Abstract
Butyrate-producing bacteria have an important role in maintaining host health. They are well studied in human and medically associated animal models; however, much less is known for other Vertebrata. We investigated the butyrate-producing community in hindgut-fermenting Mammalia (n=38), Aves (n=8) and Reptilia (n=8) using a gene-targeted pyrosequencing approach of the terminal genes of the main butyrate-synthesis pathways, namely butyryl-CoA:acetate CoA-transferase (but) and butyrate kinase (buk). Most animals exhibit high gene abundances, and clear diet-specific signatures were detected with but genes significantly enriched in omnivores and herbivores compared with carnivores. But dominated the butyrate-producing community in these two groups, whereas buk was more abundant in many carnivorous animals. Clustering of protein sequences (5% cutoff) of the combined communities (but and buk) placed carnivores apart from other diet groups, except for noncarnivorous Carnivora, which clustered together with carnivores. The majority of clusters (but: 5141 and buk: 2924) did not show close relation to any reference sequences from public databases (identity <90%) demonstrating a large ‘unknown diversity'. Each diet group had abundant signature taxa, where buk genes linked to Clostridium perfringens dominated in carnivores and but genes associated with Ruminococcaceae bacterium D16 were specific for herbivores and omnivores. Whereas 16S rRNA gene analysis showed similar overall patterns, it was unable to reveal communities at the same depth and resolution as the functional gene-targeted approach. This study demonstrates that butyrate producers are abundant across vertebrates exhibiting great functional redundancy and that diet is the primary determinant governing the composition of the butyrate-producing guild.
Introduction
Many studies investigating host–microbiota interactions have demonstrated that the intestinal microbiota has a critical role in maintaining gut homeostasis and host health. A mutualistic relationship between the host and its microbiota exists, where bacteria provide essential substrates such as vitamins or short-chain fatty acids, whereas the host delivers nutrients and a stable growth environment (Nicholson et al., 2012). Most investigations have been performed with humans and a few model organisms (mouse, rat and pig), but much less is known for other mammals or distinct vertebrate classes. In a pioneer study using 16S rRNA gene sequences, Ley et al. (2008) found an overall relationship in mammals between the structure of the fecal microbiota, diet and host phylogeny. Whereas studies based on traditional bacterial taxonomic markers can provide first insights into host and microbiota inter-relationships, functional investigations are essential in order to understand the key processes. Several studies focused on microbial functions for specific animals (c.f. Lavery et al., 2012; Tun et al., 2012), and a global metagenomic screen across Mammalia demonstrated that the bacterial functional repertoire is primarily governed by diet and does not reflect host phylogeny (Muegge et al., 2011). That study revealed that the microbiota in carnivores and herbivores are characterized by different trade-offs in amino-acid pathways and by an opposite directionality in the central phosphoenolpyruvate-pyruvate-oxaloacetate cycle. Similar differences were observed in human subjects who were exposed to diets mirroring those of carnivores or herbivores demonstrating that broad functional differences can be achieved with distinct nourishments even in the same host indicating that direct host-specific selection processes are limited (at least in humans) (David et al., 2013).
The divergence between overall bacterial function and host phylogeny raises the question whether specific physiological processes of the host that are linked to certain microbial functions, such as components of the immune system or energy requirements of the colonic epithelium, evolved with diet. If this is not the case and those processes are conserved throughout hosts, or coevolved with certain phylogenetic groups (for example, all mammals), we would expect functional redundancy of the microbiota that is adapted to the individual diets. In this context, the production of butyrate is of specific interest as it is considered essential for maintaining health in humans and usual animal models, where it serves as the main energy source for colonocytes, shows immunomodulatory effects and influences local gene expression (Hamer et al., 2008; Furusawa et al., 2013). Whereas butyrate is detected in feces of various vertebrates (Stevens and Hume, 1998), indicating that it serves important roles throughout this taxon, it is currently unknown how butyrate-producing communities evolved with distinct diets. In particular, carnivores are interesting as butyrate production is primarily linked to bacteria that feed on carbohydrates such as starch or xylan, converting pyruvate to butyrate using the ‘acetyl-CoA pathway' (Bennett and Rudolph, 1995). However, this pathway can also be fed by amino acids and additional amino-acid-based butyrate-synthesis pathways are recognized as well (Buckel and Barker, 1974; Barker et al., 1982; Gerhardt et al., 2000).
The few existing studies specifically focusing on butyrate producers in different animal taxa demonstrate that butyrate-producing communities are omnipresent; however, their composition can be considerably distinct from that of humans (c.f. Eeckhaut et al., 2011; Levine et al., 2013). Detailed investigations on these important functional communities across vertebrates are missing and it is currently unknown how they have adapted to distinct hosts.
Materials and methods
Samples and DNA extraction
An overview of all collected fecal samples is given in Table 1. The majority of samples were derived from Potter Park Zoo (Lansing, MI, USA), except for dogs (private), giant panda (Ningbo, China), guineapig, mouse, rat (commercial pet-store) and grizzly bear (Detroit Zoo, Detroit, MI, USA). One sample per animal was collected. Only hindgut fermenters were included in the analysis. Foregut fermenters were specifically excluded to avoid influences of their distinct gut composition on the structure of butyrate-producing communities that might bias diet-related signals. All samples were collected within 24 h after defecation and were stored at −80 °C. Animals were categorized into three distinct diet groups based on feeding protocols provided by individual sources (Supplementary Table S2). Categorization into carnivores and herbivores was based on the animal's predominant diet, and some taxa in those groups were additionally fed minor amounts of substances that were not strictly associated with the corresponding diet. Zoo animals were fed diets researched to be appropriate for the taxon and used by most US zoos. DNA extraction was performed using the PowerSoil kit (MoBio, Jefferson City, MO, USA) according to the manufacturer.
