The production of extracellular membrane vesicles (hereafter called MVs) is a universal cellular feature, common to the three domains of life (Deatherage and Cookson, 2012). In particular, it has been known for decades that bacteria, including marine species, produce MVs in the laboratory but also in biofilms or during infections (Schooling and Beveridge, 2006; Deatherage and Cookson, 2012). However, until recently, the presence of MVs in natural environments has been largely overlooked by molecular ecologists. This attitude is probably going to change with a recent report in Science in which Biller et al. (2014) demonstrate the abundance of bacterial vesicles in marine ecosystems. They first show that several strains from the numerically dominant marine phytoplankter Prochlorococcus produce large amounts of MVs in the laboratory, suggesting that marine phototrophic bacteria also produce MVs in their natural environment. This hypothesis has been directly tested by examining two distinct ocean water samples for the presence of MVs. As expected, the authors succeeded to isolate abundant MVs from these two samples, with concentrations ranging from 105 to 106 vesicles ml−1 of sea water. This is similar to the lower range of concentrations reported for viral particles in the oceans (105 to 109 virus-like particles/ml; Suttle, 2007). It is therefore surprising that Biller et al. (2014) observed only negligible number of apparent tailed phages (or gene transfer agents (GTA)) in their vesicle-rich ocean samples even though the methods they used to isolate MVs were similar to those traditionally employed for the isolation of viral particles. Indeed, the dimensions, morphology and molecular composition of MVs are very similar to those of some virions (Forterre et al., 2013). This observation suggests that MVs could outnumber true viral particles in some marine environments.
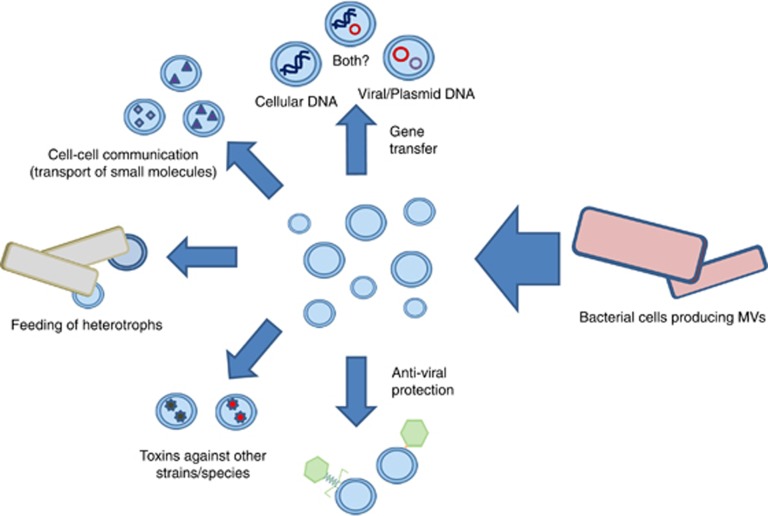
Biller et al. propose several roles for marine MVs, including cellular communication, gene transfer, phage resistance and carbon cycling (Figure 1). However, they do not consider another major consequence of their observation: the possible confusion of bacterial MVs with viral particles in ecological studies. We recently pointed out that MVs carrying DNA mimic viral particles in epifluorescence microscopy (EFM) analyses (Forterre et al., 2013). Indeed, we previously observed that MVs from hyperthermophilic archaea of the genus Thermococcus carrying cellular DNA produce fluorescent dots in EFM that cannot be distinguished from those attributed to viral particles (Soler et al., 2008). We predicted that bacterial MVs present in natural environments should also mimic viral particles in EFM studies as these MVs can also contain DNA (Yaron et al., 2000). Indeed, Hagemann et al. (2013) recently reported that MVs produced by two marine Proteobacteria, Ahrensia kielensis and Pseudoalteromonas marina, give bright spots in EFM after staining with SYBR Gold. Biller and colleagues also noticed the presence of DNA associated with MVs produced both in the laboratory by Prochlorococcus species and MVs isolated from ocean samples. Sequencing of the DNA associated with environmental MVs revealed sequences from several bacterial phyla, including Proteobacteria, Cyanobacteria and Bacteroidetes, confirming that diverse marine bacteria produce MVs containing DNA (Biller et al., 2014).
Figure 1.
Various possible physiological roles of bacterial membrane vesicles in the environment.
