Abstract
Arctic Ocean microbial eukaryote phytoplankton form subsurface chlorophyll maximum (SCM), where much of the annual summer production occurs. This SCM is particularly persistent in the Western Arctic Ocean, which is strongly salinity stratified. The recent loss of multiyear sea ice and increased particulate-rich river discharge in the Arctic Ocean results in a greater volume of fresher water that may displace nutrient-rich saltier waters to deeper depths and decrease light penetration in areas affected by river discharge. Here, we surveyed microbial eukaryotic assemblages in the surface waters, and within and below the SCM. In most samples, we detected the pronounced SCM that usually occurs at the interface of the upper mixed layer and Pacific Summer Water (PSW). Poorly developed SCM was seen under two conditions, one above PSW and associated with a downwelling eddy, and the second in a region influenced by the Mackenzie River plume. Four phylogenetically distinct communities were identified: surface, pronounced SCM, weak SCM and a deeper community just below the SCM. Distance–decay relationships and phylogenetic structure suggested distinct ecological processes operating within these communities. In the pronounced SCM, picophytoplanktons were prevalent and community assembly was attributed to water mass history. In contrast, environmental filtering impacted the composition of the weak SCM communities, where heterotrophic Picozoa were more numerous. These results imply that displacement of Pacific waters to greater depth and increased terrigenous input may act as a control on SCM development and result in lower net summer primary production with a more heterotroph dominated eukaryotic microbial community.
Introduction
Biogeography of microbial communities are governed by the interaction of deterministic and stochastic processes, with shifts in community composition ultimately reflecting changes in the influence of these community assembly processes (Dumbrell et al., 2009; Martiny et al., 2011; Hanson et al., 2012). Within the first category of deterministic, or niche-based, processes are environmental filtering (species sorting) and species interactions. Within the second category of stochastic or species-neutral processes are dispersion and genetic drift (Chase and Myers, 2011). The Arctic Ocean (AO) is an ideal environment to study these ecological processes, especially habitat filtering as it is strongly salinity stratified (Rainville et al., 2011), and consists of advective water masses with different histories. These water masses may act as selective environments, ultimately reflected in microbial community composition. A consequence of the hydrography in the Western AO is that the euphotic zone is largely nutrient limited and the majority of summer primary production occurs within a subsurface chlorophyll maximum (SCM) layer (Carmack and McLaughlin, 2011). SCM is routinely detected as peaks of chlorophyll fluorescence at depth and is clearly separated from surface chlorophyll concentrations. In the Western AO, the phytoplanktons that live within these SCM, at the interface between the euphotic zone and deeper nutrient-replete water masses (Wheeler et al., 1996; Martin et al., 2012), are exclusively microbial eukaryotes, especially diatoms and the pico-sized alga Micromonas (Martin et al., 2010; Ardyna et al., 2011). Because the depth of the SCM reflects a trade-off between light and nutrient levels capable of sustaining phytoplankton growth (Hill et al., 2013), SCM maintenance may be highly susceptible to changes in the physical structure of the water column. Any disruption of this fragile light-nutrient balance would be predicted to alter community structure (Lovejoy, 2012). In addition, less favorable light and nutrient conditions could be considered as stress factors that would influence species composition and turnover within the SCM. Marked environmental changes have occurred over the past decade across the AO (Screen and Simmonds, 2010), notably two summer sea ice minimum extent records in 2007 and 2012. Increased sea ice loss and terrestrial freshwater runoff rich in dissolved organic matter are altering the AO physical structure with impacts on vertical mixing and, in the Western Arctic, the depth of nutrient-rich Pacific waters (Carmack and McLaughlin, 2011). Evaluating the ecological consequences of multiyear sea ice loss and increased terrestrial runoff on SCM layers is essential for predicting biodiversity, productivity and potential food-web shifts in a changing AO (Lovejoy, 2012; Forest et al., 2013; Post et al., 2013).
Over the Western Arctic, SCM is associated with the inflow of nutrient-rich Pacific Ocean summer (PSW) and winter waters (PWW), which, most of the year, remain below the fresher, nutrient-depleted, polar mixed layer (PML; Carmack et al., 1989, 2004; Macdonald et al., 1989). As the AO freshens, the PML has thickened and PSW and PWW are being pushed deeper (Carmack and McLaughlin, 2011). The AO physical structure directly impacts the vertical distribution of microbial communities, which are linked to their water mass of origin (Martin et al., 2010; Ardyna et al., 2011; Lovejoy and Potvin, 2011; Monier et al., 2013). Disturbance of AO water mass depth equilibrium could thus expose SCM microbial communities to lower light levels at the nitracline, and potentially alter their taxonomic composition and distribution (Cermeno et al., 2008), ultimately affecting primary production (McLaughlin and Carmack, 2010). For example, over an 8-year period, the average phytoplankton cell size in the upper 200 m of the Canada Basin decreased, with diatoms becoming scarcer compared with small flagellates (Li et al., 2009). Furthermore, SCM microbial communities changed after the 2007 record multiyear sea-ice loss (Comeau et al., 2011).
Although oceanographic structure and microbial communities seem to be changing, there is a knowledge gap on how climate-driven oceanic processes could influence community assembly and functionality. To fully understand how microbial communities are affected by changing Arctic conditions, the underlying ecological processes responsible for their extant composition, spatial distribution and their responses to environmental changes need to be assessed (Horner-Devine and Bohannan, 2006; Stegen et al., 2012).
We used the southern Beaufort Sea, affected by mesoscale processes mediated by the Mackenzie Trough and River, as a test to gain insights into the potential effects of climate-driven changes on AO microbial biodiversity, biogeography and on the ecological processes influencing SCM establishment and maintenance. The Mackenzie Trough, a shelf canyon that cuts across the Beaufort Shelf, promotes asymmetric upwelling and gyre formation, displacing water masses by tens to hundreds of meters (Williams et al., 2006). In addition, the Mackenzie River plume, with high concentrations of colored dissolved organic matter (CDOM), can strongly attenuate photosynthetically active radiation (PAR; Matsuoka et al., 2012). Such conditions potentially mimic changes predicted to occur over the AO where the locally formed PML is thickening, driving Pacific water deeper and high CDOM river inputs are increasing (Peterson et al., 2002; Matsuoka et al., 2012).