Table 1. Diet and taxonomy of the 54 samples included in the study.
| SampleID | Diet | Class | Order | Family | Genuse/species |
|---|---|---|---|---|---|
| BaldEagle | C | Aves | Accipitriformes | Accipitridae | Haliaeetus leucocephalus |
| Duck | H | Aves | Anseriformes | Anatidae | Anseranser domestic |
| Mousebird | H | Aves | Coliiformes | Coliidae | Urocolius macrourus |
| Hawk | C | Aves | Falconiformes | Accipitridae | Buteo jamaicensis |
| Peafowl | O | Aves | Galliformes | Phasianidae | Pavo cristatus |
| KingVulture | C | Aves | Incertaesedis | Cathartidae | Sarcoramphus papa |
| Penguin | C | Aves | Sphenisciformes | Spheniscidae | Spheniscus magellanicus |
| Ostrich | O | Aves | Struthioniformes | Struthionidae | Struthio S. camelus |
| Tenrec1 | C | Mammalia | Afrosoricida | Tenrecidae | Echinops telfairi |
| Tenrec2 | C | Mammalia | Afrosoricida | Tenrecidae | Echinops telfairi |
| GuineaHog | H | Mammalia | Artiodactyla | Suidae | Sus scrofa scrofa |
| Dog1 | O | Mammalia | Carnivora | Canidae | Canis lupus |
| Dog2 | O | Mammalia | Carnivora | Canidae | Canis lupus |
| ArcticFox1 | C | Mammalia | Carnivora | Canidae | Alopex lagopus |
| ArcticFox2 | C | Mammalia | Carnivora | Canidae | Alopex lagopus |
| AfricanLion | C | Mammalia | Carnivora | Felidae | Panthera Heol krugeri |
| Tiger | C | Mammalia | Carnivora | Felidae | Panthera tigris |
| Meerkat | O | Mammalia | Carnivora | Herpestidae | Suricata suricatta |
| Ferret | C | Mammalia | Carnivora | Mustelidae | Mustela putorius furo |
| RiverOtter | C | Mammalia | Carnivora | Mustelidae | Lontra canadensis |
| RedPanda | H | Mammalia | Carnivora | Ursidae | Ailurus fulgens fulgens |
| GrizzlyBear | O | Mammalia | Carnivora | Ursidae | Ursus arctos |
| GiantPanda1 | H | Mammalia | Carnivora | Ursidae | Ailuropoda melanoleuca |
| GiantPanda2 | H | Mammalia | Carnivora | Ursidae | Ailuropoda melanoleuca |
| Binturong | O | Mammalia | Carnivora | Viverridae | Arctictis binturong |
| Armadillo | O | Mammalia | Cingulata | Dasypodidae | Tolypeutes matacus |
| Opossum | O | Mammalia | Didelphimorphia | Didelphidae | Didelphis virginiana |
| Rabbit1 | H | Mammalia | Lagomorpha | Leporidae | Oryctolagus cuniculus |
| Rabbit2 | H | Mammalia | Lagomorpha | Leporidae | Oryctolagus cuniculus |
| Donkey | H | Mammalia | Perissodactyla | Equidae | Equus asinus asinus |
| CottontopTamarin | O | Mammalia | Primates | Callitrichidae | Sagulnus oedipus |
| GoldenLionTamarin | O | Mammalia | Primates | Callitrichidae | Leontopithecus rosalia |
| Human1 | O | Mammalia | Primates | Hominidae | Homo sapiens |
| Human2 | O | Mammalia | Primates | Hominidae | Homo sapiens |
| Mandril | O | Mammalia | Primates | Cercopithecidae | Mandrillus sphinx |
| MongooseLemur | O | Mammalia | Primates | Lemuridae | Eulemur mongoz |
| RedruffeledLemur | O | Mammalia | Primates | Lemuridae | Varecia Variegatelrubra |
| RingtailedLemur | O | Mammalia | Primates | Lemuridae | Lemur catta |
| SpiderMonkey | O | Mammalia | Primates | Atelidae | Ateles fusciceps robustus |
| GuineaPig | H | Mammalia | Rodentia | Caviidae | Cavia porcellus |
| PatagonianCavy | H | Mammalia | Rodentia | Caviidae | Dolichotis patagonum |
| Chinchilla | H | Mammalia | Rodentia | Chinchillidae | Chinchilla brevicaudataus |
| Porcupine | O | Mammalia | Rodentia | Hystricidae | Hystrix cristata |
| Mouse | O | Mammalia | Rodentia | Muridae | Mus musculus |
| Rat | O | Mammalia | Rodentia | Muridae | Rattus norvegicus |
| Treeshrew | O | Mammalia | Scandentia | Tupaiidae | Tupaia belangeri |
| BeardedDragon | O | Reptilia | Squamata | Agamidae | Pogona vitticeps |
| MilkSnake | C | Reptilia | Squamata | Colubridae | Lampropeltis triangulum sinaloae |
| RatSnake | C | Reptilia | Squamata | Colubridae | Panther ophislobsoletus/spiloidesa |
| Chuckwalla | H | Reptilia | Squamata | Iguanidae | Sauromalus ater |
| Skink | O | Reptilia | Squamata | Scincidae | Tiliqua scincoides intermedia |
| BoxTurtle | O | Reptilia | Testudines | Emydidae | Terrapene carolina carolina |
| Tortoise1 | H | Reptilia | Testudines | Testudinidae | Testudo graeca traeca |
| Tortoise2 | H | Reptilia | Testudines | Testudinidae | Testudo graeca traeca |
Abbreviations: C, carnivores; H, herbivores; O, omnivores.
The sample derived from either species.
Amplification of functional genes
Primers used were described previously (Vital et al., 2013). Amplification of DNA samples was achieved with the AccuPrime PCR system (Life Technologies, Grand Island, NY, USA), where 5 ng of template DNA was combined with all individual forward and reverse primers containing the same barcode (total final concentration of 0.4 μM). The PCR protocol (35 cycles) was carried out according to the manufacturer, with annealing temperatures and MgSO4 concentrations of 51 °C, 3 mM and 48 °C, 2 mM for butyryl-CoA:acetate CoA-transferase (but) and butyrate kinase (buk), respectively. Harvesting of target bands and re-amplification are described in (Vital et al., 2013). Sequencing was performed on the 454 FLX system at the Utah State University (Logan, UT, USA) using the Lib-A kit according to the manufacturer (454 Life Sciences, Branford, CT, USA).
