Abstract
Atmospheric carbon dioxide (CO2) levels are rapidly rising causing an increase in the partial pressure of CO2 (pCO2) in the ocean and a reduction in pH known as ocean acidification (OA). Natural volcanic seeps in Papua New Guinea expel 99% pure CO2 and thereby offer a unique opportunity to explore the effects of OA in situ. The corals Acropora millepora and Porites cylindrica were less abundant and hosted significantly different microbial communities at the CO2 seep than at nearby control sites <500 m away. A primary driver of microbial differences in A. millepora was a 50% reduction of symbiotic Endozoicomonas. This loss of symbiotic taxa from corals at the CO2 seep highlights a potential hurdle for corals to overcome if they are to adapt to and survive OA. In contrast, the two sponges Coelocarteria singaporensis and Cinachyra sp. were ∼40-fold more abundant at the seep and hosted a significantly higher relative abundance of Synechococcus than sponges at control sites. The increase in photosynthetic microbes at the seep potentially provides these species with a nutritional benefit and enhanced scope for growth under future climate scenarios (thus, flexibility in symbiosis may lead to a larger niche breadth). The microbial community in the apparently pCO2-sensitive sponge species S. massa was not significantly different between sites. These data show that responses to elevated pCO2 are species-specific and that the stability and flexibility of microbial partnerships may have an important role in shaping and contributing to the fitness and success of some hosts.
Introduction
Declining seawater pH, termed ocean acidification (OA), is a direct result of increasing atmospheric carbon dioxide (CO2) that leads to higher partial pressure of CO2 (pCO2) in seawater. Rising pCO2 reduces the concentration of carbonate ions and the saturation state of calcium carbonate minerals, which are essential for calcification in carbonate-accreting invertebrates such as corals. Volcanic CO2 seeps provide a unique opportunity to investigate how organisms respond to OA in situ (Hall-Spencer et al., 2008; Fabricius et al., 2011). Within the D'Entrecasteaux Island group in Milne Bay Province, Papua New Guinea, tropical coral reef communities exist within natural volcanic seeps that continuously expel ∼99% pure CO2 into the sediments and water column (Fabricius et al., 2011). The water chemistry, temperature, currents and light of the seep environment is otherwise consistent with reef sites <500 m away, providing a unique opportunity to study organisms in an un-manipulated natural pCO2/pH experiment. The composition of reef organisms along the pCO2/pH gradient between seep and non-seep sites varies such that as pH declines, so does the richness of structurally complex corals (e.g., branching, foliose and tabulate growth forms), crustose coralline algae, foraminifera, soft corals and a range of other macro-invertebrates (Fabricius et al., 2011; Uthicke et al., 2013; Fabricius et al., 2014). Although many of the same species are found inside and outside of the CO2 seeps, the most abundant benthic organisms living at reduced pH are massive Porites spp., non-calcareous macroalgae and seagrass. Sponge communities are also common at these shallow water CO2 seeps with recent surveys revealing that some species can tolerate extreme pH conditions, whereas others are particularly vulnerable to elevated CO2 (Goodwin et al., 2013). However, at the community level, there appears to be a significant decline in sponge cover from normal to high CO2 sites (Fabricius et al., 2011; Goodwin et al., 2013).
Corals and sponges rely on intimate and dynamic associations with diverse microorganisms for their health and physiology, yet little is known about how their microbiota respond to changes in pCO2. The ability of microbes to rapidly evolve means they can shift their host range, metabolic capabilities and other essential functions in response to changing environmental conditions. Thus, to predict the response and resilience of marine organisms under future climate scenarios, we need to understand how their associated microbiota responds to increasing pCO2. An increase in CO2 or reduction in pH can have consequences for microbially driven nutrient cycling, including carbon and nitrogen fixation (Hutchins et al., 2007), nitrification (Beman et al., 2011) and iron availability (Shi et al., 2010). CO2 enrichment studies in microcosms have documented distinct and seasonal changes in pelagic microbial communities (Krause et al., 2012), with some bacteria demonstrating enhanced growth efficiency and increased CO2 fixation at elevated pCO2 (Teira et al., 2012). Bacteria may even profit energetically if the homeostatic difference between external and internal pH (7.4 to 7.8) is reduced (Padan et al., 2005). Most previous studies were conducted on pelagic bacteria and it is unknown whether such microbial shifts and physiological changes also occur within host-associated communities. Furthermore, whether such changes compromise host health or infer an adaptive advantage has not yet been explored. Aquarium-based studies have shown that increased pCO2 or reduced pH can cause microbial shifts in communities associated with corals, foraminifera and crustose coralline algae (Meron et al., 2011; Webster et al., 2013a, 2013b). However, to date, no study has explored microbial communities in organisms that have spent their entire lives at elevated CO2, with the potential for their communities to adapt or physiologically acclimatize over large timescales (>3 years).
Coral-associated microbial communities are composed of a core population of species-specific bacteria, in addition to transient associates that can vary in response to geographic location, climate and other environmental factors (Ritchie, 2006). Some coral species support highly specific associations, such as the coral Porites astreoides in the Caribbean and Stylophora pistillata in the Red Sea, which both host microbiomes largely composed of bacteria in the family Hahellaceae (Endozoicomonas sp.; (Morrow et al., 2012; Bayer et al., 2013a, 2013b), whereas other corals have more diverse associations (Bourne et al., 2013). The significance of these diversity differences has not yet been fully elucidated although it is believed that this has an important role in host fitness and stability in changing environmental conditions. Marine sponges are also known to host dense, diverse and highly stable microbial communities with many of the microorganisms being specific to sponge hosts (Simister et al., 2012). Environmental disturbances such as temperature, bleaching, disease, nutrient enrichment and changes in pH can all cause dramatic changes in both coral and sponge microbiota (Webster et al., 2008; Vega Thurber et al., 2009; Meron et al., 2011; Fan et al., 2012; Vega Thurber et al., 2014). Symbiotically conferred stress tolerance has been demonstrated in a number of systems (Rodriguez et al., 2008) and may provide a strategy for withstanding the effects of climate change and/or OA. The current study uses 16S rRNA gene amplicon pyrosequencing to assess the pCO2/pH sensitivity of microbial associations within two coral and three sponge species and explore whether host tolerance to OA stress could be enabled via habitat-adapted microbial associations.
Materials and methods
Sample collection and processing
Sponge and coral abundance was surveyed at the volcanic shallow water CO2 seep system (Upa Upasina) within the D'Entrecasteaux Islands, Milne Bay Province, Papua New Guinea (PNG; 9°49.45' S, 150°49.07' E), and at a nearby ambient pCO2 ‘control' site not exposed to elevated pCO2 (<500 m from the seeps, 9°49.68' S, 150°49.23' E; (Fabricius et al., 2011; Uthicke et al., 2013). Sponge abundance was surveyed across five transects (20 m × 1 m) within each site in 2011 (depth 3–4 m). Coral abundance was surveyed across 15 transects (10 m × 0.5 m) at each site in 2014 to determine the abundance of Acropora spp. and other CO2-sensitive coral species, including P. cylindrica.
Replicate samples of Acropora millepora (n=10) and Porites cylindrica (n=5) coral colonies and Coelocarteria singaporensis, Cinachyra sp. and Stylissa massa sponge individuals (n=8–10 per species per site) were collected on SCUBA from both the high pCO2 seep site and the ambient control site. Seawater carbonate chemistry varies in response to bubble activity and water motion at the seep; thus, corals experience a pH range of 7.91–8.09 (Avg. pCO2 346 μatm) at the control site and pH 7.28–8.01 (Avg. pCO2 624 μatm) at the seep site (Fabricius et al., 2014), which is within the range of future predictions for the year 2100 (Moss et al., 2010). Coral and sponge tissue samples were transported to the surface in individual plastic bags and immediately preserved in 95% ethanol within 15 ml falcon tubes and stored at −20 °C for transport back to the Australian Institute of Marine Science (AIMS).
Tissues from sampled coral colonies were removed with sterile tips and pressurized air into the original preservation ethanol, then transferred to Beckman Ultra-clear centrifuge tubes (Beckman-Coulter, Brea, CA, USA), and pelleted in a swinging bucket rotor (20 000 g for 15 min at 4 °C). Each pellet was resuspended using PowerPlant Pro Microbead solution (MoBio Laboratories, Carlsbad, CA, USA), transferred to MoBio microbead tubes and frozen at −20 °C. Samples were later thawed at 65 °C for 10 min and extracted using the PowerPlant Pro DNA Isolation Kit as per the manufacturer's instructions (MoBio Laboratories). DNA was also extracted from all sponge samples using MoBio PowerPlant DNA Isolation Kit as per the manufacturer's instructions.
16S gene-targeted pyrosequencing
Extracted DNA was quantified using a NanoDrop 2000 UV–vis spectrophotometer (Wilmington, DE, USA), and aliquoted to the same concentration (20 ng μl−1). All samples were PCR amplified with universal bacterial primers targeting the hypervariable region V1-V3 of the 16S ribosomal rRNA gene with 28 F (5′GAGTTTGATCNTGGCTCAG3′) and 519 R (5′GTNTTACNGCGGCKGCTG3′) and sequenced at the Molecular Research Laboratories (Shallowater, TX, USA). Briefly, a single-step 30-cycle PCR using the HotStarTaq Plus Master Mix Kit (Qiagen, Valencia, CA, USA) was used under the following conditions: 94 °C for 3 min, followed by 28 cycles of 94 °C for 30 s; 53 °C for 40 s and 72 °C for 1 min; after which a final elongation step at 72 °C for 5 min was performed. Following PCR, all amplicon products were mixed in equal concentrations and purified using Agencourt AMPure beads (Beckman Coulter), followed by sequencing using the Roche (Basel, Switzerland) 454 FLX+ platform and Titanium reagents.
