Abstract
Sea ice is one of the most frigid environments for marine microbes. In contrast to other ocean ecosystems, microbes in permanent sea ice are space confined and subject to many extreme conditions, which change on a seasonal basis. How these microbial communities are regulated to survive the extreme sea ice environment is largely unknown. Here, we show that filamentous phages regulate the host bacterial community to improve survival of the host in permanent Arctic sea ice. We isolated a filamentous phage, f327, from an Arctic sea ice Pseudoalteromonas strain, and we demonstrated that this type of phage is widely distributed in Arctic sea ice. Growth experiments and transcriptome analysis indicated that this phage decreases the host growth rate, cell density and tolerance to NaCl and H2O2, but enhances its motility and chemotaxis. Our results suggest that the presence of the filamentous phage may be beneficial for survival of the host community in sea ice in winter, which is characterized by polar night, nutrient deficiency and high salinity, and that the filamentous phage may help avoid over blooming of the host in sea ice in summer, which is characterized by polar day, rich nutrient availability, intense radiation and high concentration of H2O2. Thus, while they cannot kill the host cells by lysing them, filamentous phages confer properties advantageous to host survival in the Arctic sea ice environment. Our study provides a foremost insight into the ecological role of filamentous phages in the Arctic sea ice ecosystem.
Introduction
Sea ice, covering 13% of the earth's surface (Parkinson and Gloersen, 1993), is one of the most frigid environments for marine microbes. It critically impacts the productivity of polar oceans, global energy budgets and atmosphere–ocean interactions in the Arctic and Antarctic zones (Dieckmann and Hellmer, 2003). Sea ice is characterized by perennially low temperatures ranging from −35 °C to 0 °C and poor nutrient supplies, with great fluctuations on many other factors such as pH, salinity and dissolved gas concentrations in different seasons (Mock and Thomas, 2005). Despite the extreme conditions, there is still a high variety and abundance of cold-adapted microorganisms including bacteria and bacteriophages (Steward et al., 1996; Thomas and Dieckmann, 2002). Different from the other ocean ecosystems, microbes in sea ice are space confined and face heavy pressure to survive under extreme conditions. How these microbes are adapted to survive the vast environmental changes in a confined space is largely unknown.
Bacteriophages far outnumber the prokaryotic cells in the ocean (Whitman et al., 1998; Suttle, 2007), which suggests the important roles of bacteriophages in the ocean ecosystems. Previous studies show that bacteriophages are also widespread in polar regions (Säwström et al., 2007, 2008a, 2008b) including the sea ice environment (Smith et al., 1992; Bird et al., 1993; Marchant et al., 2000; Guixa-Boixereu et al., 2002). How the widespread bacteriophages in sea ice regulate the microbial community is still unclear. Among bacteriophages, filamentous phages infect a large number of Gram-negative bacteria without lysing them (Rakonjac et al., 2011). Filamentous phages exist as a circular, double-stranded plasmid-like replicative form (RF) or an integrated form (IF) in the chromosome of the host cells (Rakonjac et al., 2011; Jian et al., 2012). Mature filamentous virions contain single-stranded DNA (ssDNA) and are packaged on the cell surface and secreted to the extracellular space (Rakonjac et al., 1999). Homologous filamentous phages usually reside in hosts of the same species, for example, f237 and VCYΦ are widely distributed in Vibrio parahaemolyticus and V. cholerae, respectively (Nasu et al., 2000; Xue et al., 2012). Lateral gene transfer conducted by filamentous phages among these bacteria belonging to the same genus is helpful for bacterial evolution in the ocean (Faruque et al., 2005). Filamentous phages have been isolated from different hosts inhabiting various environments, such as phage SW1 from the deep sea bacterium Shewanella piezotolerans WP3 (Wang et al., 2007), phage VCYΦ from V. cholerae from a coastal brackish pond (Xue et al., 2012), phage Φ Lf from Xanthomonas campestris pv. campestris from crucifers (Tseng et al., 1990) and phage CTXΦ from V. cholerae from cholera patients (Waldor and Mekalanos, 1996). However, filamentous phages have never been observed in or isolated from sea ice to date.
During the Second Chinese National Arctic Research Expedition cruise of the Chinese icebreaker Xue Long into the Canada Basin in August 2003, permanent sea ice samples were collected from seven sites in the area of 74°41′N–80°12′N and 149°06′W–164°04′W. A total of 356 aerobic heterotrophic bacterial strains were isolated from the ice samples. Phylogenetic analysis shows that more than 50% of the isolated strains are Pseudoalteromonas (Yu et al., 2009), which suggests that Pseudoalteromonas is a predominant group in the culturable diversity within the sea ice ecosystem. Study of the relationship between Pseudoalteromonas and derived bacteriophages will help us understand the role of bacteriophages in the sea ice ecosystem. In this article, a filamentous phage, termed as f327, was isolated from Pseudoalteromonas sp. BSi20327 from Arctic sea ice and characterized. Then, the distribution of this type of phage in the Arctic sea ice Pseudoalteromonas strains from different sites and its ecological role in sea ice ecosystem were studied. The results showed that the phage confers different physiologic properties on the host that may be advantageous to host survival in the Arctic sea ice environment. Our results provide evidence for filamentous phage's impact on the bacterial community in Arctic sea ice.
Materials and methods
Collection of sea ice samples and isolation of bacterial strains and plasmid-like RF
Sea ice samples (150–340 cm core length with 9 cm diameter) were collected at the seven sites using a MARK II ice auger (Kovacs Enterprises Inc., Lebanon, NH, USA) during the Second Chinese National Arctic Research Expedition cruise of the Chinese icebreaker Xue Long into the Canada Basin in August 2003. Sterile conditions were maintained during sampling and processing. The ice cores were cut into 10–20 cm sample sections using a sterile saw. Each ice section was melted at 4 °C in the same amount of pre-filtered (0.2 μm pore size) and autoclaved natural seawater from 5 m below the ice. Heterotrophic bacteria strains were isolated from the samples with three different media, including marine R2A (Suzuki et al., 1997), Difco marine 2216 (BD, Franklin Lakes, NJ, USA) and natural seawater. All media contained 15 g of agar per liter. Ice-melt samples were diluted in natural seawater and 100 μl of each dilution was plated on the agar plates. The plates were incubated in the dark at 4 °C for up to 8 weeks. Colonies from various agar plates were picked on the basis of differing colony morphologies and re-plated at least three times before further identification and study. The isolated Pseudoalteromonas strains (Supplementary Table S1) were grown at 15 °C in a marine Luria-Bertani (LB) broth (10 g peptone, 5 g yeast extract, 1 L artificial seawater, pH 7.5) (Zhao et al., 2011) for plasmid-like RF isolation with a plasmid extraction kit (BioTeke, Beijing, China).
