leaf area (see asterisk in Supplementary Figure S6), likely because the gross infectious microbial cell mass is located in the leaf interior. If the MALDI-MS nucleotide-phosphate signal was used to estimate bacterial density and the disaccharide signal for substrate presence at a given position on the leaf (which corresponds to a single scan of a MALDI-MS measurement), spatial correlations at the scale of the scan raster size can be investigated. Calculation of the scan-wise (that is, spatially resolved) sum of the average-normalized sucrose over ADP ratio resulted in a lower value for the potentially glycolytic strains (S. melonis and P. syringae) compared with the non-glycolytic M. extorquens (Supplementary Figure S7). This result is in accordance with the hypothesis that the ratio of the normalized substrate to nucleotide ion count (that is, relative to the respective average intensities) should be approximately one if there is no localized substrate depletion by the resident microbial cell mass and lower than one if the microbial cell mass depletes the present substrate locally by glycolytic activity. Additionally, we performed a statistical test for the observed difference, in which the measured disaccharide to nucleotide ratio of an individual sample was compared with the analogously calculated ratio from randomly joined substrate and nucleotide intensities derived from the entire measured area, which would correspond to random co-localization. This statistical approach confirmed the measured ratios for S. melonis and P. syringae colonized leaves to be lower than expected for random co-localization, but not for M. extorquens colonized leaves (Supplementary Figure S7b).
Colonization-dependent compound pool changes in central and secondary plant metabolism
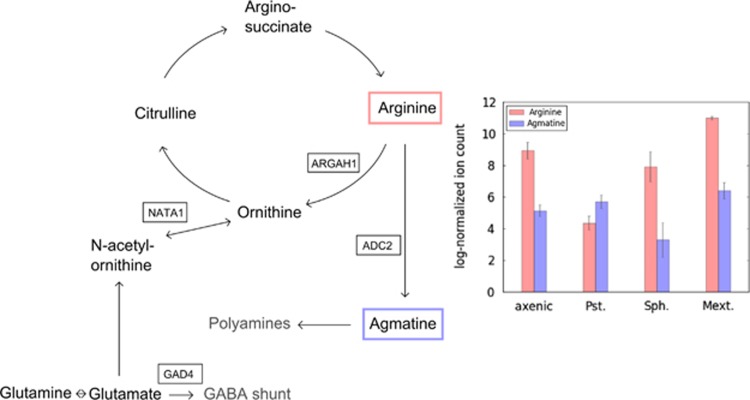
Striking differences in dependence on leaf inoculation were found for certain pathways of amino acid metabolism. In particular, intermediates of arginine degradation pathways showed high normalized ion count changes when individual treatments were compared; P. syringae infection led to a pronounced decrease in arginine with increased levels of agmatine when compared with axenic ion counts (Figure 4). This observation correlated with the transcript-levels of arginine amido-hydrolase (ARGAH1) and the stress-inducible arginine decarboxylase (ADC2), which were highly upregulated after A. thaliana infection with phytopathogenic pseudomonads in 'Genevestigator' (www.genevestigator.com, Zimmermann et al. (2008)). In addition, polyamine biosynthesis, for which agmatine is a precursor, has been shown to be highly beneficiary for the in planta performance of various epiphytes (Park et al., 2010; Ward et al., 2010). Notably, S. melonis Fr1-colonized leaves showed the largest arginine-to-agmatine ratio of all treatments, suggesting metabolic adaptations in agmatine metabolism of A. thaliana in response to Sphingomonas.
Figure 4.
Phylloplane compounds implicated in arginine metabolism, which show colonization specific pool changes as determined by IMS. Genes shown in boxes have been reported as strongly upregulated after P. syringae infection in literature (Zimmermann et al., 2008; Adio et al., 2011). The bar plot on the right hand side shows the varying levels of arginine and agmatine for all the colonization treatments. Bars represent the mean of three replicates each with standard errors of the mean shown as error bars. Abbreviations used here for the different bacterial strains are: Pst., P. syringae pv. tomato; Sph., S. melonis; Mext., M. extorquens.