The presence of bacterial DNA in MVs isolated from ocean samples, together with the fact that MVs appear much more abundant than bona fide viral particles in these samples, validates our prediction that MVs present in various environments could lead to an overestimation of viral titers by EFM and even suggest the presence of viruses when there are none (Forterre et al., 2013). EFM has been widely used for the last two decades by microbial ecologists to quantify viral particles (Patel et al., 2007), leading to the conclusion that viruses are usually 10 times more abundant than bacteria in environmental samples and have major impact on global biogeochemical cycles (Danovaro et al., 2011). If MVs are indeed more abundant than true viral particles in natural environments, these conclusions should be reconsidered.
The two ocean samples studied by Biller and co-workers were possibly especially rich in MVs (or poor in true viruses). However, a recent analysis by Sullivan and co-workers suggests that this might not be the case. These authors noticed that non-tailed particles numerically dominate over tailed particles in 2600 ocean and sea water samples from six different sites all over the planet (Brum et al., 2013). In their analysis, non-tailed particles correspond to 50–90% of the total ‘viral-like particles' observed by electron microscopy. Importantly, they took all precautions to prevent tail loss during preparation and indicated that they never observed viral tails separated from capsids in any of the analyzed samples. The authors assume in their discussion that all observed non-tailed particles were of bona fide viral origin, produced by bacterial or eukaryotic hosts and belong to unknown viral families. However, many of the observed particles could correspond to MVs. Indeed, it is impossible to discriminate with confidence between bona fide tailless virions and MVs by electron microscopy.
All these data call for a revision of all quantitative studies dealing with enumeration of viruses in natural ecosystems, and introduction of new methods to evaluate their impact on global biogeochemical cycles. For instance, one of us has previously suggested focusing on new methods enumerating infected cells (virocells) corresponding to the active form of viruses, rather than viral particles that are passive vehicles of viral genome dissemination (Forterre, 2013) and can be confused with MVs. However, the identification of virocells in natural environments is not an easy task and can be a subject to various pitfalls (discussed in Forterre et al., 2013). In general, greater attention should be given to the preparation of samples: some environmental samples were treated with chloroform (Dinsdale et al., 2008), possibly eliminating MVs (along with some true viral particles), whereas such treatment was not performed in other analyses (keeping MVs within viral samples). However, the efficiency of such treatment to discriminate between MVs and bona fide viral particles remains to be evaluated and is also prone to various pitfalls (discussed in Forterre et al., 2013). Discriminating between virions and vesicles (some of them containing viral DNA, see below) appears to be a major challenge for future studies in viral ecology.
A priori, the presence of abundant MVs in marine environments should also be a problem for metagenomic analyses of viral communities as the methods used to enrich samples for viruses also enrich for MVs containing cellular DNA (Forterre et al., 2013). In fact, contamination of viromes by MVs could nicely explain the rather high percentage of bacterial reads in marine viromes (Kristensen et al., 2010; Roux et al. 2013). In a recent computational analysis of 67 viromes, Roux et al. (2013) found that most viromes were contaminated, albeit to a varying extent, with cellular sequences. However, when comparing microbiomes and viromes that are only mildly contaminated with cellular sequences, one can observe a significant enrichment of these viromes with viral sequences and sequences encoding for hypothetical proteins (Dinsdale et al., 2008; Kristensen et al., 2010; Roux et al., 2013; Enav et al., 2014). This suggests that cellular DNA associated to MVs might not be a major problem when analyzing marine viromes.
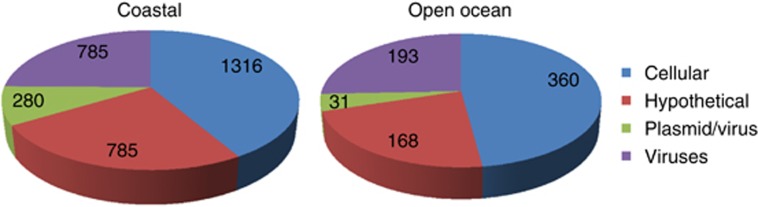
At first, the latter conclusion appears to contradict Biller and co-workers who assumed that most tailless particles in their ocean samples were MVs carrying cellular DNA, since only 4% to 8% of DNA associated to marine MVs could be clearly affiliated to tailed and other marine phages (Biller et al., 2014). However, we think that this inference probably underestimates the ‘contamination' of MV-associated DNA with sequences from viruses or other mobile elements, such as plasmids. Indeed, functional affiliations of presumably cellular proteins encoded by MV-associated DNA (Table S6 in Biller et al.) reveal very few ribosomal proteins (the hallmark of cellular organisms) but instead many sequences typical of viruses or plasmids, such as various capsid proteins, integrases, DNA methyltransferases or else plasmid replication proteins. Furthermore, a very high percentage of sequences with cellular affiliation encode for hypothetical proteins of unknown functions, another typical feature of viral genomes. According to our analysis, viral/plasmid DNA could represent from 30% to 50% of DNA from MVs in the two ocean samples analyzed by Biller and co-workers (Figure 2). These DNA sequences could have originated from genuine tailless viruses of unknown families observed by Brum et al. (2013). However, another (non exclusive) possibility to explain the presence of so much ‘viral' DNA in samples that do not contain viral particles visible by electron microscopy is that many MVs present in the ocean samples in fact contain viral (or plasmid) DNA instead of bona fide cellular DNA (Figure 1).
