Abstract
Soil microorganisms found in the root zone impact plant growth and development, but the potential to harness these benefits is hampered by the sheer abundance and diversity of the players influencing desirable plant traits. Here, we report a high level of reproducibility of soil microbiomes in altering plant flowering time and soil functions when partnered within and between plant hosts. We used a multi-generation experimental system using Arabidopsis thaliana Col to select for soil microbiomes inducing earlier or later flowering times of their hosts. We then inoculated the selected microbiomes from the tenth generation of plantings into the soils of three additional A. thaliana genotypes (Ler, Be, RLD) and a related crucifer (Brassica rapa). With the exception of Ler, all other plant hosts showed a shift in flowering time corresponding with the inoculation of early- or late-flowering microbiomes. Analysis of the soil microbial community using 16 S rRNA gene sequencing showed distinct microbiota profiles assembling by flowering time treatment. Plant hosts grown with the late-flowering-associated microbiomes showed consequent increases in inflorescence biomass for three A. thaliana genotypes and an increase in total biomass for B. rapa. The increase in biomass was correlated with two- to five-fold enhancement of microbial extracellular enzyme activities associated with nitrogen mineralization in soils. The reproducibility of the flowering phenotype across plant hosts suggests that microbiomes can be selected to modify plant traits and coordinate changes in soil resource pools.
Introduction
Recent studies have highlighted the ability of plant-associated microbiomes to influence plant traits including disease resistance, growth and abiotic stress tolerance (Swenson et al., 2000; Mendes et al., 2011; Lau and Lennon, 2012; Bainard et al., 2013; Sugiyama et al., 2013). When a fast-growing plant is studied in conjunction with its microbiome across multiple generations, new forms of interactions can be observed between plants and microorganisms shaping plant development. Similar experimental designs using a multi-generational approach have been used to document rapid evolution in plant–insect interactions (Züst et al., 2012; Agrawal et al., 2013). In the rhizosphere specifically, two recent publications have focused on soil microbiomes to address drought tolerance and disease resistance (Mendes et al., 2011; Lau and Lennon, 2012). An earlier study by Swenson et al. (2000) indicates that the microbially mediated mechanisms of plant growth can be passed on through multiple generations of experimental evolution to modulate plant biomass levels. Here, we merge these approaches to examine how selection at the community level (soil microorganisms) for the plant host trait (flowering time) has consistent and reproducible effects on plant function across multiple hosts.
The ability to assemble trait-associated microbiomes into new plant hosts is a critical step in developing an effective microbiome component to plant production systems, particularly for high-value crops that rely on transplantation of young plants into field or greenhouse settings. The ability to progressively enrich for microbiota associated with a specific plant trait facilitates the use of more complex communities instead of a single microbial strain. Related research on disease-suppressive soils illustrates how the concept of multiple plantings of a plant species can enrich for beneficial microbiota that enhance disease resistance in successive generations of plant hosts (Mazzola, 2004; Kinkel et al., 2011). Transfer of the microbiota into fumigated or sterilized soil containing the young developing plant could help establish persistent plant-associated microbiomes after transplantation to field settings.
In this study, we used a multi-generation approach to generate enriched microbiomes that induce flowering time as the targeted plant trait. Applying community selection on microbiomes through observable changes on a plant trait can demonstrate the potential for complex communities of microorganisms to shape rapid change in plant population traits. We examined the ability of soil microbiomes selected over 10 plantings for progressively earlier or later flowering in Arabidopsis thaliana genotype Col to induce the same early (EF)- and late-flowering (LF) times in four novel plant hosts (Supplementary Figure S1). The soils received low fertilizer inputs to maintain nutrient limitation throughout the study and the soils were steam-sterilized to facilitate establishment of the inoculating microbiome into new soils. The central focus of this study features microbiomes from the tenth generation of plantings inoculated into the soils of novel plant hosts that included Brassica rapa (BR) and three A. thaliana genotypes: Rld, Landsberg erecta (Ler) and Bensheim (Be). We hypothesized that the community selection of microbiomes across 10 generations of earlier or later flowering times in A. thaliana Col would result in EF- vs LF plastic responses across all A. thaliana hosts and the related B. rapa upon inoculation into these novel host soils, and that these microbiomes would differ in phylogenetic composition by flowering time responses.