Sequence analysis of RF327
RF327 was isolated from BSi20327 and treated with various restriction endonucleases, which showed that it contained restriction sites for HindIII (Supplementary Figure S1). Then, the nucleotide sequence of the plasmid was determined by using a combination of subcloning and primer walking. The whole sequence was obtained with at least 3 × coverage. The protein-coding regions were predicted with GeneMarkS for phage at http://topaz.gatech.edu/genemarks.cgi (Besemer et al., 2001). Similarity searches were carried out using the BLAST program at http://www.ncbi.nlm.nih.gov/blast (Tatusova and Madden, 1999). Signal peptide prediction was implemented using the SignalP 3.0 program at http://www.cbs.dtu.dk/services/SignalP-3.0/ (Bendtsen et al., 2004). The nucleotide sequence of RF327 was submitted to GenBank under the accession number GU198194. The phylogenetic trees based on different filamentous phage proteins were constructed with Molecular Evolutionary Genetics Analysis software (MEGA) (Tamura et al., 2011) by neighbor joining using the p-distance method. Bootstrap values were calculated based on 1000 computer-generated trees.
Phage isolation and observation by electron microscopy
A BSi20327 culture grown in marine LB broth at 15 °C overnight was centrifuged, and the supernatant was filtered through a 0.22 μm pore size membrane. The filtered solution was precipitated with 40% saturated ammonium sulfate overnight, and the precipitate was suspended in SM buffer (0.1 M NaCl, 0.01% gelatin, 8 mM MgSO4·7H2O, 50 mM Tris-HCl, pH 7.5) (Nasu et al., 2000). After dialysis against ddH2O, 3 μl of the resuspension was subjected to negative staining using a 1% (wt vol−1) uranyl formate solution on glow-discharged continuous carbon-coated copper grids and was examined under an electron microscope (Tecnai G2 F30; FEI, Hillsboro, OR, USA) with a field emission gun operated at 300 kV. Images of the samples were captured on a charge-coupled device camera (UltraScan1000XP, 2 k × 2 k; Gatan, Pleasanton, CA, USA) at magnifications of × 23 000 with the defocus ranging from −3 to −5 μm. The samples were also examined at tilt angles of ±60° using the same image conditions.
Characterization of phage f327
Phage f327 was isolated from a culture of BSi20327 grown in marine LB broth at 15 °C overnight, and the genomic DNA of f327 was extracted by phenol–chloroform extraction and ethanol precipitation. The phage genomic DNA was treated individually with RNase A, S1 nuclease, DNase I and restriction endonuclease HindIII followed by agarose gel electrophoresis. For N-terminal sequencing of the phage protein, the purified filamentous phage was separated on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to a polyvinylidene fluoride transfer membrane. The N-terminal amino-acid sequence was analyzed by Edman degradation on an Applied Biosystems PROCISE491 protein sequencer (Applied Biosystems, Foster City, CA, USA) at Beijing University (Beijing, China).
Phage distribution analysis
To examine the phage distribution in the Pseudoalteromonas strains isolated from different sites of Arctic sea ice, PCR was performed on the 53 isolated Pseudoalteromonas strains to amplify a phage DNA fragment from ORF386 to ORF448 using primers 386F and 448R (Supplementary Table S5) to detect the phage-containing Pseudoalteromonas strains. The PCR assay was performed using Fastpfu DNA polymerase (TransGen, Beijing, China) following the manufacturer's instruction. To confirm the phage-containing Pseudoalteromonas strains, the amplified DNA fragments were sequenced, and plasmids were extracted from the phage-containing Pseudoalteromonas strains by using a plasmid extraction kit (BioTeke). The extracted plasmids were further verified by PCR.
To analyze the phylogenetic relationship of the phage-containing Pseudoalteromonas strains, the rpoD genes (the structural gene for RNA polymerase σ70 factor) were amplified from the Arctic sea ice Pseudoalteromonas strains using primers 70F and 70R (Yamamoto et al., 1999) with Ex Taq DNA polymerase (TaKaRa, Tokyo, Japan) and sequenced. Based on the sequences of the rpoD genes, a phylogenetic tree was constructed by the neighbor-joining method (Saitou and Nei, 1987). Genomic DNA for the DNA–DNA hybridization assay was extracted from the Pseudoalteromonas strains using a previously described method (Brenner et al., 1982). DNA–DNA hybridization was performed according to renaturation rates (De Ley et al., 1970).
Curing of RF327 from Pseudoalteromonas sp.BSi20327
RF327 was cured from BSi20327 by elevating the culture temperature (Ghosh et al., 2000), and a derivative harboring no RF327 was screened and named BSi20327A. Southern blot hybridization was performed to verify whether BSi20327A produced the phage ssDNA. The genomic DNA from BSi20327 and BSi20327A were extracted and each was divided into two aliquots; one aliquot was digested with HindIII and the other was treated with S1 nuclease before digestion with HindIII. Southern blot hybridization was performed using the treated genomic DNA with ORF386 as the probe.
Growth, stress tolerance and swimming motility assays for BSi20327 and BSi20327A
BSi20327 and BSi20327A were grown at 15 °C overnight in marine LB broth. When they reached similar OD600 values indicative of late-exponential growth phase, 500 μl of each culture was transferred into 200 ml of modified marine LB broth (10 g peptone, 5 g yeast extract, 1 g MgSO4, 1 g (NH4)2SO4, 2 g K2HPO4, 1 L distilled water (pH 7.5)) containing different concentrations of NaCl (0.5%, 3%, 6% and 8%), which were cultured at 15 °C with shaking. The OD600 value was measured at different time intervals. The colony-forming unit counts of BSi20327 and BSi20327A in stationary growth phase was estimated after the cultures were serially diluted and spread onto marine LB plates.
To test the H2O2 tolerance of BSi20327 and BSi20327A, different concentrations (250 nM, 500 nM and 1.5 μM) of H2O2 were added to BSi20327 and BSi20327A cultures with an OD600 of 0.6. The cultures were then incubated at 0 °C for 16 h. After incubation, the cultures were serially diluted and spread on marine LB agar. Survival rates of BSi20327 and BSi20327A treated with H2O2 were measured and calculated using the previously described method (Watson et al., 1998).