Similarly, N-acetyl-ornithine could be fragmented using Flowprobe-MS2 with highest intensities on P. syringae-infected plants. Ornithine N-δ-acetyltransferase NATA-1 has been reported to be implicated in P. syringae infection of Arabidopsis (Adio et al., 2011), although its influence on pathogen physiology has not been fully elucidated yet. Note that, owing to direct injection mass spectrometry from the phylloplane conducted here (that is, without chromatographic separation and derivatization), the resulting fragment spectrum would be a mixture of N-α- and N-δ-acetyl-ornithine if the latter was indeed present. It is therefore no surprise that the fragment ion C6H15N2O+ for N-δ-acetyl-ornithine (published in Adio et al. (2011)) that was found with 10 milli mass units deviation from its theoretical mass was overshadowed by fragment ions belonging either to the ubiquitous N-α-acetyl-ornithine or to both mass isomers (Supplementary Figure S8).
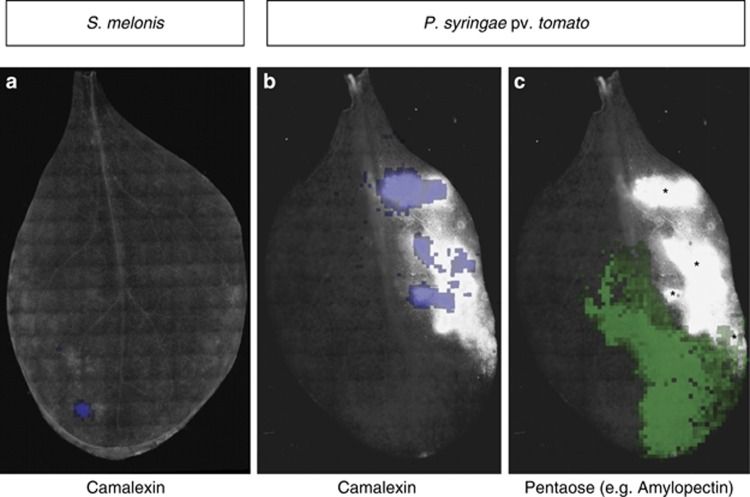
Oligosaccharides also showed treatment-specific ion count patterns (Figure 5). Pentaose levels were clearly upregulated after P. syringae infection compared with almost zero ion counts in axenic samples. Possible explanations for this phenomenon involve degradation of structural plant polysaccharides or upregulated enzymes of storage oligosaccharides, for example, the P. syringae-induced plant starch synthase (SS2), which increases amylopectin levels (although more efficiently at higher degrees of polymerization than pentaoses) (Zhang et al., 2008). Oligosaccharides have also been implicated in phytoalexin biosynthesis as a possible regulatory molecule in phytopathogen sensing (Glawischnig, 2007). Related to that, the secretion of camalexin was clearly detectable on P. syringae-treated plant leaf surfaces (Figure 5). Camalexin production in response to pathogen infection has been previously described (Schuhegger et al., 2006; Glawischnig, 2007; Chassot et al., 2008); here, we were able to visualize camalexin directly confined to pathogen-elicited lesions by imaging mass spectrometry in a spatially resolved manner. We noted that camalexin was not found on the entire lesioned area of a leaf with comparable intensity, and it is tempting to speculate that camalexin accumulated to highest levels in the lesions that had progressed the farthest. Camalexin is a growth inhibitor for a variety of phytopathogens, such as Gram-negative bacteria (Rogers et al., 1996), which is why it would be important to accelerate its production if periods of high bacterial inoculation frequency is at risk, by building a first line of chemical defense against phytopathogen infections. Low but detectable levels of camalexin were also found on some, but not all, S. melonis-colonized leaf surfaces, at loci that were devoid of lesions or other forms of apparent tissue damage (Figure 5a). Camalexin biosynthesis genes have been shown to be elicited in leaves by the commensal rhizobacterium Pseudomonas fluorescens SS101 (van de Mortel et al., 2012), with no significant expression in roots. Commensal-induced camalexin production at the site of bacterial colonization has not been previously demonstrated, but is interesting in view of the plant protection effect of S. melonis Fr1 against phytopathogens (Innerebner et al., 2011; Vogel et al., 2012), for which proof for the mode of action at the molecular level is currently lacking. It must be stressed that more data are needed to elucidate the mechanism of plant protection; however, S. melonis-elicited camalexin production might be one element involved in the reported plant protection from P. syringae pv. tomato infection.