Materials and methods
Growth chamber conditions
All plants were grown at 22 °C on a 16/8 h day/night cycle. Relative humidity was set to 75%, and light level was set at 250 μE. (Percival-Cornell University Weill Hall Life Sciences Growth Chamber Facility, Ithaca, NY, USA)
Multi-generation selection of microbiome inoculants
Inoculants for EF- and LF-associated microbiomes were generated through an iterative selection process adapted from Swenson et al. (2000). Approximately 100 A. thaliana seeds were placed in each of 14 replicate microcosms (7.6 cm diameter × 8 cm height pots) containing 1:1 mixture of field soil: potting mix soil (LM-3 General Purpose Mix; Lambert Peat Moss Inc., Rivière-Ouelle, QC, Canada). The field soil was obtained from a collection of sites across Ithaca, NY, USA (42.456583, −76.368822; 42.452265, −76.369477; and 42.414913, −76.442272) representative of agricultural, forest and grassland ecosystems. The intention was to include a diversity of soil microorganisms for the initial generation. The potting mix was autoclaved for each generation, and became the growing media for the experimental selection. The EF- and LF-associated treatments were established with 14 replicate units each per planting and a control group included 7 units paired with each flowering treatment (14 control units). In each generation, four microcosms were selected based on the highest degree of the plant trait desired. This corresponded with progressively later flowering or earlier flowering as determined by uniform flower bolting in 90% of the individuals in a unit. Controls were paired with each flowering time microbiome treatment to examine plant traits and soil extracellular enzyme activity results relative to plant phenology. The controls consisted of the plants and steam-sterilized soils, but the units were not inoculated with EF- or LF-associated microbiomes.
Biomass and soils were harvested immediately following flowering of all pots within a group. Loose soil was separated from roots of the four earliest vs four latest flowering replicate units of each treatment group, pooled and mixed with sterile water to form the EF and LF inoculants. Soil slurry inoculants were prepared with 180 ml of sterile deionized water and 30 g of fresh rhizosphere soil, and then shaken vigorously for 60 s upon preparation and periodically during inoculation to make a soil suspension. Each unit for the subsequent generation received 12 ml of the corresponding treatment inoculant. The control group did not receive inoculants of the microbiomes. All seeds across the multi-generation planting were derived from a static seed pool of a highly inbred line, A. thaliana Col (Lehle Seed Co., Round Rock, TX, USA). Seeds were derived from this common seed pool to maintain consistent allelic frequencies across all generations and to ensure that any changes in plant traits are the result of microbiome selection. For example, the same pool of seeds was used across generations 1 through 10 and in the EF, LF and control treatments. All microcosms were watered through capillary action using individual reservoirs for each unit. A low level of available nutrients in the potting medium, as well as in the watering regime ensured that the plants were under nutrient limitation, providing a strong filter to impose microbiome effects on soil nutrient mineralization. Fertilizer requirements for A. thaliana are high (200 ppm nitrogen (N) every other day) to achieve optimal growing conditions (Eddy et al., 2008), but we used a fraction of the amount comprising applications of 10 ppm N for generations 1 through 5 for each watering event and three applications of 10 ppm N per generation for generations 6 through 10. As the genetic pool of the plants was held constant, the only adaptive traits to evolve over the iterative generations were derived from the soil inoculation (soil microbial community). This selection process continued for 10 successive generations (plantings) to develop distinct, trait-associated soil microbiomes associated with EF/LF time.
Transfer of microbiomes to novel plant hosts
Approximately 100 seeds of A. thaliana Col, Ler, RLD and Be (Lehle Seed Co.) and six seeds of rapid cycling B. rapa (Carolina Biological Supply Co., Burlington, NC, USA) were placed in separate units with 14 replicates measuring 7.6 cm diameter × 8 cm height. Each unit contained the same potting mix used in the multi-generation microbiome selection. The soil mix was autoclaved prior to adding seeds. The EF- and LF microbiomes were inoculated separately into their corresponding replicated units. Each flowering time treatment was paired with a control with seven replicate units. Microorganisms were excluded in the control group inoculants. Each plant host was arranged in the growth chamber in a randomized block design. All units of a plant host received the same amount of fertilizer consisting of a 10% solution (10ppm N, 10.5% nitrate/89.5% urea) of 20-10-20 Jack's Professional General Purpose Fertilizer (J.R. Peters, Inc., Allentown, PA, USA). Plant hosts Be, Col and Ler received three equal doses of fertilizer during growth for a total of 0.9 mg added N, whereas RLD received two doses for a total of 0.65 mg added N, and B. rapa received no fertilizer. The difference in fertilizing regimes was due to the rapid flowering, and completion of life cycle, in the EF group for RLD and B. rapa in advance of the fertilization schedule and the need to keep nutrient addition constant between treatments.
Plant biomass
Plant aboveground biomass was harvested after flower bolting had begun in 90% of the individuals of each replicate microcosm. Biomass was harvested in two separate portions, reproductive structure and leaf tissue, for the A. thaliana genotypes, and whole for B. rapa. Harvested tissue was dried at 50 °C until constant weight.
Soil collection and storage
To maximize rhizosphere soil yield, loose soil was removed and soil adhering to the roots was collected. The soil was homogenized and a portion was immediately frozen at −80 °C for DNA analysis. The other half was analyzed for extracellular enzyme potential activity.