To examine processes driving phylogenetic composition and structure of microbial communities, we sampled the principal water masses and their microbial eukaryotic communities, from the Beaufort Sea and Mackenzie Trough. We targeted stations with a range of in situ SCM fluorescence profiles, from sharp, intense peaks to very weak, diffuse fluorescence signals. We then used high-throughput 18S rDNA amplicon sequencing to assess phylogenetic and taxonomic composition of microbial communities across the ecological landscape. In a previous study focusing on Beaufort Sea stations with clear SCM peaks, we showed that the heterotrophic flagellate component of the eukaryotic communities were clearly partitioned based on their water mass of origin (Monier et al., 2013). Here, we investigated stations with remarkably low SCM peaks, and analyzed the distribution of phototrophs and smaller heterotrophs that made up microbial eukaryotic communities to determine: (i) the effect of AO physical oceanography on microbial diversity and biogeography; (ii) the influence of ecological processes and microbial species turnover in SCM formation and maintenance, and (iii) whether specific taxa are associated with community changes.
Materials and methods
Microbial sampling and high-throughput sequencing
Sampling was from the Canadian research icebreaker CCGS Amundsen in August 2009, as part of the International Polar Year, project MALINA. Detailed methods are given in Supplementary Information and all ancillary data for the MALINA project are available at http://malina.obs-vlfr.fr. Samples for microbial analysis were selected on the downward conductivity-temperature–salinity (CTD) cast, based on salinity, temperature, relative nitrate and chlorophyll fluorescence traces (Supplementary Table S1). For this study, six stations, three east of the Mackenzie Canyon (stations 430, 460 and 540), one west of the canyon (station 760) and two within the canyon (620 and 670) were sampled (Figure 1). Four depths were sampled at each station: surface (ZSURF), the SCM (ZSCM) and waters from above and below the SCM (Za-SCM and Zb-SCM, respectively).
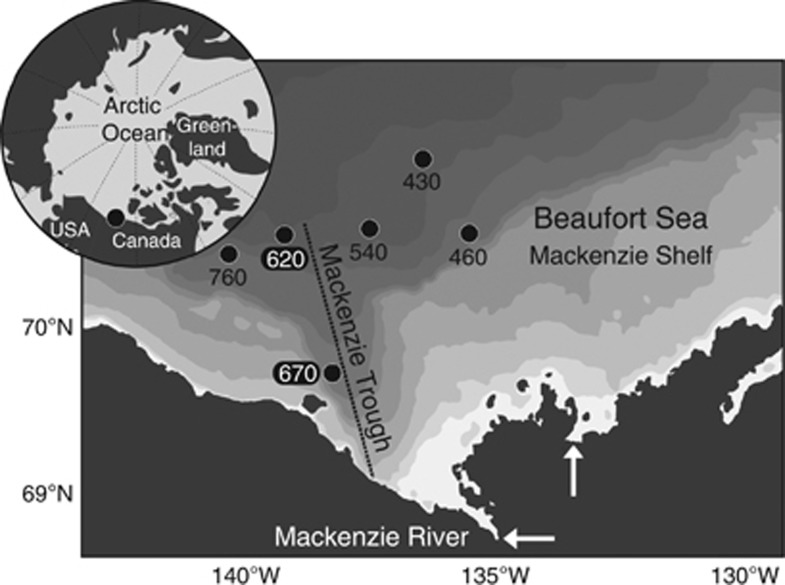
Figure 1.
Location of the Beaufort Sea stations sampled in August 2009. White arrows indicate the Mackenzie River main outlets. The Mackenzie Trough is marked with a dashed line and the shaded contours denotes the bathymetry retrieved from the International Bathymetric Chart of the Arctic Ocean (IBCAO v.3; Jakobsson et al., 2012). Station latitude and longitude coordinates are: St 430 (71.217,−136.720); St 460 (70.678,−136.047); St 540 (70.751,−137.892); St 620 (70.680,−139.627); St 670 (69.798,−138.441); St 760 (70.554,−140.798).
Filtration and sequencing protocols were as in Monier et al. (2013). Briefly, 6 l of water were filtered through 3 μm pore size filters (47 mm diameter polycarbonate) followed by 0.2 μm filters (Sterivex; Millipore, Billerica, MA, USA). For this study, the 0.2–3 μm size fraction was used to enrich for smaller cells. Sterivex units were stored at −80 °C with a buffer (1.8 ml of 40 mmol l−1 EDTA; 50 mmol l−1 Tris (pH=8.3); 0.75 mol l−1 sucrose). DNA was extracted from the Sterivex units using a modified salt extraction technique (Harding et al., 2011). The V4 region of the 18S rRNA gene was amplified using specific 454 primers that included Roche/454 tags specific for each sample as described previously (Comeau et al., 2011). The same quantity of DNA product, from each of the 24 samples (6 stations across 4 depths), was mixed and run on a 454-GS-FLX Titanium platform (Roche/454 Life Sciences, Branford, CT, USA) at the IBIS/Université Laval Plateforme d'Analyses Génomiques (Quebec, QC, Canada). All sequences are available at the NCBI Sequence Read Archive (SRA), under the accession number SRA063446.
Sequence processing and microbial community analyses
Care was given to avoid generation of spurious operational taxonomic units (OTUs), leading to inflation in microbial biodiversity. To this end, we combined QIIME v.1.6 (Caporaso et al., 2010b) and USEARCH v.6 (Edgar, 2010, 2011) sequence preprocessing pipelines. Sequences shorter than 150 nucleotides were removed and remaining sequences were ‘denoized' using the QIIME built-in ‘denoizer' program (Supplementary Table S2). The resulting reads were then aligned using pyNAST (Caporaso et al., 2010a) with the Silva 18S alignment (Pruesse et al., 2007) as a template; aligned sequences were manually inspected and 5′/3′-trimmed. Additional sequence quality controls were performed through the USEARCH quality-filtering pipeline (Edgar, 2010); putative chimeras were detected with UCHIME (Edgar et al., 2011) using the Silva 18S database (release 108) as reference. Remaining sequences were clustered in OTUs using a 98% sequence identity cutoff with UCLUST (Edgar, 2010) and singleton OTUs (i.e., cluster composed of a single sequence) were discarded from this study. OTU sequence representatives were selected as the most abundant sequence within the cluster. OTU representatives were taxonomically classified via the mothur Bayesian classifier (Schloss et al., 2009) with a custom-curated 18S sequence database as the reference data set, which was an updated version of Comeau et al. (2011) as in Monier et al. (2013). Sequences from OTUs classified as multicellular organisms (Opisthonkonta or Streptophyta) were discarded from the study as we chose to focus on microbial diversity.