Figure 2.
Evaluation of the origin of MV-associated DNA sequences from coastal (left) and open ocean (right) samples. The original data on the best-blast hits and functional annotation of the DNA sequences recovered from marine MVs were obtained from the publication of Biller et al (2014). Sequences were categorized into four different groups according to the proteins they encode: (i) cellular proteins (bacterial, archaea and eukaryotic proteins that are rarely found in viruses and plasmids); (ii) hypothetical proteins (proteins with uncertain provenance; hypothetical proteins are typically enriched in mobile genetic elements); (iii) plasmid/viral proteins (sequences that have closest homologs in cellular organisms but are annotated as typical viral or plasmid proteins, such as structural components of virions, diverse methyltransferases, integrases, invertases, transposases and so on); (iv) viral proteins (encoded in genuine viral genomes).
In agreement with this possibility, it is well known that bacterial and archaeal MVs can be enriched in plasmid and/or viral DNA (Yaron et al., 2000; Renelli et al., 2004; Soler et al., 2011; Gaudin et al., 2014). Yaron et al. (2000) first detected phage DNA in MVs produced by Escherichia coli O157:H7. More recently, we found that MVs produced by the archaeon Thermococcus nautili (formerly T. nautilus) selectively incorporate the genome of a defective virus belonging to the PRD1/Adenovirus lineage as well as a small plasmid (Gaudin et al., 2014). Viral/plasmid genomes could have been directly packaged during the formation of MVs by unknown mechanisms or, in some case, injected directly into MVs in the course of ‘mistaken' infection (Biller et al., 2014). MVs containing either plasmid or viral genomes are distinct from true virions, because they do not contain proteins encoded by the viral genome, but a subset of cellular membrane proteins, testifying for their production by budding from the cell envelope (Gaudin et al., 2013). We suggest calling these unique biological entities viral membrane vesicles (vMV), considering that virus and plasmid sequence spaces often overlap (see Cortez et al., 2009; Koonin and Dolja, 2014).
vMVs that are built from cellular proteins but carry viral genomes or plasmids are the mirror image of GTA that are built from viral proteins but carry cellular genomes (Lang et al., 2012). However, similar to GTA, microbial MVs can be involved in the transfer of genetic material from cell to cell (Renelli et al., 2004; Velimirov and Hagemann, 2011; Gaudin et al., 2013). We have thus suggested that vMVs can serve as vehicles for the intercellular transport of viral genomes thereby facilitating recombination between viral, plasmid and/or cellular chromosomes (Gaudin et al., 2014). If a significant percentage of MVs present in the environmental samples corresponds in fact to vMVs, it is possible that vMVs represent major, presently unrecognized players in the gene fluxes between viruses and cellular organisms in the biosphere.
All these observations call for large-scale studies of MVs in various environments to fully understand their biological significance and complex interplay with viral particles. In eukaryotes, some virus-infected cells produce MVs harboring viral proteins and/or mRNA and microRNA (Pegtel et al., 2010; Meckes and Raab-Traub, 2011). We suspect that some human MVs (microparticles, ectosomes or exosomes) probably also carry viral genomes. It should be extremely important to verify if such vMVs indeed exist in eukaryotes, and if they can be vectors for the long distance transmission of viral genomes in multicellular organisms. Such mechanisms could help viral information escaping immune system in vertebrates.
Acknowledgments
PF was supported by an ERC grant from the European Union's Seventh Framework Programme (FP/2007-2013)/Project EVOMOBIL—ERC Grant Agreement no.340440.
The authors declare no conflict of interest.