Soil extracellular enzyme activity
We sought to assess microbiome influence on soil processes by measuring the potential activities of soil extracellular enzymes involved in N mineralization. The inclusion of potential extracellular enzyme activity measurements in this study is based on observations of soil nutrient variables established over generations of selection for changes in plant biomass (Swenson et al., 2000). The enzymes include N-acetyl glucosaminidase, leucine aminopeptidase and phenol oxidase. They function in depolymerizing organic matter and facilitate microbial access to N sequestered within the complex structures (Sinsabaugh, 2010). N-acetyl glucosaminidase and leucine aminopeptidase were measured by fluorometric quantification and phenol oxidase was quantified by absorption. We used 4-methylumbelliferone- and 7-amino-4-methylcoumarin-labeled substrates (200 μM), and L-3,4-dihydroxyphenylalanine (25 mM) substrate to provide quantifiable fluorescence and color for quantification of oxidation (Saiya-Cork et al., 2002; German et al., 2011). Soil slurries were prepared from 5 g fresh soil in 150 ml sodium bicarbonate buffer (50 mM, pH 7) and homogenized with an immersion blender for 1 min. Hydrolytic enzyme assays were conducted in black 96-well microplates and oxidative assays were carried out in transparent-bottom 96-well microplates. Standard curves were made for each soil sample (soil slurry+4-methylumbelliferone or 7-amino-4-methylcoumarin standard of 0, 2.5, 5, 10, 25, 50 μM). A 200 μl volume of soil slurry and 50 μl of 4-methylumbelliferone or 7-amino-4-methylcoumarin standards were added into wells of standard plate, and 200 μl of soil slurry and 50 μl of the labeled substrate into wells of substrate plate. Plates were incubated in the dark at 25 °C for 3 h and fluorescence was measured immediately after removal from the incubator with a BioTek Synergy HT microplate reader (BioTek Industries, Inc., Winooski, VT, USA) (ex: 365 nm, em: 450 nm). The oxidative enzyme plate contained a buffer blank (250 μl buffer), a L-3,4-dihydroxyphenylalanine blank (200 ul buffer+50 μl L-3,4-dihydroxyphenylalanine), sample blank (200 μl slurry+50 μl buffer) and the sample wells (200 μl slurry+50 μl L-3,4-dihydroxyphenylalanine). Oxidative plates were incubated in the dark at 25 °C for 3 h and absorbance was measured at 460 nm with the BioTek microplate reader. We calculated activity based on equations from previous work (Saiya-Cork et al., 2002; German et al., 2011).
Microbiome 16 S rRNA gene sequencing
Soil DNA was extracted from frozen samples using the PowerSoil DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA) according to the recommended protocol for highly organic soil. Approximately 0.1 g of soil from each sample was used for isolation of soil DNA. We normalized isolated samples to a concentration of 10 ng ul−1 by dilution with PCR-grade water. Quantification was performed with the standard dsDNA quantification protocol for Picogreen (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Samples with concentrations below 10 ng ul−1 were extracted again at lower elution volume and pooled until a concentration above 10 ng ul−1 was reached for normalization. All pipetting for DNA extraction and normalization was conducted with an Eppendorf epMotion 5075 pipetting robot (Eppendorf AG, Hamburg, Germany).
We amplified 16 S rRNA gene sequences in duplicate from the extracted DNA. PCR primers used are described in Caporaso et al. (2012) that target the bacterial/archaeal 16 S rRNA gene variable region 4 (515 F/806 R) for downstream paired-end Illumina (Illumina, Inc., San Diego, CA, USA) barcoded sequencing (Caporaso et al., 2012). We pooled duplicate amplified samples and purified them with the desalting protocol of the Qiagen QiaQuick spin filter purification kit (QIAGEN Inc., Valencia, CA, USA). An epMotion 5075 was used to combine equal concentrations of all barcoded samples and then to dilute the pooled, barcoded amplicons for submission to the Cornell Life Sciences Sequencing Core with the custom sequencing primers as detailed in Caporaso et al. (2012) for multiplexed paired-end sequencing on the Illumina MiSeq platform.
Statistics
We used the R statistical package (Rproject.org) and JMP (SAS Institute, Inc., Cary, NC, USA) for all statistical modeling. All manipulations and calculations on 16 S rRNA gene sequence data were conducted in the R statistical package. We modeled biomass, flowering, tissue nutrient and enzyme activity data by standard least squares linear regression with control group values for each response variable included as a covariate to control for the effect of being grown at separate times. The analysis of covariance evaluates each dependent variable across our treatment groups while controlling for covariates. Treatment means adjusted to account for covariates are what are presented in figures to compare differences between the divergent treatment groups. Statistical significances of these comparisons are from the application of a post hoc Fisher's test of each plant host, and dependent variable, individually.
Multivariate statistics included multiple linear regression, correlation and covariance matrices to understand the data structure and interactions, and were conducted not only on the biomass, enzyme potential activity and flowering data but also on the relative abundance data of the major phyla/classes. We determined the significance of differences in abundance data by ANOVA (False Discovery Rate-corrected) and significant differences between community composition across groups (LF, EF and Control) were assessed by a nonparametric statistical method, adonis, which identifies relevant centroids, calculates squared deviations and determines significance by F-tests on sequential sums of squares from permutations of data (Quantitative Insights into Microbial Ecology (QIIME) Documentation).