Sequencing and statistical analyses
Raw.sff sequence reads from all samples were denoized and processed through the MOTHUR software package (www.mothur.org) and the analysis protocol was based on the 454 standard operating procedure (Schloss et al., 2011). Briefly, sequences of length<200 bp, ambiguous base calls or homopolymer runs exceeding 8 bp were removed. Sequences were aligned with the SILVA 16 S rRNA alignment (Pruesse et al., 2007) as a reference and operational taxonomic units (OTUs) were defined using Pre.cluster after removal of singleton sequences, clustering at 3% divergence (97% similarity). Chimeric artefacts were also removed using UCHIME (Edgar et al., 2011). OTUs were taxonomically classified using a BLAST-based method against the May 2013 curated Greengenes database ((Altschul et al., 1997; DeSantis et al., 2006), http://greengenes.secondgenome.com/), and compiled at each taxonomic level into a counts file. Any sequences that were classified as Mitochondria, Eukaryotic or Chloroplast as well as any sequences of unknown origin were filtered out of the dataset. Sequences without hits were rechecked against the NCBI-nr database (http://www.ncbi.nlm.nih.gov/) for 16S rRNA gene sequences retrieved from uncultured bacteria using a BLAST search (Altschul et al., 1997) with the E-value threshold (0.001). Data from each coral and sponge species were analysed separately and each rarefied to account for variation in sampling depth. The raw pyrosequencing reads were submitted to the NCBI Sequence Read Archive under accession numbers PRJNA253930 for corals and SRP029239 for sponges.
Alpha diversity statistics including total observed and predicted Chao1 (Chao, 1984) OTUs and the Shannon H' diversity statistic were calculated in MOTHUR and plotted. OTU tables were built in Microsoft Excel from two MOTHUR output files (SHARED and TAXONOMY), and pivot tables were used to condense tables by phyla and family for graphical interpretation. OTU data were square-root-transformed and similarity percentage (SIMPER) analyses were performed to examine which OTUs contributed most to the dissimilarity between sites. Bray-Curtis distance matrices were built to examine additional patterns of community structure and visualized using principal coordinate analyses. Pearson correlation vectors were overlayed to demonstrate which taxa have strong positive or negative correlations with either PCO axis, indicative of site differences. A permutational analysis of variance (PERMANOVA, using 10 000 permutations) determined whether spatial separation between sites was statistically significant. All multidimensional statistical analyses were performed in PRIMER V6 with the PERMANOVA+ add-on (PRIMER-E Ltd., Devon, UK).
Functional profile prediction for key symbionts
To better understand the potential functional contributions of the observed shifts in microbial diversity, we imputed functional profiles for two key microbial symbionts in the PICRUSt software package ((Langille et al., 2013), http://picrust.github.io/picrust/). This technique estimates the genomic copy number of each gene family (KO) in an environmental microorganism based on the position of that organism in a reference phylogeny with regard to microorganisms for which finished genomes are available. Hidden state prediction (Zaneveld and Vega Thurber, 2014) is then used to model changes in each gene family over time, and estimate mean and 95% confidence intervals for the abundance of each gene family.
Functional profile imputation was performed on partial 16S rRNA gene sequences for the Endozoicomonas (Gammaproteobacteria:Oceanospirillales) symbiont of A. millepora (Otu_0002; 256 bp) and the Synechococcus (Cyanobacteria: Synechococcophycideae) symbiont of C. singaporensis (Otu_0001; 125 bp). These sequences were mapped to the Greengenes 13_8 reference phylogeny (Mcdonald et al., 2012) using pick_closed_reference_otus.py script of the QIIME 1.8 software package (Caporaso et al., 2010), running in the MacQIIME environment. Functional profiles, Nearest Sequenced Taxon Index (NSTI) scores and 95% confidence intervals were calculated using the PICRUSt's predict_metagenomes.py script, treating each OTU as a separate sample. Following functional imputation, the obtained profiles of gene family abundance across samples were summarized into KEGG Pathways using the categorize_by_function.py PICRUSt script (PICRUSt counts genes that participate in multiple KEGG pathways once per pathway).
Results
Community abundance surveys
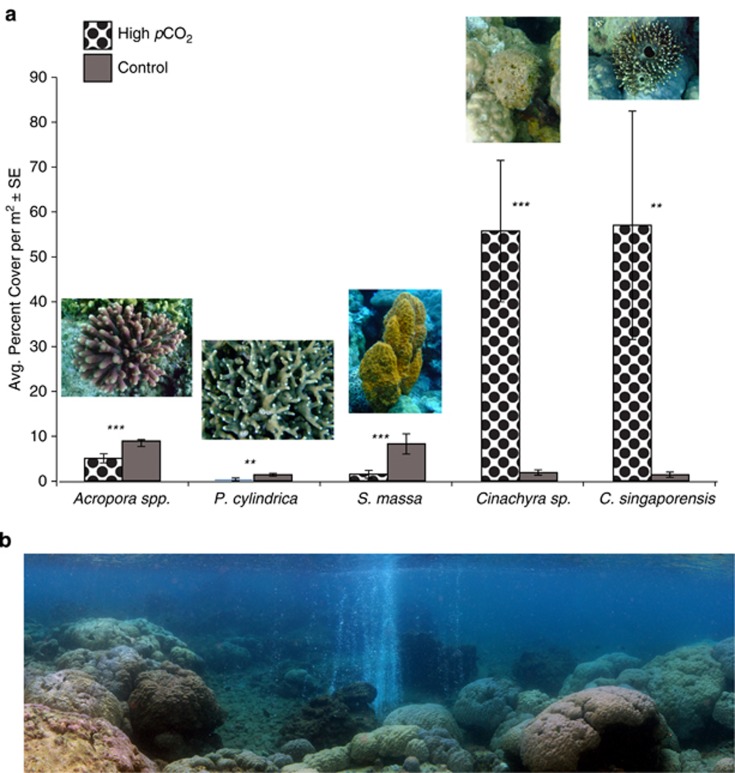
Previous coral abundance surveys conducted at the volcanic CO2 seep in Milne Bay reported that both coral species, A. millepora and P. cylindrica, were significantly less abundant at the seep than at the adjacent control site (see Fabricius et al., 2011). Surveys repeated in March 2014 confirm a significant reduction in Acropora spp. (∼ two times less abundant) and P. cylindrica (∼five times less abundant) at the seep site in comparison with the control site (ANOVA, F(1,33)=20.6, P<0.001 and F(1,33)=11.4, P=0.002, respectively; Figure 1). Sponge surveys showed that S. massa was ∼six times less abundant at the seep than at the control site (ANOVA, F(1,10)=12.0, P=0.006; Figure 1). In contrast, both C. singaporensis and Cinachyra sp. were almost 40 times more abundant at the CO2 seep than at the control site (ANOVA, F(1,10)=8.6, P=0.015; F(1,10)=17.5, P= 0.002, respectively; Figure 1).
Figure 1.
(a) Sponge and coral percent cover per m2. (b) Panorama of the PNG seep site. Photo: S. Noonan.
Coral-associated bacterial communities
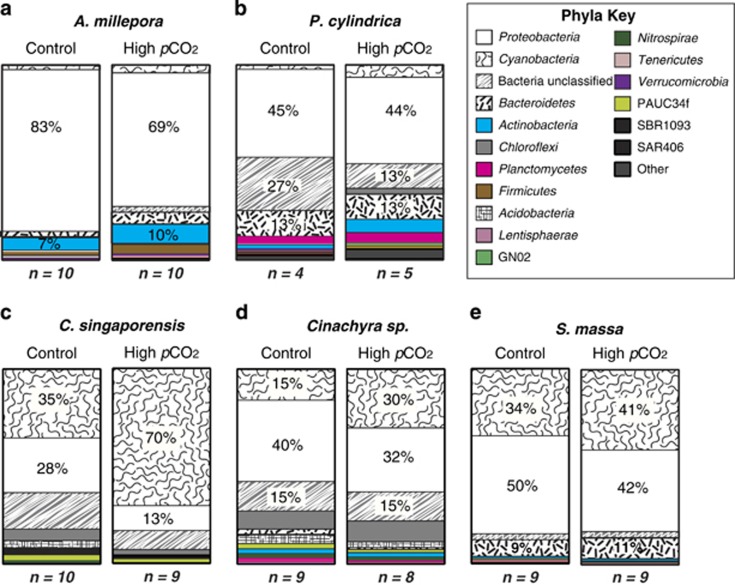
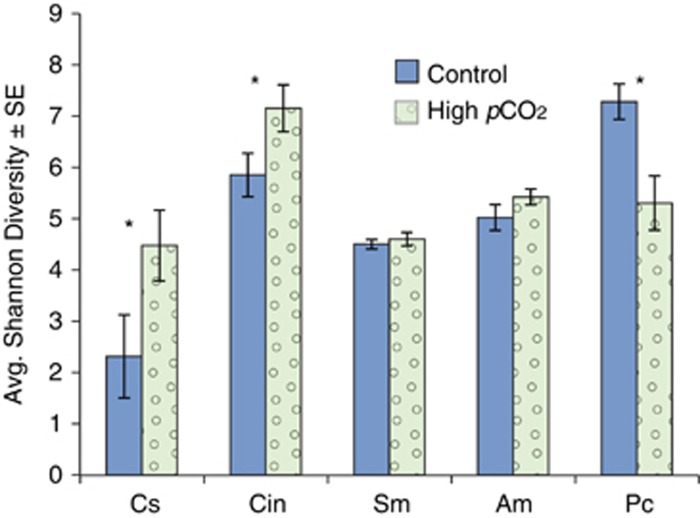
A total of 193 724 high-quality 16S rRNA gene pyrosequencing reads were recovered for both coral species from the seep and control sites, with an average of 6457±1213 reads per sample. Each coral species was analysed separately and retrieved sequences were rarefied to the sample with the least reads (1426 reads for A. millepora and 1850 reads for P. cylindrical) (Table 1). Both coral species hosted a number of unique bacterial phyla, including 27 phyla represented in A. millepora and 30 in P. cylindrica. The most abundant and diverse phylum recovered from both coral species was the Proteobacteria (Figure 2). P. cylindrica hosted higher bacterial diversity than A. millepora, with a significant reduction in microbial diversity at the phylum level at the high pCO2 site (H'=7.2±0.34 s.e.) compared with the control site (H'=5.3±0.5 s.e.; F (1,7)=10.7, P=0.01; Figure 3).