Phylogenetic analyses
All ecological analyses based on phylogenetic information (UniFrac, phylogenetic diversity (PD) and net relatedness index (NRI)) were based on a phylogenetic tree reconstructed with an approximate maximum-likelihood approach as in FastTree v.2.1 (Price et al., 2010) using the ‘accurate' mode (i.e., -slownni –mlacc 4; with pseudocounts). The phylogenetic reconstruction was based on an alignment of final OTUs, aligned with pyNAST and manually curated.
UniFrac dW5000 (weighted) and dUW5000 (unweighted) distances (Lozupone and Knight, 2005) were computed on data sets subsampled at an even depth of 5000 sequences using generalized UniFrac procedure (Chen et al., 2012) implemented in the R package ‘GUniFrac' v.1. Clusters based on dW5000 were generated using hierarchical clustering. One thousand jackknife replicates (with 3750 sequences, i.e., 75% of the subsampled data sets) were used to assess clustering robustness.
PD rarefactions analyses were also conducted on samples rarefied to 5000 sequences; PD values were measured following Faith's definition (i.e., total branch length of the OTU phylogenetic tree) and computed every 20 sequences using 100 subsampling iterations. PD values were computed using the R package ‘picante' v.1.6 (Kembel et al., 2010). Other α-diversity measures, Chao1 estimator and Shannon index, were computed with QIIME. NRI values were computed using ‘picante', through the commands ses.mpd (−1 × NRI) on abundance weighted data, and tips of the original phylogenic tree were shuffled to generate randomized phylogenetic relationships (null.model=‘taxa.labels'). We also used an alternative null model, based on the independent swap algorithm (null.model=‘independentswap' Gotelli and Entsminger, 2003; Kembel, 2009), to compute NRI indexes. Results similar to those obtained with the null model based on taxa label shuffling were observed; for example weak SCM (wSCM) communities exhibited higher NRI indexes than SCM ones (analyses of variance (ANOVA) and Tukey's honestly significant difference (HSD) test; P=0.004).
Phylogenetic placements of Picozoa-like OTUs were performed using the RAxML v.7.2 (Stamatakis, 2006) evolutionary placement algorithm (Berger and Stamatakis, 2011). We first built the Picozoa reference tree with sequences and clade information from Cuvelier et al. (2008) and Seenivasan et al. (2013). Picozoa reference sequences (‘long' sequences from clone libraries) were clustered at 99% using CD-HIT v.4.6 (Fu et al., 2012) to avoid redundant sequence information that could affect phylogenetic placements. Resulting sequence cluster representatives were aligned with MAFFT v.7 (in accurate mode with global homology and 1PAM scoring matrix; Katoh and Standley, 2013). The resulting alignment was then manually inspected, curated and trimmed. The maximum-likelihood phylogenetic tree was reconstructed with RAxML using GTR+G model, which was selected by ModelGenerator v.0.85 (Keane et al., 2006) as the nucleotide substitution model best-fitting our data. Node support was computed from 100 bootstrap replicates. Reads representing Picozoa-like OTUs were aligned to our reference Picozoa alignment with MAFFT v.7 using 1PAM matrix and —addfragments mode (Katoh and Frith, 2012). These sequences were then placed on the Picozoa reference phylogenetic tree using RAxML evolutionary placement algorithm.
Statistical analyses
β-Diversity significance was assessed using UniFrac Monte Carlo significance test on dW5000 with 10 000 randomizations, as implemented in QIIME. All subsequent statistical analyses were computed in the R environment v.3 (http://www.r-project.org). Mantel tests for assessing the correlation between dW5000 and depth or geographical distances were computed with the Pearson's correlation method and 10 000 permutations using the R package ‘vegan' v.2 (Oksanen et al., 2007). To determine the contextual variables driving the microbial community compositions, we first followed a methodology similar to Martiny et al. (2011). We removed redundant environmental and biological variables (Supplementary Table S1) by using varclus (R package ‘Hmisc' v.3.12; Harrell, 2008) with Spearman's correlation (ρ2 cutoff: 0.8; PO4 and salinity had ρ2>0.8 but were retained for subsequent analyses). Non-redundant variables best explaining community dissimilarity matrix (dW5000) were then identified using the BEST procedure (i.e., best subset of environmental variables with maximum rank correlation with community dissimilarities; Clarke and Ainsworth, 1993). The resulting nine variables best explaining the dW5000 matrix were fitted as vectors onto the nonmetric multidimensional scaling ordination (NMDS) ordination and used for permutational analyses of variance on distance matrix analysis, which were computed using ‘vegan' (envfit and adonis functions, respectively, both using 10 000 permutations). Overall significance of the model composed of the nine BEST-selected variables in explaining dW5000 was assessed using multiple regressions on matrices (‘ecodist' R package v.1.2; Goslee and Urban, 2007; 10 000 permutations and Spearman's correlations). Differences in PD, NR and taxonomic composition mean among community clusters were tested using ANOVA and pairwise differences were evaluated with Tukey's HSD test. Statistical analyses of sample taxonomic enrichments were computed with Metastats (White et al., 2009) using 1000 bootstrap permutations and a 0.05 significance threshold.