Sequence analysis
Paired-end sequences were truncated at the first low-quality base and quality filtered to remove those with an average quality score below 25, fewer than 200 nt, greater than 700 nt, ambiguous bases, primer mismatches, erroneous barcodes and homopolymer runs exceeding six bases. Paired-end reads were joined and then demultiplexed within the QIIME software package (Qiime.org) (Caporaso et al., 2010b). We analyzed 16 S rRNA gene sequences in the QIIME software tool with the default parameters for each step. De novo operational taxonomic unit (OTU) picking was performed with the uclust option in QIIME (Edgar, 2010). Representative OTU sequences were aligned using the PyNAST algorithm with a minimum percent identity of 80% (Caporaso et al., 2010a). Assignment of taxonomy to representative OTUs was carried out with the Ribosomal Database Project (RDP) classifier (Lan et al., 2012) at the default 97% sequence identity and 80% confidence level with the bundled RDP-assigned taxonomies. Sequences matching plant chloroplast or mitochondrial 16 S rRNA were filtered from the dataset.
We determined the optimal sampling depth through examination of exploratory rarefaction curves of observed species plotted against sampling depth and the dataset was rarefied to 12 000 sequences per sample. Samples with fewer reads were removed. Alpha diversity metrics (chao1, PD, observed species) were computed within QIIME. Distance matrices were generated with the unweighted and weighted UniFrac methods to compare the relative abundance and presence/absence patterns between treatment groups. Our beta diversity measures (between-sample diversity) were computed with QIIME and jackknifed by repeatedly sampling at 3000 sequences per sample. Beta diversity was then plotted by principal coordinates analysis with confidence ellipses generated from the jackknifing procedure.
The heatmap was created from the log abundance of all genera and classified by the Prediction Analysis for Microarrays for the R package, which uses the least shrunken centroid method (Tibshirani et al., 2002). The ternary plot was created with ggplot2 in R.
Results
Soil microbiome composition
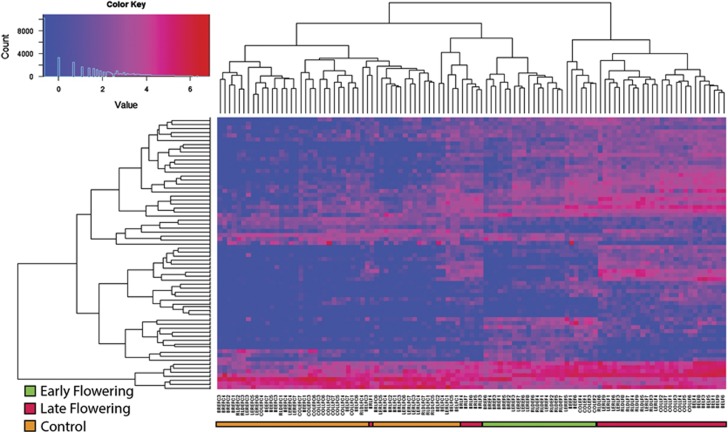
Soil microbiotas obtained from the root zone of B. rapa and four A. thaliana genotypes grouped together primarily by flowering time treatment and controls. Figure 1 shows a heatmap of log absolute abundance for all taxa. The samples grouped specifically by EF, LF and control (C) treatments. The ‘control' serves as a profile of the surviving and residual microbiota endemic in the soils after steam-sterilization and without inoculation of additional microbiota. Although the heatmap showed strong clustering by treatment, eight samples were misclassified representing an error rate of 0.075.
Figure 1.
Soil microbiota group together primarily by flowering time treatment and controls. Heatmap of log absolute abundance of all taxa. Classification, dendrograms and order of samples and taxa were determined by the Prediction Analysis for Microarrays in the R statistical package. The color key at the top left includes a frequency histogram of number of OTUs at each expression level. Vertical columns represent samples mapping primarily into ‘Control', EF and LF treatment groups. The ‘control' serves as a profile of the surviving and residual microbiota endemic in the soils after steam-sterilization and without inoculation of additional microbiota. Although the heatmap showed strong clustering by treatment, eight samples were misclassified representing an error rate of 0.075.
Distribution of OTUs across samples revealed a core microbiome with 60% of all OTUs shared across flowering time and control treatment groups (Figure 2a). The center of the ternary plot shows the core microbiome (high density of circles) across the EF, LF and control treatments. The OTUs uniquely associated with a specific treatment (where more than 80% of the total abundance of a particular OTU is uniquely associated with only one group) corresponded to the points within the corners of the ternary plot. The genera assigned to these OTUs fall into a handful of key families (Figure 2b), with more specific associations in Supplementary Table 1. The bacteria most strongly associated with the EF treatments include genera within two families with many known plant pathogens (Xanthomonadaceae and Pseudomonadaceae) and genera within three families with members associated with nutrient mineralization and substrate depolymerization (Moraxellaceae, Cellulomonadaceae and Saprospiraceae) (Sarkar and Guttman, 2004; Xia et al., 2008; Dodds and Rathjen, 2010; Rokhbakhsh-Zamin et al., 2011). In contrast, the LF treatments were associated with families that include plant-growth-promoting bacteria (Iamiaceae, Alcaligenaceae and Corynebacteriaceae) and a family of bacteria (Verrucomicrobiaceae) that are ubiquitous in soil but are poorly represented through culturing methods (Altman and Lawlor, 1966; Bertrand et al., 2000; Kurahashi et al., 2009; Da Rocha et al., 2013.