Table 1. Alpha diversity statistics based on sequences derived from two coral species and three sponge species in the control and high pCO2 seep sites.
|
A. millepora |
P. astreoides |
C. coelocarteria |
Cinachyra sp. |
S. massa |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Control (10)a | High pCO2 (10) | Control (4) | High pCO2 (5) | Control (10) | High pCO2 (9) | Control (9) | High pCO2 (8) | Control (9) | High pCO2 (9) | |
| Avg # Trimmed Seqs | 6488±2462 | 3463±534 | 10 274±3954 | 5848±1863 | 5520±961 | 7393±1116 | 9478±753 | 5320±431 | 5399±680 | 6043±1703 |
| # Rarefied Seqs | 1426 | 1850 | 2420 | 2568 | 2148 | |||||
| Greengenes OTUs | 1528 | 2794 | 3279 | 6348 | 2744 | |||||
| Unique OTUsb | 431 | 449 | 140 | 448 | 267 | |||||
| Chao1 | 208±20 | 216±21 | 595±70 | 789±66 | 389±89 | 558±61 | 1322±130 | 1394±198 | 395±26 | 446±33 |
| Observed | 145±9 | 141±10 | 261±40 | 439±48 | 135±33 | 209±22 | 431±44 | 602±86 | 191±11 | 202±8 |
Abbreviations: OTUs, operational taxonomic units; pCO2, partial pressure of CO2.
Average alpha diversity statistics±s.e. calculated with alpha_diversity.py in qiime_VirtualBox.
Number in parentheses indicates samples included in analyses (n=#).
Number of unique OTUs after all identical OTUs exported from Mothur were combined.
Figure 2.
Average percent contribution to total abundances of most dominant phyla across all species replicates. (a) A. millepora, (b) P. cylindrica, (c) C. singaporensis, (d) Cinachyra sp. and (e) S. massa. n=the total number of individual sponges or coral colonies sampled for each species at each site.
Figure 3.
Shannon Diversity index: means±s.e. The asterisk indicates significant difference at alpha<0.05.
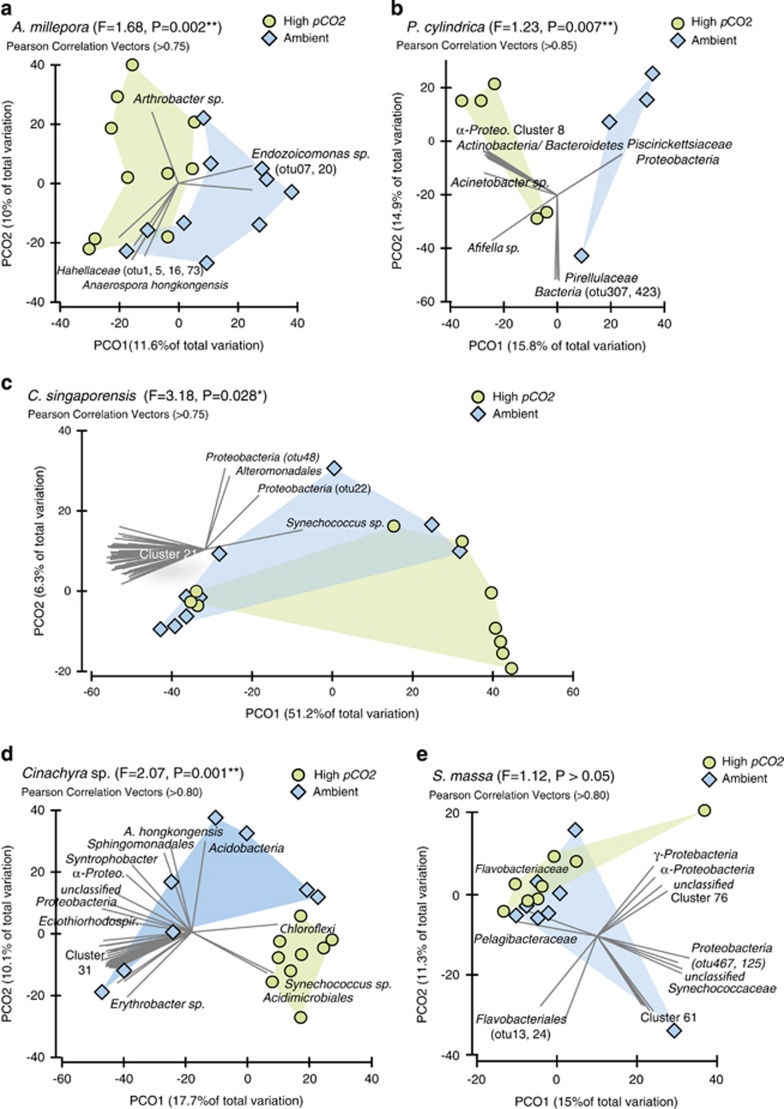
At the bacterial species level (97% sequence similarity), significant differences in community composition were observed between sites in both A. millepora (PERMANOVA Pseudo-F1,19=1.68, P=0.002) and P. cylindrica (Pseudo-F1,8=1.23, P=0.007; Figure 4). Significant differences in A. millepora microbes between sites were the direct result of changes in the relative abundance of OTUs affiliated with members of the Proteobacteria (Figure 4, Table 2). The relative abundance of most Proteobacteria sequences increased in corals at the CO2 seeps, except for sequences associated with the class Gammaproteobacteria. In particular, sequences associated with the genus Endozoicomonas sp. (class Gammaproteobacteria, family Hahellaceae) were two times less abundant in A. millepora at the seep compared with the control site (26.4 vs 51.6% Table 2). SIMPER analysis and Pearson correlation vectors associated with principal coordinate analyses plots also indicate that members of the Endozoicomonas sp. were particularly affected by the differences in environmental and host-related conditions between seep and control sites (Figure 4, Table 3). Most other Gammaproteobacteria affiliated sequences from A. millepora also decreased at the seep site in comparison with the control, except for sequences within the family Halomonadaceae, which increased in relative abundance from 1.2 to 4.8% of the total microbiome at high pCO2 (Table 2). In contrast, the relative abundance of sequences associated with many other coral-associated bacterial phyla increased at the CO2 seep in comparison with the adjacent control site (e.g., Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes; Table 2).
Figure 4.
Principal coordinate analysis plots for each coral and sponge species based on square-root transformed Bray-Curtis distances. (a) A. millepora, (b) P. cylindrica, (c) C. singaporensis, (d) Cinachyra sp. and (e) S. massa. Green circles indicate bacterial communities sampled at the seep sites (high pCO2 and low pH), and blue diamonds represent samples from adjacent control locations. Pearson correlation vectors represent the most resolved taxonomy for bacterial OTUs. PERMANOVA. Pseudo-F and P-values are reported. The list of taxa composing the OTU clusters is available in Supplementary Material.