Results
Oceanographic context
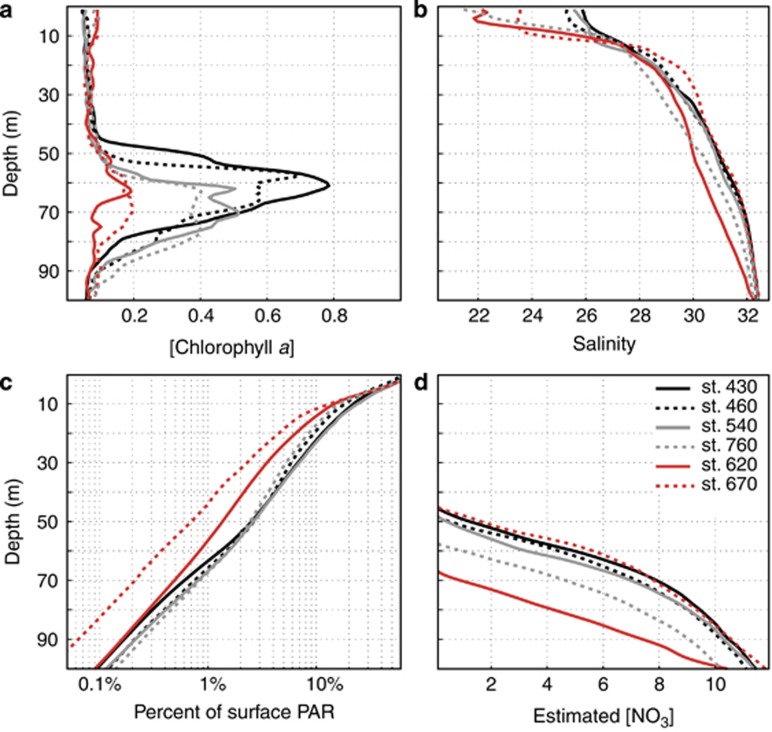
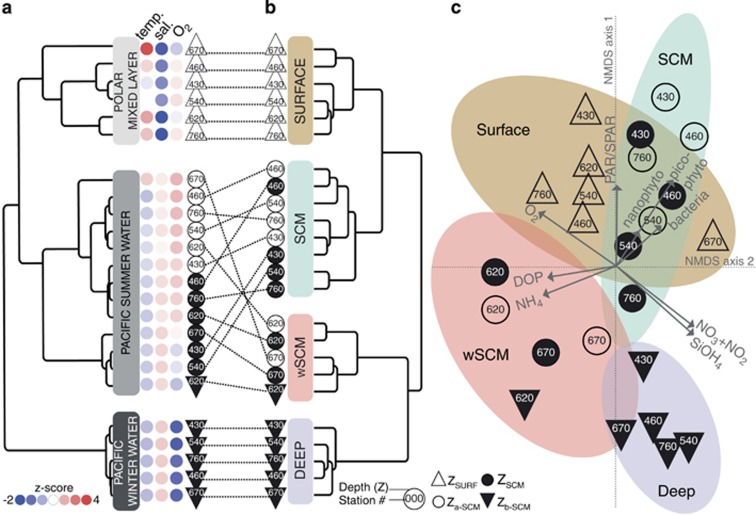
Six stations were sampled (Figure 1) at four depths (Z): surface (ZSURF), the SCM (ZSCM), just above the SCM (Za-SCM) and below the SCM (Zb-SCM). Four stations had pronounced in situ chlorophyll a (Chl a) fluorescence peaks at ZSCM (0.4–0.8 mg Chl a m−3); however, this peak was barely discernible (<0.2 mg Chl a −3) for two stations; 620 and 670 along line 600 in the Mackenzie Trough (Figure 2a). Chl a concentrations at ZSCM from discrete water samples followed the same trends, with lowest concentrations at stations 620 and 670. Surface Chl a concentrations were ⩽0.1 mg m−3, except at station 620 with a value of 0.13 mg m−3. All stations were salinity stratified (Matsuoka et al., 2012). The depth where salinity is equal to 31 indicates PSW, and this ranged from 50 to 60 m for all stations except station 620, where the PSW indicator was at 70 m (Figure 2b). The euphotic zone (1% of surface PAR) ranged from 59 to 69 m at all stations except station 670 with a value of 45 m (Figure 2c). Nutrient concentrations, especially nitrate, were depleted in the upper waters (Supplementary Table S1) and the nitracline occurred between 45 and 57 m for all stations except station 620 where nitrate was depleted to 67 m (Figure 2d). Surface temperatures were also greater in the Mackenzie Trough stations (Supplementary Figure S1A). The beam attenuation coefficient (cp) was greatest for Mackenzie Trough surface waters (Supplementary Figure S1B). A hierarchical clustering of the seawater samples using temperature, salinity and dissolved O2 was used to further discriminate the different water masses (Figure 3a). The samples themselves grouped into three distinct clusters, which matched the three well-defined AO water masses. Samples collected at ZSURF corresponded to the PML; Za-SCM plus ZSCM, corresponded to PSW and with one exception, Zb-SCM had origins in the PWW, the one deeper sample station 620 Zb-SCM grouped with the PSW cluster rather than with PWW. Although branching correctly within the PML cluster, the ZSURF sample from station 670 was an outlier of this cluster.
Figure 2.
Profiles from the Beaufort Sea selected stations. (a) Chlorophyll a concentration, from calibrated in situ fluorescence profiles (mg Chl a m−3). (b) Salinity, which is unitless but often referred to as practical salinity units (PSU). (c) Percentage of surface PAR. (d) Nitrate concentration from calibrated in situ profiles (μM). The red lines indicate weak SCM stations; depth indicated on Y axis.
Figure 3.
Water mass and microbial community partitioning across the Beaufort Sea water column. (a) Hierarchical clustering on temperature, salinity and dissolved O2 distances (centered-scaled; z-score scale denoted by color gradient) on 24 seawater samples collected for microbial sampling. The water column partitioned into the PML, PSW and PWW water masses. The station number and depth classifications are indicated by the given symbols. (b) Hierarchical clustering on weighted UniFrac distances (dW5000) of OTUs. Communities were classified into four distinct clusters: Surface (brown; support, 65% of 1000 jackknife replicates), SCM (cyan, 58%), wSCM (pink, 91%) and Deep (lavender, 93%). (c) NMDS ordination on dW5000 distances (2D stress=0.0971). For each community cluster, ellipses represent 95% confidence intervals. Arrows represent best explanatory environmental and biological variables fitted onto the ordination space (environmental factors as in Supplementary Table S3).
