Abstract
A large soil CO2 pulse is associated with rewetting soils after the dry summer period under a Mediterranean-type climate, significantly contributing to grasslands' annual carbon budget. Rapid reactivation of soil heterotrophs and a pulse of available carbon are both required to fuel the CO2 pulse. Understanding of the effects of altered summer precipitation on the metabolic state of indigenous microorganisms may be important in predicting changes in carbon cycling. Here, we investigated the effects of extending winter rainfall into the normally dry summer period on soil microbial response to a controlled rewetting event, by following the present (DNA-based) and potentially active (rRNA-based) soil bacterial and fungal communities in intact soil cores (from a California annual grassland) previously subjected to three different precipitation patterns over 4 months (full summer dry season, extended wet season and absent dry season). Phylogenetic marker genes for bacteria and fungi were sequenced before and after rewetting, and the abundance of these genes and transcripts was measured. After having experienced markedly different antecedent water conditions, the potentially active bacterial communities showed a consistent wet-up response. We found a significant positive relation between the extent of change in the structure of the potentially active bacterial community and the magnitude of the CO2 pulse upon rewetting dry soils. We suggest that the duration of severe dry summer conditions characteristic of the Mediterranean climate is important in conditioning the response potential of the soil microbial community to wet-up as well as in framing the magnitude of the associated CO2 pulse.
Introduction
In systems that are characterised by fluctuating soil water availability over time, rewetting events trigger intense microbial activity that releases soil CO2 efflux pulses (Birch, 1958; Borken and Matzner, 2009; Inglima et al., 2009). For example, in Mediterranean climate grasslands, soil microbial activity includes a strong seasonal component that is related to the extreme soil desiccation during the summer followed by a wet fall and winter season (Waldrop and Firestone, 2006a; Cruz-Martinez et al., 2009). Carbon substrates accumulated during the dry hot summer period fuel a large microbial-driven mineralisation pulse upon soil rewetting, initiating the wet period of the annual cycle. The large soil CO2 efflux associated with this rewetting accounts for a large part of the annual C budget of these annual grasslands under Mediterranean climates (Xu et al., 2004; Jarvis et al., 2007).
The decoupling between C cycling and microbial community that takes place during dry periods is followed by an abrupt reconnection between soil microorganisms and C substrates upon rewetting. Our understanding of the processes underlying the dry–wet transition is based in part on its effects on the availability of the carbon consumed to fuel the CO2 pulse and in part on the capacity of the indigenous community to rapidly reactivate and resume full metabolic activity. Although there has been substantial interest in identifying the C sources consumed upon rewetting (Miller et al., 2005; Xiang et al., 2008; Boot, 2011; Parker and Schimel, 2011; Kakumanu et al., 2013), that question is to a large degree distinct from that of understanding the physiological state of the soil community before and during the wet-up process. From the microbial activity point of view, microbial communities indigenous to California annual grasslands have been shown to react extremely rapidly to rewetting (Fierer and Schimel, 2003; Placella et al., 2012; Barnard et al., 2013). In a recent study, we found that different bacterial phyla responded differently to rewetting in California grasslands, displaying contrasting life strategies (Barnard et al., 2013), resulting from long-term evolutionary adaptation.
The response of soil communities to changes in water resources after exposure to altered climatic conditions has recently been the topic of intense interest (Bouskill et al., 2012; Evans and Wallenstein, 2012; de Vries et al., 2012b; Evans and Wallenstein, 2013; Evans et al., 2014). Climate change models predict changes in precipitation patterns for California, with the wet winter season extending further into spring (Field et al., 1999; Suttle et al., 2007). Several studies have shown an effect of climate change scenarios, including altered precipitation, on the structure and function of microbial communities in soils under a Mediterranean climate (Fierer and Schimel, 2002, 2003; Fierer et al., 2003; Waldrop and Firestone, 2006b; Cruz-Martinez et al., 2009). Microbial communities in Mediterranean-type systems are the result of long-term evolutionary adaptation to an annual repeatable climatic pattern of wet fall season following a long period of soil desiccation during the summer. Drought, that is, water deficit that is outside the normally experienced environmental envelope, has different adaptative implications, since microbial communities experience it as a stochastic stress. Recent insights have been gained from studies of microbial community response to drought in a mesic continental climate ecosystems using BrdU approaches to assess bacterial response (Evans et al., 2014), as well as from pulsed rainfall events in arid and semi-arid systems (Angel and Conrad, 2013). Although Mediterranean systems are quite distinct from continental and arid systems, results from these three system types help construct a more general framework of the microbial response to changes in water availability. These results help us understand the importance of antecedent water conditions to soil microbes more generally but they do not directly assess the impacts of a changing annual pattern of summer desiccation in Mediterranean climates on indigenous communities highly adapted to a relatively regular annual climate pattern. The effects of changes in the duration of the summer dry period on the metabolic capacity of the microbial effectors of the rewetting mineralisation pulse remain unclear. A better understanding of the effects of antecedent conditions on the activity of the soil microbial community as it transitions from extreme desiccation to moist conditions may be crucial in predicting the future changes in carbon and nutrient cycling in Mediterranean climate grasslands, likely to result from changing patterns of precipitation.
The primary objective of the present study was to elucidate how changes in the length of the Mediterranean-type summer dry period before rewetting cause shifts in (a) bacterial and fungal communities and (b) the associated soil CO2 release. By analysing the soil ribosomal RNA (rRNA) pool, it is possible to identify which organisms are primed to synthesise proteins in response to rewetting and the substrates that thereby become available, and thus can be considered ‘potentially active' (Schippers et al., 2005, e.g. Jones and Lennon, 2010, see Blazewicz et al., 2013 for a review). Although these results do not establish a direct causal link between microbial metabolic state and the ensuing CO2 pulse, they allow phylogenetic characterisation of the potentially active bacterial and fungal groups that are present and respond to changing environmental conditions.
The present study investigated the wet-up response of the present and potentially active soil bacterial and fungal communities using intact soil cores taken from a California annual grassland and subjected to three different precipitation patterns over a 4 month summer dry-down. The precipitation patterns were (i) weekly water inputs, (ii) weekly water inputs for 2 months followed by no inputs or (iii) no water input for 4 months. Ribosomal RNA transcripts and genes from bacteria, fungi and archaea were extracted at the end of the summer period (before wet-up) and 2 h after wet-up, over which time soil CO2 efflux was monitored. Genes and transcripts from bacteria and fungi were analysed by 454 Titanium pyrosequencing, and the abundance of selected phylogenetic marker genes and transcripts was measured using quantitative PCR.
Materials and methods
Experimental setup
A homogeneous 10 × 10 m plot was selected in a California grassland (University of California Hopland Research and Extension Center, 39°0'N, 123°3'W, 400 m a.s.l.). This Sutherland series soil is a loam soil over clay (texture 46% sand, 35% silt, 19% clay), pH was 6.2, cation exchange capacity was 20.0 cmol kg−1, organic matter was 3.9%, total N 0.23%, total C 2.56%. The vegetation cover of the plot is grass, dominated by the introduced annual Avena barbata.