Table 2. Relative abundance of OTUs affiliated with dominant phyla.
| a. A. millepora | Control | High pCO2 | a. C. singaporensis | Control | High pCO2 | % of Total |
|---|---|---|---|---|---|---|
| Proteobacteria (% of Total) | 83.5 | 68.8 | Proteobacteria (% of Total) | 23.3 | 10.8 | 0.1–0.5 |
| Alphaproteobacteria | 14.4 | 19.8 | Alphaproteobacteria | 2.8 | 2.3 | 0.6–1.0 |
| Caulobacteraceae | 3.5 | 2.6 | Pelagibacteraceae | 0.3 | 0.2 | 1.1–2.5 |
| Rhodobacteraceae | 2.4 | 2.9 | unclassified | 2.3 | 2.0 | 2.5–5.0 |
| Sphingomonadaceae | 1.5 | 0.8 | Deltaproteobacteria | 3.6 | 1.3 | 5.1–7.5 |
| unclassified | 3.7 | 6.9 | Syntrophobacteraceae | 0.9 | 0.4 | 7.6–10 |
| Betaproteobacteria | 2.2 | 4.4 | Entotheonellaceae | 2.6 | 0.8 | 10.1–25 |
| Comamonadaceae | 1.4 | 3.4 | Gammaproteobacteria | 0.6 | 0.3 | >25% |
| Gammaproteobacteria | 66.2 | 43.2 | HTCC2089 | 2.9 | 1.6 | |
| Hahellaceae | 51.6 | 26.4 | HTCC2188 | 0.5 | 0.4 | |
| Enterobacteriaceae | 1.8 | 0.6 | unclassified | 11.2 | 5.2 | |
| Halomonadaceae | 1.2 | 4.8 | Cyanobacteria (% of Total) | 35.0 | 70.1 | |
| Moraxellaceae | 3.7 | 2.9 | Synechococcaceae | 35.0 | 70.0 | |
| Pseudoalteromonadaceae | 1.5 | 0.5 | unclassified | 0.1 | 0.1 | |
| Pseudomonadaceae | 2.7 | 3.1 | ||||
| Actinobacteria | 6.55 | 9.82 | b. Cinachyra sp. | Control | High pCO2 | |
| Bacteroidetes | 2.72 | 5.72 | Proteobacteria (% of Total) | 39.6 | 31.8 | |
| Cyanobacteria | 2.05 | 4.19 | Alphaproteobacteria | 15.9 | 7.2 | |
| Firmicutes | 1.82 | 4.65 | Rhodobacteraceae | 6.6 | 2.8 | |
| unclassified | 1.48 | 2.90 | Rhodospirillaceae | 0.4 | 0.2 | |
| Pelagibacteraceae | 0.7 | 0.8 | ||||
| b. P. cylindrica | Control | High pCO2 | unclassified | 6.5 | 3.5 | |
| Proteobacteria (% of Total) | 45.0 | 43.8 | Deltaproteobacteria | 0.5 | 0.2 | |
| Alphaproteobacteria | 13.1 | 20.7 | unclassified | 0.1 | 0.1 | |
| Rhodobacteraceae | 1.6 | 4.1 | Gammaproteobacteria | 19.2 | 16.3 | |
| Rhodobiaceae | 1.4 | 1.1 | HTCC2089 | 1.3 | 2.6 | |
| unclassified | 7.6 | 11.1 | Piscirickettsiaceae | 0.9 | 0.2 | |
| Betaproteobacteria | 0.4 | 1.8 | OM60 | 0.3 | 0.2 | |
| Deltaproteobacteria | 0.5 | 1.3 | Alteromonadaceae | 0.3 | 0.1 | |
| Gammaproteobacteria | 29.9 | 16.1 | Colwelliaceae | 0.1 | 0.1 | |
| Alteromonadaceae | 11.4 | 1.1 | unclassified | 15.2 | 12.5 | |
| Chromatiaceae | 0.2 | 4.2 | Cyanobacteria (% of Total) | 14.7 | 30.0 | |
| Pseudoalteromonadaceae | 8.1 | 0.7 | Synechococcophycideae | 9.9 | 25.8 | |
| Vibrionaceae | 2.0 | 0.8 | Synechococcaceae | 7.2 | 25.7 | |
| unclassified | 1.5 | 8.9 | Pseudanabaenaceae | 2.6 | 0.1 | |
| Bacteroidetes (% of Total) | 13.3 | 12.8 | Oscillatoriophycideae | 0.6 | 0.4 | |
| Bacteroidia | 0.3 | 0.4 | Cyanobacteriaceae | 0.0 | 0.2 | |
| Cytophagia | 6.0 | 3.0 | Phormidiaceae | 0.0 | 0.1 | |
| Flavobacteriia | 1.3 | 7.0 | unclassified | 0.3 | 0.1 | |
| Saprospirae | 1.9 | 0.2 | ||||
| unclassified | 3.0 | 2.0 | c. S. massa | Control | High pCO2 | |
| Actinobacteria | 2.22 | 7.18 | Proteobacteria (% of Total) | 46.7 | 38.8 | |
| Cyanobacteria | 2.47 | 6.64 | Alphaproteobacteria | 16.0 | 15.3 | |
| Planctomycetes | 3.70 | 5.39 | Pelagibacteraceae | 12.7 | 10.6 | |
| Chloroflexi | 0.76 | 3.02 | Rhodobacteraceae | 2.6 | 4.2 | |
| unclassified | 3.00 | 2.00 | AEGEAN_112 | 0.3 | 0.2 | |
| unclassified | 0.1 | 0.1 | ||||
| Deltaproteobacteria | 6.3 | 2.0 | ||||
| unclassified | 6.2 | 1.9 | ||||
| Gammaproteobacteria | 23.1 | 20.7 | ||||
| Halomonadaceae | 1.8 | 3.6 | ||||
| unclassified | 20.5 | 16.5 | ||||
| Cyanobacteria (% of Total) | 33.9 | 40.5 | ||||
| Synechococcophycideae | 31.4 | 38.0 | ||||
| Synechococcaceae | 31.4 | 37.9 | ||||
| Bacteroidetes (% of Total) | 8.2 | 10.0 | ||||
| Cytophagia | 1.6 | 2.3 | ||||
| Flammeovirgaceae | 1.6 | 2.3 | ||||
| Flavobacteriia | 2.9 | 3.5 |
Abbreviation: OTUs, operational taxonomic units.
Table 3. Similarity Percentage Analysis (SIMPER) for 10 most significant OTUs driving differences between the CO2 seep and control site.
| Phylum | Most resolved taxonomy (Blastn) | “Otu” |
Higher abundance |
Fold difference | Rank | |
|---|---|---|---|---|---|---|
| High pCO2 | Ambient | |||||
| A. millepora dissimilarity=85.17% | ||||||
| Actinobacteria | Corynebacterium vitaeruminis | Otu0008 | x | 1.02 | 9 | |
| Proteobacteria | ||||||
| Alphaproteobacteria | Brevundimonas sp. | Otu0010 | x | 2.46 | 10 | |
| Gammaproteobacteria | ||||||
| Hahellaceae | unclassified | Otu0001 | x | 0.53 | 1 | |
| Hahellaceae | unclassified | Otu0005 | x | 0.24 | 7 | |
| Hahellaceae | Endozoicomonas sp. | Otu0002 | x | 6.65 | 2 | |
| Hahellaceae | Endozoicomonas sp. | Otu0003 | x | 4.39 | 3 | |
| Hahellaceae | Endozoicomonas sp. | Otu0007 | x | 5.12 | 4 | |
| Hahellaceae | Endozoicomonas numazuensis | Otu0006 | x | 3.89 | 5 | |
| Halomonadaceae | Chromohalobacter salexigens | Otu0013 | x | 3.88 | 8 | |
| Bacteria | unclassified | Otu0004 | x | 0.99 | 6 | |
| P. cylindrica dissimilarity=94.8% | ||||||
| Actinobacteria | Corynebacterium vitaeruminis | Otu0007 | x | 1.6 | 6 | |
| Bacteroidetes | Flavobacteriaceae | Otu0006 | x | 3.83 | 7 | |
| Cyanobacteria | Oscillatoriophycideae | Otu0001 | x | 3.66 | 1 | |
| Proteobacteria | ||||||
| Alphaproteobacteria | unclassified | Otu0018 | x | 3.97 | 9 | |
| Gammaproteobacteria | ||||||
| Alteromonadaceae | Candidatus Endobugula | Otu0004 | x | 5.23 | 4 | |
| Chromatiaceae | Rheinheimera sp.NG3 | Otu0013 | x | 4.04 | 10 | |
| Pseudoalteromonadaceae | Pseudoalteromonas sp. RKVR13 | Otu0005 | x | 5.22 | 3 | |
| Pseudoalteromonadaceae | Pseudoalteromonas sp.EF1A-B164 | Otu0009 | x | 4.47 | 5 | |
| Bacteria | unclassified | Otu0003 | x | 5.37 | 2 | |
| Bacteria | unclassified | Otu0002 | x | 4.48 | 8 | |
| C. singaporensis avg. dissimilarity=71.85% | ||||||
| Cyanobacteria | Synechococcus sp. | Otu0001 | x | 13.68 | 1 | |
| Proteobacteria | ||||||
| Deltaproteobacteria | Entotheonellaceae | Otu0008 | x | 3.63 | 6 | |
| Gammaproteobacteria | unclassified | Otu0004 | x | 4.54 | 2 | |
| unclassified | Otu0005 | x | 3.74 | 5 | ||
| unclassified | Otu0010 | x | 3.41 | 8 | ||
| Bacteria | unclassified | Otu0002 | x | 5.69 | 3 | |
| SBR1093 | Otu0003 | x | 4.64 | 4 | ||
| unclassified | Otu0006 | x | 3.27 | 7 | ||
| PAUC34f | Otu0007 | x | 2.57 | 9 | ||
| unclassified | Otu0009 | x | 2.96 | 10 | ||
| Cinachyra sp. avg. dissimilarity=79.42% | ||||||
| Actinobacteria | Acidimicrobiales | Otu00030 | x | 3.76 | 10 | |
| Cyanobacteria | Synechococcus | Otu00001 | x | 16.13 | 1 | |
| Synechococcus | Otu00002 | x | 2.07 | 6 | ||
| Proteobacteria | unclassified | Otu00005 | x | 8.45 | 2 | |
| Gammaproteobacteria | unclassified | Otu00010 | x | 7.07 | 3 | |
| unclassified | Otu00003 | x | 2.64 | 4 | ||
| HTCC2089 | Otu00006 | x | 3.19 | 8 | ||
| Nitrospirae | Nitrospiraceae | Otu00007 | x | 1.1 | 5 | |
| Bacteria | unclassified | Otu00016 | x | 3.91 | 7 | |
| Bacteria | PAUC34f | Otu00012 | x | 2.67 | 9 | |
| S. massa avg. dissimilarity=47.06% | ||||||
| Alphaproteobacteria | Pelagibacteraceae | Otu0006 | x | 2.01 | 5 | |
| Pelagibacteraceae | Otu0004 | x | 0.75 | 10 | ||
| Cyanobacteria | Synechococcus sp. | Otu0001 | x | 2.57 | 3 | |
| Synechococcaceae | Otu0012 | x | 0.64 | 6 | ||
| Synechococcaceae | Otu0030 | x | 0.45 | 9 | ||
| Proteobacteria | ||||||
| Deltaproteobacteria | unclassified | Otu0005 | x | 4.59 | 2 | |
| Gammaproteobacteria | Candidatus Portiera | Otu0017 | x | 1.44 | 8 | |
| Thiohalorhabdales | Otu0003 | x | 0.97 | 4 | ||
| unclassified | Otu0002 | x | 4.40 | 1 | ||
| unclassified | Otu0009 | x | 1.05 | 7 | ||
Abbreviation: OTUs, operational taxonomic units.