Functional genes' data analysis
To estimate gene abundances of but and buk (as percentage of total bacteria exhibiting the gene), target bands after the first PCR amplification were categorized into six distinct brightness groups (5: above 20%. 4: 10%–20%, 3: 4%–10%, 2: 1%–4%, 1: below 1% and 0: not detectable; category 1 was omitted for but gene analysis because of a higher detection threshold compared with buk) based on standard curves. The following concentrations from reference organisms (but: mix of Roseburia inulinivorans/Faecalibacterium prausnitzii and buk: Clostridium perfringens) were used to establish standard curves: 5 ng (100%), 2 ng (40%), 1 ng (20%), 0.5 ng (10%), 0.2 ng (4%), 0.1 ng (2%), 0.05 ng (1%), 0.02 ng (0.4%) and 0.01 ng (0.2%). Band brightness was measured using the software imageJ (http://imagej.nih.gov/ij/). Two independent experiments were performed and average values were used as the final result for statistical analyses. 16S rRNA was included as an amplification control (PCR conditions and primers as presented in Kozich et al. (2013) but increase of cycle numbers to 35). This method was used as qPCR analysis was not possible for quantification of these genes because of the formation of primer dimers as well as some nonspecific amplification (Vital et al., 2013). Processing of raw reads was carried out according to Vital et al. (2013) with some alterations. Raw reads were processed through Ribosomal Database Project (RDP) pyrosequencing pipeline (http://rdp.cme.msu.edu) yielding 491 000 but and 286 761 buk sequences after trimming. Sequences were then frameshift-corrected with FrameBot (Wang et al., 2013). During these initial pipelining steps all sequences that display ⩾93% coverage to seeding sequences from FunGene (Fish et al., 2013) were included. This selection does contain many sequences of distinct functionality/substrate specificity and sequences considered as real but/buk genes (ref. seq.) form a separate clade (on a neighbor joining tree) that includes mainly well-known butyrate producers and where function of several sequences was biochemically proven (Vital et al., 2013). FunGene sequences that encode enzymes with distinct functions/substrate specificities were intentionally included here as they encompass the most closely related sequences to but and buk genes, respectively, and amplicons matching those genes were omitted from further analyses (filtered-out). Thus, amplicons were not forced to match a specific ref. seq. in order to ensure that amplicons displaying low identities to a specific ref. seq. are not originating from genes encoding distinct functionality. This exclusion criterion was previously not applied for buk ref. seq. (Vital et al., 2013) but is included in this study because several sequences previously considered as possible buk are most likely of distinct functionality/substrate specificity (namely methyl-butyrate kinase, Vital et al., 2014). For buk, a few sequences linked to butyrate producers expected to exhibit that gene namely Acidaminococcus sp. 21, Anaerotruncus colihominis and Halanaerobium praevalens fell outside the main clade and were manually included as real buk. The percentage of retained sequences after the pipeline process were 66.5% and 71.1% for but and buk, respectively, excluding the samples that produced too few sequences (see below). Subsequently, chimeras were removed using UCHIME (www.drive5.com) in de novo mode. A very low percentage of but (0.2%) and buk (0.1%) sequences, respectively, were identified as chimeric. Remaining protein sequences were aligned to Hidden Markov Models of each individual gene (hmmalign), trimmed from both ends and clustered using RDP's complete-linkage-clustering applying a 5% cutoff. It should be noted that closest match assignments of amplicons during FrameBot analysis do not imply that the carrier exhibiting an amplicon/gene does necessarily fall into the same taxonomic unit associated with the closest matching ref. seq. All pipeline steps and clustering were performed using Michigan State University High Performance Computing Center (MSU HPCC). The number of clusters for individual diet groups including identity of representative sequences to a ref. seq. was calculated excluding global singletons. For ordination analysis, operational taxonomic unit tables were subsampled to equal depth for each gene (1093 sequences per sample) using the function rrarefy in R's (R Development Core Team, 2008) package vegan and, subsequently, combined. Some samples contained too few sequences as most (⩾97%) were filtered-out during pipeline processing (but: Bald Eagle, GiantPanda 1, GiantPanda 2, Penguin, Tenrec 1; buk: CottontopTamarin). For GrizzlyBear and Chuckwalla, only 747 but genes (88.5% filtered-out) and 743 buk genes (74% filtered-out), respectively, were retrieved. Furthermore, a few samples produced no band after re-amplification (but: RedPanda; buk: MilkSnake, Mousebird, GiantPanda 1, GiantPanda 2, Peafowl). For all those samples the corresponding gene was treated as absent during ordination analysis.
Nonmetric multidimensional scaling analysis (vegan's metaMDS) using Bray–Curtis dissimilarities on Hellinger-transformed data (vegan's decostand) was performed. Statistical analysis (permutational ANOVA) was calculated with vegan's adonis. For FrameBot closest match analysis, samples were normalized to equal depth (but: 1093 and buk: 1142) before analysis. Samples exhibiting values below those thresholds were not considered for analysis (the same samples as for ordination analysis were excluded). Results were separated into distinct categories based on their percent identity to a reference gene, where each category encompassed a 10% identity range. Results originating from the same genome as well as genes from closely related strains (see (Vital et al., 2013)) were merged. For statistical analysis of FrameBot closest match data, the functions mt (test=f, R package labdsv) and vegan's sim (simper analysis) were used. Noncarnivorous Carnivora as well as taxa that exhibited <1% of the total gene communities in all samples were excluded from the analysis.
Amplification and analysis of 16S rRNA genes
Primers, barcoding strategy and amplification was performed according to (Kozich et al., 2013). Samples were individually amplified, gel-extracted (QIAquick gel extraction kit; Qiagen, Valencia, CA, USA) and purified (QIAquick gel purification kit; Qiagen). All samples produced bright bands indicating that the extract was not inhibitory to amplification. Samples were pooled to equal concentrations and sequenced on the Illumina MiSeq platform (Illumina, San Diego, CA, USA) at the Research Technology Support Facility at the Michigan State University. Raw paired-end reads were merged using RDP's APE (Cole et al., 2014). Before read taxonomic classifications (RDP's classifier), chimeras were removed using UCHIME (www.drive5.com) using the gold database. All steps were performed using the MSU HPCC. The data were harvested for genera linked to butyrate producers according to Supplementary Table S1 (based on Vital et al., 2014). Pearson correlations and corresponding statistical analyses were calculated in excel and results were visualized using cytoscape (http://www.cytoscape.org). Ordination analysis and statistics on entire communities are based on a supervised approach at the genus level (Sul et al., 2011) and was carried out in R as described above for the functional genes. Shannon diversity was calculated using R's package phyloseq. Data were subsampled to equal depth (42 270 sequences) before analysis. All sequences (but, buk, 16S rRNA) are available via SRA (PRJEB6978).
Results
The aim of the present study was to reveal the butyrate-producing communities in a broad set of hindgut-fermenting mammals, and members of the related classes birds and reptiles, to investigate their association with diet and host phylogeny. A list of all animals including their diet and taxonomy is given in Table 1. Butyrate-producing communities were explored in depth using a functional gene-based pyrosequencing approach targeting the terminal genes of the main butyrate-synthesis pathways namely but and buk (Vital et al., 2013). 16S rRNA gene analysis served as a reference method.