On 12 April 2010, 25 intact soil cores were taken by inserting polyvinyl chloride (PVC) tubes (7.5 cm internal diameter, 15 cm long) 15 cm deep in the soil, ∼1 m apart, and removing them carefully. The vegetation present on the cores was cut flush with the soil and subsequent regrowth was cut. The cores were transported to a greenhouse at University of California, Berkeley. The soils were subjected to three different spring-summer dry-down precipitation pattern treatments over 4 months: (i) weekly water inputs, (ii) weekly water inputs for 2 months followed by no inputs and (iii) no water input (closest approximation of normal). The cores were weighed weekly to monitor soil water content. The weekly water inputs were calculated for each core based on its weight, to compensate the water loss of the cores during the week preceding watering.
Soil cores were destructively sampled at the end of the treatment period (31 August 2010, four cores per treatment), and 2 h after controlled wet-up (1 September 2010, four other cores per treatment, see below for wet-up and soil sampling details). Total soil DNA and RNA were extracted (see below) and the present (DNA-based) and potentially active (RNA-based) soil bacterial and fungal communities were assessed by 454 pyrosequencing, and the abundance of selected genes determined by quantitative polymerase chain reaction (qPCR) analysis (see below).
Wet-up and soil CO 2 efflux rate
At the end of the treatment period, four cores per treatment were subjected to a controlled wet-up simulating the first rainfall event after summer, during which soil CO2 efflux rates were determined. In the treatment subjected to weekly water inputs, the controlled wet-up event took place instead of a weekly water input, that is, after 6 days without precipitation. The bottoms of the cores were sealed, and a 1-l PVC chamber was fitted airtight on top of the core. Immediately after installing the chamber, distilled water was dispensed onto the soil surface through a septa over 2 min, whereas a needle through another septa ensured no pressure build-up in the system. The amount of water given to each core was calculated to approximate the amount of water lost over the treatment period and amounted to a ∼25 mm rain event. The cores submitted to weekly water inputs were subjected to wet-up after 6 days without water inputs, in order to impose the most conservative conditions of comparison with the other treatments. Headspace gas samples (3 ml) were taken just before adding water, then 5, 10, 20, 30, 45, 65, 90 and 115 min after the end of water addition. The volume removed was replaced at each sampling time by the same volume of synthetic air (20:80 O2:N2). Headspace samples were injected into vials that had been previously evacuated and filled with N2. Soil CO2 efflux rate and CO2 amount released were calculated after measuring CO2 concentrations in the vials using a model 6890 series gas chromatograph (Agilent Technologies, Wilmington, DE, USA) fitted with a pulsed-discharge detector (VICI Valco Instruments, Houston, TX, USA). Soil in the cores that had been subjected to wet-up was sampled 120 min after the end of water addition (see below), before nucleic acid extraction.
Soil sampling
To avoid edge effects, smaller cores (5 cm diameter, 10 cm depth) were taken from the intact cores. Soil was sieved and a subsample immediately frozen (−80 °C) before nucleic acid extraction (see below). Soil water content was determined gravimetrically by comparing fresh and 105 °C dried soil weights on another subsample. A soil water retention curve was established based on soil water potential measured using a constant preset atmospheric pressure potential (four points between −0.03 and −1 MPa), and an isopiestic method based on four points between −4.6 and −88 MPa, using saturated solutions of copper sulfate, sucrose, sodium chloride and magnesium nitrate (Tokunaga et al., 2003).
Soil nucleic acid extraction and purification
Both DNA and RNA were extracted from the soil samples, using a protocol adapted from Griffiths et al. (2000) and Brodie et al. (2002). All solutions and glassware were rendered RNase-free by diethyl pyrocarbonate treatment. Briefly, for each soil sample, three 0.4 g dry weight subsamples were extracted separately. Each aliquot was transferred to a 2-ml Lysing Matrix E tube (MP Biomedicals, Solon, OH, USA) and extracted twice as follows. 500 μl extraction buffer (5% CTAB, 0.5 M NaCl, 240 mM K2HPO4, pH 8.0) and 500 μl 25:24:1 phenol:chloroform:isoamyl alcohol were added before shaking (FastPrep24, MP Biomedicals; 30 s, 5.5 m s−1). After spinning down the debris (16 100 g, 5 min, 4 °C), residual phenol was removed using pre-spun 2 ml Phase Lock Gel tubes (5 Prime, Gaithersburg, MD, USA) with an equal volume of 24:1 chloroform:isoamyl alcohol, mixed and centrifuged (16 100 g, 2 min, 4 °C). The aqueous phases from both extractions were pooled, mixed with 1 ml 40% polyethylene glycol 6000 dissolved in 1.6 M NaCl and 3 μl linear acrylamide (5 mg ml−1; Ambion, Grand Island, NY, USA), and incubated for 1 h at room temperature. After centrifugation (16 100 g, 20 min, room temperature), the pellet was rinsed with 1 ml ice-cold 70% ethanol, air-dried, resuspended in 20 μl RNase-free water and stored at −80 °C. For each sample, the three subsample extracts were pooled during purification (AllPrep DNA/RNA Mini Kit, Qiagen, Valencia, CA, USA) according to manufacturer's instructions.
Abundance of genes and transcripts
The extracted DNA and RNA were quantified using Quant-iT PicoGreen dsDNA and and RiboGreen RNA reagents, respectively (Invitrogen, Grand Island, NY, USA) with a CFX96 thermocycler (Bio-Rad Laboratories, Hercules, CA, USA). cDNA was generated from 50 ng of the extracted RNA from each subsample using the QuantiTect Reverse Transcription Kit (Qiagen), then pooled.
The abundance of genes and transcripts was assessed by qPCR. The genes encoding bacterial 16S, archaeal 16S and fungal 28S ribosomal components were selected as phylogenetic markers. The bacterial rpoB gene was also selected, since this single-copy gene encodes a sub-unit of the bacterial RNA polymerase and thus provides an estimation of transcriptional activity. The primers used were: EUB338 and EUB518 for bacterial 16S (Fierer et al., 2005), A364aF and A934b for archaeal 16S (Kemnitz et al., 2005), NL1f and LS2r for fungal 28S (Bates and Garcia-Pichel, 2009) and rpoB-f-4 and rpoB-r-2 for bacterial rpoB genes (Silkie and Nelson, 2009).
DNA and cDNA quantification was performed with a CFX96 thermocycler (Bio-Rad) in a total volume of 12 μl including 2–4 ng DNA or 2.5 ng cDNA, 10 μl SsoFast EvaGreen Supermix (Bio-Rad) and 300 μM of each primer. All qPCR assays were run for 30 s at 95 °C, and then 40 cycles, with plate-reading, of 95 °C for 5 s and 60 °C for 15 s, with a final melt-curve step from 75 to 95 °C. Standard curves were obtained using serial dilutions of plasmids containing the cloned genes and the efficiencies ranged between 91.9 and 108.7% (minimum R2: 0.985).