References
- Biller SJ, Schubotz F, Roggensack SE, Thompson AW, Summons RE, Chisholm SW. (2014). Bacterial vesicles in marine ecosystems. Science 343: 183–186. [DOI] [PubMed] [Google Scholar]
- Brum R, Schenck RO, Sullivan MB. (2013). Global morphological analysis of marines viruses shows minimal regional variation and dominance of non-tailed viruses. ISME J 7: 1738–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Forterre P, Gribaldo S. (2009). A hidden reservoir of integrative elements is the major source of recently acquired foreign genes and ORFans in archaeal and bacterial genomes. Genome Biol 10: R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danovaro R, Corinaldesi C, Dell'anno A, Fuhrman JA, Middelburg JJ, Noble RT et al. (2011). Marine viruses and global climate change. FEMS Microbiol Rev 35: 993–1034. [DOI] [PubMed] [Google Scholar]
- Deatherage BL, Cookson BT. (2012). Membrane vesicle release in bacteria, eukaryotes, and archaea: A conserved yet underappreciated aspect of microbial life. Infect Immun 80: 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM et al. (2008). Functional metagenomic profiling of nine biomes. Nature 452: 629–632. [DOI] [PubMed] [Google Scholar]
- Enav H, Mandel-Gutfreund Y, Béjà O. (2014). Comparative genomic analyses reveal viral-induced shifts of host metabolism toward nucleotide biosynthesis. Microbiome 2: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P. (2013). The virocell concept and environmental microbiology. ISME J 7: 233–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P, Soler N, Krupovic M, Marguet E, Ackermann HW. (2013). Fake virus particles generated by fluorescence microscopy. Trends Microbiol 21: 1–5. [DOI] [PubMed] [Google Scholar]
- Gaudin M, Gauliard E, Schouten S, Houel-Renault L, Lenormand P, Marguet E et al. (2013). Hyperthermophilic archaea produce membrane vesicles that can transfer DNA. Environ Microbiol Rep 5: 109–116. [DOI] [PubMed] [Google Scholar]
- Gaudin M, Krupovic M, Marguet E, Gauliard E, Cvirkaite-Krupovic V, Le Cam E et al. (2014). Extracellular membrane vesicles harbouring viral genomes. Environ Microbiol 16: 1167–1175. [DOI] [PubMed] [Google Scholar]
- Hagemann S, Stöger L, Kappelmann M, Hassl I, Ellinger A, Velimirov B. (2013). DNA-bearing membrane vesicles produced by Ahrensia kielensis and Pseudoalteromonas marina. J Basic Microbiol doi:10.1002/jobm.201300376. [DOI] [PubMed]
- Koonin EV, Dolja VV. (2014). Virus world as an evolutionary network of viruses and capsidless selfish elements. Microbiol Mol Biol Rev 78: 278–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen DM, Mushegian AR, Dolja VV, Koonin EV. (2010). New dimensions of the virus world discovered through metagenomics. Trends Microbiol 18: 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AS, Zhaxybayeva O, Beatty JT. (2012). Gene transfer agents: phage-like elements of genetic exchange. Nat Rev Microbiol 10: 472–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckes Jr DG, Raab-Traub N. (2011). Microvesicles and viral infection. J Virol 85: 12844–12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Noble RT, Steele JA, Schwalbach MS, Hewson I, Fuhrman JA. (2007). Virus and prokaryote enumeration from planktonic aquatic environments by epifluorescence microscopy with SYBR Green I. Nat Protoc 2: 269–276. [DOI] [PubMed] [Google Scholar]
- Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA,. (2010) 107: 6328–6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renelli M, Matias V, Lo RY, Beveridge TJ. (2004). DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology 150: 2161–2169. [DOI] [PubMed] [Google Scholar]
- Roux S, Krupovic M, Debroas D, Forterre P, Enault F. (2013). Assessment of viral community functional potential from viral metagenomes may be hampered by contamination with cellular sequences. Open Biol 3: 130160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooling SR, Beveridge TJ. (2006). Membrane vesicles: an overlooked component of the matrices of biofilms. J Bacteriol 188: 5945–5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler N, Marguet E, Verbavatz JM, Forterre P. (2008). Virus-like vesicles and extracellular DNA produced by hyperthermophilic archaea of the order Thermococcales. Res Microbiol 159: 390–399. [DOI] [PubMed] [Google Scholar]
- Soler N, Gaudin M, Marguet E, Forterre P. (2011). Plasmids, viruses and virus-like membrane vesicles from Thermococcales. Biochem Soc Trans 39: 36–44. [DOI] [PubMed] [Google Scholar]
- Suttle CA. (2007). Marine viruses – major players in the global ecosystem. Nat Rev Microbiol 5: 801–812. [DOI] [PubMed] [Google Scholar]
- Velimirov B, Hagemann S. (2011). Mobilizable bacterial DNA packaged into membrane vesicles induces serial transduction. Mob Genet Elements 1: 80–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron S, Kolling GL, Simon L, Matthews KR. (2000). Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl Environ Microbiol 66: 4414–4420. [DOI] [PMC free article] [PubMed] [Google Scholar]