Estimation of but and buk gene abundances in individual diet groups
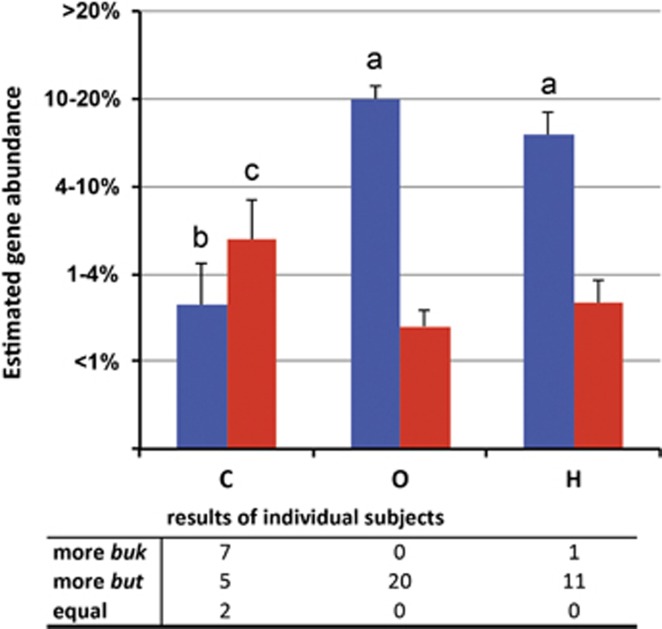
Gene abundance estimations based on band intensities of amplified genes revealed abundant butyrate-producing communities in almost all fecal samples. A clear diet-specific pattern was observed, where estimated but gene abundances were significantly enriched in omni- and herbivores with an average closest to the category representing 10%–20% of the total community compared with an estimated average abundance positioned between categories of 1%–4% and<1% in carnivores (Figure 1). Estimated average percentages of the communities harboring buk trended to be increased in carnivores compared with omnivores (P=0.051), although not so compared with herbivores, P=0.27. Overall, but dominated the butyrate-producing community in omni- and herbivores (P « 0.01) where 100% and 92% showed a positive but to buk ratio, respectively, whereas buk was more abundant in many carnivorous animals (50% Figure 1). However, a few carnivorous animals, especially the two arctic foxes and ferret, exhibited high but gene abundances. Noncarnivorous Carnivora followed the overall results of carnivores with significantly reduced estimated but gene abundances (P<0.01; data not shown) and dominance of buk in several subjects. Individual results for each sample are presented in Supplementary Figure S1. No significant estimated abundance differences were found for total butyrate-producing genes and individual but and buk genes, respectively, between vertebrate classes, whereas within the Mammalia but was significantly reduced in Carnivora (P<0.01) compared with primates and rodents (data not shown).
Figure 1.
Average gene abundance estimates for butyryl-CoA:acetate CoA-transferase (blue) and butyrate kinase (red) for individual diet groups (C: carnivores, O: omnivores, H: herbivores) are shown. Abundances were estimated by categorizing band intensities of PCR products into six distinct brightness groups (0–5) that were related to standard curves established with reference genomes. a: Differ significantly within diet groups P « 0.01, b: significantly reduced in carnivores (P<0.01), c: different to omnivores (P=0.051) and to herbivores (P=0.27). The s.e.m. is indicated. At the bottom the dominant gene for each subject of individual diet groups is shown.
Many unknown but and buk gene sequences were detected
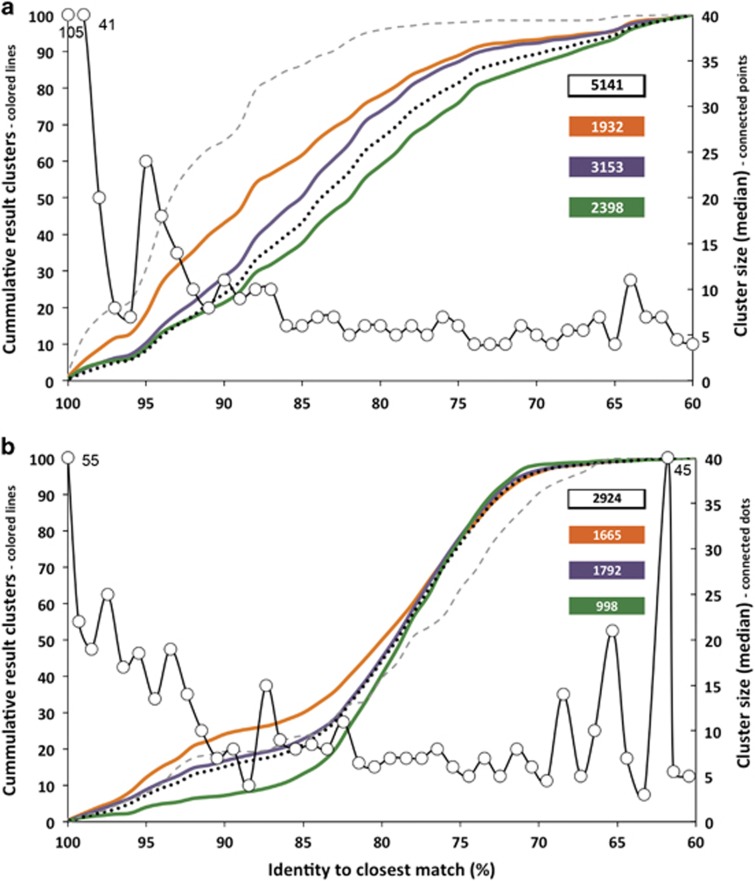
Diversity analysis based on complete-linkage-clustering of protein sequences applying a 5% cutoff revealed a large (unknown) gene diversity (Figure 2). For but, 5141 different clusters were detected. Analysis of representative sequences (rep. seq.) for each cluster demonstrated that the majority of but-linked clusters were not closely related to any known sequences from the FunGene database. For example, in carnivorous communities only 43.1% of rep. seq. showed ⩾90% identity to a reference, followed by omnivores (38.4%) and herbivores (21.3% Figure 2a). Human-derived communities exhibit many more clusters closely related to known sequences—65.6% of rep. seq. showed ⩾90% identity to a reference. For the buk gene, 2924 clusters were detected (Figure 2b). Individual diet groups displayed a similar pattern as but; however, even less clusters had a close identity to a reference and many exhibited a rep. seq. that displayed <80% identity to database sequences. Buk clusters linked to humans followed the overall trend of other categories. However, the median cluster size was not constant but considerably larger for clusters with a rep. seq. exhibiting ⩾95% identity for both but and buk (Figure 2). Nevertheless, whereas analysis based on each gene sequence indeed displayed steeper curves, a large amount of obtained sequences are not closely related to a reference (Supplementary Figure S2).