Pyrosequencing of bacterial and fungal communities
DNA and cDNA from three replicate samples per precipitation pattern treatment and for both pre- and post-wet-up sampling time points was sequenced by 8 bp tag-encoded FLX amplicon pyrosequencing. In total, 18 cDNA samples and 18 DNA samples were sequenced (Research and Testing Laboratory, Lubbock, TX, USA; Roche 454 FLX Titanium, Branford, CT, USA). Initial generation of the sequencing libraries utilised a one-step PCR with a total of 30 cycles, a mixture of Hot Start and HotStar High Fidelity Taq polymerases (Qiagen). The primers used were 939F (TTGACGGGGGCCCGCAC) and 1492R (TACCTTGTTACGACTT) to sequence the V6–V9 regions of the bacterial 16S, and the LR0R (ACCCGCTGAACTTAAGC) and LR3 (CCGTGTTTCAAGACGGG) to sequence the D1 domain of the fungal 28S. The sequence data was submitted to the NCBI Sequence Read Archive database (BioSample accession No. SAMN03016010). The pyrotag analysis was run using PyroTagger (Kunin and Hugenholtz, 2010) as follows. Sequences were denoised, quality-filtered by removing reads with ⩾3% low-quality bases over a given length, trimmed at 200 nucleotides, clustered (uclust algorithm, Edgar, 2010) with a 95% similarity threshold, using the most abundant unique sequence as cluster representative. Taxonomy was assigned using the greengenes and Silva databases for bacteria and fungi, respectively.
Sequencing depth for rRNA and rRNA genes of both bacterial and fungal datasets was analysed by analysis of variance, using wet-up, precipitation pattern and their interaction as explanatory variables. In case at least one variable was significant, the corresponding dataset was rarefied.
Phylogenetic community structure
Phylogenetic pairwise dissimilarity matrices were calculated using Fast UniFrac (Hamady et al., 2010), based on sequence abundance, without normalising for differing mutation rates. Phylogenetic trees for bacterial 16S rRNA and fungal 28S rRNA gene sequences (4559 and 804 clusters, respectively) were constructed using a generalised time-reversible model of the nucleotide sequences in FastTree 2.1.1 (Price et al., 2009), after aligning the representatives sequences against the greengenes core set and the silva database, respectively, using the Nearest Alignment Space Termination algorithm (75% minimum sequence similarity, 200 nt minimum length, no gap character removal).
Statistical analyses
Statistical analyses were performed in R 2.15.0 (R Development Core Team, 2012). DNA and RNA data were analysed separately. The log-transformed abundances of RNA genes and transcripts were analysed by analysis of variance, using precipitation pattern, wet-up and their interaction as explanatory variables. The effect of wet-up was determined by comparing the pre-wet-up time point (taken the day before wet-up) and a time point 2 h after wet-up.
UniFrac distances were compared by non-parametric permutational multivariate analysis of variance (perMANOVA, adonis function of vegan package, Anderson, 2001), nested by precipitation pattern. The explanatory variables were precipitation pattern and wet-up, and their interaction.
Relative abundances of bacterial and fungal groups were analysed by aggregating all taxa at the phylum and class levels. The analysis targeted the most abundant phyla and classes, by using the lowest relative abundance threshold that ensured that all taxa in the analysis were present in all the samples. For bacteria, this threshold corresponded to the phyla and class accounting for more than 1% of the total number of operational taxonomic units sequenced in at least one sample at one sampling time and site. For fungal phyla and classes, the threshold was 0.5% and 1%, respectively. The relative abundance of the most abundant bacterial and fungal taxa was analysed by analysis of variance, after log-transformation of the data. The explanatory variables were precipitation pattern, wet-up, taxonomic identity (phylum or class) and their interactions. The statistical model respected the hierarchical structure of the experiment: the taxa identity level was nested within the soil core level.
Wet-up-related changes in the structure of the present and the potentially active bacterial and fungal communities were measured by differences in the abundance-weighted UniFrac phylogenetic distance (see above) between the respective pre- and post-wet-up communities.
Results
Precipitation patterns
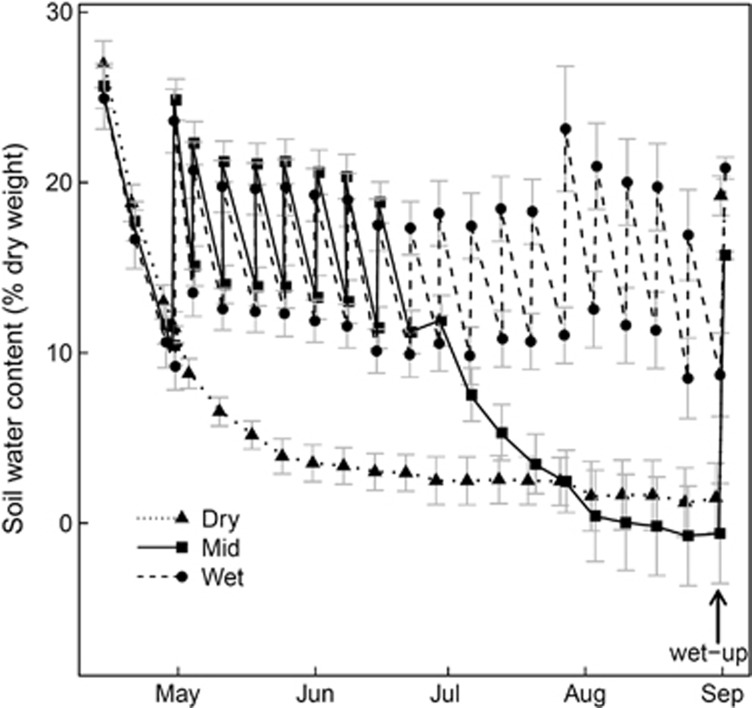
Soil water content responded clearly to the different summer precipitation pattern treatments. Under weekly water inputs, soil water content fluctuated between 10 and 25% (Figure 1). In contrast, no water inputs resulted in a sharp decrease over a month, followed by stabilisation of soil water content at around 2% (corresponding to a water potential of around −30 MPa). Weekly water inputs for 2 months followed by a dry period also resulted in a sharp decrease in soil water content after water inputs were ceased, the soil reaching water content levels that were comparable with those in the unwatered treatment in less than 1 month.
Figure 1.
Dynamics of soil water content in the precipitation pattern treatments (Dry: no water input, triangles; Mid: weekly water inputs for 2 months followed by no input, squares; Wet: weekly water inputs, circles) and controlled wet-up. Bars indicate ±1s.e.