Figure 5.
Overlays of whole leaf epifluorescent images of Arabidopsis leaves inoculated with the S. melonis mCherry or the P. syringae pv. tomato YFP reporter strain, analogous to the ones shown in Supplementary Figure S6. Asterisk in panel (c) (pentaose) indicate P. syringae-elicited lesions. The camalexin distribution is strictly limited to these regions on Pseudomonas infected leaves, but do not cover them completely with comparable concentrations (see panel b), whereas Sphingomonas-colonized leaves can show camalexin production in apparently healthy regions (see panel a). Pentaoses (shown in panel c) can be found in close proximity to lesions, but do not overlap with the latter.
Conclusions
In this study, we showed that the developed analytical workflows capture novel aspects of bacterial and plant physiology in the phyllosphere. Colonization of Arabidopsis leaf surfaces by bacterial model strains was found to have a profound effect on the phylloplane exometabolome, which is similar in magnitude to changes in a mammalian host–microbe interaction study (Wikoff et al., 2009). Some of these effects might be explained by conversion of plant-derived compounds for bacterial growth and includes carbohydrates and amino acids such as arginine. Other changes like the in situ production of phytoalexins in dependence on colonization treatments point toward a recognition of and response to the epiphyte, discriminating not only pathogens from commensals but also different commensal strains.
Acknowledgments
Whole-leaf epifluorescence microscopy was performed at the ScopeM facility (ETH Zurich) together with Justine Kusch. We thank Mitja N. P. Remus-Emsermann (ETH Zurich) for providing reporter strains, Jean-Pierre Both (CEA, Saclay, France) for MALDI-MS visualization tools and Christine Vogel (ETH Zurich) for assistance in statistical analysis. This work was supported by ETH Zurich (ETH-41 14-2).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abanda-Nkpwatt D, Müsch M, Tschiersch J, Boettner M, Schwab W. (2006). Molecular interaction between Methylobacterium extorquens and seedlings: growth promotion, methanol consumption, and localization of the methanol emission site. J Exp Bot 57: 4025–4032. [DOI] [PubMed] [Google Scholar]
- Adio AM, Casteel CL, De Vos M, Kim JH, Joshi V, Li BH et al. (2011). Biosynthesis and defensive function of N-δ-acetylornithine, a jasmonate-induced Arabidopsis metabolite. Plant Cell 23: 3303–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atamna-Ismaeel N, Finkel O, Glaser F, von Mering C, Vorholt JA, Koblížek M et al. (2012). Bacterial anoxygenic photosynthesis on plant leaf surfaces. Env Microbiol Rep 4: 209–216. [DOI] [PubMed] [Google Scholar]
- Beattie GA, Lindow SE. (1999). Bacterial colonization of leaves: a spectrum of strategies. Phytopathology 89: 353–359. [DOI] [PubMed] [Google Scholar]
- Bodenhausen N, Horton MW, Bergelson J. (2013). Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. Plos ONE 8: e56329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch AY, Browne PJ, Dunlap CA, Price NP, Lindow SE. (2011). Comparison of biosurfactant detection methods reveals hydrophobic surfactants and contact-regulated production. Environ Microbiol 13: 2681–2691. [DOI] [PubMed] [Google Scholar]
- Chassot C, Buchala A, Schoonbeek HJ, Metraux JP, Lamotte O. (2008). Wounding of Arabidopsis leaves causes a powerful but transient protection against Botrytis infection. Plant J 55: 555–567. [DOI] [PubMed] [Google Scholar]