Figure 2.
Diversity analysis of gene clusters (5% cutoff on protein level) of butyryl-CoA:acetate CoA-transferase (a) and butyrate kinase (b) is shown. Individual diet groups are indicated as orange (carnivores including noncarnivorous Carnivora), violet (omnivores), green (herbivores), gray-dashed line (human) and black-dotted line (all clusters). The data are displayed as a cumulative percentage of representative sequence identities to a reference gene in our database. Numbers of clusters per group are shown in boxes (individual diet groups associated with corresponding color, white: total clusters) and the median cluster sizes are indicated (numbers associated with certain points indicate the median cluster sizes that were above the axis-maximum of 40 sequences). Global singletons were not considered for analysis.
Diet-specific clustering
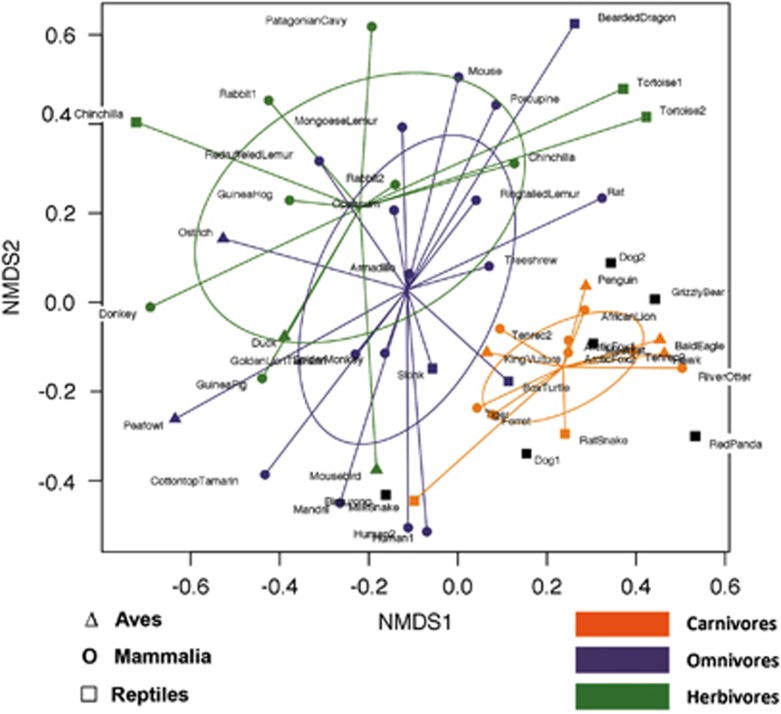
Ordination analysis based on the combined butyrate-producing community (but and buk results were merged) revealed a diet-specific clustering (P<0.01 based on PERMANOVA analysis), where carnivores formed a separate group from omnivores and herbivores, which did not robustly separate from each other (Figure 3). No significant differences in diversity (Shannon) were obtained between diet groups (data not shown). Noncarnivorous Carnivora (n=6) clustered closer to carnivores than to members of their corresponding diet group. No significant grouping was found for individual vertebrate classes (data not shown); however, Carnivora formed a separate cluster from primates and rodents within the Mammalia (Supplementary Figure S3).
Figure 3.
Nonmetric multidimensional scaling (NMDS) of combined gene data (butyryl-CoA:acetate CoA-transferase and butyrate kinase) based on multilinkage clustering (5% cutoff on the protein level) is shown (stress=0.27). Individual diet groups are indicated as orange (carnivores), violet (omnivores) and green (herbivores). Distinct Vertebrate classes are specified with symbols. Noncarnivorous Carnivora are presented as black squares. Ellipses represent the s.d.'s of points. Diet was revealed as a significant factor (P<0.01 based on permutational ANOVA analysis).
Closest match analysis also revealed distinct communities associated with different diets (Figure 4; individual results for each sample are presented in Supplementary Figure S4). All groups exhibited high percentages of but gene communities linked to several Roseburia sp. In carnivores, but genes associated with Acidaminococcus sp. D21 were enriched, whereas genes related to 80_Ruminococcacaea bacterium D16 and 60_Eubacterium hallii L2-7 were specifically abundant in herbivores and omnivores. Simper analysis revealed several Roseburia sp., Clostridium sp. SS2/1 and 80_E. rectale (both down in herbivors), E. limosum (up in omnivores), 80_Ruminococcacaea bacterium D16 (down in carnivores), Oscillibacter valericigenes (up in herbivores) as well as Clostridium symbiosum WAL-14163 and 60_Megasphaera micronuciformis F0359 as the main contributors for dissimilarities between groups (Supplementary Figure S5). For buk, a clear separation of carnivores and herbivores was observed, where genes associated with C. perfringens and 80_C. difficile were significantly abundant in the former and the latter predominantly exhibited buk sequences related to 70/80_Lachnospiracae bacterium 3 1 57FAA CT1 and 70/80_Coprococcus sp. ART55/1, which were both detected as the most dissimilar taxa (next to C. perfringens) based on simper analysis. Omnivores were characterized by an ‘in-between' pattern displaying abundant gene communities that were specific for the two other diets.
Figure 4.
FrameBot closest match analysis of butyryl-CoA:acetate CoA-transferase (blue) and butyrate kinase (red) for individual diet groups (C: carnivores and noncarnivorous Carnivora, O: omnivores, H: herbivores) is shown. Closest match results were separated into distinct categories based on their percent identity to a reference gene (bins containing 10% identity ranges were created, where the number preceding the reference name specifies the lowest identity in that group; for example, ‘70_reference name' is the combined percentage of all sequences that show 70%–79% identity to that reference; no number indicates categories from 90% to 100% identity). Only taxa accounting for more than one percent of the entire gene community are shown. A black star indicates taxa significantly difference (P<0.05) between diet groups.