Bacterial and fungal sequencing
Since wet-up significantly affected the number of bacterial DNA and cDNA pyrosequences, these datasets were rarefied to 1800 and 1620 pyrotag sequences per sample, respectively, representing a total of 4647 clusters (or operational taxonomic unit) for each dataset. The fungal DNA and cDNA dataset had 18 921 and 9633 pyrotag sequences remaining after curation, representing 1347 clusters for each. Fungal DNA and cDNA samples consisted of an average±s.e. of 1051±75 and 536±66 pyrosequences, respectively. Since none of precipitation pattern, wet-up and their interaction had a significant effect on the number of fungal DNA or cDNA pyrosequences, these datasets were not rarefied.
Bacterial and fungal community structure
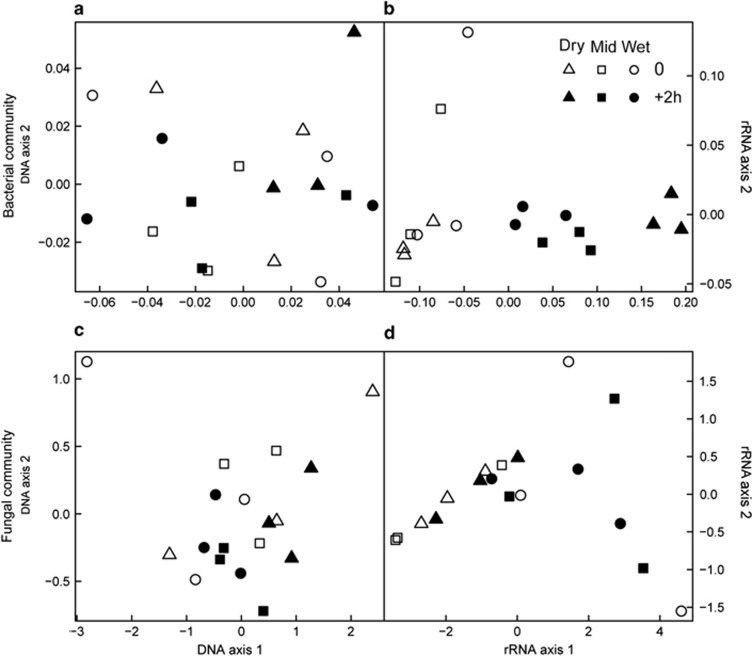
Before wet-up, neither the DNA-based or RNA-based soil bacterial community structures were detectably different between treatments (P=0.70 and 0.33, respectively, open symbols in Figures 2a and b). We found no detectable wet-up-related changes in the present soil bacterial community structure (Figure 2a, Supplementary Table S1). In contrast, both wet-up and the preceding precipitation pattern resulted in significant differences in the structure of the potentially active soil bacterial community. Potentially active bacterial communities that had been subjected to different antecedent precipitation treatments tended to respond differently to rewetting (Figure 2b, Supplementary Table S1: P=0.098 for precipitation pattern × wet-up interaction). Indeed, adding a contrast term to the anova model showed that the communities in the soils that had received weekly precipitation inputs differed significantly in their wet-up response from those subjected to the dry and mid treatments (P=0.035).
Figure 2.
Principal coordinates analysis (PCoA) of the UniFrac pairwise dissimilarity of the relative abundance of bacterial and fungal pyrotag sequences, based on 16S rRNA gene (a) and 16S rRNA (b) for bacteria and on 28S rRNA gene (c) and 28S rRNA (d) for fungi, before (open symbols) and 2 h after wet-up (closed symbols) in the three precipitation pattern treatments (Dry: no water input, triangles; Mid: weekly water inputs for 2 months followed by no input, squares; Wet: weekly water inputs, circles).
Before wet-up, the structure of the DNA-based soil fungal community was not detectably different between treatments (P=0.38, open symbols in Figure 2c). The RNA-based fungal community structure was significantly different between treatments before wet-up (P=0.032). Over wet-up, we found no detectable differences in either the present or the potentially active soil fungal community structure due to wet-up or the preceding precipitation pattern treatment (Figures 2b and c, Supplementary Table S2).
Relative abundance of bacterial groups
Immediately before wet-up, the relative abundance of most abundant bacterial groups present (DNA-based) or potentially active (RNA-based) did not detectably differ between precipitation patterns (Supplementary Table S3). Soil bacterial communities were dominated in relative abundance by the Acidobacteria phylum (mostly iii1-15 class), followed by the Actinobacteria (mostly Rubrobacteridae class) and Proteobacteria (primarily alpha, delta and gamma) phyla (Supplementary Figure S1).
Rewetting and its interaction with precipitation pattern showed no detectable effect on the relative abundance of the present bacterial phyla and classes (Supplementary Figure S3, Supplementary Table S4).
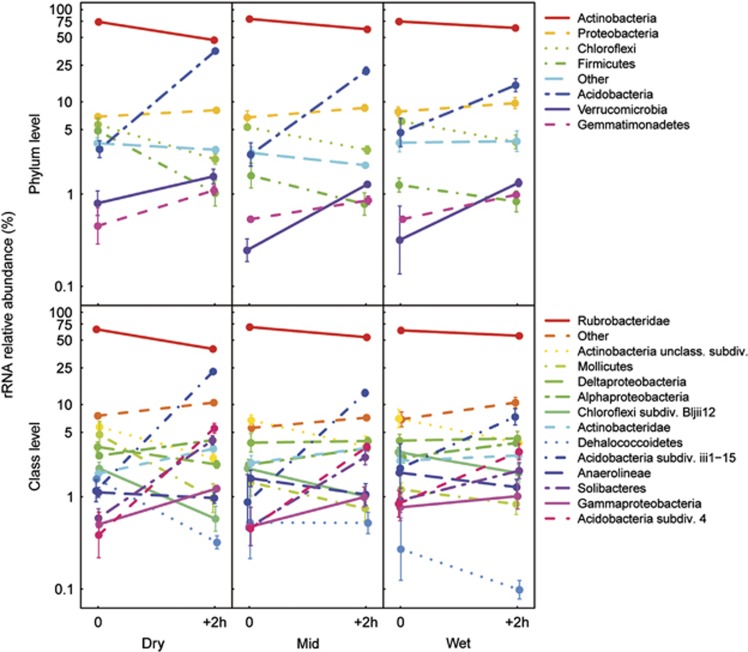
The relative abundance of the potentially active bacterial phyla and classes was significantly affected by wet-up (Figure 3, Supplementary Table S5). The different phyla and classes responded differently to wet-up (significant wet-up × bacterial phylum or class interactions). The three-way interaction wet-up × precipitation pattern × phylum or class evaluates differences in the wet-up response of the different phyla or classes between precipitation patterns. It was significant at both the phylum and class level, but explained only 1.0% and 2.0%, respectively, of the variance of the wet-up dataset. This amounts to 1.1% and 2.8% of the variance explained by wet-up-related differences in relative abundance of the different phyla and classes, respectively. Adding a contrast term based on taxa or groups of taxa to the anova models did not enable us to relate wet-up-related differences to the response of individual taxa or groups of taxa. Nevertheless, our data shows that within the most abundant bacterial taxa, the phyla that responded most to wet-up were the Acidobacteria, which increased from 3.8 to 24.4%, and the Actinobacteria that decreased from 75.9 to 57.2% with wet-up. The most responsive classes were the iii1-15 subdivision of the Acidobacteria, which increased from 1.8 to 14.7%, and the Rubrobacteria (Actinobacteria phylum), which decreased from 66.3 to 50.0%.