- Choi YJ, Bourque D, Morel L, Groleau D, Míguez CB. (2006). Multicopy integration and expression of heterologous genes in Methylobacterium extorquens ATCC 55366. Appl Environ Microb 72: 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuppels DA. (1986). Generation and characterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato. Appl Environ Microbiol 51: 323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B, Schlapbach R et al. (2009). Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc Natl Acad Sci USA 106: 16428–16433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert B, Zöller D, Erban A, Fehrle I, Hartmann J, Niehl A et al. (2010). Metabolic profiling of Arabidopsis thaliana epidermal cells. J Exp Bot 61: 1321–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effantin G, Rivasseau C, Gromova M, Bligny R, Hugouvieux-Cotte-Pattat N. (2011). Massive production of butanediol during plant infection by phytopathogenic bacteria of the genera Dickeya and Pectobacterium. Mol Microbiol 82: 988–997. [DOI] [PubMed] [Google Scholar]
- Fiala V, Glad C, Martin M, Jolivet E, Derridj S. (1990). Occurrence of soluble carbohydrates on the phylloplane of maize (Zea mays L.): variations in relation to leaf heterogeneity and position on the plant. New Phytol 115: 609–615. [Google Scholar]
- Folkers A, Hüve K, Ammann C, Dindorf T, Kesselmeier J, Kleist E et al. (2008). Methanol emissions from deciduous tree species: dependence on temperature and light intensity. Plant Biology 10: 65–75. [DOI] [PubMed] [Google Scholar]
- Gibon Y, Usadel B, Blaesing OE, Kamlage B, Hoehne M, Trethewey R et al. (2006). Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol 7: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glawischnig E. (2007). Camalexin. Phytochemistry 68: 401–406. [DOI] [PubMed] [Google Scholar]
- Goudeau DM, Parker CT, Zhou YG, Sela S, Kroupitski Y, Brandl MT. (2013). The Salmonella transcriptome in lettuce and cilantro soft rot reveals a niche overlap with the animal host intestine. Appl Environ Microb 79: 250–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourion B, Francez-Charlot A, Vorholt JA. (2008). PhyR is involved in the general stress response of Methylobacterium extorquens AM1. J Bacteriol 190: 1027–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivet JP, Delort AM, Portais JC. (2003). NMR and microbiology: from physiology to metabolomics. Biochimie 85: 823–840. [DOI] [PubMed] [Google Scholar]
- Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K et al. (2010). MassBank: A public repository for sharing mass spectral data for life sciences. J Mass Spectrom 45: 703–714. [DOI] [PubMed] [Google Scholar]
- Innerebner G, Knief C, Vorholt JA. (2011). Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl Environ Microbiol 77: 3202–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner DC, Lindow SE. (2000). Heterogeneity of iron bioavailability on plants assessed with a whole-cell GFP-based bacterial biosensor. Microbiology 146: 2435–2445. [DOI] [PubMed] [Google Scholar]
- Kaczmarczyk A, Hochstrasser R, Vorholt JA, Francez-Charlot A. (2014). Complex two-component signaling regulates the general stress response in Alphaproteobacteria. Proc Natl Acad Sci USA 111: E5196–E5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi K, Yurimoto H, Oku M, Sakai Y. (2011). Yeast methylotrophy and autophagy in a methanol-oscillating environment on growing Arabidopsis thaliana leaves. Plos ONE 6: e25257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer P, Buchhaupt M, Christen P, Kaup B, Schrader J, Vorholt JA. (2009). Metabolite profiling uncovers plasmid-induced cobalt limitation under methylotrophic growth conditions. Plos ONE 4: e7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer P, Schmitt U, Vorholt JA. (2013). eMZed: An open source framework in python for rapid and interactive development of LC/MS data analysis workflows. Bioinformatics 29: 963–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knief C, Frances L, Vorholt JA. (2010. a. Competitiveness of diverse Methylobacterium strains in the phyllosphere of Arabidopsis thaliana and identification of representative models, including M. extorquens PA1. Microb Ecol 60: 440–452. [DOI] [PubMed] [Google Scholar]