Microbial community partitioning
Across all 24 samples, ∼200 000 quality controlled 18S rRNA gene reads clustered into 1411 OTUs (Supplementary Table S2). β-Diversity, assessed using weighted UniFrac distances (phylogenetic-based β-diversity measure; dW5000), indicated that communities recovered from the same water masses (Figure 3a) were more similar, even among geographically distant stations, as highlighted by hierarchical clustering (Figure 3b) and NMDS (Figure 3c) on dW5000. The samples significantly partitioned into four community clusters based on their phylogenetic composition (permutational analysis of variance on dW5000; R2=0.72; P=0.001). The communities from PML and PWW water masses grouped into two distinct clusters, hereafter referred to as ‘Surface' and ‘Deep' communities (Figure 3b). In the dW5000 NMDS ordination space, the only Surface outlier was from station 670, a sample influenced by the Mackenzie River plume (Figure 3a). In contrast to PML and PWW water masses with single coherent microbial communities, PSW harbored two distinct assemblages: those with substantial Chl a concentrations grouped into an ‘SCM' cluster (stations 430, 460, 540 and 760), whereas PSW samples with low Chl a concentrations (620 and 670) formed a separate cluster, termed ‘wSCM' for weak SCM.
Defining community composition using unweighted UniFrac distances (i.e., presence–absence; dUW5000), instead of dW5000, had a larger influence on Surface and SCM communities (Supplementary Figure S2A), even though dW5000 and dUW5000 NMDS ordinations were significantly correlated (procrustes test m12=0.46; P=0.001; Supplementary Figure S2B). In contrast, wSCM and Deep community compositions were much less affected. Discrepancies between dW5000 and dUW5000 for Surface and SCM communities, suggest the presence of very abundant taxa. Rank abundance curves (Supplementary Figure S2C), and sequence abundances of the 50 dominant OTUs (Supplementary Figure S2D) confirmed that Surface and SCM communities were dominated by a few OTUs, each represented by more than 1000 sequences. Most of these sequences were classified as Arctic Micromonas, exemplified by isolate CCMP2099 (Lovejoy et al., 2007).
Environmental and biological drivers
To identify which local environmental and biological variables drove microbial community composition, we determined the set of variables best explaining the dW5000 distances among communities. The resulting nine variables significantly explained a large fraction of the community phylogenetic compositions (multiple regression model: R2=0.507; P=0.0001) and were projected as vectors onto the corresponding NMDS ordination space (environmental factor fitting; Figure 3c). Light (percentage of surface PAR) matched the first ordination axis, correctly oriented with Surface and SCM communities that were subject to higher irradiance levels. Higher nutrient levels (SiOH4 and NO3+NO2) characterized Deep samples, whereas Surface ones were nutrient depleted but O2 rich. SCM community compositions were also characterized by higher bacterial, nano- and picophytoplankton cell concentrations (Supplementary Table S1). Light and nutrient vectors were identified as likely driving the composition of wSCM assemblages. PSW assemblages from station 670 (with turbid surface water; Za-SCM and ZSCM) and deeper waters from station 620 (Zb-SCM) were under a low-light regime (Figure 2c). In contrast, for the other wSCM communities (station 620 Za-SCM and ZSCM), PAR was higher but nutrient concentrations were lower. All communities classified as wSCM were associated with higher NH4 and PO4 concentrations (Supplementary Table S1). To assess the contribution of each of the nine environmental and biological variables in explaining the compositional differences among microbial communities, we retrieved their environmental factor fitting statistics as well as those from permutational analyses of variance on distance matrices. For both environmental factor fitting and permutational analyses of variance on distance matrix analyses, the variables most influencing the community phylogenetic compositions were nutrient and picophytoplankton concentrations and O2 (Supplementary Table S3).
Phylogenetic diversity
To determine variations at the α-diversity level, we next measured PD (here, the total sum of branch lengths for a given OTU phylogenetic tree). For each community there was substantial microbial biodiversity (Supplementary Figure S3A) with large and significant variations (ANOVA; P=0.0001; Supplementary Table S2). Deep communities were the most diverse (PD mean and s.d.: 35.8±2.2), whereas Surface ones were the least diverse (22.1±2.5). PD of SCM and wSCM communities did not significantly differ (Tukey's HSD test; P=0.81). Community clusters also exhibited distinct diversity levels when using non-phylogenetic measures of α-diversity (e.g., Chao1; ANOVA; P=0.011; Supplementary Table S2). There was also a positive relationship between depth and PD (R2=0.511, P<0.001; Supplementary Figure S3B).
Ecological processes
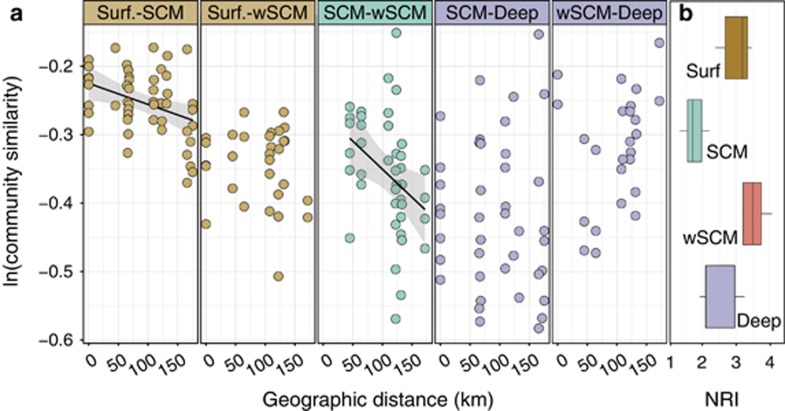
We evaluated the degree of compositional specificity among microbial communities and found that compositions of Surface, wSCM and Deep communities were significantly specific (UniFrac Monte Carlo significance test; P<0.05) but SCM ones were not (P=0.25). To determine whether the lack of compositional specificity from SCM communities resulted from historical contingencies, such as past dispersions originating from other water masses, we used distance–decay relationships (i.e., the effect of geographical distance on community similarity) to infer community assembly processes, while controlling for the concomitant influences of dispersion and environmental filtering. Specifically, we compared communities from segregated environments (contiguous water masses; dispersal-limited) and from within the same environment (PSW water mass; dispersal non-limited). The physicochemical separation between water masses (Figure 3a) represented environmental barriers hindering microbial dispersion and colonization between water masses. Distant–decay curves with slopes significantly different from zero were found between SCM and Surface communities (Figure 4a; adjusted P=0.028) and, to a lesser extent, between SCM and wSCM communities (adjusted P=0.058). In the case of non-dispersal limitation, the negative relationship between phylogenetic similarity and geographic distances across SCM and wSCM communities suggests that environmental filtering was mainly operating. In the case of dispersal limitation, colonization of microbes from one water mass could be hampered between contiguous water masses. As such, no relationship between community similarity and geographic distance between Deep and PSW (either SCM or wSCM) communities would be expected. In contrast, a significant distance–decay curve was observed between Surface and SCM assemblages. This distance–decay relationship implies that Surface and SCM communities were more compositionally similar when geographically closer, despite being from distinct water masses.