16S rRNA gene analysis
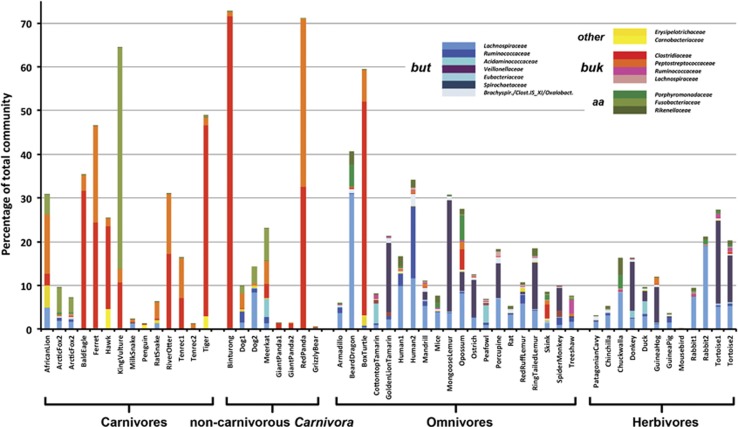
Overall, 16S rRNA analysis grouped samples according to diet confirming results on Mammalia reported earlier by (Ley et al., 2008; Supplementary Figure S6A). Similar to the functional gene results, noncarnivorous Carnivora grouped closer to Carnivora than to their individual dietary partners (result not clear for the two dog samples). A reduced bacterial diversity (Shannon) for carnivores compared with other two diet groups was observed (Supplementary Figure S6B). Results on the phylum level are presented in Supplementary Figure S7. Specific analysis for butyrate-producing taxa on the genus level showed a similar overall pattern as the functional gene-targeted approach. Genera linked to buk were enriched in carnivores, whereas but-linked taxa were much more abundant in omnivores and herbivores (a list of butyrate-producing genera found in the data is presented in Supplementary Table S1). But gene communities primarily consisted of Lachnospiraceae (in particular Clostridium XIVa sp., Lachnospiracea incertae sedis sp. and Roseburia sp.), Ruminococcaceae (mainly Faecalibacterium sp.) and Spirochaetaceae (only Treponema sp.), whereas Clostridium sensu stricto sp. (Clostridiaceae) and Clostridium XI sp. (Peptostreptococcaceae) were the main buk-associated taxa. A few abundant genera contain members exhibiting either but or buk, such as Clostridium sensu stricto sp. or Coprococcus sp., and the applied gene association can not be regarded as strict for those taxa. Furthermore, not always all members of a certain genus are butyrate producers; this is especially true for bacteria derived from Clostridium XIVa and Lachnospiraceae_insertae_sedis. Functional predictions shown in Figure 5 should, hence, be regarded as an estimate. Genera linked to alternative transferases (Carnobacterium sp. and Erysipelotrichaceae incertae sedis sp.) were detected in a few samples as well. Furthermore, all diet groups displayed several samples that contained taxa that are specifically known to produce butyrate via amino-acid fermentation namely Fusobacterium sp., which were primarily detected in several carnivores (and noncarnivorous Carnivora), as well as Porphyromonadaceae (mainly Porphyromonas sp. and Odoribacter sp.) and Alistipes sp. (Rikenellaceae), which were enriched in omnivores and herbivores (Figure 5).
Figure 5.
16S rRNA gene analysis for potential butyrate-producing taxa is shown as percentages of the total 16S rRNA gene sequences. Data were searched on a genus level and subsequently categorized into butyryl-CoA:acetate CoA-transferase (but: bluish colors) and butyrate kinase (buk; reddish colors)-linked taxa. The applied gene association should not be regarded as strict for those taxa as several genera contain both but and buk as well as non-butyrate-producing members (see main text). Thus, this type of analysis cannot substitute for a functional gene-targeted approach but is used here for global comparisons. Taxa associated with alternative transferases (other: yellowish colors) or amino-acid-fed butyrate-producing pathways (aa; greenish colors) are shown. Individual genera from the same family were combined (see Supplementary Table S1).
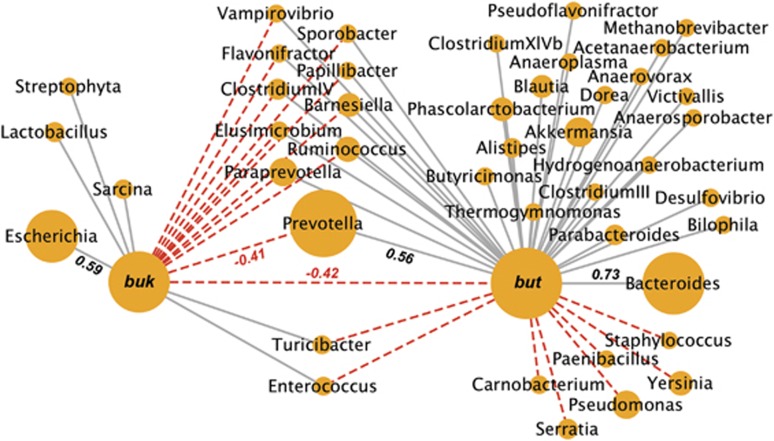
Pearson correlation analysis between the entire but- and buk-associated communities and other genera showed a clear separation between the two groups (negative correlation of r2: −0.42) and unique correlations with other taxa (Figure 6). Most obvious was a positive correlation of but-related communities with the abundant Bacteroides sp. (r2: 0.73) and Prevotella sp. (r2: 0.56), whereas Escherichia sp. was associated with buk-linked taxa (r2: 0.59).
Figure 6.
Pearson correlations of butyryl-CoA:acetate CoA-transferase communities (but) and butyrate kinase communities (buk) with other genera based on the 16S rRNA results is presented. Genera linked to individual genes (but or buk) were combined before analysis. The type of correlation (gray line: positive, red-dashed line: negative) is indicated. The size of nodes is proportional to their overall abundance in our samples. Only significant correlations (P<0.01) exhibiting an r2 of >0.4 were considered. r2 values for most important correlations are shown.
Discussion
We detected abundant butyrate-producing communities in most samples analyzed, demonstrating that they are omnipresent across Mammalia and adjunct vertebrate classes. The enormous functional redundancy (distinct microbial taxa that perform the same function) of the microbiota, which is adapted to individual host diets, enables bacteria to produce butyrate under various nutritional conditions. Owing to the volatile nature of butyrate and the different masses/densities between samples as well as differences in times of sample collection after defecation (1–24 h), accurate measurements of butyrate were not feasible. However, short-chain fatty acid measurements in fecal content of various vertebrates under controlled conditions found in literature support our results of a consistent butyrate-producing potential across members of this taxon. Stevens and Hume (1998) report high hindgut short-chain fatty acid concentrations in a series of comparative studies for carnivores, omnivores and herbivores with similar ratios between acetate-to-propionate-to-butyrate. Comparable concentrations were found in birds and reptiles (Stevens and Hume, 1998). Furthermore, in vitro incubation experiments of fecal matter showed similar butyrate-synthesis rates for species across dietary groups (Sunvold and Hussein, 1995). This strong conservation of butyrate synthesis across host taxa indicates that microbiota host interactions via butyrate are not specific to certain mammalian groups, but are, potentially, ubiquitous in vertebrates. Additional studies investigating several vertebrate taxa from distinct classes, specifically focusing on how butyrate affects host physiology, are needed in order to gain more insights into this apparent symbiosis.