Figure 3.
Relative abundance of potentially active bacterial phyla (>1%), based on 16S rRNA sequenced before and 2 h after wet-up in the three precipitation pattern treatments (Dry: no water input, Mid: weekly water inputs for 2 months followed by no input, Wet: weekly water inputs). Bars indicate ±1s.e.
Relative abundance of fungal groups
Immediately before wet-up, the relative abundance of the most abundant fungal groups present (DNA-based) or potentially active (RNA-based) did not differ between precipitation patterns, besides a marginal difference (P=0.057) between present fungal classes (Supplementary Table S6). Soil fungal communities were dominated in relative abundance by the Ascomycota and, to a lesser extent, by the Basidiomycota phyla, while the distribution of fungal classes, dominated by Sordariomycetes and Agaricomycetes, Eurotiomycetes was more even and variable between treatments (Supplementary Figure S2).
The relative abundance of the present fungal phyla and classes was overall unaffected by wet-up. The different present fungal classes showed a differential response to wet-up (Supplementary Figure S4, Supplementary Table S7).
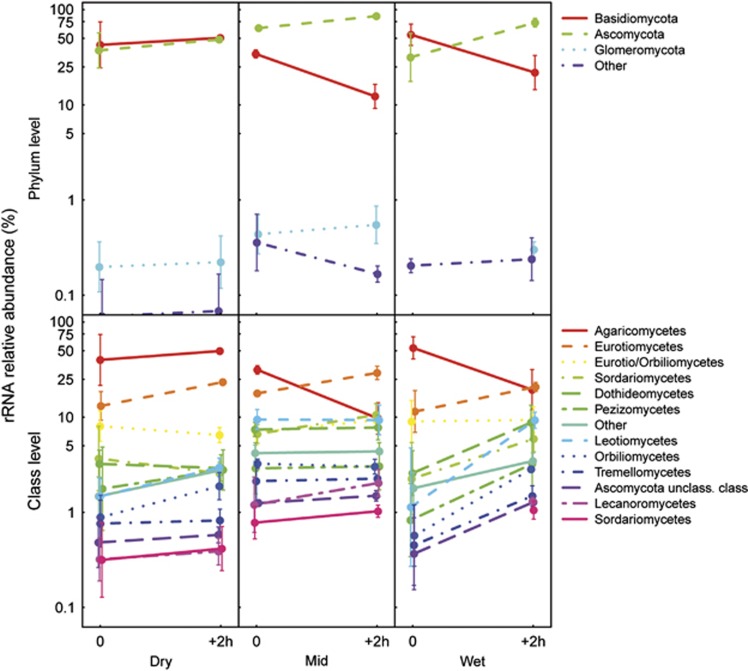
The relative abundance of the potentially active fungal classes was marginally significantly affected by wet-up (P=0.055, Figure 4, Supplementary Table S8). The different potentially active phyla responded differently to wet-up, both at the phylum and the class level. Adding a contrast term based on taxa or groups of taxa to the anova models did not enable us to relate wet-up-related differences to the response of individual taxa or groups of taxa.
Figure 4.
Relative abundance of potentially active fungal phyla (>1%), based on 28S rRNA sequenced before and 2 h after wet-up in the three precipitation pattern treatments (Dry: no water input, Mid: weekly water inputs for 2 months followed by no input, Wet: weekly water inputs). Bars indicate ±1s.e.
Quantification of bacterial, fungal and archaeal genes and transcripts
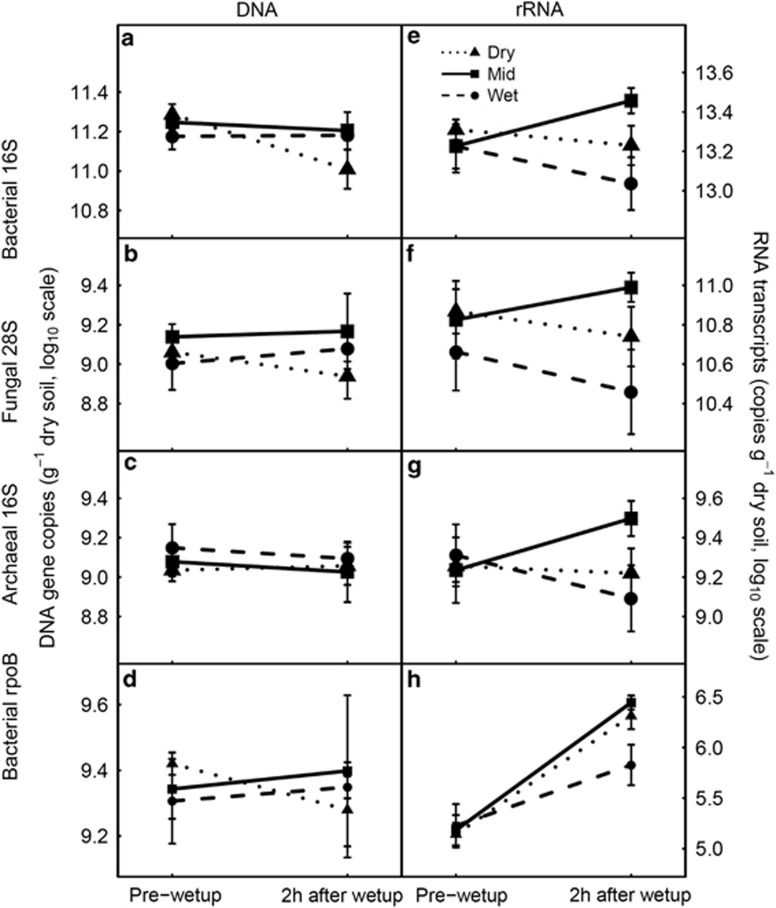
Wet-up tended to decrease the overall abundance of bacterial 16S rRNA genes (Supplementary Table S9, P=0.061), mostly driven by a reduction of 45% by wet-up in the dry treatment (Figure 5a). Indeed, adding a contrast term to the anova model showed that the effect of wet-up was significantly different between the dry treatment and the other treatments (P=0.036). The abundances of fungal 28S and archaeal 16S rRNA genes as well as of the rpoB gene (Figures 5b–d) were not significantly affected by wet-up (Supplementary Table S9). Abundances of the bacterial 16S, fungal 28S and archaeal 16S transcripts were not significantly affected by wet-up (Figures 5e–g). Wet-up significantly increased rpoB transcript abundance in all treatments (Figure 5h, Supplementary Table S9, overall +1026%). In addition, the dry and mid treatments responded more strongly than the wet treatment (P=0.43 for the interaction between wet-up and a contrast term grouping dry and mid against wet in the anova model).