Figure 4.
Community phylogenetic similarities across the selected Beaufort Sea stations. (a) Distance–decay curves for microbial communities from contiguous water masses (Surface vs SCM or wSCM; Deep vs SCM or wSCM) or from the same water mass (SCM vs wSCM). Each dot represents a pairwise comparison of community phylogenetic similarity (expressed as ln(1−dW5000)) and geographic distance between microbial communities. Lines represent best linear fit, with a gray-shaded 95% confidence interval, for which the slope was significantly different than zero (SCM vs Surface: slope=−0.0003 and Bonferroni adjusted P=0.028; SCM vs wSCM: slope=−0.0008 and adjusted P=0.058). (b) NRI (in units of standard deviation) for each community cluster. NRIs were based on standardized mean pairwise distances from community cluster OTU phylogenetic tree. To generate null models to compute NRIs, tips of this phylogenic tree were shuffled to create randomized OTU phylogenetic relationships. An alternative null model yielded similar results (see Materials and methods).
Next, community phylogenetic structure was tested using null models. We calculated the NRI, a measure of community phylogenetic relatedness for each community (Webb et al., 2002) that determines if OTUs are more closely related to co-occurring relatives than expected by chance), which can assess environmental filtering inferred from phylogenetic clustering (Kembel, 2009; Stegen et al., 2012). All microbial community clusters had significant phylogenetic structure (positive NRI values; one sample t-test, P⩽0.0005; Figure 4b; Supplementary Table S2), indicating that communities of microbial eukaryotes were more closely related to each other than expected by chance. However, significant differences among community clusters were also observed (ANOVA; P<0.001), reflecting differences in their phylogenetic structure. wSCM communities showed the highest level of phylogenetic clustering (NRI mean and s.d.: 3.5±0.3) followed by surface communities (3±0.4). In contrast, SCM communities were the least phylogenetically clustered (1.7±0.3), and were significantly more overdispersed than wSCM communities (Tukey's HSD test; P<0.001).
Taxonomic compositions
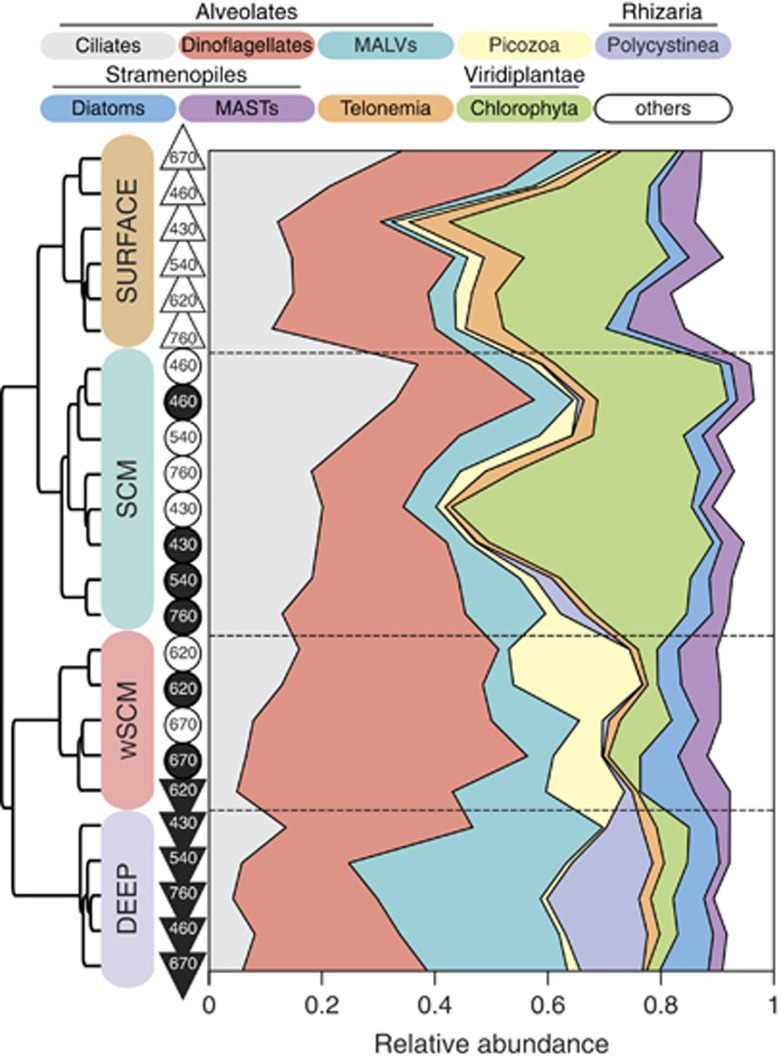
Among the groups with easily defined trophic and size categories, green algae largely dominated the SCM assemblages at the phylum level (Figure 5), with relative abundance mean and s.d. of 31.4±1% (P=0.002, Metastats; White et al., 2009), and were also abundant in Surface communities. Sequences classified as green algae were mostly assigned to Arctic Micromonas. In contrast to SCM communities, green algae were less abundant in wSCM communities (4.9±0.4% P=0). SCM samples were also particularly rich in sequences classified as ciliates (Ciliophora; 26.1±0.9% P=0.02), which were also less abundant in wSCM assemblages (12.1±0.5% P=0.003). Dinoflagellates were the most abundant microbial eukaryotes in wSCM assemblages (30.1±1.1% P=0.03). wSCM samples were also enriched in sequences classified as Picozoa (18.2±1% P=0.007), a heterotrophic lineage (Seenivasan et al., 2013).
Figure 5.
Taxonomic composition differences among Beaufort Sea microbial communities. Areas colored according to the top legend denote the relative abundances of the main eukaryotic lineages. Markers represent each sample as in Figure 3. Left dendrogram represents the hierarchical clustering output on dW5000, as displayed in Figure 3b.