Both methods, the functional gene approach as well as 16S rRNA analysis, revealed similar overall results and complement each other well. However, specifically targeting but and buk allowed deeper insights into the butyrate-producing community at fine resolutions as certain genera include both producers and non-producers and some abundant genera such as Clostridium sensu stricto or Coprococcus exhibit members linked to but and others to buk. Furthermore, a few bacteria, which were frequently detected by the functional gene approach, in particular Lachnospiraceae bacterium 3_1_57FAA_CT1 and Ruminococcaceae bacterium D16, are not classified to the genus level. In total, a considerable percentage of sequences were not classifiable at the genus level (median: 20.4%, range: 0.51%–66.2%, data not shown). Those caveats of the 16S rRNA-based analysis are most probably the reason why this method revealed much less buk-linked communities in both omni- and herbivores compared with the functional gene-based results.
The gene diversity analysis presented in Figure 2 (as well as Figure 4, Supplementary Figure S4) demonstrates a huge gap between isolated, sequenced organisms and actual in vivo communities in animal-derived samples. Most intestinal isolates and associated genome sequences in public databases are anthropocentric (many derived from the Human Microbiome Project (HMP, 2012)), leaving the microbiota of other hosts unknown. This was reflected in our data, where the majority of human-derived but-clusters, which is the predominant terminal butyrate-synthesis gene in this taxon (Louis et al., 2004), display much higher identity to known references compared with other samples. However, even in human samples the majority of the less prevalent buk gene communities are not closely related to any reference. Our data suggest that additional sequencing efforts specifically targeting bacterial genomes from distinct host taxa are needed to close the current gap.
Communities differed considerably between diets where in particular carnivores displayed a unique pattern distinct from omnivores and herbivores. Buk gene communities linked to taxa that are associated with protein-rich environments and require several amino acids for growth, namely C. difficile and C. perfringens (Haslam et al., 1986; Shimizu et al., 2002), were prevalent in this group suggesting how butyrate production of the microbiota is adapted to a carnivorous diet. The low amount of but genes in the majority of carnivores might stem from negative selection because of a lack of a consistent external acetate supply that is required for this gene to perform (Duncan et al., 2004). Pearson correlations suggest that the strict anaerobic, abundant, acetate producers Bacteroides sp. and Prevotella sp. are strongly positively correlated to but gene communities supporting the proposed but-external acetate link (Figure 6). Although acetate production is also known for Proteobacteria that are abundantly found in carnivores (in particular Escherichia sp.), environmental conditions can change their physiologies towards consumption (Kleman and Strohl, 1994). This inconsistency renders functional co-evolution based on acetate production with other taxa unlikely. Thus, in this context the predominance of buk genes in carnivores makes sense. However, additional experiments specifically investigating the link between acetate concentration and but gene abundance as a possible co-evolutionary process are needed in order to verify our hypothesis. Interestingly, a study analyzing the short-chain fatty acid contents in Grizzly Bear feces showed high butyrate concentrations and only minimal concentrations of acetate indicating non-but butyrate synthesis in those individuals (Schwab et al., 2009).
Although the acetyl-CoA pathway was abundant in most analyzed samples based on both methods, 16S rRNA analysis additionally revealed certain genera that are known to exhibit alternative, protein-fed, butyrate-synthesis pathways (Barker et al., 1982, Vital et al., 2014; Figure 5). Interestingly, they were not more abundant in carnivores but are detected at similar levels across members of all diet groups. This is in accordance with metagenomic results presented earlier (Muegge et al., 2011), where the increased abundance of amino-acid degradation pathways in carnivores did not result in enrichments of genes specifically associated with amino-acid-fed butyrate-synthesis pathways (based on EC numbers) and suggests that the distinct strategy in the terminal step of the acetyl-CoA pathway favoring either using but or buk is indeed the predominant adaptation process of the microbiota between animals of individual diet groups. However, those samples were not sequenced very deeply and investigations using new sequencing techniques could add additional information. In a recent study, human subjects who were exposed to diets composed of animal- or plant-derived products displayed functional communities that mirrored differences between herbivorous and carnivorous mammals (based on Muegge et al. (2011)) namely a distinct trade-off between carbohydrate and amino-acid degradation (David et al., 2013). From a butyrate-producing perspective, it is, however, unclear whether the human microbiota has the potential to adapt to those extreme diets in a similar way as found for different animals in this study. This is in particular in question for a strict carnivorous diet as butyrate production was significantly reduced in those human subjects (David et al., 2013). Our study revealed specific buk-containing Clostridia species as the key for butyrate synthesis in carnivorous animals and, although OTUs linked to Clostridium sp. were indeed reported to increase in individuals consuming the animal-based products, it is unknown whether those taxa are linked to butyrate production and whether they particularly exhibit the buk gene. Furthermore, abundant butyrate-producing taxa in carnivorous animals such as C. difficile or C. perfingens are associated with disease in humans (Rood and Cole, 1991; Kelly and LaMont, 2008) and negative selection processes of the host might hinder their establishment.
Most noncarnivorous Carnivora exhibited communities that were closer to those of carnivores than to those of their dietary partners (Figure 3), which is analogous to the observed pattern of entire microbial fecal communities (Supplementary Figure S6A and Ley et al., 2008). This illustrates that the governance of diet over the composition of butyrate-producing communities is not all-inclusive but that phylogenetic factors have a certain role as well. Carnivora are hindgut fermenters; however, they exhibit a distinct gut anatomy namely a so-called simple gut that might create additional selective environments for specific microbes explaining the non-diet-related similarities of their microbiota. Giant panda was the only animal that did not exhibit any butyrate-producing genes indicating that the extreme combination of a carnivorous gut anatomy with short retention times and a strict herbivorous diet (Dierenfeld et al., 1982) does not allow the establishment of a butyrate-producing community. However, the related herbivorous red panda, which also belongs to the Carnivora, exhibited abundant buk-associated butyrate producers that are typical for that group. Escherichia sp. overwhelmingly dominated the microbial flora in our giant panda samples (data not shown). It was previously reported that captive animals display Proteobacteria-enriched communities at the expense of Clostridia (containing potential butyrate-producing bacteria), which prevail in wild individuals (Zhu et al., 2011). This hints that specific unknown factors possibly associated with their captivity are biasing results and indicates that the observed absence of butyrate producers may not be normal for giant pandas.