Figure 2.
Family-level taxa uniquely associated with EF/LF time groups and controls. (a) Ternary plot of OTUs showing the percent of each OTU's observations present in each group (EF, LF and Control) across different plant hosts. For example, a point's position within the ‘0.8' triangle at the ‘EF' corner of the ternary plot indicates that 80% of all observations of that OTU occur within the EF group. Diameter of plotted points corresponds to relative abundance of the OTU. Compartments of the dotted grid correspond to 20% increments. (b) List of taxonomy at the family level corresponding to OTUs of points falling within the 80% compartment of each group.
Principal coordinates analysis of the unweighted UniFrac distances showed separation of the trait-associated microbiome treatments (EF vs LF) and the control by microbial taxa (Figure 3). In contrast, the weighted UniFrac analysis indicated no separation of flowering time and control microbiomes in this study (Supplementary Figure S2). Although the majority of soil studies place emphasis on the relative abundance of taxa, to which weighted UniFrac is sensitive, the multiple generations of selection in this study may have led to the enrichment of trait-associated rare taxa—meaning abundant taxa alone may not drive the observed differences in flowering time. Furthermore, patterns of presence/absence can be obscured by the high relative abundances of core microbiome taxa, making unweighted UniFrac (insensitive to relative abundance) better for elucidating these patterns (Lozupone et al., 2011).
Figure 3.
Unweighted UniFrac distances show separation of the EF/LF-associated microbiome treatments and controls by microbial taxa. Principle coordinates analysis of unweighted UniFrac distances generated from 16 S rRNA sequence data obtained from the rhizosphere soils of the plant hosts. Unweighted UniFrac distances are insensitive to relative abundance of observed OTUs and instead reveal patterns and differences in the presence/absence of taxa. Samples were rarefied to an even sampling depth of 12 000 seqs per sample. The orange points refer to LF microbiomes, the blue points are the EF microbiomes and the red points are the control microbiomes. Percentages on each axis represent the percent variation explained by the PCs.
Effect of selected microbiomes on plant host traits
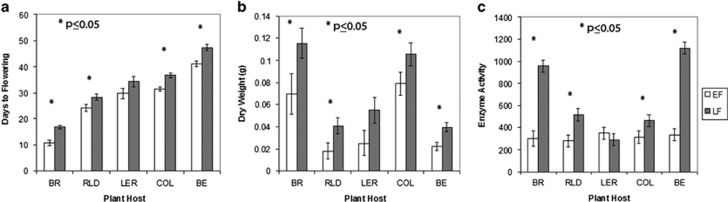
When the EF- and LF-associated microbiomes were inoculated into soils containing novel plant hosts, we found consistent responses in differences between flowering times. All A. thaliana hosts grown with LF-associated microbiomes flowered 15–17% later than plants containing the EF-associated microbiomes. The related crucifer, B. rapa, flowered 56% later in the LF treatments than in EF treatments. The significant delays in flowering were associated with increases in inflorescence biomass of three of the four A. thaliana genotypes Col, RLD and Be. Similarly, B. rapa, also showed delayed reproduction and an increase in total aboveground biomass in the LF-associated microbiome treatment (Figures 4a and b).
Figure 4.
Flowering time, reproductive biomass and potential extracellular enzyme activity show consistent changes across plant hosts. (a) Days to flowering of each plant host after inoculation with EF and LF microbiomes. (b) Reproductive biomass for the A. thaliana genotypes and total biomass for B. rapa. (c) Potential extracellular enzyme activity in soils across plant hosts. Enzyme activity associated with N mineralization is represented by the sum of leucine aminopeptidase, N-acetyl glucosaminidase and phenol oxidase (Sinsabaugh, 2010). Enzyme activity is measured in nmol per gram soil per hour. Values reported are from a standard least squares regression model including control values as a covariate (analysis of covariance). Plant host abbreviations correspond to B. rapa (BR) and the four A. thaliana genotypes Rld (RLD), Ler (LER), Col-0 (COL) and Be (BE). Asterisks denote statistical significance at P<0.05. Error bars represent s.e.m.
Potential microbiome-mediated shifts in soil environment
Soil microbial communities have a strong role in biogeochemical processes that determine soil environmental parameters such as pH, mineralization and nutrient availability (Burns, 1982; Allison and Vitousek, 2005). We observed no significant changes in soil pH between treatments and plant hosts, which indicates that pH is not responsible for the observed differences in plant growth and phenology. Soil inorganic NH4+ and NO3− concentrations did not differ across treatments, but any differences generated from mineralization could be explained by rapid immobilization in soil microorganisms and plants. Although aboveground plant tissue N did not differ across flowering treatment groups, it is plausible that root tissue N differed (but root tissue N was not measured in this study). The potential soil extracellular enzyme activities associated with N mineralization increased two- to fivefold in the LF microbiome treatment over the EF microbiome treatment (Figure 4c). Enhanced extracellular enzyme activity can indicate increased microbial coordination in the depolymerization of complex substrates and release of bioavailable N or phosphorus (Schimel and Bennett, 2004; Allison, 2005).