Porites cylindrica also demonstrated a decrease in OTUs affiliated with the Gammaproteobacteria at the seep compared with the control site (Table 2), especially in the three families Alteromonadaceae (11.4–1.1% of the total microbiome), Pseudoalteromonadaceae (8.1–0.7%), and Vibrionaceae (2.0–0.8%, Table 2). The only group that increased at the seep site were purple sulphur bacteria in the family Chromatiaceae (0.2–4.2% of the total microbiome), mostly composed of bacteria associated with the genus Rheinheimera sp. (Table 2). SIMPER analyses indicate that members of the Alteromonadaceae and Pseudoalteromonadaceae influence site differences in the community structure for P. cylindrica (Table 3). Members of the phylum Bacteroidetes composed approximately 13% of the P. cylindrica microbiome at both sites, but they varied in composition (Table 2). For example, bacteria in the class Flavobacteria increased in relative abundance (1.3–7.0% of the total microbiome), and Cytophaga decreased in relative abundance (6.0–3.0%) at the seep site in comparison with the control (Table 2). Furthermore, members of the Bacteroidetes phylum are indicated by Pearson correlation vectors and SIMPER analyses as influencing site differences (Table 3, Figure 4). The relative abundance of several other bacterial phyla (e.g., Actinobacteria, Cyanobacteria, Planctomycetes and Chloroflexi) increased in abundance at the CO2 seep in comparison with the control site (Table 2). Interestingly, both corals demonstrate a twofold increase in Cyanobacteria-affiliated OTUs from approximately 2.5% of the total microbiome at the control site to 6% at the seep site (Table 2).
Sponge-associated bacterial communities
A total of 373 494 high-quality 16S rRNA reads were recovered from all three sponge species with an average of 6163±279 s.e. reads per sample (Table 1). Samples from each sponge species were rarefied to the lowest retrieved number of sequences, ranging from 2148 to 2568 reads (Table 1). Each sponge species hosted a number of unique bacterial phyla including 15 hosted by C. singaporensis, 30 hosted by Cinachyra sp. and 18 hosted by S. massa. Like in corals, Proteobacteria were the most abundant and diverse phylum in Cinachyra sp and S. massa, although C. singaporensis was dominated by Cyanobacteria, particularly at the seep site (70%, Figure 2). Among the three sponge species, Cinachyra sp. hosted the highest bacterial diversity at the high pCO2 site (H'=7.2±0.5) in comparison with the control (Shannon H'=5.9±0.4; ANOVA F(1,15)=4.3 P=0.05; Figure 3).
At the bacterial species level (97% sequence similarity), significant differences in community composition between sites were detected for the two pCO2-tolerant sponge species C. singaporensis (PERMANOVA F=3.18, P=0.03) and Cinachyra sp. (F=2.07, P=0.001; Figure 4). The only species examined that did not differ in microbial community composition between the seep and control site was the apparently pCO2-sensitive sponge species S. massa (F=1.12, P>0.05; Figure 4). OTUs affiliated with Synechococcus sp. and Proteobacteria were responsible for much of the between-site differences in both C. singaporensis and Cinachyra sp. (Figure 4) and these Synechococcus sp. OTUs were clearly distinct from the cyanobacterial OTUs observed within the two corals (Table 3). A twofold increase in Cyanobacteria (Synechococcus spp.) from the control to the seep site was observed in both C. singaporensis (35–70% of the total microbiome) and Cinachyra sp. (1–30%), whereas Cyanobacteria showed only a marginal increase in S. massa (34–41% Figure 2, Table 2). SIMPER analyses also demonstrated that Synechococcus sp. was the major driver of differences between seep and non-seep bacterial communities for C. singaporensis and Cinachyra sp. (Table 3). In addition to Synechococcus spp., a large number of less-discriminating bacterial OTUs also contributed to the between-site variation (Figure 4). The increase in Cyanobacteria corresponded to a relative decrease in classes Alpha-, Beta-, Delta- and Gammaproteobacteria in all sponges at the seep site in comparison with the control (Table 2). In addition, concomitant with the elevated abundance of Cyanobacteria in C. singaporensis and Cinachyra sp. at the seep was a generally lower relative abundance of phyla containing known sponge symbionts. For example, in C. singaporensis, Chloroflexi declined from 5.8 to 2.7%, Acidobacteria declined from 3.8 to 1.1%, Deltaproteobacteria declined from 3.6 to 1.3% and Nitrospirae declined from 1.3 to 0.4%. (Figure 2, Table 2). Similarly, in Cinachyra sp., Acidobacteria declined from 4.6 to 2.9%, Planctomycetes declined from 1.9 to 0.6% and Bacteroidetes declined from 2.6 to 1.1% (Figure 2, Table 2). In contrast, with the exception of Cyanobacteria and Bacteroidetes, which both showed a marginal but non-significant increase at the seep, the distribution of OTUs within phyla in S. massa were consistent across both sites (Figure 2).
Functional implications of community shifts
To summarize some of the potential functional contributions of key symbionts, functional profiles were predicted for the major Endozoicomonas symbiont lost in A. millepora near the seep site and Synechococcus that increased in C. singaporensis using the PICRUSt software package. This approach uses evolutionary modelling to put bounds on the gene content of environmental organisms based on their sequenced relatives. The accuracy of the method is highest when closely related reference genomes are available. The validity of reference genomes can be quantified using the NSTI score, which is simply the average branch length on the reference phylogeny between the organism(s) to be predicted and an organism for which a genome sequence is available (Langille et al., 2013). Sufficiently close reference genomes were available for the C. singaporensis Synechococcus symbiont (NSTI 0.056), whereas available genomic data were at the outer limits of accurate prediction for the Endozoicomonas symbiont of A. millepora (NSTI 0.156). On the basis of previous cross-validation studies of all sequenced genomes, these NSTI scores suggest that a balanced accuracy of ∼0.95 should be expected for prediction of gene family presence/absence (for genes in KEGG KOs) in the Synechococcus and ∼0.85 for the Endozoicomonas (Langille et al., 2013, Supplementary Figure 1). Dominant KEGG Pathways for Synechococcus included photosynthesis, membrane transport, and DNA repair and recombination (Supplementary Figure 1). Dominant KEGG Pathways in the Endozoicomonas from A. millepora included pathways for membrane transporters, ABC transporters, two-component signalling systems, cell motility and secretion (Supplementary Figure 1).
These imputed functional profiles largely reflected the known roles of these organisms. The Synechococcus functional profile included many genes involved in photosynthesis pathways, whereas these were absent or at very low abundance in the heterotrophic Endozoicomonas. Conversely, Endozoicomonas were marked by high abundances of membrane transporters (∼1.7 × their abundance in Synechococcus), motility genes and two-component signalling systems. Both profiles included poorly characterized gene families within their top three KEGG Pathways. Although not atypical in bacteria, the abundance of gene families with poorly characterized function in both abundant symbionts highlights the potential for unexplored functional novelty in these groups.