- Knief C, Ramette A, Frances L, Alonso-Blanco C, Vorholt JA. (2010. b). Site and plant species are important determinants of the Methylobacterium community composition in the plant phyllosphere. ISME J 4: 719–728. [DOI] [PubMed] [Google Scholar]
- Knief C. (2014). Analysis of plant microbe interactions in the era of next generation sequencing technologies. Front Plant Sci 5: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambin EF, Meyfroidt P. (2011). Global land use change, economic globalization, and the looming land scarcity. Proc Natl Acad Sci USA 108: 3465–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveau JH, Lindow SE. (2001). Appetite of an epiphyte: Quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc Natl Acad Sci USA 98: 3446–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg M, Cunnac S, Collmer A. (2012). Pseudomonas syringae type III effector repertoires: last words in endless arguments. Trends Microbiol 20: 199–208. [DOI] [PubMed] [Google Scholar]
- Lindow SE, Leveau JH. (2002). Phyllosphere microbiology. Curr Opin Biotech 13: 238–243. [DOI] [PubMed] [Google Scholar]
- Lindow SE, Brandl MT. (2003). Microbiology of the phyllosphere. Appl Environ Microb 69: 1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13: 614–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier J, Lindow SE. (2000). Role of leaf surface sugars in colonization of plants by bacterial epiphytes. Appl Environ Microbiol 66: 369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier JM, Lindow SE. (2003). Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. Proc Natl Acad Sci USA 100: 15977–15982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Mirabella R, Bronstein PA, Preston GM, Haring MA, Lim CK et al. (2010). Mutations in γ-aminobutyric acid (GABA) transaminase genes in plants or Pseudomonas syringae reduce bacterial virulence. Plant J 64: 318–330. [DOI] [PubMed] [Google Scholar]
- Peyraud R, Kiefer P, Christen P, Massou S, Portais JC, Vorholt JA. (2009). Demonstration of the ethylmalonyl-CoA pathway by using 13C metabolomics. Proc Natl Acad Sci USA 106: 4846–4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan VV, Liu WT, Pogliano K, Dorrestein PC. (2012). Microbial metabolic exchange—the chemotype-to-phenotype link. Nat Chem Biol 8: 26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, Zamioudis C, Berendsen RL, Weller DM, Van Wees SC, Bakker PA. (2014). Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol 52: 347–375. [DOI] [PubMed] [Google Scholar]
- Reisberg EE, Hildebrandt U, Riederer M, Hentschel U. (2013). Distinct phyllosphere bacterial communities on Arabidopsis wax mutant leaves. Plos ONE 8: e78613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus-Emsermann MN, Tecon R, Kowalchuk GA, Leveau JHJ. (2012). Variation in local carrying capacity and the individual fate of bacterial colonizers in the phyllosphere. ISME J 6: 756–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus-Emsermann MN, Lücker S, Müller DB, Potthoff E, Daims H, Vorholt JA. (2014). Spatial distribution analyses of natural phyllosphere-colonizing bacteria on Arabidopsis thaliana revealed by fluorescence in situ hybridization. Environ Microbiol 16: 2329–2340. [DOI] [PubMed] [Google Scholar]
- Rico A, Preston GM. (2008). Pseudomonas syringae pv. tomato DC3000 uses constitutive and apoplast-induced nutrient assimilation pathways to catabolize nutrients that are abundant in the tomato apoplast. Mol Plant Microbe Interact 21: 269–282. [DOI] [PubMed] [Google Scholar]
- Rico A, McCraw SL, Preston GM. (2011). The metabolic interface between Pseudomonas syringae and plant cells. Curr Opin Microbiol 14: 31–38. [DOI] [PubMed] [Google Scholar]
- Rogers EE, Glazebrook J, Ausubel FN. (1996). Mode of action of the Arabidopsis thaliana phytoalexin camalexin and its role in Arabidopsis-pathogen interactions. Mol Plant Microbe Interact 9: 748–757. [DOI] [PubMed] [Google Scholar]