The swimming motility of strains BSi20327 and BSi20327A was assessed using semisolid modified marine LB broth supplemented with 3% or 8% NaCl and 0.3% agar. The plates were seeded with 5 μl BSi20327 or BSi20327A in the exponential phase with the same OD600 and were incubated at 15 °C. The diameter was measured after 24 h for the colonies on the plates containing 3% NaCl or 72 h for the colonies on the plates containing 8% NaCl using at least five plates for each condition.
RNA extraction, whole-transcriptome sequencing and real-time qPCR
Total RNA was extracted from BSi20327 or BSi20327A grown to exponential phase (OD600≈1.0) at 15 °C in modified marine LB broth containing 3% or 8% NaCl with the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). Whole-transcriptome sequencing was performed by Beijing Genomics Institute Co., Ltd. (Shenzhen, China), where removal of rRNA, fragmentation, cDNA synthesis and PCR amplification were carried out. The constructed cDNA libraries were sequenced with Illumina HiSeq 2000 (Illumina Inc., San Diego, CA, USA). The RPKM (reads per kb per million reads) method (Mortazavi et al., 2008) was used to calculate the expression levels of genes. A rigorous algorithm described previously (Audic and Claverie, 1997) with a few modifications was used to identify differently expressed genes between two samples. Genes with false discovery rate ⩽0.001 and ratio between two samples >2 were identified as differently expressed genes. Relative expression level was indicated as fold change=log2(BSi20327/BSi20327A). Several genes that differently expressed in BSi20327 and BSi20327A were subjected to real-time quantitative PCR (qPCR) to verify the results of the whole-transcriptome sequencing. Reverse transcription was performed using PrimeScript RT Reagent Kit with gDNA Eraser (Perfect Real Time; TaKaRa). Real-time qPCR was performed using LightCycler 480 Real-Time PCR System (Roche, Basel, Switzerland) with the SYBR Premix EX Taq (TaKaRa) according to the manufacturer's instructions. Fold change was calculated using the LightCycler480 software (Roche). Each sample for real-time qPCR was performed in triplicate and a mean value and standard deviation were calculated. The rpoD gene of Bsi20327 amplified with the primers of rpoDF/rpoDR was used as the reference gene. Primers used for real-time qPCR were listed in Supplementary Table S5.
Results
Isolation and characterization of the filamentous phage f327 from Pseudoalteromonas sp. BSi20327
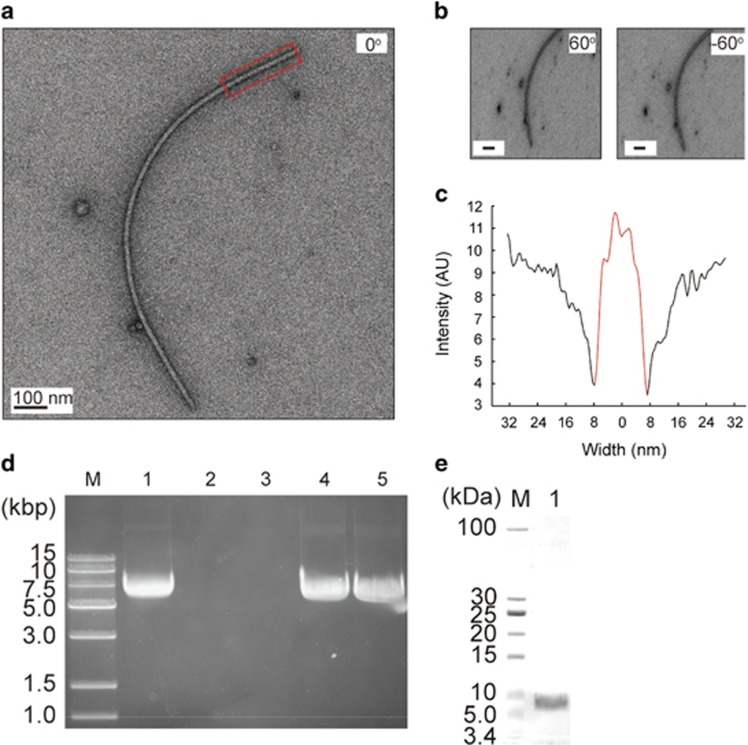
A plasmid-like structure, designated pSM327, was isolated from strain Pseudoalteromonas sp. BSi20327 (hereafter called BSi20327) from a sea ice sample collected at 78°23′14′′N and 149°06′55′′W. pSM327 is 6114 bp in length and is composed of nine open reading frames (ORFs). Sequence analysis reveals that each pSM327 ORF shows the greatest homology to ORFs at the corresponding positions in well-characterized filamentous phages (Waldor and Mekalanos, 1996; Wang et al., 2007), suggesting that pSM327 may an RF of a filamentous phage. We isolated the filamentous phage from a BSi20327 culture, which was named f327. High-resolution electron microscopy (Figures 1a–c) revealed that phage f327 exhibits a filament-like structure 14 nm in width and ∼1.5 μm in length, agreeing with the morphologic characteristics shared by filamentous phages. The genomic type of f327 is ssDNA, because the genome of f327 was resistant to RNase A and HindIII but sensitive to S1 nuclease and DNase I (Figure 1d). The purified f327 phage has a major protein band visible on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Figure 1e), and the corresponding protein is encoded by ORF74 (where the number represents the number of amino acids), based on N-terminal amino-acid sequencing. All of these data support the existence of a filamentous phage in Pseudoalteromonas sp. isolated from sea ice. In addition, the filamentous morphology and the ssDNA genome of f327 clearly indicate that it belongs to the Inoviridae family (http://ictvdb.bio-mirror.cn/Ictv/fs_inovi.htm).
Figure 1.
Morphology and basic characters of f327. (a and b) Representative images of phage f327. The tilting angles were set at 0° (a) and ±60° (b), respectively. No obvious width change was observed at different tilting angles. (c) A straight phage filament (red, boxed region in panel a) was chosen for width analysis. The phage component was colored in red. (d) Agarose gel electrophoresis analysis of the genomic DNA of phage f327. Lanes: M, DNA marker; 1, f327 genomic DNA treated with RNase A; 2, f327 genomic DNA treated with S1 nuclease; 3, f327 genomic DNA treated with DNase I; 4, f327 genomic DNA treated with HindIII; 5, untreated f327 genomic DNA. (e) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of f327 phage. Lanes: M, protein ladder; 1, purified f327 phage.