Figure 5.
Abundance of selected genes (left panels) and RNA transcripts (right panels) over wet-up. Bars indicate ±1s.e. (n=3).
Soil CO 2 efflux rate
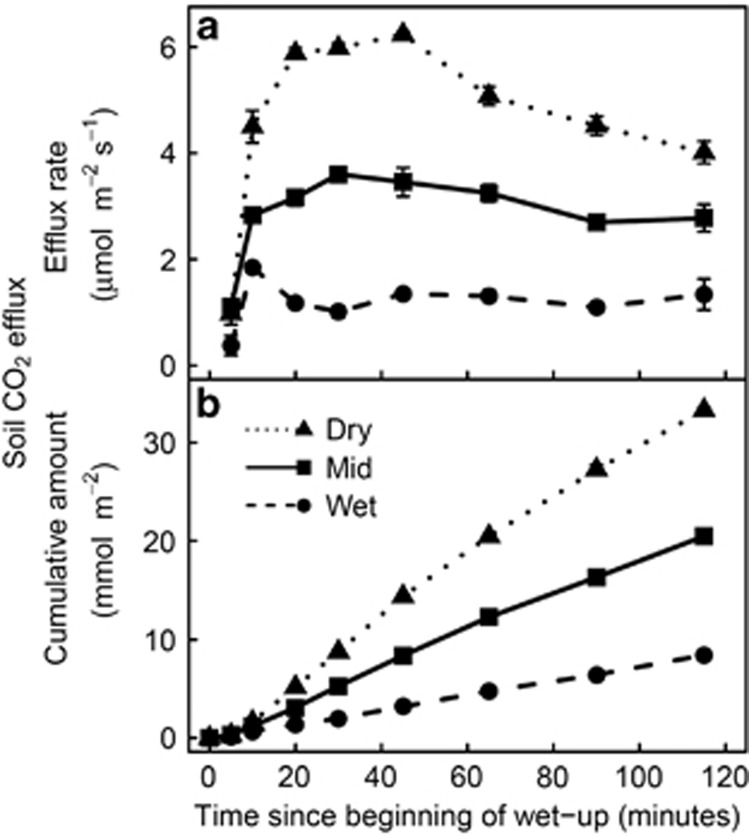
Wet-up resulted in an immediate CO2 pulse from the soil, as expected. The preceding precipitation pattern significantly affected the amount of CO2 released during the short-term measurement. 115 min after the beginning of the wet-up, the dry treatment had released on average 1.6 times more CO2 than the mid treatment, the latter having released 2.4 times more CO2 than the wet treatment (Figure 6).
Figure 6.
Soil CO2 efflux rate (a) and cumulative emissions (b) over two hours after rewetting, in the three precipitation pattern treatments (Dry: no water input; Mid: weekly water inputs for 2 months followed by no input; Wet: weekly water inputs). Bars indicate ±1s.e. (n=3).
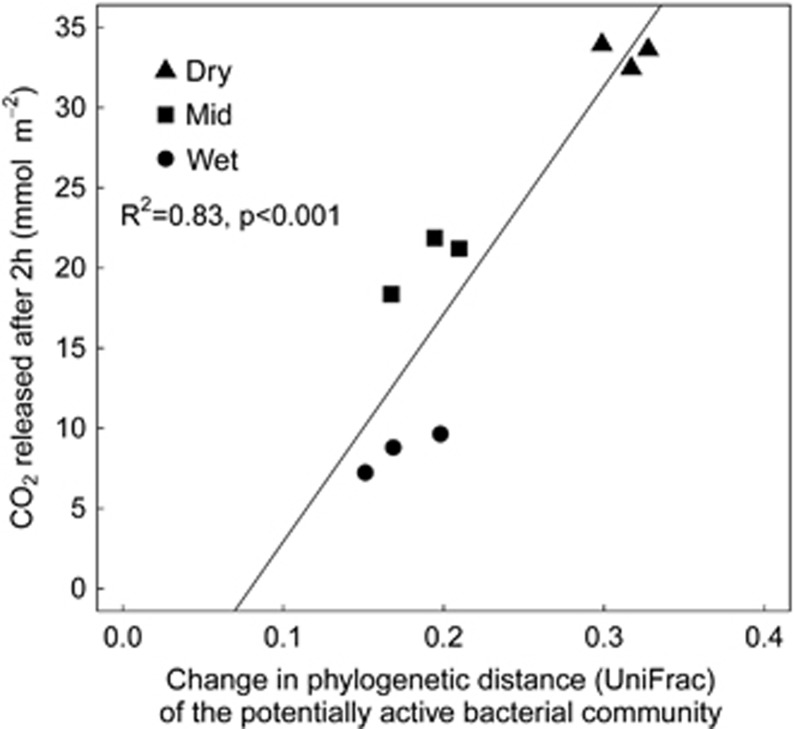
The cumulative amount of CO2 released during 2 h after rewetting was significantly positively correlated with the magnitude of wet-up-related change in the rRNA-based soil bacterial community structure (R2=0.83, P<0.001, Figure 7).
Figure 7.
Relation between changes in community structure (abundance-weighted UniFrac phylogenetic distance) of the potentially active soil bacteria and the amount of CO2 emitted from the soil, two hours after rewetting soil from the three precipitation pattern treatments (Dry: no water input, triangles; Mid: weekly water inputs for 2 months followed by no input, squares; Wet: weekly water inputs, circles).
Discussion
By the end of the dry-down period, the contrasting dry-down patterns resulting from the different precipitation treatments had not shaped different present or potentially active bacterial communities. In contrast, the rewetting event significantly altered the structure of the potentially active (rRNA-based) bacterial communities. The precipitation patterns experienced during 4 months before wet-up resulted in limited differences in the responses of the potentially active bacterial communities to rewetting, mainly driven by soils that were exposed to a pronounced dry-down period before rewetting. This pattern is consistent with the large wet-up-related increase in the abundance of rpoB transcripts, indicating that transcription resumed very rapidly after rewetting; the increase in the abundance of rpoB transcripts was more pronounced in the soils that had experienced a dry-down period before wet-up. At the phylum level, soil rewetting triggered a consistent change of the rRNA-based community structure, mainly characterised by large changes in relative abundance of only a few bacterial groups, with a strong increase of Acidobacteria (driven by the iii1-15 class) and Verrucobacteria, and a large decrease of Actinobacteria (driven by the Rubrobacteria class) and Firmicutes. Across three Mediterranean climate grasslands that differed in soil, climate and microbial community, we previously reported that the rRNA-based bacterial community shared a similar wet-up response pattern which likely resulted from contrasting bacterial life-strategies related to water availability (Barnard et al., 2013). Our current results are consistent with this previous work, in that the wet-up response patterns of the most abundant phyla were indistinguishable between communities that had experienced markedly different antecedent water conditions. In contrast with bacteria, the potentially active soil fungal community showed little response to differing precipitation treatments and to subsequent wet-up. This is consistent with the expected larger resistance of fungi than bacteria to drought (Bapiri et al., 2010; Barnard et al., 2013) and the generally more stable properties of fungal food webs compared to bacterial food webs (de Vries et al., 2012a).