- Ryu CM, Farag MA, Hu CH, Reddy MS, Kloepper JW, Paré PW. (2004). Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol 134: 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm T, Hester A, Klinkert I, Both JP, Heeren RM, Brunelle A et al. (2012). imzML - A common data format for the flexible exchange and processing of mass spectrometry imaging data. J Proteomics 75: 5106–5110. [DOI] [PubMed] [Google Scholar]
- Schreiber L, Schönherr J. (2009) Water and Solute Permeability of Plant Cuticles: Measurement and Data Analysis. Springer: Berlin, New York. [Google Scholar]
- Schuhegger R, Nafisi M, Mansourova M, Petersen BL, Olsen CE, Svatoš A et al. (2006). CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol 141: 1248–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnewald U, Brauer M, Vonschaewen A, Stitt M, Willmitzer L. (1991). Transgenic tobacco plants expressing yeast-derived invertase in either the cytosol, vacuole or apoplast: a powerful tool for studying sucrose metabolism and sink/source interactions. Plant J 1: 95–106. [DOI] [PubMed] [Google Scholar]
- Steeghs M, Bais HP, de Gouw J, Goldan P, Kuster W, Northway M et al. (2004). Proton-transfer-reaction mass spectrometry as a new tool for real time analysis of root-secreted volatile organic compounds in Arabidopsis. Plant Physiol 135: 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy A, Timmers AC, Knief C, Vorholt JA. (2005). Methylotrophic metabolism is advantageous for Methylobacterium extorquens during colonization of Medicago truncatula under competitive conditions. Appl Environ Microb 71: 7245–7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey HB. (1970). Leaching of Substances from Plants. Ann Rev Plant Physio 21: 305. [Google Scholar]
- van de Mortel JE, de Vos RC, Dekkers E, Pineda A, Guillod L, Bouwmeester K et al. (2012). Metabolic and transcriptomic changes induced in Arabidopsis by the rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol 160: 2173–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wal A, Leveau JH. (2011). Modelling sugar diffusion across plant leaf cuticles: the effect of free water on substrate availability to phyllosphere bacteria. Environ Microbiol 13: 792–797. [DOI] [PubMed] [Google Scholar]
- Vogel C, Innerebner G, Zingg J, Guder J, Vorholt JA. (2012). Forward genetic in planta screen for identification of plant-protective traits of Sphingomonas sp strain Fr1 against Pseudomonas syringae DC3000. Appl Environ Microb 78: 5529–5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorholt JA. (2012). Microbial life in the phyllosphere. Nat Rev Microbiol 10: 828–840. [DOI] [PubMed] [Google Scholar]
- Ward JL, Forcat S, Beckmann M, Bennett M, Miller SJ, Baker JM et al. (2010). The metabolic transition during disease following infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Plant J 63: 443–457. [DOI] [PubMed] [Google Scholar]
- Wessling R, Epple P, Altmann S, He YJ, Yang L, Henz SR et al. (2014). Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe 16: 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC et al. (2009). Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA 106: 3698–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M, Lindow SE. (1994). Coexistence among epiphytic bacterial-populations mediated through nutritional resource partitioning. Appl Environ Microb 60: 4468–4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XL, Szydlowski N, Delvallé D, D'Hulst C, James MG, Myers AM. (2008). Overlapping functions of the starch synthases SSII and SSIII in amylopectin biosynthesis in Arabidopsis. BMC Plant Biol 8: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Laule O, Schmitz J, Hruz T, Bleuler S, Gruissem W. (2008). Genevestigator transcriptome meta-analysis and biomarker search using rice and barley gene expression databases. Mol Plant 1: 1088–1088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.