Genomic analysis of phage f327 and its evolutionary relationships to other filamentous phages
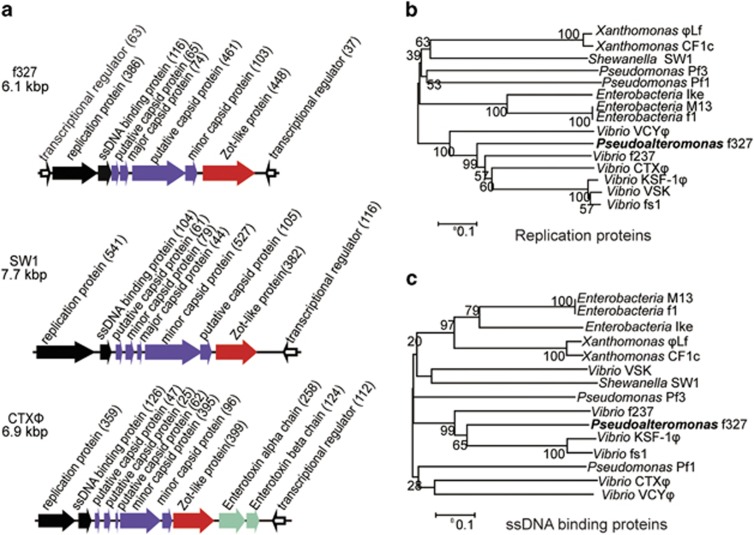
Detailed sequence alignment and genetic organization analysis of f327 with other reported phages, such as CTXΦ and SW1 (Waldor and Mekalanos, 1996; Wang et al., 2007), revealed that the genes of f327 can be classified into four functional modules as follows: replication, structure, assembly and regulation (Figure 2a). The regulation module consists of ORF37 and ORF63. ORF37, which is oriented in the opposite direction compared with the other eight ORFs, is similar to RstR of Vibrio phage CTXΦ (Supplementary Table S2), and ORF63 is similar to the transcriptional regulator of Marinobacter. The replication module contains ORF386 and ORF116. ORF386 encodes a protein similar to the replication proteins of Vibrio phages, and ORF116 encodes a protein similar to the ssDNA-binding proteins of Vibrio phages. The structural module comprises ORF65, ORF74, ORF461 and ORF103 based on their map positions. ORF74, ORF461 and ORF103 are predicted to have signal peptides and therefore may function as capsid proteins. ORF103 shares similarity with the minor coat protein of Ralstonia solanacearum and the integral membrane protein of Neisseria flavescens; ORF74 shows similarity with the coat protein of Haloplasma contractile; ORF65 and ORF461 have no obvious similarity to any functional proteins from previously described filamentous phages. The major coat protein of f327 is translated from ORF74 as revealed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis (Figure 1e) and N-terminal sequence analysis mentioned above. The assembly module contains only ORF448, which encodes a protein similar to the phage assembly protein of Ralstonia syzygii and the Zot protein of CTXΦ (a homolog to the gene I product of phage M13 involved in filamentous phage assembly; Waldor and Mekalanos, 1996). Based on the phage replication protein (Figure 2b) and the ssDNA-binding protein (Figure 2c) sequences, f327 is phylogenetically closely related to other known Inoviridae phages, further confirming that f327 belongs to the Inoviridae family.
Figure 2.
The genomic structural map of f327 and phylogenetic relationships among the amino-acid sequences of different proteins. (a) The genomic structural maps of f327 and related filamentous phages. The length of the arrow represents the size of the protein. The direction of the arrow represents the transcription direction of the ORF. The number within parentheses indicates the number of amino-acid residues. ORFs in black represent the replication module; the purple arrows represent the structural module; the assembly modules are shown in red; the hollow arrows represent the regulation module. (b) Phylogenetic relationships among amino-acid sequences of replication proteins. (c) Phylogenetic relationships among amino-acid sequences of ssDNA-binding proteins.
Distribution of f327 in the Arctic sea ice Pseudoalteromonas
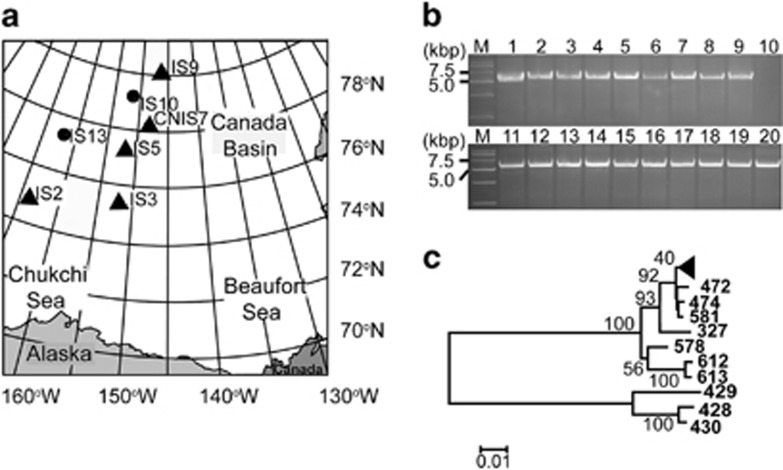
To investigate the distribution of this filamentous phage in Arctic sea ice Pseudoalteromonas strains, attempts were made to amplify a fragment between ORF386 and ORF448 of f327 (shown in Figure 2a) from 53 Pseudoalteromonas strains isolated from different sites in the Canada Basin (Figure 3a and Supplementary Table S1). The expected DNA fragments were amplified from 19 Pseudoalteromonas strains. Respective RFs were also detected in these 19 strains by plasmid isolation (Supplementary Figure S3). Therefore, 36% of the Pseudoalteromonas strains (19/53) were found to contain f327 or f327-like genes (Figure 3b). Further genomic analysis on one typical phage-containing strain designated BSi20439 (DDBJ/GenBank/EMBL accession number: BADW00000000) showed that BSi20439 contains all orthologous genes of f327 (Bian et al., 2011) (Supplementary Table S3). These data indicate a remarkably high prevalence of f327 or similar phage types in the Arctic sea ice Pseudoalteromonas population. It is important to emphasize here that phages f327 and f439 are not 100% identical (identities from 68% to 100% Supplementary Table S3), which indicates that different phages may preferentially target different Pseudoalteromonas strains. We selected 19 phage-containing strains and 9 strains without phage to analyze the phylogenetic relationship of the phage-containing strains. The phylogenetic tree based on the rpoD gene sequence of the Arctic sea ice Pseudoalteromonas strains revealed that all of the phage-containing strains are in the same branch (Figure 3c). To further analyze whether the phage-containing strains belong to the same species of Pseudoalteromonas, BSi20327, BSi20439, BSi20514 (three phage-containing strains) and BSi20612 (a strain without phage) were chosen to perform DNA–DNA hybridization. BSi20327 had a relatedness of 91.7% with BSi20439 and 93.2% with BSi20514. Both percentages are higher than 70%, a standard for bacterial species delineation (Johnson, 1973), which indicates that these phage-containing strains belong to the same species of Pseudoalteromonas. In contrast, BSi20612 (no phage) belongs to another species of Pseudoalteromonas, as it only exhibited a relatedness of 51.6% to BSi20327. These results indicated that f327 or f327-like filamentous phages are prevalent in strains of one Pseudoalteromonas species, which is widely distributed in Arctic sea ice samples from 74°N to 80°N (Figure 3a).