Discussion
Although the mechanisms underlying the apparent microbiome-driven shifts in flowering time are unknown in this study, we speculate that microbial modification of the soil altered a suite of environmental cues controlling flowering time. Regulation of flowering time is primarily driven by abiotic factors, such as vernalization and photoperiod, but it is well known that stress and nutrient availability also influence flowering time (Simpson and Dean, 2002; Amasino, 2010). The increase in A. thaliana reproductive biomass and B. rapa total biomass associated with the LF microbiomes points to several possibilities that include changes in soil resource pools altering flowering time, delayed reproduction altering soil resource pools and accumulated pathogens that contribute to early stress flowering. In this study, the delay in flowering corresponded to a 50–100% increase in host reproductive or total biomass. Minor increases in bioavailable N or other limiting nutrients could result in the biomass gains observed in the plant hosts particularly because the plants in this experiment were grown under nutrient limitation.
The production of extracellular enzymes provides a major mechanism by which microorganisms gain access to limiting nutrients bound in soil organic matter. Under N- or phosphorus-limiting conditions, groups of microorganisms capable of producing extracellular enzymes are able to capture N or phosphorus that would otherwise be inaccessible for biological uptake (Burns, 1982; Sinsabaugh, 1994; Sinsabaugh, 2010). These groups of microorganisms may include both bacteria and fungi, although fungi were not specifically examined in this study because of the lack of mycorrhizal association in A. thaliana and less robust community profiling methods. Plant rhizodeposition and root exudates represent a potential catalyst needed to prime the breakdown of complex polymers that release mineralized N and phosphorus (Haichar et al., 2008). Given this beneficial association, the production of extracellular enzymes and their value to the many organisms inhabiting the rhizosphere represent a unique situation in which selective pressures may encourage higher level coordination between plants and their microbiome (Wilson, 1975; Kerr and Godfrey-Smith, 2002; Okasha, 2009).
It is conceivable that the multi-generational approach to microbiome assembly we used may have led to the development of microbiomes in the LF treatment that enhance N mineralization via extracellular enzyme production. The resulting increase in mineralized N could modulate nutrient stress responses thereby favoring delays in bolting. Reproductive delay in A. thaliana grown in low phosphorus soils has been shown to increase biomass by 30% presumably by allowing greater time for soil phosphorus mineralization and root exploration (Nord and Lynch, 2008). Similarly in the study by Swenson et al. (2000), continuous selection for high vs low biomass A. thaliana plants showed changes in soil chemistry. By generations 13 and 14, phosphorus was one of the major factors explaining the separation of soil nutrients by low vs high host biomass selection lines.
The ability to reproduce microbiome function in novel plant hosts suggests that microbiome composition is also reproducible. However, inoculation of a plant's root-associated microbiome into the soils of novel plant hosts does not necessarily lead to a reassembly of microbial communities representative of the inoculant. For example, legumes inoculated with a mixture of rhizobial strains showed that nodule formation with the effective strain was not achieved uniformly across legume genotypes (Kiers et al., 2007). In this study, we showed that the plant trait-associated microbiomes developed over multiple generations were able to assemble into distinct community profiles by flowering time treatment across novel plant hosts. Although the soils were steam-sterilized to reduce viable microorganisms, a small fraction of the community is still able to persist as found in other studies (Lau and Lennon, 2011). In spite of the persistence, the inoculated microbiomes were able to populate the soils of novel hosts and induce plastic responses in flowering phenology and soil function. While bacterial sequencing was emphasized in this study as in other plant microbiome-focused papers (Lundberg et al., 2012; Bulgarelli et al., 2012; Peiffer et al., 2013), fungi could have had a significant role in modulating flowering time, altering extracellular enzyme activities and enhancing reproductive biomass. Root-colonizing endophytic fungi and root-associated fungi are able to modulate stress and enhance plant growth in Arabidopsis and other hosts (McLellan et al., 2007; Sherameti et al., 2008). Although the multi-generation approach to enriching microbiomes is likely to favor bacterial populations, it is conceivable that certain fungi are enriched across plantings assuming that fungal hyphae were able to persist through the inoculation procedure we used and establish in host tissues or rhizospheres within the short lifecycle of the rapidly cycling Brassicas.