At a finer taxonomic resolution (i.e., species, genus or class depending on the taxonomic resolution available), we identified 155 out of 227 taxa that exhibited significant differential abundances across the four community clusters (Supplementary Figure S4). When comparing SCM with wSCM communities, 30 taxa differed significantly (Supplementary Table S4). Thirteen were under-represented in wSCM communities, including Arctic Micromonas, which was most abundant in SCM samples (24.9±1% P=0.01) and most depleted in wSCM ones (9.4±2.1% P=0.03 vs SCM). Several ciliate taxa were also less abundant in wSCM assemblages, in particular Novistrombidium and Pseudotontonia (P<0.01) compared with SCM communities. Telonema, a heterotrophic nanoflagellate, was also less abundant in wSCM communities (P=0.03). Within the wSCM communities, dinoflagellates from the Gyrodinium group were significantly over-represented (Dinophyceae; 10.5±0.7% P=0.009) along with sequences matching the Picozoan environmental sequence NW617.02 (11.6±0.7% P=0.01).
Picozoa were represented by 22 OTUs, which could be phylogenetically placed onto a Picozoa 18S rRNA gene reference tree. The Picozoa OTUs mapped to four previously established clades (Seenivasan et al., 2013) with placement likelihoods ⩾0.75: BP2, P5, P8 and P9 (Supplementary Figure S5; clade BP3.1 attracted four OTUs but the likelihood of their placements was low). Picozoa assemblages from wSCM were dominated by two OTUs, from clades P5 and P8, but only the latter OTU was significantly enriched compared with other community clusters (P⩽0.05). Surface, SCM and Deep communities were also populated with distinct Picozoa sequences, but in much lower abundance than for wSCM. Outside of wSCM, only Deep communities were significantly enriched with a Picozoa OTU (clade P8).
Discussion
Oceanographic processes affect microbial biogeography
Station 670 ZSURF waters were fresh (Figure 2b), warm (Supplementary Figure S1A) and had high CDOM concentrations (Matsuoka et al., 2012), characteristic of the Mackenzie River plume. In those waters, cp (beam attenuation) was high (Supplementary Figure S1B), the CDOM and particulate material in the plume reduced water column light penetration (Figure 2c), and the communities in PSW were light limited. In the surface waters of station 620, cp was also high but light still reached depths normally corresponding to PSW, indicating that lower CDOM and low surface Chl a concentrations offset the attenuation signal allowing light to penetrate. However, although light was available at the usual depth of PSW, the PSW itself was displaced by ∼10 m, and there was a lack of nutrients at the (weak) SCM, detected at 65 m (Figure 2d). Physical oceanographic transect data along the Mackenzie Trough (Matsuoka et al., 2012) were consistent with the presence of a cyclonic eddy triggered by the Mackenzie Trough (Williams et al., 2006) at the time of sampling. This eddy activity would have deepened the PSW and nitracline. In summary, our results indicate that the weak SCM signals were due to oceanographic processes affecting the light–nutrient balance required for phytoplankton growth and maintenance (Martin et al., 2010). Both processes resulted in low Chl a SCM layers. Communities with similar phylogenetic compositions were present in both the low-light and the low-nutrient ZSCM systems, and the wSCM communities were distinct from high Chl a SCM ones. Mesoscale oceanographic processes have also been implicated in compositional changes in Pacific Ocean diatom populations (Chappell et al., 2013) and Sargasso Sea bacterioplankton (Nelson et al., 2013), and appear to be important drivers of microbial species turnover in marine environments, for both bacterial and eukaryotic communities.
Ecological processes influencing SCM microbial community compositions
Using phylogenetic information, we found that the different Beaufort Sea water masses harbored distinct microbial eukaryotic communities. Vertical partitioning of microbial assemblages in this region has been reported previously (Monier et al., 2013). Here we used a larger and more varied data set to address an important goal of microbial ecology, to unveil ecological and evolutionary mechanisms beyond biogeographic patterns (Hanson et al., 2012). In contrast to heterotrophic flagellate communities from AO stations with well-defined SCM, which were partitioned based on their water mass of origin (Monier et al., 2013), we found that this vertical β-diversity of microbial eukaryotic communities was disrupted by Arctic Ocean processes. To identify the processes driving the β-diversity among AO microbial eukaryotic communities, we investigated distance–decay relationships (Figure 4a) and community phylogenetic structures (Figure 4b). Distance–decay curves arise from the negative relationship between community composition similarity and spatial distances, and can reflect the influences of environmental filtering and historical contingencies (Chase, 2003; Martiny et al., 2006, 2011; Chase and Myers, 2011; Hanson et al., 2012). Although the use of distance–decay relationships to investigate community assembly mechanisms is frequently applied, local environmental conditions and geographic location may be autocorrelated and caution is required in interpreting the results (Peres-Neto and Legendre, 2010). Dispersal limitation may also be influenced by microbial dispersal routes and capabilities (Leibold et al., 2004; Martiny et al., 2006; Soininen et al., 2011).
Although most microbial biogeography studies have addressed the contribution of environmental filtering and dispersal limitation, historical contingencies may also affect species distributions (Ricklefs, 2007; Leibold et al., 2010). Here, mixing and turbulent processes between PML and PSW water masses have the potential to allow the dispersion, colonization and survival of microbial species able to persist under conditions found in both water masses. The Surface-SCM distance–decay curve suggests ecological drift between the two contiguous PML and PSW water masses as a result of dispersal limitation. The significant differences between their corresponding NRI indexes suggest distinct phylogenetic structures, and the extent of the niche breadth of the dominant phylotype inhabiting the surface and SCM layers. The AO euphotic zone is an ecological continuum for Micromonas, which is widely distributed throughout the AO, likely due to its plasticity in light requirements and small cell size (Lovejoy et al., 2007). However, Micromonas was mostly absent from the PSW of stations 620 and 670; no distance–decay relationship connecting Surface and wSCM communities was detected at those two stations. The absence of a trend in this case indicated that environmental filtering overwhelmed the effect of historical events. In the case of non-limited dispersion, the distance–decay between SCM and wSCM communities indicates that environmental filtering is predominantly structuring PSW microbial communities in the disturbed conditions, defined as conditions modified by the mesoscale processes.