Figure 3.
Distribution of f327 or f327-like phages in the Arctic sea ice Pseudoalteromonas population. (a) Location of the sampling sites in the Arctic sea ice of the Canada Basin where the 53 Pseudoalteromonas strains were isolated. The sites where f327 or f327-like phages were detected are indicated by black triangles. (b) Agarose gel electrophoresis analysis of the PCR product of 19 phage gene-containing strains. Lanes 1–20 were BSi20327, BSi20335, BSi20381, BSi20465, BSi20401, BSi20514, BSi20407, BSi20357, BSi20436, BSi20670, BSi20366, BSi20340, BSi20455, BSi20447, BSi20462, BSi20451, BSi20464, BSi20341, BSi20437 and BSi20439, respectively. Strain BSi20670 harboring no phage gene was used as the negative control. (c) Phylogenetic tree of Pseudoalteromonas based on the rpoD sequences. The black triangles represent the 18 phage-containing strains, which are BSi20357, BSi20341, BSi20407, BSi20439, BSi20447, BSi20401, BSi20437, BSi20381, BSi20366, BSi20514, BSi20462, BSi20465, BSi20451, BSi20455, BSi20335, BSi20436, BSi20340 and BSi20464. ‘BSi20' was omitted from the identification numbers in the figure.
Influence of phage f327 on the growth, cell density, stress tolerance and motility of the host BSi20327
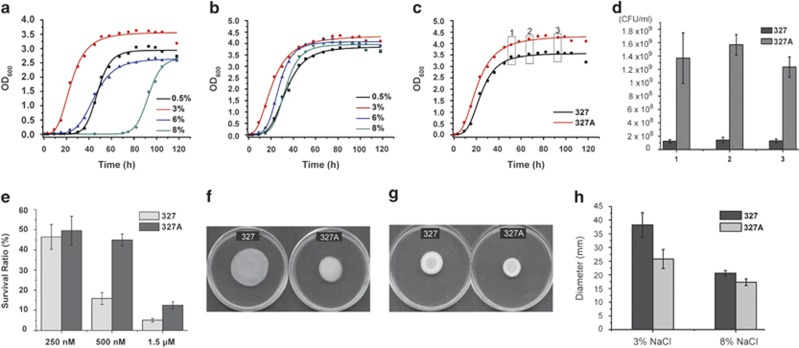
To investigate the role of f327 in the sea ice ecosystem, the effect of f327 on the growth, cell density, stress tolerance and motility of the host BSi20327 strain was examined. An RF327-cured derivative of BSi20327 was obtained and named Pseudoalteromonas sp. BSi20327A (hereafter called BSi20327A) (Supplementary Figure S2). Considering that the salinity of sea ice can range from 0 to 20% during seasonal shifts (Mock and Thomas, 2005), strains BSi20327 and BSi20327A were subjected to growth rate analysis at different salt concentration conditions. Both strains showed the highest growth rates at 3% NaCl (Figures 4a and b). However, BSi20327A had a higher growth rate than BSi20327 under 3% NaCl (Figure 4c). Moreover, the cell density of BSi20327A obtained was also higher than that of BSi20327. The colony-forming units of BSi20327A were ∼7–10-fold more abundant than those of BSi20327 at the stationary phase under 3% NaCl (Figure 4d). In addition, BSi20327 and BSi20327A differed significantly in NaCl tolerance (Figures 4a and b). NaCl in the range of 0.5–8% only had a slight influence on the BSi20327A growth rate (Figure 4b), but the higher NaCl concentrations within this range markedly decreased the growth rate of BSi20327 (Figure 4a). Taken together, these results indicate that the presence of phage f327 can reduce the host growth rate, impair the host tolerance against NaCl and lower the cell density of the host community.
Figure 4.
Growth, stress tolerance and motility of BSi20327 and BSi20327A. (a) Growth curves of BSi20327 in different NaCl concentrations. (b) Growth curves of BSi20327A in different NaCl concentrations. (c) Growth curves of BSi20327 and BSi20327A cultured in 3% NaCl at 15 °C. The points indicated by the three hollow boxes were the sampling times for the colony-forming unit (CFU) estimation. (d) CFU of BSi20327 and BSi20327A in the stationary phase. (e) Survival ratios of BSi20327 and BSi20327A treated with 250 nM, 500 nM and 1.5 μM H2O2. (f) Swimming motility of BSi20327 and BSi20327A on motility agar plates containing 3% NaCl. (g) Swimming motility of BSi20327 and BSi20327A on motility agar plates containing 8% NaCl. (h) Comparison of the diameters of BSi20327 and BSi20327A on motility agar plates containing 3% or 8% NaCl. The P-value is 5.08 × 10−4 at 3% NaCl and 0.011 at 8% NaCl.
In addition to the variation in salt concentration, H2O2 is photochemically produced under high irradiance levels in polar zones, especially in the summer time (Cooper and Zirka, 1983; Sigg and Neftel, 1988), and can reach concentrations up to 500 nM in sea ice (Mock and Thomas, 2005). We tested the tolerance of BSi20327 and BSi20327A to H2O2. Although both strains had a similar survival rate in 250 nM H2O2, the survival rate of BSi20327A was at least two times as high as that of BSi20327 in 500 nM or 1.5 μM H2O2 (Figure 4e). Therefore, the presence of f327 seems to impair the H2O2 tolerance of the host.