Discussion
Compared with preindustrial levels, global oceans have already experienced a 30% increase in acidity, and are predicted to undergo a further 170% increase in acidity by 2100 (IGBP, IOC, SCOR, 2014), leading to reduced biogenic calcification (Anthony et al., 2008), increased bio-erosion (Fang et al., 2013) and significant changes to coral reef community structure (Hall-Spencer et al., 2008; Fabricius et al., 2011, 2014). This study assessed how the microbiomes of key reef invertebrates might respond to OA in situ, by surveying corals and sponges that have had a post-larval lifetime of exposure to elevated CO2 within a volcanic CO2 seep. The composition of invertebrate-associated microbial communities is known to be intimately linked to host health (Bourne and Webster, 2013), and we therefore predicted that OA-sensitive species would host microbial communities that are clearly distinct from the same species under ambient conditions, including a potential loss of putative symbionts and the appearance of microorganisms from taxa commonly associated with stress and disease. Additionally, not all tropical reef organisms are detrimentally impacted by elevated pCO2 (Fabricius et al., 2011); therefore, we also hypothesized that tolerance to OA stress in some species may be enabled via habitat-acclimated microbial associations. Here we have shown that distinct microbial communities have developed in two sponge species that appeared to benefit from high pCO2 / low pH conditions as well as one sponge species and two coral species that were apparently sensitive to this environment. The primary driver of microbial variation in the two sponge species that were more abundant at the seeps was an increased relative abundance of photosynthetic Synechococcus, whereas the major driver of variation in the apparently sensitive coral species was a loss of Gammaproteobacteria at the seep site, in particular the putatively endosymbiotic Endozoicomonas in A. millepora. The development of distinct microbial communities between sites may influence the fitness and success of the host, although the functional implications of these divergent microbial populations are still to be fully elucidated.
Our understanding of how OA will directly affect marine microbial communities is limited (Joint et al., 2011), although microbial shifts in response to elevated pCO2 have previously been reported from CO2 mesocosm experiments on bacterioplankton and picoplankton (Allgaier et al., 2008; Meakin and Wyman, 2011), studies of surface-associated biofilms (Witt et al., 2011) and experimental analyses of host-associated microbes (Meron et al., 2011; Webster et al., 2013a, 2013b; Kerfahi et al., 2014). Manipulative experiments with corals exposed to high pCO2 / low pH have demonstrated an increase in the relative abundance of disease-associated bacteria within the class Flavobacteria in the coral Porites compressa (Vega Thurber et al., 2009). This is largely consistent with findings from the CO2 seep where we also observed an increase in bacterial OTUs affiliated with Bacteroidetes (A. millepora microbiome), specifically within the class Flavobacteria (P. cylindrica microbiome). However, when two Mediterranean coral species were transplanted for 7 months into a CO2 seep in the Gulf of Naples, Italy, and assessed by 16S rRNA gene clone sequencing, there was no detectable impact on the composition of their associated microbial communities in comparison with control sites (Meron et al., 2012). Microbial community shifts reported in these previous experimental studies tend to be host- and study-specific with no microbial taxa consistently reported as present or absent from specific pH/pCO2 treatments. It is likely that experiments conducted over extended time periods are required, particularly to enable host-associated microbial communities to adapt and reflect consistent patterns associated with environmental change. In most cases, these studies indicate that marine microbial communities can shift in response to high pCO2, yet the speed and stability of these changes, as well as the implications for host health, adaptation, acclimation or ecosystem function, have not yet been determined.
Both coral species were less abundant and hosted significantly different microbial communities at the site of active CO2 discharge compared with the adjacent control site. Within the A. millepora microbiome, a notable ∼50% reduction in the family Hahellaceae (order Oceanospirillales) was observed, largely as a result of loss of sequences related to Endozoicomonas sp. The Endozoicomonas are commonly found in close association with marine invertebrates, including the deep-sea bone-eating polychaete Osedax frankpressi (Goffredi et al., 2005), the Pacific sea slug Elysia ornata (Kurahashi and Yokoto, 2007), the hydrothermal vent snail Alviniconcha (Beinart et al., 2014), in addition to gorgonians (La Rivière et al., 2013; Bayer et al., 2013b) and sponges (Bourne et al., 2013). Associated with corals specifically, members of the genus Endozoicomonas have been isolated from Montipora aequituberculata in the Pacific (Yang et al., 2010), comprise up to 80% of the microbiome of Porites astreoides from the Caribbean (Morrow et al., 2012; Rodriguez-Lanetty et al., 2013) and 70 to 95% of the microbiome in Red Sea corals Stylophora pistillata, Pocillopora damicornis and Acropora humilis (Bayer et al., 2013a). Importantly, a meta-analysis also revealed that Oceanospirillales-related sequences (which include the Endozoicomonas genus) were among the most common ribotypes found in healthy corals (Mouchka et al., 2010), suggesting that the loss of this important taxon at high pCO2 would have significant implications for coral health, and may contribute to the inability of these species to thrive at the CO2 seep.
Hypotheses about the function of the Endozoicomonas-related symbionts have ranged from parasite consumers of host tissue to beneficial mutualists that assist in the metabolism, nutrient acquisition and/or cycling of organic compounds. The Endozoicomonas are members of the order Oceanospirillales, a group widely known to be heterotrophic and capable of degrading complex organic compounds (Garrity et al., 2005), including the production of extracellular hydrolytic enzymes that can degrade various complex organic substrates such as whale bones (Goffredi et al., 2007). Previous studies have also found that Endozoicomonas aggregate and may live endosymbiotically, including within the coral endoderm (Bayer et al., 2013b), and within membrane-bound vesicles inside the gill cells of the hydrothermal vent snail Alviniconcha (Beinart et al., 2014).
Isolates of Endozoicomonas-related sequences from A. millepora coral tissues possessed the ability to metabolise the organosulphur compound dimethylsulphoniopropionate, which suggests a role in sulphur cycling (Raina et al., 2009). Furthermore, Endozoicomonas isolates from sponges were shown to produce the quorum-sensing metabolites N-acyl homoserine lactones (Mohamed et al., 2008), and demonstrate antimicrobial activity against Bacillus subtilis (Rua et al., 2014). In the present study, the Endozoicomonas-related OTUs associated with A. millepora that were significantly reduced at the seep site were ∼96% similar to Endozoicomonas numazuensis (NCBI strain HC50), a strain isolated from marine sponges in Japan (Nishijima et al., 2013) and similar to E. montiporae (Pike et al., 2013), a strain isolated from the coral Montipora aquetuberculata (Yang et al., 2010). The E. numazuensis strain is facultatively anaerobic and capable of fermenting carbohydrates, reducing nitrate and hydrolysing DNA (Nishijima et al., 2013). Consistent with the proposed roles in nutrient acquisition and cycling of organic compounds, KEGG pathway prediction for the A. millepora Endozoicomonas OTU using PICRUSt revealed the categories of membrane transport, amino acid and carbohydrate metabolism, replication and repair, and cell motility to be the most abundant known pathways (genes of unknown function were also abundant). Taken together, these several lines of evidence suggest that host-associated Endozoicomonas may have important ecological roles in host health by providing otherwise unavailable macromolecular nutrients, contributing to nitrogen and/or sulphur cycling, and shaping the associated microbial community via signalling molecules and antimicrobial activity.
In contrast to the observed decrease in Endozoicomonas at the seep site, another related bacterial genus within the family Halomonadaceae increased in abundance within A. millepora. The Halomonadaceae are considered coral ‘residents' (Ritchie, 2006), are known for their successful growth over a wide range of temperature and pH levels and may produce enzymes capable of catabolizing dimethylsulphoniopropionate and acrylate produced by Symbiodinium within the coral tissues (Todd et al., 2012). In the present study, the OTU responsible for this shift in community structure was ∼99% similar to Chromohalobacter salexigens based on 16S rRNA gene identity (NCBI strain DSM3043), a strain isolated from high saline environments and capable of H2S production, nitrate reduction and anaerobic growth in the presence of nitrate. Thus, although the seep environment may be suboptimal for Endozoicomonas sp., it may still be suitable for other putatively symbiotic members of the coral microbial community. These results suggest that coral-associated microbial communities are dynamic and responsive to changes in pCO2 and/or pH. Although the microbial shifts reported here are not entirely consistent with previous mesocosm and laboratory studies, they do reflect host–microbial relationships that have stabilized given a lifetime of acclimation to an environment with high pCO2.
Sponge species that are abundant at shallow water CO2 seeps in Papua New Guinea host significantly different microbial communities to individuals of the same species at control sites less than 500 m away. Although many bacteria contribute to these differences, the variation is largely influenced by the dominance of Synechococcus (phylum Cyanobacteria) within C. singaporensis and Cinachyra sp. at the CO2 seep. Although carbon metabolism in sponges is typically based on filter feeding of bacterioplankton, some species can obtain >50% of their carbon demand from cyanobacterial photosymbionts (Wilkinson, 1983; Cheshire and Wilkinson, 1991; Freeman and Thacker, 2011). Hosting a higher abundance of cyanobacteria may therefore lead to higher carbon fixation, but nutritional benefits may also extend to nitrogen fixation as the products of nitrogen metabolism can be transferred from the symbionts to the sponge host in some species (Freeman and Thacker, 2011). An elevated abundance of cyanobacteria likely provides at least some sponge species with enhanced scope for growth in these seep environments. Consistent with this role as providers of photosynthates, the three primary KEGG level 2 pathways identified from PICRUSt analysis of the Synechococcus OTU were energy metabolism (in this case, primarily pathways involved in photosynthesis), carbohydrate metabolism and amino acid metabolism.