Our findings that drier antecedent conditions resulted in a larger rewetting CO2 pulse are consistent with results from field studies (Xu et al., 2004; Cable et al., 2008) as well as controlled rewetting experiments (Göransson et al., 2013). Further, we found a strong, significant positive relation between the magnitude of the CO2 pulse upon rewetting dry soils and the magnitude of change in the community structure of the potentially active bacterial groups. Although this strong correlation could result from a direct cause and effect, it could also result from both the magnitude of the CO2 pulse and the magnitude of change in the community structure being dependent on antecedent conditions. Non-biological sources of soil CO2 upon rewetting are expected to have contributed only marginally to the amount of CO2 released over a 2 h period after wet-up, as these soils (pH 6.2) are not carbonate-rich (Inglima et al., 2009). In these soils, the main inorganic sources should include CO2 displaced from soil pores and dissolved in rain water. Theoretical calculation shows that given a CO2 concentration of 500 mmol mol−1 in soil pores and assuming that all the rain degasses and displaces all the CO2 out of the soil pores, a 25 mm rain event can release 0.52 mmol CO2 per m2 from soil pores and 0.40 mmol CO2 dissolved in rain, that is, a total of 0.92 mmol CO2 per m2, amounting to 0.13 μmol CO2 per m2 s1 (assuming a homogeneous efflux over 2 h). These numbers represent only a small fraction of the CO2 efflux amounts and rates measured in our experiment.
The contrasting bacterial strategies that emerge from the main potentially active bacterial groups indicate that abrupt rewetting benefits the activity of quick responders that likely rapidly metabolise readily available carbon substrates that accumulated during the dry period. Both the resulting rapid change in the structure of the RNA-based bacterial community and the associated CO2 pulse would be expected to be smaller in soils in which C substrates are likely to have accumulated over less time, as found in our study. In our experiment, soil water content dropped to similar extremely low levels (below 5%) in the dry and mid treatments, but remained at this level for twice as long in the former. Shortening the duration of the extreme dry conditions resulted in dampening the effect of rewetting on both the magnitude of the soil CO2 pulse and the changes in the structure of the potentially active soil bacterial community, which could be independent phenomena or could both result from a lesser release of C resources due to a shorter dry period.
Our results suggest that the duration of dry conditions also affects cell mortality. We found that wet-up decreased bacterial 16S rRNA gene abundance in soils subjected to the longest dry summer period. Since 16S rRNA-gene-based bacterial community structure was not detectably affected by wet-up, decreased bacterial 16S rRNA gene abundance likely reflected a generalised decline in live cell abundance. In addition to C substrates that become bio-available upon rewetting, dead bacterial material might potentially contribute to the CO2 pulse observed in our study. Indeed, recent isotope probing analysis of the actual death and growth of microbial cells indicates that the release of carbon from cell death due to desiccation and/or wetting could account for a significant portion of the CO2 produced on wet-up (Blazewicz et al., 2014).
Our study shows that predicted changes in summer precipitation pattern will likely affect the metabolic potential of the soil bacterial community and the related magnitude of the rainfall-induced CO2 pulse upon rewetting. Although we found that a shorter dry summer season may reduce the contribution of the rewetting CO2 pulse to yearly C losses, we did not assess the integrated impact on the yearly C budget. In addition to impacts on C cycling, rewetting frequency can also significantly impact soil nitrogen cycling dynamics (Miller et al., 2005). Based on our results, it appears that the duration of severe dry conditions may be very important in conditioning the response of the potentially active bacterial community to wet-up. Our results suggest that the predicted extension of the wet season into summer will dampen the magnitude of the response of potentially active soil bacteria in response to rewetting after the dry season.
Acknowledgments
We thank Maria Patanwala for nucleic acid extractions, James Cleaver for help in the field, Barbara Rotz for help in the greenhouse, Pascal Niklaus for statistical advice, Chris Jones for help with processing sequencing data and Laurent Philippot for constructive comments on the manuscript. This work was funded by the Kearney Soil Science Foundation. RB was funded by the European Community's Seventh Framework Programme under grant agreement PIOF-GA-2008-219357.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Anderson MJ. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol 26: 32–46. [Google Scholar]
- Angel R, Conrad R. (2013). Elucidating the microbial resuscitation cascade in biological soil crusts following a simulated rain event. Environ Microbiol 15: 2799–2815. [DOI] [PubMed] [Google Scholar]
- Bapiri A, Bååth E, Rousk J. (2010). Drying–rewetting cycles affect fungal and bacterial growth differently in an arable soil. Microb Ecol 60: 419–428. [DOI] [PubMed] [Google Scholar]
- Barnard RL, Osborne CA, Firestone MK. (2013). Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J 7: 2229–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates ST, Garcia-Pichel F. (2009). A culture-independent study of free-living fungi in biological soil crusts of the Colorado Plateau: their diversity and relative contribution to microbial biomass. Environ Microbiol 11: 56–67. [DOI] [PubMed] [Google Scholar]
- Birch HF. (1958). The effect of soil drying on humus decomposition and nitrogen availability. Plant Soil 10: 9–31. [Google Scholar]
- Blazewicz SJ, Barnard RL, Daly RA, Firestone MK. (2013). Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J 7: 2061–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazewicz SJ, Schwartz E, Firestone MK. (2014). Growth and death of bacteria and fungi underlie rainfall-induced carbon dioxide pulses from seasonally dried soil. Ecology 95: 1162–1172. [DOI] [PubMed] [Google Scholar]
- Boot CM. (2011). Microbial response to drying and rewetting: osmotic and matric effects. Plant Soil 348: 99–102. [Google Scholar]
- Borken W, Matzner E. (2009). Reappraisal of drying and wetting effects on C and N mineralization and fluxes in soils. Global Change Biol 15: 808–824. [Google Scholar]
- Bouskill NJ, Lim HC, Borglin S, Salve R, Wood TE, Silver WL et al. (2012). Pre-exposure to drought increases the resistance of tropical forest soil bacterial communities to extended drought. ISME J 7: 384–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie E, Edwards S, Clipson N. (2002). Bacterial community dynamics across a floristic gradient in a temperate upland grassland ecosystem. Microb Ecol 44: 260–270. [DOI] [PubMed] [Google Scholar]