Swimming motility is another factor we chose to test. BSi20327 and BSi20327A were assayed on semisolid plates. The diameter of BSi20327 colonies was ∼48% larger than those of BSi20327A in 3% NaCl (Figures 4f and h). Although BSi20327 demonstrated a much lower growth rate than BSi20327A in 8% NaCl (Figures 4a and b), BSi20327 colonies were ∼19% larger than those of BSi20327A (Figures 4g and h). This result suggests that the presence of f327 can increase the motility of the host, thereby enabling the bacterial community to access rapidly favorable environments.
Transcriptome analysis of the molecular mechanism of f327 affecting the host
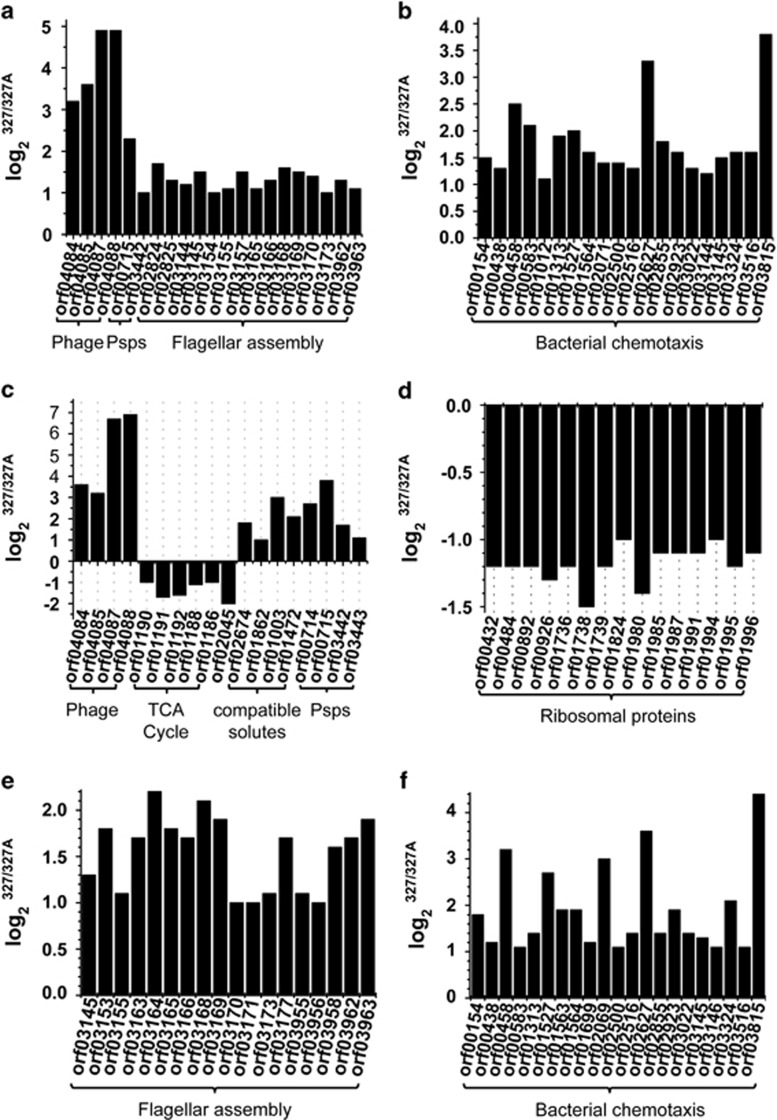
To determine the mechanism underlying how f327 affects its host BSi20327, the whole transcriptomes of BSi20327 and BSi20327A grown in 3% and 8% NaCl were analyzed and compared. At 3% NaCl, the only upregulated genes in BSi20327 were the genes encoded by phage f327 and genes related to phage shock, flagellar assembly and bacterial chemotaxis (Figures 5a and b). No significant difference in transcription was observed between BSi20327 and BSi20327A for all other genes. As the processes of phage and flagella assembly are energy intensive, the reduction in the growth rate of BSi20327 in 3% NaCl may be mainly because of these two energy-intensive processes. At 8% NaCl, in addition to those upregulated at 3% NaCl, five genes encoding succinyl-CoA synthetase and succinate dehydrogenase (key enzymes in the tricarboxylic acid cycle) are downregulated in BSi20327 (Figure 5c), which would result in a reduction in energy production. One transcription elongation factor gene (Figure 5c) and 15 genes related to ribosome assembly (Figure 5d) are also downregulated in BSi20327, which likely decreases the rate of gene transcription and translation. Therefore, at 8% NaCl, overexpression of f327 in BSi20327 not only consumes energy but also affects energy production and gene transcription and translation of the host, which will severely reduce the host growth rate as we observed (Figure 4a).
Figure 5.
Analysis of the differentially expressed genes in BSi20327 and BSi20327A grown at 3% NaCl (a and b) and 8% NaCl (c and d). (a) Phage genomic genes and genes related to phage shock proteins and flagellar assembly. (b) Genes related to bacterial chemotaxis. (c) Phage genomic genes and genes related to the tricarboxylic acid cycle, gene transcription (orf02045), amino-acid transport, glutamate and proline synthesis and phage shock proteins. (d) Genes related to ribosome assembly. (e) Genes related to flagellar assembly. (f) Genes related to bacterial chemotaxis. The verification of the results of the whole-transcriptome sequencing by real-time qPCR is shown in Supplementary Figure S4 and the detailed gene annotation is listed in Supplementary Table S4.
Bacteria usually respond to hyperosmolarity by accumulating compatible solutes, such as some amino acids (glutamate, proline) and sugars, by absorbing or synthesizing them (McLaggan et al., 1994; Sleator and Hill, 2002). Based on transcriptome analysis, two genes involved in amino-acid transport, as well as the glutamate synthase gene and the glutamate-5-semialdehyde dehydrogenase gene (both taking part in the glutamate and proline synthesis), are upregulated in both BSi20327 and BSi20327A in 3–8% NaCl. This suggests that these genes are likely involved in the salt tolerance of BSi20327 and BSi20327A. However, these genes are upregulated to a greater extent in BSi20327 compared with BSi20327A, especially at 8% NaCl (Figure 5c). Because these four genes are related to the accumulation of compatible solutes to cope with the hyperosmotic environment, this result suggests that 8% NaCl is more stressful to BSi20327 than that to BSi20327A; therefore, BSi20327 needs to accumulate more compatible solutes to resist the hyperosmolarity.