Although research has not yet explored how symbiotic cyanobacteria respond to elevated pCO2, recent studies of planktonic cyanobacteria have revealed species-specific responses to CO2 conditions projected for the end of this century (Hutchins et al., 2009). For instance, at elevated pCO2, increased rates of N2 and CO2 fixation occur in Trichodesmium (Hutchins et al., 2007; Lomas et al., 2012), whereas carbon assimilation in Prochlorococcus and Synechococcus is relatively unaffected by increased pCO2 except at elevated temperatures (Fu et al., 2007; Lomas et al., 2012). Although pelagic Synechococcus spp. show an immediate increase in carbon fixation (productivity) at elevated pCO2, this effect appears short term with acclimation of cellular physiology occurring within 1–3 days (Lomas et al., 2012). However, it is important to consider that, individual strains of Cyanobacteria may respond differently to altered CO2 concentrations, and these short-term experiments are unable to account for how microbes adapt and evolve in response to higher pCO2 environments. Potential variation in photosynthetic performance of different cyanobacterial types is particularly relevant to this study where Cyanobacteria increased in all three sponge species and both coral species at the seep, yet only two of these sponge species were more abundant at the seep site.
In addition to improved host fitness and enhanced competitive advantage, microbial symbionts have been shown to infer environmental stress tolerance to their plant and animal hosts. For example, in the aphid Buchnera aphidicola symbiosis, a single nucleotide mutation of the symbiont governs the thermal tolerance of the host (Dunbar et al., 2007), and in grass species from coastal and geothermal habitats, fungal endophytes confer salt and heat tolerance, respectively (Rodriguez et al., 2008). On the basis of these findings, it is plausible to consider whether specific members of the microbial community in C. singaporensis and Cinachyra sp. confer low pH/high pCO2 tolerance to the populations that occupy the CO2 seep habitat. For instance, the internal pH of the sponge tissue may be dramatically influenced by utilisation or production of CO2 by symbionts (e.g., Cyanobacteria) as well as other microbial processes such as nitrification. Transplantation experiments (potentially coupled with experimental addition of cultured symbionts) or targeted experimental analysis with microsensors would further elucidate whether these symbiont populations enhance or hinder the survival of organisms in such extreme pH environments. Furthermore, future studies at shallow CO2 seep environments would benefit from a combination of isotope labelling experiments and metagenomic/metatranscriptomic sequencing to enhance our ability to tease apart the relative contribution of bacterial symbionts to the growth and nutrition of ‘sensitive' and ‘tolerant' CO2 species.
Although the vulnerability of corals to OA is well established (Orr et al., 2005), it has previously been proposed that sponges are one taxon that could benefit from projected climate change scenarios (Bell et al., 2013), potentially by acquiring microbially mediated competitive advantages that enable them to thrive under future conditions of OA. Here we have shown that distinct microbial communities have developed in two sponge species that were tolerant of the high pCO2 conditions occurring within a natural volcanic CO2 seep. Conversely, the third sponge species was significantly less abundant at the CO2 seep and showed no flexibility in microbial symbiosis. Both coral species were also significantly less abundant at the CO2 seep. The proposed coral symbiont Endozoicomonas sp. was significantly reduced in corals at the CO2 seep, whereas the Cyanobacteria prominent in sponge microbiomes at the seep were present in only low abundance in the coral microbiomes. In addition, distinct microbial communities were observed in both coral species at the seep site including an overrepresentation of opportunistic bacteria previously linked with diseased and stressed corals (e.g., Flavobacteria). Although these results emphasise how some non-calcifying coral reef organisms (i.e., C. singaporensis and Cinachyra sp.) can benefit under reduced pH/elevated pCO2 conditions, we also showed that these benefits do not extend to all species and symbiotic partnerships. Organisms that lack the flexibility (i.e., S. massa) to alter the composition of their symbiotic microbial community or have particularly sensitive symbiont populations (i.e., A. millepora, P. cylindrica) may be more vulnerable to the effects of OA.
Acknowledgments
NSW was funded through an Australian Research Council Future Fellowship (FT120100480). The expedition to conduct the field work was funded by the Australian Institute of Marine Science.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allgaier M, Riebesell U, Vogt M, Thyrhaug R, Grossart HP. (2008). Coupling of heterotrophic bacteria to phytoplankton bloom development at different pCO2 levels: a mesocosm study. Biogeosciences 5: 1007–1022. [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony KRN, Kline DI, Diaz-Pulido G, Dove S, Hoegh-Guldberg O. (2008). Ocean acidification causes bleaching and productivity loss in coral reef builders. Proc Natl Acad Sci USA 105: 17442–17446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer T, Arif C, Ferrier-Pagès C, Zoccola D, Aranda M, Voolstra CR. (2013. a). Bacteria of the genus Endozoicomonas dominate the microbiome of the Mediterranean gorgonian coral Eunicella cavolini. Mar Ecol Prog Ser 479: 75–84. [Google Scholar]
- Bayer T, Neave MJ, Alsheikh-Hussain A, Hughen A, Aranda M, Yum LK et al. (2013. b). The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Appl Environ Microb 79: 4759–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinart RA, Nyholm SV, Dubilier N, Girguis PR. (2014). Intracellular Oceanospirillales inhabit the gills of the hydrothermal vent snail Alviniconcha with chemosynthetic, γ-Proteobacterial symbionts. Environ Microbiol Rep doi: 10.1111/1758-2229.12183. [DOI] [PubMed]
- Bell JJ, Davy SK, Jones T, Taylor MW, Webster NS. (2013). Could some coral reefs become sponge reefs as our climate changes? Glob Change Biol 19: 2613–2624. [DOI] [PubMed] [Google Scholar]
- Beman JM, Chow C-E, King AL, Feng Y, Fuhrman JA, Andersson AJ et al. (2011). Global declines in oceanic nitrification rates as a consequence of ocean acidification. Proc Natl Acad Sci USA 108: 208–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne DG, Dennis PG, Uthicke S, Soo RM, Tyson GW, Webster NS. (2013). Coral reef invertebrate microbiomes correlate with the presence of photosymbionts. ISME J 7: 1459–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne DG, Webster NS. (2013). Coral Reef Bacterial Communities. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, (eds) The Prokaryotes. Springer: Berlin Heidelberg, pp 163–187. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A. (1984). Non-parametric estimation of the number of classes in a population. Scand J Stat 11: 265–270. [Google Scholar]
- Cheshire AC, Wilkinson CR. (1991). Modelling the photosynthetic production by sponges on Davies Reef, Great Barrier Reef. Mar Biol 109: 13–18. [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microb 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar HE, Wilson ACC, Ferguson NR, Moran NA. (2007). Aphid thermal tolerance is governed by a point mutation in bacterial symbionts. PLoS Biol 5: e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinfomatics 27: 2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabricius KE, De'ath G, Noonan S, Uthicke S. (2014). Ecological effects of ocean acidification and habitat complexity on reef-associated macroinvertebrate communities. Proc. Biol Sci 281: 20132479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabricius KE, Langdon C, Uthicke S, Humphrey C, Noonan S, De'ath G et al. (2011). Losers and winners in coral reefs acclimatized to elevated carbon dioxide concentrations. Nat Climate Change 1: 165–169. [Google Scholar]
- Fan L, Liu M, Simister R, Webster NS, Thomas T. (2012). Marine microbial symbiosis heats up: The phylogenetic and functional response of a sponge holobiont to thermal stress. ISME J 7: 991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang JKH, Mello-Athayde MA, Schönberg CHL, Kline DI, Hoegh-Guldberg O, Dove S. (2013). Sponge biomass and bioerosion rates increase under ocean warming and acidification. Glob Change Biol 19: 3581–3591. [DOI] [PubMed] [Google Scholar]
- Freeman CJ, Thacker RW. (2011). Complex interactions between marine sponges and their symbiotic microbial communities. Limnol Oceanogr 56: 1577–1586. [Google Scholar]
- Fu F-X, Warner ME, Zhang Y, Feng Y, Hutchins DA. (2007). Effects of increased temperature and CO2 on photosynthesis, growth and elemental ratios of marine Synechococcus and Prochlorococcus (cyanobacteria). J Phycol 43: 485–496. [Google Scholar]
- Garrity GM, Bell JA, Lilburn T. (2005). Oceanospirillales ord. nov. In Brenner DJ, Krieg NR, Garrity GM, Boone DR, De Vos P, et al (eds) Bergey's Manual® of Systematic Bacteriology. Springer: New York, USA, pp 270–323. [Google Scholar]
- Goffredi SK, Orphan VJ, Rouse GW, Jahnke L, Embaye T, Turk K et al. (2005). Evolutionary innovation: a bone-eating marine symbiosis. Environ Microbiol 7: 1369–1378. [DOI] [PubMed] [Google Scholar]
- Goffredi SK, Johnson SB, Vrijenhoek RC. (2007). Genetic diversity and potential function of microbial symbionts associated with newly discovered species of Osedax polychaete worms. Appl Environ Microbiol 73: 2314–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin C, Rodolfo-Metalpa R, Picton B, Hall-Spencer JM. (2013). Effects of ocean acidification on sponge communities. Mar Ecol 35: 41–49 doi:10.1111/maec.12093. [Google Scholar]
- Hall-Spencer JM, Rodolfo-Metalpa R, Martin S, Ransome E, Fine M, Turner SM et al. (2008). Volcanic carbon dioxide vents show ecosystem effects of ocean acidification. Nature 454: 96–99. [DOI] [PubMed] [Google Scholar]
- Hutchins DA, Fe F-X, Zhang Y, Warner ME, Feng Y, Portune K et al. (2007). CO2 control of Trichodesmium N2 fixation, photosynthesis, growth rates, and elemental ratios: implications for past, present, and future ocean biogeochemistry. Limnol Oceanogr 52: 1293–1304. [Google Scholar]
- Hutchins DA, Mulholland MR, Fu F. (2009). Nutrient cycles and marine microbes in a CO2-enriched ocean. Oceanography 22: 128–145. [Google Scholar]
- IGBP, IOC, SCOR. (2014). Ocean acification summary for policymakers-third symposium on the ocean in a high-CO2 world. International Geosphere-Biosphere Programme, Stockholm, Sweden.