Irreproducibility of microbiome function in Landsberg Erecta
The A. thaliana genotype Ler showed microbiome profiles consistent with the other plant hosts, but was unable to show the same significant shifts in flowering time, biomass and soil extracellular enzyme activities. Genotypic variability within a species can influence the composition of plant-associated microorganisms. For A. thaliana, a study conducted on eight genotypes in two different soil types showed that genotype explained a small but significant fraction of variation in the composition of the endophytic microbiome (Lundberg et al., 2012). Similarly, a study conducted on maize genotypes showed that a similar fraction of variation in rhizosphere microbial diversity was explained by plant host genetics (Peiffer et al., 2013). In our study, the lack of a significant response found in Ler to the inoculated microbiomes could be related to variation in host genetics. In particular, Ler shows unique genetic traits relevant to flowering regulation that could contribute to a reduced ability to delay flowering. For example, LF associated with the FRIGIDA (FRI) gene is partially suppressed in Ler and the suppressor allele found in Ler (FLC-Ler) may constrain the expression of the LF phenotype through inhibiting increases in Flowering Locus C (FLC) expression (Michaels and Amasino, 2001).
Conclusion
We have shown that experimental selection on soil microbial communities can alter major plant traits, including flowering time. These trait-associated microbiomes can then populate the soils of novel hosts and reproduce their intended functions. The ability of microbiomes to reproduce their effects on soil processes and host plant traits is critical to advancing the use of soil microbiomes in plant production systems. Our findings from the sequencing analysis indicate that rare taxa may have important roles in plant trait development. Accordingly, our results suggest that selection based on diverse microbial communities holds strong potential for using microbiomes to address key agronomic and environmental concerns.
Acknowledgments
We thank Stacey Lee, Biao Zhu, Danielle Park, Michael Grome, Lillian Aoki, Xiaoyun Gong, Sarah Carver, Grant Thompson, Dan Vandam and Iffie Ikem for their assistance with the research. We thank James Booth for help with statistical analysis and Anurag Agrawal for helpful comments on the paper. This project was funded by a grant from the President's Council of Cornell Women (PCCW), the USDA-CSREES and by a fellowship through NSF Integrative Graduate Education and Research Traineeship (IGERT) to KPB.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Agrawal AA, Johnson MTJ, Hastings AP, Maron JL. (2013). A field experiment demonstrating plant life-history evolution and its eco-evolutionary feedback to seed predator populations. Am Nat 181: S35–S45. [DOI] [PubMed] [Google Scholar]
- Allison SD. (2005). Cheaters, diffusion and nutrients constrain decomposition by microbial enzymes in spatially structured environments. Ecol Lett 8: 626–635. [Google Scholar]
- Allison SD, Vitousek PM. (2005). Responses of extracellular enzymes to simple and complex nutrient inputs. Soil Biol Biochem 37: 937–944. [Google Scholar]
- Altman J, Lawlor S. (1966). The effects of some chlorinated hydrocarbons on certain soil bacteria. J Appl Microbiol 29: 260–265. [DOI] [PubMed] [Google Scholar]
- Amasino R. (2010). Seasonal and developmental timing of flowering. Plant J 61: 1001–1013. [DOI] [PubMed] [Google Scholar]
- Bainard LD, Koch AM, Gordon AM, Klironomos JN. (2013). Growth response of crops to soil microbial communities from conventional monocropping and tree-based intercropping systems. Plant Soil 363: 345–356. [Google Scholar]
- Bertrand H, Plassard C, Pinochet X, Touraine B, Normand P, Cleyet-Marel JC. (2000). Stimulation of the ionic transport system in Brassica napus by a plant growth-promoting rhizobacterium (Achromobacter sp.). Can J Microbiol 46: 229–236. [DOI] [PubMed] [Google Scholar]
- Bulgarelli D, Rott M, Schlaeppi K, van Themaat EVL, Ahmadinejad N, Assenza F et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488: 91–95. [DOI] [PubMed] [Google Scholar]
- Burns RG. (1982). Enzyme activity in soil - location and a possible role in microbial ecology. Soil Biol Biochem 14: 423–427. [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. (2010. a). PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010. b). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6: 1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11: 539–548. [DOI] [PubMed] [Google Scholar]
- Eddy R, Hahn DT, Aschenbeck L. (2008). 101 ways to try to grow Arabidopsis: What fertilizer strength and frequency was required? Purdue Methods for Arabidopsis Growth. Paper 10http://docs.lib.purdue.edu/pmag/10.