Phage shock proteins (Psps) can be induced during the secretion of filamentous phages (Joly et al., 2010). Several genes in the phage shock protein operon, pspA, pspB and pspC, and a transcriptional activator gene are upregulated in BSi20327 (Figure 5c). It has been reported that the expression of phage shock proteins can affect chemotaxis and motility of the host (Darwin, 2005; Joly et al., 2010). Correspondingly, flagella-related genes (Figure 5e) and chemotaxis-related genes (Figure 5f) are also upregulated in BSi20327, which may enhance the cell motility of BSi20327. These results are consistent with the swimming motility assay of BSi20327 and BSi20327A on semisolid plates (Figure 4). It can be concluded that the upregulation of phage shock protein genes caused by the assembly and extrusion of f327 increases the motility and chemotaxis of the host organism.
Discussion
Although several bacteriophages have been isolated from Pseudoalteromonas (Wichels et al., 1998; Kivelä et al., 2002), no filamentous phage has yet been described in Pseudoalteromonas. In this study, a filamentous phage f327 was isolated from Pseudoalteromonas sp. BSi20327 from the Arctic sea ice of Canada Basin. f327 is a member of the Inoviridae family. This filamentous phage is prevalent in the strains of a Pseudoalteromonas species widely distributed in the Arctic sea ice. As Pseudoalteromonas is probably a dominant group in Canada Basin sea ice (Yu et al., 2009), the effect of this filamentous phage on the host would influence the sea ice ecosystem.
Although some researches show that the mortality and community structure of the host bacteria in polar areas are regulated by lytic viruses (Guixa-Boixereu et al., 2002; López-Bueno et al., 2009), the ecological role of filamentous phages in sea ice is still unknown. Sea ice is a confined space permeated by a highly connected network of channels and pores, which are filled with brine formed from expelled salts during the freezing of the ice crystals (Thomas and Dieckmann, 2002). Owing to its closed or semiclosed characteristics, exchange of inorganic nutrients is greatly restricted in the sea ice ecosystem (Thomas and Dieckmann, 2002). In winter, the bacteria in sea ice are confronted with high salinity and nutrient deficiency because of the low temperatures and long polar nights. In summer, although the salinity in the sea ice decreases with increased temperature, the sea ice bacteria are faced with rich nutrients and a high concentration of H2O2 due to the polar day and intense sunlight (Mock and Thomas, 2005). Thus, in winter, the reduction in the growth rate and NaCl tolerance of BSi20327 caused by f327 would diminish the community scale of BSi20327 and thereby reduce nutrient consumption; in the meantime, motility enhancement caused by f327 can help BSi20327 access environments suitable for growth. These regulatory effects of phage f327 on the host cells favor the survival of the host community during the long nutrient-deficient polar night winter days. In summer, because of the presence of f327, the H2O2 tolerance of BSi20327 decreases, which would lead to a high mortality rate of BSi20327 because of the high concentration of H2O2. In addition, the presence of f327 also reduces the cell density of the BSi20327 community. These mechanisms may help avoid the over blooming of BSi20327 during the months of round-the-clock sunlight and the nutrient-rich summer. Thus, as a filamentous phage that cannot kill the host cells by lysing them, f327 can moderately regulate its host community. Although f327 has some negative effects on the metabolism and growth of the host cells under different environmental conditions in Arctic sea ice, our results suggest that f327 is helpful in maintaining the host community at an appropriate scale, which may be advantageous to host survival in Arctic sea ice.
Acknowledgments
We thank Aharon Oren (the Hebrew University of Jerusalem, Israel) for helpful suggestions. The work was supported by National Natural Science Foundation of China (31290231, 41106161, 31025001, 31070061 and 31000034), Hi-Tech Research and Development program of China (2011AA09070303, 2012AA092105 and 2012AA092103) and Program of Shandong for Taishan Scholars (2008BS02019).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Audic S, Claverie JM. (1997). The significance of digital gene expression profiles. Genome Res 7: 986–995. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. (2004). Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795. [DOI] [PubMed] [Google Scholar]
- Besemer J, Lomsadze A, Borodovsky M. (2001). GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29: 2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian F, Xie BB, Qin QL, Shu YL, Zhang XY, Yu Y et al. (2011). Genome sequences of six Pseudoalteromonas strains isolated from Arctic sea ice. J Bacteriol 194: 908–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird DF, Maranger R, Karl DM. (1993). Palmer LTER: aquatic virus abundances near the Antarctic Peninsula. Antarct J US 28: 234–235. [Google Scholar]
- Brenner DJ, McWhorter AC, Leete Knutson JK, Steigerwalt AG. (1982). Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J Clin Microbiol 15: 1133–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper WJ, Zirka RG. (1983). Photochemical formation of hydrogen peroxide in surface and ground waters exposed to sunlight. Science 220: 711–712. [DOI] [PubMed] [Google Scholar]
- Darwin AJ. (2005). The phage-shock-protein response. Mol Microbiol 57: 621–628. [DOI] [PubMed] [Google Scholar]
- De Ley J, Cattoir H, Reynaerts A. (1970). The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12: 133–142. [DOI] [PubMed] [Google Scholar]
- Dieckmann GS, Hellmer HH. (2003) Sea Ice: An Introduction to its Physics, Chemistry, Biology and Geology. Blackwell Science Press: Oxford, UK. [Google Scholar]
- Faruque SM, Naser IB, Fujihara K, Diraphat P, Chowdhury N, Kamruzzaman M et al. (2005). Genomic sequence and receptor for the Vibrio cholerae phage KSF-1Φ: evolutionary divergence among filamentous vibriophages mediating lateral gene transfer. J Bacteriol 187: 4095–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Mahapatra NR, Ramamurthy T, Banerjee PC. (2000). Plasmid curing from an acidophilic bacterium of the genus Acidocella. FEMS Microbiol Lett 183: 271–274. [DOI] [PubMed] [Google Scholar]