- Kerfahi D, Hall-Spencer JM, Tripathi B, Milazzo M, Lee J, Adams JM. (2014). Shallow water marine sediment bacterial community shifts along a natural CO2 gradient in the Mediterranean Sea off Vulcano, Italy. Microb Ecol 67: 819–828. [DOI] [PubMed] [Google Scholar]
- Krause E, Wichels A, Giménez L, Lunau M, Schilhabel MB, Gerdts G. (2012). Small changes in pH have direct effects on marine bacterial community composition: A microcosm approach. PLoS One 7: e47035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi M, Yokoto A. (2007). Endozoicomonas elysicola gen. nov., sp. nov., a γ-proteobacterium isolated from the sea slug Elysia ornate. Syst Appl Microbiol 30: 202–206. [DOI] [PubMed] [Google Scholar]
- Langille MG, Zaneveld J, Caporaso JG, Mcdonald D, Knights D, Reyes JA et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31: 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rivière M, Roumagnac M, Garrabou J, Bally M. (2013). Transient shifts in bacterial communities associated with the temperate Gorgonian Paramuricea clavata in the Northwestern Mediterranean Sea. PLoS One 8: e57385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomas MW, Hopkinson BM, Losh JL, Ryan DE, Shi DL, Xu Y et al. (2012). Effect of ocean acidification on cyanobacteria in the subtropical North Atlantic. Aquat Microb Ecol 66: 211–222. [Google Scholar]
- Mcdonald D, Price MN, Goodrich J, Nawrocki EP, Desantis TZ, Probst A et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meakin NG, Wyman M. (2011). Rapid shifts in picoeukaryote community structure in response to ocean acidification. ISME J 5: 1397–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meron D, Atias E, Iasur Kruh L, Elifantz H, Minz D, Fine M et al. (2011). The impact of reduced pH on the microbial community of the coral Acropora eurystoma. ISME J 5: 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meron D, Rodolfo-Metalpa R, Cunning R, Baker AC, Fine M, Banin E. (2012). Changes in coral microbial communities in response to a natural pH gradient. ISME J 6: 1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed N, Cicirelli EM, Kan J, Chen F, Fuqua C, Hill RT. (2008). Diversity and quorum-sensing signal production of Proteobacteria associated with marine sponges. Environ Microbiol 10: 75–86. [DOI] [PubMed] [Google Scholar]
- Morrow KM, Moss AG, Chadwick NE, Liles MR. (2012). Bacterial associates of two Caribbean coral species reveal species-specific distribution and geographic variability. Appl Environ Microbiol 78: 6438–6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss RH, Edmonds JA, Hibbard KA, Manning MR, Rose SK, van Vuuren DP et al. (2010). The next generation of scenarios for climate change research and assessment. Nature 463: 747–756. [DOI] [PubMed] [Google Scholar]
- Mouchka ME, Hewson I, Harvell CD. (2010). Coral-associated bacterial assemblages: current knowledge and the potential for climate-driven impacts. Integr Comp Biol 50: 662–674. [DOI] [PubMed] [Google Scholar]
- Nishijima M, Adachi K, Katsuta A, Shizuri Y, Yamasato K. (2013). Endozoicomonas numazuensis sp. nov., a gammaproteobacterium isolated from marine sponges, and emended description of the genus Endozoicomonas Kurahashi and Yokota 2007. Int J Syst Evol Microbiol 63: 709–714. [DOI] [PubMed] [Google Scholar]
- Orr JC, Fabry VJ, Aumont O, Bopp L, Doney SC, Feely RA et al. (2005). Anthropogenic ocean acidification over the twentyfirst century and its impact on calcifying organisms. Nature 437: 681–686. [DOI] [PubMed] [Google Scholar]
- Padan E, Bibi E, Ito M, Krulwich TA. (2005). Alkaline pH homeostasis in bacteria: New insights. Biochim Biophys Acta 1717: 67–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike RE, Haltli B, Kerr RG. (2013). Description of Endozoicomonas euniceicola sp. nov. and Endozoicomonas gorgoniicola sp. nov., bacteria isolated from the octocorals Eunicea fusca and Plexaura sp., and an emended description of the genus Endozoicomonas. Int J Syst Evol Microbiol 63: 4294–4302. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina JB, Tapiolas D, Willis B, Bourne DG. (2009). Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl Enviro Microbiol 75: 3492–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie KB. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol Prog Ser 322: 1–14. [Google Scholar]
- Rodriguez-Lanetty M, Granados-Cifuentes C, Barberan A, Bellantuono AJ, Bastidas C. (2013). Ecological inferences from a deep screening of the complex bacterial consortia associated with the coral, Porites astreoides. Mol Ecol 22: 4349–4362. [DOI] [PubMed] [Google Scholar]
- Rodriguez RJ, Henson J, Van Volkenburgh E, Hoy M, Wright L, Beckwith F et al. (2008). Stress tolerance in plants via habitat-adapted symbiosis. ISME J 2: 404–416. [DOI] [PubMed] [Google Scholar]
- Rua CPJ, Trindade-Silva AE, Appolinario LR, Venas TM, Garcia GD, Carvalho LS et al. (2014). Diversity and antimicrobial potential of culturable heterotrophic bacteria associated with the endemic marine sponge Arenosclera brasiliensis. PeerJ 2: e419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Gevers D, Westcott SL. (2011). Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6: e27310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D, Xu Y, Hopkinson BM, Morel FMM. (2010). Effect of ocean acidification on iron availability to marine phytoplankton. Science 327: 676–679. [DOI] [PubMed] [Google Scholar]
- Simister RL, Deines P, Botté ES, Webster NS, Taylor MW. (2012). Sponge-specific clusters revisited: a comprehensive phylogeny of sponge-associated microorganisms. Environ Microbiol 14: 517–524. [DOI] [PubMed] [Google Scholar]
- Teira E, Fernández A, Álvarez-Salgado XA, García-Martín EE, Serret P, Sobrino C. (2012). Response of two marine bacterial isolates to high CO2 concentration. Mar Ecol Prog Ser 453: 27–36. [Google Scholar]
- Todd J, Curson ARJ, Nikolaidou-Katsaraidou N, Brearley CA, Watmough NJ, Chan Y et al. (2012). Molecular dissection of bacterial acrylate catabolism -unexpected links with dimethylsulfoniopropionate catabolism and dimethyl sulfide production. Environ Microbiol 12: 327–343. [DOI] [PubMed] [Google Scholar]
- Uthicke S, Momigliano P, Fabricius KE. (2013). High risk of extinction of benthic foraminifera in this century due to ocean acidification. Scientific Rep 3: 1769. [Google Scholar]
- Vega Thurber R, Willner-Hall D, Rodriguez-Mueller C, Desnues C, Edwards RA, Angly F et al. (2009). Metagenomic analysis of stressed coral holobionts. Environ Microbiol 11: 2148–2163. [DOI] [PubMed] [Google Scholar]
- Vega Thurber RL, Burkepile DE, Fuchs C, Shantz AA, McMinds R, Zaneveld JR. (2014). Chronic nutrient enrichment increases prevalence and severity of coral disease and bleaching. Glob Change Biol 20: 544–554. [DOI] [PubMed] [Google Scholar]
- Webster NS, Cobb RE, Negri AP. (2008). Temperature thresholds for bacterial symbiosis with a sponge. ISME J 2: 830–842. [DOI] [PubMed] [Google Scholar]
- Webster NS, Negri AP, Flores F, Humphrey C, Soo R, Botté ES et al. (2013. a). Near-future ocean acidification causes differences in microbial associations within diverse coral reef taxa. Environ Microbiol Rep 5: 243–251. [DOI] [PubMed] [Google Scholar]
- Webster NS, Uthicke S, Botté ES, Flores F, Negri AP. (2013. b). Ocean acidification reduces induction of coral settlement by crustose coralline algae. Glob Change Biol 19: 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson CR. (1983). Net primary productivity in coral reef sponges. Science 219: 410–412. [DOI] [PubMed] [Google Scholar]
- Witt V, Wild C, Anthony KRN, Diaz-Pulido G, Uthicke S. (2011). Effects of ocean acidification on microbial community composition of, and oxygen fluxes through, biofilms from the Great Barrier Reef. Environ Microbiol 13: 2976–2989. [DOI] [PubMed] [Google Scholar]
- Yang CS, Chen MH, Arun AB, Chen CA, Wang JT, Chen WM. (2010). Endozoicomonas montiporae sp. nov., isolated from the encrusting pore coral Montipora aequituberculata. Int J Syst Evol Micr 60: 1158–1162. [DOI] [PubMed] [Google Scholar]
- Zaneveld JR, Vega Thurber RL. (2014). Hidden State Prediction: a modification of classic ancestral state reconstruction algorithms helps unravel complex symbioses. Front Microbiol 5: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.