- Edgar RC. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- German DP, Weintraub MN, Grandy AS, Lauber CL, Rinkes ZL, Allison SD. (2011). Optimization of hydrolytic and oxidative enzyme methods for ecosystem studies. Soil Biol Biochem 43: 1387–1397. [Google Scholar]
- Haichar F, el Z, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J et al. (2008). Plant host habitat and root exudates shape soil bacterial community structure. ISME J 2: 1221–1230. [DOI] [PubMed] [Google Scholar]
- Kerr B, Godfrey-Smith P. (2002). Individualist and multi-level perspectives on selection in structured populations. Biol Philos 17: 477–517. [Google Scholar]
- Kiers ET, Hutton MG, Denison RF. (2007). Human selection and the relaxation of legume defences against ineffective rhizobia. Proc R Soc B-Biological Sci 274: 3119–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkel LL, Bakker MG, Schlatter DC. (2011). A coevolutionary framework for managing disease-suppressive soils. Annual Review of Phytopathology 49: 47–67. [DOI] [PubMed] [Google Scholar]
- Kurahashi M, Fukunaga Y, Sakiyama Y, Harayama S, Yokota A. (2009). Lamia majanohamensis gen. nov., sp nov., an Actinobacterium isolated from sea cucumber Holothuria edulis, and proposal of Lamiaceae fam. nov. Int J. Syst Evol Microbiol 59: 869–873. [DOI] [PubMed] [Google Scholar]
- Lan Y, Wang Q, Cole JR, Rosen GL. (2012). Using the RDP classifier to predict taxonomic novelty and reduce the search space for finding novel organisms. PLoS One 7: e32491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau JA, Lennon JT. (2011). Evolutionary ecology of plant-microbe interactions: soil microbial structure alters selection on plant traits. New Phytol 192: 215–224. [DOI] [PubMed] [Google Scholar]
- Lau JA, Lennon JT. (2012). Rapid responses of soil microorganisms improve plant fitness in novel environments. Proc Natl Acad Sci USA 109: 14058–14062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Lladser ME, Knights D, Stombaugh J et al. (2011). UniFrac: an effective distance metric for microbial community comparison. ISME J 5: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzola M. (2004). Assessment and management of soil microbial community structure for disease suppression. Annu Rev Phytopathol 42: 35–59. [DOI] [PubMed] [Google Scholar]
- McLellan CA, Turbyville TJ, Wijeratne EK, Kerschen A, Vierling E, Queitsch C et al. (2007). A rhizosphere fungus enhances Arabidopsis thermotolerance through production of an HSP90 inhibitor. Plant Physiol 145: 174–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332: 1097–1100. [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13: 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord EA, Lynch JP. (2008). Delayed reproduction in Arabidopsis thaliana improves fitness in soil with suboptimal phosphorus availability. Plant Cell Environ 31: 1432–1441. [DOI] [PubMed] [Google Scholar]
- Okasha S. (2009). Individuals, groups, fitness and utility: multi-level selection meets social choice theory. Biol Philos 24: 561–584. [Google Scholar]
- Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL et al. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci USA 110: 6548–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Rocha UN, Plugge CM, George I, van Elsas JD, van Overbeek LS. (2013). The rhizosphere selects for particular groups of acidobacteria and verrucomicrobia. PLoS One 8: e82443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokhbakhsh-Zamin F, Sachdev D, Kazemi-Pour N, Engineer A, Pardesi KR, Zinjarde S et al. (2011). Characterization of plant-growth-promoting traits of Acinetobacter species isolated from rhizosphere of Pennisetum glaucum. J Microbiol Biotechnol 21: 556–566. [PubMed] [Google Scholar]
- Saiya-Cork KR, Sinsabaugh RL, Zak DR. (2002). The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biol Biochem 34: 1309–1315. [Google Scholar]
- Sarkar SF, Guttman DS. (2004). Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl Environ Microbiol 70: 1999–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimel JP, Bennett J. (2004). Nitrogen mineralization: Challenges of a changing paradigm. Ecology 85: 591–602. [Google Scholar]
- Sherameti I, Tripathi S, Varma A, Oelmüller R. (2008). The root-colonizing endophyte Pirifomospora indica confers drought tolerance in Arabidopsis by stimulating the expression of drought stress-related genes in leaves. Mol Plant Microbe Interact 21: 799–807. [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dean C. (2002). Flowering - Arabidopsis, the rosetta stone of flowering time? Science 296: 285–289. [DOI] [PubMed] [Google Scholar]
- Sinsabaugh RL. (1994). Enzymatic analysis of microbial pattern and process. Biol Fertil Soils 17: 69–74. [Google Scholar]
- Sinsabaugh RL. (2010). Phenol oxidase, peroxidase and organic matter dynamics of soil. Soil Biol Biochem 42: 391–404. [Google Scholar]
- Sugiyama A, Bakker MG, Badri D V, Manter DK, Vivanco JM. (2013). Relationships between Arabidopsis genotype-specific biomass accumulation and associated soil microbial communities. Botany-Botanique 91: 123–126. [Google Scholar]
- Swenson W, Wilson DS, Elias R. (2000). Artificial ecosystem selection. Proc Natl Acad Sci USA 97: 9110–9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R, Hastie T, Narasimhan B, Chu G. (2002). Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA 99: 6567–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DS. (1975). Theory of group selection. Proc Natl Acad Sci USA 72: 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Kong YH, Thomsen TR, Nielsen PH. (2008). Identification and ecophysiological characterization of epiphytic protein-hydrolyzing Saprospiraceae (‘Candidatus epiflobacter' spp.) in activated sludge. Appl Environ Microbiol 74: 2229–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst T, Heichinger C, Grossniklaus U, Harrington R, Kliebenstein DJ, Turnbull LA. (2012). Natural enemies drive geographic variation in plant defenses. Science 338: 116–119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.