- Guixa-Boixereu N, Vaqué D, Gasol JM, Sánchez-Cámara J, Pedrós-Alió C. (2002). Viral distribution and activity in Antarctic waters. Deep-Sea Res 49: 827–845. [Google Scholar]
- Jian HH, Xu J, Xiao X, Wang FP. (2012). Dynamic modulation of DNA replication and gene transcription in deep-sea filamentous phage SW1 in response to changes of host growth and temperature. PLoS One 7: e41578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL. (1973). Use of nucleic-acid homologies in the taxonomy of anaerobic bacteria. Int J Syst Bacteriol 23: 308–315. [Google Scholar]
- Joly N, Engl C, Jovanovic G, Huvet M, Toni T, Sheng X et al. (2010). Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol Rev 34: 797–827. [DOI] [PubMed] [Google Scholar]
- Kivelä HM, Kalkkinen N, Bamford DH. (2002). Bacteriophage PM2 has a protein capsid surrounding a spherical proteinaceous lipid core. J Virol 76: 8169–8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bueno A, Tamames J, Velázquez D, Moya A, Quesada A, Alcamí A. (2009). High diversity of the viral community from an Antarctic lake. Science 326: 858–861. [DOI] [PubMed] [Google Scholar]
- Marchant H, Davidson A, Wright S, Glazebrook J. (2000). The distribution and abundance of viruses in the Southern Ocean during spring. Antarct Sci 12: 414–417. [Google Scholar]
- McLaggan D, Naprstek J, Buurman ET, Epstein W. (1994). Interdependence of K+ and glutamate accumulation during osmotic adaptation of Escherichia coli. J Biol Chem 269: 1911–1917. [PubMed] [Google Scholar]
- Mock T, Thomas DN. (2005). Recent advances in sea-ice microbiology. Environ Microbiol 7: 605–619. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Nasu H, Iida T, Sugahara T, Yamaichi Y, Park KS, Yokoyama K et al. (2000). A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J Clin Microbiol 38: 2156–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson CL, Gloersen P. (1993) Atlas of Satellite Observations Related to Global Change. Cambridge University Press: Cambridge, UK. [Google Scholar]
- Rakonjac J, Bennett NJ, Spagnuolo J, Gagic D, Russel M. (2011). Filamentous bacteriophage: biology, phage display and nanotechnology applications. Curr Issues Mol Biol 13: 51–76. [PubMed] [Google Scholar]
- Rakonjac J, Feng JN, Model P. (1999). Filamentous phage are released from the bacterial membrane by a two-step mechanism involving a short C-terminal fragment of pIII. J Mol Biol 289: 1253–1265. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Sigg A, Neftel A. (1988). Seasonal variations in hydrogen peroxide in polar ice cores. Ann Glaciol 10: 157–162. [Google Scholar]
- Sleator RD, Hill C. (2002). Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol Rev 26: 49–71. [DOI] [PubMed] [Google Scholar]
- Smith DC, Steward GF, Azam F, Hollibaugh JT. (1992). Virus and bacteria abundance in the Drake Passage during January and August 1991. Antarct J US 27: 125–127. [Google Scholar]
- Steward GF, Smith DC, Azam F. (1996). Abundance and production of bacteria and viruses in the Bering and Chukchi Seas. Mar Ecol Prog Ser 131: 287–300. [Google Scholar]
- Suttle CA. (2007). Marine viruses—major players in the global ecosystem. Nat Rev Microbiol 5: 801–812. [DOI] [PubMed] [Google Scholar]
- Suzuki MT, Rappe MS, Haimberger ZW, Winfield H, Adair N, Ströbel J et al. (1997). Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol 63: 983–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Säwström C, Granéli W, Laybourn-Parry J, Anesio AM. (2007). High viral infection rates in Antarctic and Arctic bacterioplankton. Environ Microbiol 9: 250–255. [DOI] [PubMed] [Google Scholar]
- Säwström C, Lisle J, Anesio AM, Priscu JC, Laybourn-Parry J. (2008. a). Bacteriophage in polar inland waters. Extremophiles 12: 167–175. [DOI] [PubMed] [Google Scholar]
- Säwström C, Pearce I, Davidson AT, Rosén P, Laybourn-Parry J. (2008. b). Influence of environmental conditions, bacterial activity and viability on the viral component in 10 Antarctic lakes. FEMS Microbiol Ecol 63: 12–22. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. (1999). Blast 2 sequences—a new tool for comparing proteins and nucleotide sequences. FEMS Microbiol Lett 174: 247–250. [DOI] [PubMed] [Google Scholar]
- Thomas DN, Dieckmann GS. (2002). Antarctic sea ice—a habitat for extremophiles. Science 295: 641–644. [DOI] [PubMed] [Google Scholar]
- Tseng YH, Lo MC, Lin KC, Pan CC, Chang RY. (1990). Characterization of filamentous bacteriophage phi Lf from Xanthomonas campestris pv. campestris. J Gen Virol 71: 1881–1884. [DOI] [PubMed] [Google Scholar]
- Waldor MK, Mekalanos JJ. (1996). Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272: 1910–1914. [DOI] [PubMed] [Google Scholar]
- Wang F, Wang FP, Li Q, Xiao X. (2007). A novel filamentous phage from the deep-sea bacterium Shewanella piezotolerans WP3 is induced at low temperature. J Bacteriol 189: 7151–7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson SP, Clements MO, Foster SJ. (1998). Characterization of the starvation-survival response of Staphylococcus aureus. J Bacteriol 180: 1750–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. (1998). Prokaryotes: the unseen majority. Proc Natl Acad Sci USA 95: 6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichels A, Biel SS, Gelderblom HR, Brinkhoff T, Muyzer G, Schütt C. (1998). Bacteriophage diversity in the North Sea. Appl Environ Microbiol 64: 4128–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H, Xu Y, Boucher Y, Ploz MF. (2012). High frequency of a novel filamentous phage, VCYΦ, within an environmental Vibrio cholerae population. Appl Environ Microbiol 78: 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Bouvet PJM, Harayama S. (1999). Phylogenetic structures of the genus Acinetobacter based on gyrB sequences: comparison with the grouping by DNA–DNA hybridization. Int J Syst Bacteriol 49: 87–95. [DOI] [PubMed] [Google Scholar]
- Yu Y, Li HR, Zeng YX, Chen B. (2009). Extracellular enzymes of cold-adapted bacteria from Arctic sea ice, Canada Basin. Polar Biol 32: 1539–1547. [Google Scholar]
- Zhao DL, Yu ZC, Wu ZY, Chen XL, Shi M, Yu Y et al. (2011). Characterization of a cryptic plasmid pSM429 and its application for heterologous expression in psychrophilic Pseudoalteromonas. Microb Cell Fact 10: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.