- Cable JM, Ogle K, Williams DG, Weltzin JF, Huxman TE. (2008). Soil texture drives responses of soil respiration to precipitation pulses in the Sonoran desert: implications for climate change. Ecosystems 11: 961–979. [Google Scholar]
- Cruz-Martinez K, Suttle KB, Brodie EL, Power ME, Andersen GL, Banfield JF. (2009). Despite strong seasonal responses, soil microbial consortia are more resilient to long-term changes in rainfall than overlying grassland. ISME J 3: 738–744. [DOI] [PubMed] [Google Scholar]
- de Vries FT, Liiri ME, Bjornlund L, Bowker MA, Christensen S, Setala HM et al. (2012. a). Land use alters the resistance and resilience of soil food webs to drought. Nat Climate Change 2: 276–280. [Google Scholar]
- de Vries FT, Liiri ME, Bjornlund L, Setala HM, Christensen S, Bardgett RD. (2012. b). Legacy effects of drought on plant growth and the soil food web. Oecologia 170: 821–833. [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Evans SE, Wallenstein MD, Burke IC. (2014). Is bacterial moisture niche a good predictor of shifts in community composition under long-term drought? Ecology 95: 110–122. [DOI] [PubMed] [Google Scholar]
- Evans SE, Wallenstein MD. (2012). Soil microbial community response to drying and rewetting stress: does historical precipitation regime matter? Biogeochemistry 109: 101–116. [Google Scholar]
- Evans SE, Wallenstein MD. (2013). Climate change alters ecological strategies of soil bacteria. Ecol Lett 17: 155–164. [DOI] [PubMed] [Google Scholar]
- Field CB, Daily GC, Davis FW, Gaines S, Matson PA, Melack J et al. (1999) Confronting climate change in California: ecological impacts on the golden state. Union of Concerned Scientists, Cambridge, MA and Ecological Society of America: Washington, DC. [Google Scholar]
- Fierer N, Jackson JA, Vilgalys R, Jackson RB. (2005). Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl Environ Microbiol 71: 4117–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Schimel JP, Holden PA. (2003). Influence of drying-rewetting frequency on soil bacterial community structure. Microb Ecol 45: 63–71. [DOI] [PubMed] [Google Scholar]
- Fierer N, Schimel JP. (2002). Effects of drying-rewetting frequency on soil carbon and nitrogen transformations. Soil Biol Biochem 34: 777–787. [Google Scholar]
- Fierer N, Schimel JP. (2003). A proposed mechanism for the pulse in carbon dioxide production commonly observed following the rapid rewetting of a dry soil. Soil Sci Soc Am J 67: 798–805. [Google Scholar]
- Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ. (2000). Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol 66: 5488–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göransson H, Godbold DL, Jones DL, Rousk J. (2013). Bacterial growth and respiration responses upon rewetting dry forest soils: Impact of drought-legacy. Soil Biol Biochem 57: 477–486. [Google Scholar]
- Hamady M, Lozupone C, Knight R. (2010). Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglima I, Alberti G, Bertolini T, Vaccari FP, Gioli B, Miglietta F et al. (2009). Precipitation pulses enhance respiration of Mediterranean ecosystems: the balance between organic and inorganic components of increased soil CO2 efflux. Global Change Biol 15: 1289–1301. [Google Scholar]
- Jarvis P, Rey A, Petsikos C, Wingate L, Rayment M, Pereira J et al. (2007). Drying and wetting of Mediterranean soils stimulates decomposition and carbon dioxide emission: the ‘Birch effect'. Tree Physiol 27: 929–940. [DOI] [PubMed] [Google Scholar]
- Jones SE, Lennon JT. (2010). Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci USA 107: 5881–5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakumanu ML, Cantrell CL, Williams MA. (2013). Microbial community response to varying magnitudes of desiccation in soil: a test of the osmolyte accumulation hypothesis. Soil Biol Biochem 57: 644–653. [Google Scholar]
- Kemnitz D, Kolb S, Conrad R. (2005). Phenotypic characterization of Rice Cluster III archaea without prior isolation by applying quantitative polymerase chain reaction to an enrichment culture. Environ Microbiol 7: 553–565. [DOI] [PubMed] [Google Scholar]
- Kunin V, Hugenholtz P. (2010). PyroTagger: A fast, accurate pipeline for analysis of rRNA amplicon pyrosequence data. Open J Available at: http://www.theopenjournal.org/toj_articles/1.
- Miller AE, Schimel JP, Meixner T, Sickman JO, Melack JM. (2005). Episodic rewetting enhances carbon and nitrogen release from chaparral soils. Soil Biol Biochem 37: 2195–2204. [Google Scholar]
- Parker SS, Schimel JP. (2011). Soil nitrogen availability and transformations differ between the summer and the growing season in a California grassland. Appl Soil Ecol 48: 185–192. [Google Scholar]
- Placella SA, Brodie EL, Firestone MK. (2012). Rainfall-induced carbon dioxide pulses result from sequential resuscitation of phylogenetically clustered microbial groups. Proc Natl Acad Sci USA 109: 10931–10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. (2009). FastTree: Computing large minimum-evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. (2012). R: A language and environment for statistical computing: Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org/.
- Schippers A, Neretin LN, Kallmeyer J, Ferdelmna TG, Cragg BA, Parkes RJ et al. (2005). Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature 433: 861–864. [DOI] [PubMed] [Google Scholar]
- Silkie SS, Nelson KL. (2009). Concentrations of host-specific and generic fecal markers measured by quantitative PCR in raw sewage and fresh animal feces. Water Res 43: 4860–4871. [DOI] [PubMed] [Google Scholar]
- Suttle KB, Thomsen MA, Power ME. (2007). Species interactions reverse grassland responses to changing climate. Science 315: 640–642. [DOI] [PubMed] [Google Scholar]
- Tokunaga TK, Olson KR, Wan JM. (2003). Moisture characteristics of Hanford gravels: bulk, grain-surface, and intragranular components. Vadose Zone J 2: 322–329. [Google Scholar]
- Waldrop MP, Firestone MK. (2006. a). Seasonal dynamics of microbial community composition and function in oak canopy and open grassland soils. Microb Ecol 52: 470–479. [DOI] [PubMed] [Google Scholar]
- Waldrop MP, Firestone MK. (2006. b). Response of microbial community composition and function to soil climate change. Microb Ecol 52: 716–724. [DOI] [PubMed] [Google Scholar]
- Xiang S-R, Doyle A, Holden PA, Schimel JP. (2008). Drying and rewetting effects on C and N mineralization and microbial activity in surface and subsurface California grassland soils. Soil Biol Biochem 40: 2281–2289. [Google Scholar]
- Xu L, Baldocchi DD, Tang J. (2004). How soil moisture, rain pulses, and growth alter the response of ecosystem respiration to temperature. Global Biogeochem Cy 18: GB4002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.