This study presents the first assessment on butyrate-producing communities over a broad range of vertebrates (n=54) and adds substantial new information on the diversity of this important microbial function as well as its adaptation strategies to distinct diets and hosts. It demonstrates that functional properties can be conserved in bacterial communities, despite substantial differences in its structure illustrating the need of functional-based investigations to reveal specific host–microbiota interactions.
Acknowledgments
We thank the entire Potter Park Zoo team working with Dr Tara Harrison for providing access to samples, helping with sample collection and providing dietary details for all animals analyzed. Qiong Wang, Jordan Fish and Beiting Huang are specifically acknowledged for their assistance with data analysis. This study was financially supported by the NIH Human Microbiome Project Demonstration Project (UH3 DK083993). Microbial informatics support was provided by RDP under grant DE-FG02-98ER62678 from the Office of Science, US Department of Energy.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Barker H, Kahn J, Hedrick L. (1982). Pathway of Lysine Degradation in Fusobacterium nucleatum. J Bacteriol 152: 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett G, Rudolph F. (1995). The central metabolic pathway from acetyl-CoA to butyryl-CoA in Clostridium acetobutylicum. FEMS Microbiol Rev 17: 241–249. [Google Scholar]
- Buckel W, Barker H. (1974). Two pathways of glutamate fermentation by anaerobic bacteria. J Bacteriol 117: 1248–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Fish J, Chai B, McGarrell DM, Sun Y et al. (2014). Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42: D633–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE et al. (2013). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505: 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierenfeld ES, Hintz HF, Robertson JB, Van Soest PJ, Oftedal OT. (1982). Utilization of bamboo by the giant panda. J Nutr 112: 636–641. [DOI] [PubMed] [Google Scholar]
- Duncan SH, Holtrop G, Lobley GE, Calder G, Stewart CS, Flint HJ. (2004). Contribution of acetate to butyrate formation by human faecal bacteria. Br J Nutr 91: 915–923. [DOI] [PubMed] [Google Scholar]
- Eeckhaut V, Van Immerseel F, Croubels S, De Baere S, Haesebrouck F, Ducatelle R et al. (2011). Butyrate production in phylogenetically diverse Firmicutes isolated from the chicken caecum. Microb Biotechnol 4: 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish J, Chai B, Wang Q, Sun Y. (2013). FunGene: the functional gene pipeline and repository. Front Microbiol 4: 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo T, Nakato G, Takahashi D et al. (2013). Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504: 446–450. [DOI] [PubMed] [Google Scholar]
- Gerhardt A, Cinkaya I, Linder D, Huisman G, Buckel W. (2000). Fermentation of 4-aminobutyrate by Clostridium aminobutyricum: cloning of two genes involved in the formation and dehydration of 4-hydroxybutyryl-CoA. Arch Microbiol 174: 189–199. [DOI] [PubMed] [Google Scholar]
- Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. (2008). Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther 27: 104–119. [DOI] [PubMed] [Google Scholar]
- Haslam SC, Ketley JM, Mitchell TJ, Stephen J, Burdon DW, Candy DC. (1986). Growth of Clostridium difficile and production of toxins A and B in complex and defined media. J Med Microbiol 21: 293–297. [DOI] [PubMed] [Google Scholar]
- HMP. (2012). A framework for human microbiome research. Nature 486: 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly C, LaMont J. (2008). Clostridium difficile—more difficult than ever. N Engl J Med 359: 1932–1940. [DOI] [PubMed] [Google Scholar]
- Kleman GL, Strohl WR. (1994). Acetate metabolism by Escherichia coli in high-cell-density fermentation. Appl Environ Microbiol 60: 3952–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79: 5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery TJ, Roudnew B, Seymour J, Mitchell JG, Jeffries T. (2012). High nutrient transport and cycling potential revealed in the microbial metagenome of Australian sea lion (Neophoca cinerea) faeces. PLoS One 7: e36478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine UY, Looft T, Allen HK, Stanton TB. (2013). Butyrate-producing bacteria, including mucin degraders, from the swine intestinal tract. Appl Environ Microbiol 79: 3879–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS et al. (2008). Evolution of mammals and their gut microbes. Science 320: 1647–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P, Duncan SH, McCrae SI, Millar J, Jackson MS, Flint HJ. (2004). Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J Bacteriol 186: 2099–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L et al. (2011). Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332: 970–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W et al. (2012). Host-gut microbiota metabolic interactions. Science 336: 1262–1267. [DOI] [PubMed] [Google Scholar]
- Rood JI, Cole ST. (1991). Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol Rev 55: 621–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab C, Cristescu B, Boyce MS, Stenhouse GB, Gänzle M. (2009). Bacterial populations and metabolites in the feces of free roaming and captive grizzly bears. Can J Microbiol 55: 1335–1346. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T et al. (2002). Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc Natl Acad Sci USA 99: 996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens CE, Hume ID. (1998). Contributions of microbes in vertebrate gastrointestinal tract to production and conservation of nutrients. Physiol Rev 78: 393–427. [DOI] [PubMed] [Google Scholar]
- Sul WJ, Cole JR, Jesus EDC, Wang Q, Farris RJ, Fish J et al. (2011). Bacterial community comparisons by taxonomy-supervised analysis independent of sequence alignment and clustering. Proc Natl Acad Sci USA 108: 14637–14642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunvold G, Hussein H. (1995). In vitro fermentation of cellulose, beet pulp, citrus pulp, and citrus pectin using fecal inoculum from cats, dogs, horses, humans, and pigs and ruminal fluid from cattle. J Anim Sci 73: 3639–3648. [DOI] [PubMed] [Google Scholar]
- Tun HM, Brar MS, Khin N, Jun L, Hui RKH, Dowd SE et al. (2012). Gene-centric metagenomics analysis of feline intestinal microbiome using 454 junior pyrosequencing. J Microbiol Methods 88: 369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vital M, Howe Chuang A, Tiedje JM. (2014). Revealing the bacterial butyrate-synthesis pathways from (meta)genomic data. MBio 5: e00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vital M, Penton CR, Wang Q, Young VB, Antonopoulos DA, Sogin ML et al. (2013). A gene-targeted approach to investigate the intestinal butyrate-producing bacterial community. Microbiome 1: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Quensen J, Fish J, Lee T. (2013). Ecological patterns of nifH genes in four terrestrial climatic zones explored with targeted metagenomics Uuing FrameBot, a new informatics tool. MBio 4: e00592-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Wu Q, Dai J, Zhang S, Wei F. (2011). Evidence of cellulose metabolism by the giant panda gut microbiome. Proc Natl Acad Sci USA 108: 17714–17719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.