Although there is some evidence of link between microbial community diversity and productivity (Waide et al., 1999; Chase and Leibold, 2002), the shape and strength of this relationship is highly variable (Horner-Devine et al., 2003; Smith, 2007). We found that low Chl a wSCM communities were as diverse as the high Chl a SCM (Supplementary Figure S3), suggesting no relationship. Interestingly, the similar diversity of both communities may indicate that wSCM communities are a stable alternative to SCM communities, as low diversity may render communities more sensitive to rapid changes compared with more diverse communities (Eisenhauer et al., 2012).
Environmental filtering contributions to SCM communities
Although all community clusters had significant phylogenetic structure (NRI indices significantly >0), significant differences were found among them; SCM communities were the most phylogenetically overdispersed (lowest NRIs) and wSCM were the most clustered of all communities (highest NRI values). Low NRI values indicate that assemblages were overall less phylogenetically related than ones with high NRI values (Webb et al., 2002), suggesting that SCM taxa were less ecologically similar to each other than those from wSCM. More favorable conditions support a wide range of niches, which can be filled by organisms that differ in their functional capacities, and would permit diversification within the SCM. In contrast, conditions experienced by wSCM communities were more constrained (low light or low nutrients), supporting a narrower range of traits, leading to the recruitment or retention of more specialized organisms. Similarly decreasing phylogenetic relatedness among Betaproteobacteria across a productivity gradient has also been reported (Horner-Devine and Bohannan, 2006), and increased phylogenetic clustering was induced experimentally in disturbed marine bacterioplankton (Pontarp et al., 2013) and rain forests (Ding et al., 2011). Likewise, zooplankton populations are reported to be phylogenetically more related in environmentally disturbed lakes than in more stable ones (Helmus et al., 2010). Alternatively, we note that competitive exclusion can also increase species relatedness and phylogenetic clustering (Mayfield and Levine, 2010); however, the relatively low cell concentrations in wSCM (Supplementary Table S1) suggest that species interactions would be less important in that environment than in the SCM where niche differentiation (i.e., environmental filtering, taxon specialization and competition) were acting. It is likely that these factors, rather than competitive exclusion, led to the differences in phylogenetic relatedness across the SCM/wSCM layers.
Phototrophic vs heterotrophic systems
The SCM was striking because of the high proportion of Arctic Micromonas reads (Figure 5 and Supplementary figure S4). The pan-Arctic Micromonas, represented in culture by isolate CCMP2099, has been reported from flow cytometry (Balzano et al., 2012a) and genetic surveys (Comeau et al., 2011; Balzano et al., 2012b). Interestingly, the proportion of Micromonas reads was much lower in wSCM assemblages, potentially related to low-light, low-nutrient levels or from top-down pressure, where Micromonas might have been exposed to higher concentrations of grazers or viral infection. Both light and nutrients are required for SCM to form (Klausmeier and Litchman, 2001) and given their interconnectivity, it is challenging to identify their relative contributions to community assembly. Here we addressed this question by combining phylogenetic β-diversity with multivariate analyses. The NMDS ordination (Figure 3c) revealed SCM communities distributed along a narrow space, orthogonal to the nutrient vector (NO3+NO2, SiOH4), but stretched across the irradiance vector. This spatial ordination suggested that nutrient levels mainly influenced phylogenetic compositions of SCM communities. Multivariate statistical analyses on best explanatory environmental variables also suggested the predominant role of nutrient concentrations over light (Supplementary Table S3). The arctic isolate of Micromonas maintains growth under a large range of light conditions (Lovejoy et al., 2007) and arctic phytoplankton in general seem able to grow under low-light levels, consistent with summer production being nutrient, rather than light-limited (Tremblay and Gagnon, 2009). Overall, the lack of nutrients would more likely have been the main exclusionary factor operating in wSCM systems. Such nutrient limitation may become more marked in the summer if under-ice phytoplankton blooms were to become widespread (Arrigo et al., 2012), as they would have the effect of drawing down euphotic zone nutrients earlier in the season.
The lower proportion of Micromonas in the wSCM was countered by sequences of heterotrophic or mixotrophic dinoflagellates and heterotrophic Picozoa (Figure 5 and Supplementary Figure S4 and Supplementary Table S4). Although Micromonas and Picozoa are in the same size category (1–2 μm), they are very different functionally. To date, we know only that the members of the recently described Picozoa phylum are exclusively marine and heterotrophs (Yoon et al., 2011; Seenivasan et al., 2013). The ultrastucture of the isolate, Picomonas judraskeda, precludes phagotrophic feeding on bacteria and it is thought to actively seek out colloidal material (Seenivasan et al., 2013). The Arctic ecotype of Micromonas was reported to be capable of bacterivory (Sherr et al., 2003; McKie-Krisberg and Sanders, 2014). In a low production environment, we hypothesize that the ability to scavenge colloids may give the Picozoa an advantage over bacterivores, including small mixotrophic species such as Micromonas. The Picozoa-related sequences grouped into several clades (Supplementary Figure S5), indicating that a particular trait shared among Picozoa taxa was selected for in the wSCM. Picozoa themselves are almost universally recovered from open ocean Arctic 18S rRNA gene surveys (Comeau et al., 2011; Monier et al., 2013), and the genetic diversity appears to be relatively high in the Arctic, as shown by phylogenetic reconstruction (Supplementary Figure S5), especially in comparison with Arctic Micromonas (Balzano et al., 2012a). The genetic diversity within Arctic Picozoa suggests that this eukaryotic lineage has specific traits that allow persistence in regions of periodic low productivity, such as the summer Arctic.
Conclusions
Oceanographic processes that affected nutrient concentrations and light levels influenced microbial communities in the PSW layer that supports much of the summer biological production in the Western Arctic Ocean. We identified taxonomic, phylogenetic and ecological drivers associated with a shift from picophytoplankton to heterotrophic microbial eukaryotes in SCM communities. The microbial eukaryotic communities were sensitive to relatively moderate oceanographic changes, for example, a 10 m depth displacement of PSW, suggesting the usefulness of microbial observations over different regions and seasons. The results here show that changes in AO physical forcing have the potential to modify the ecology of the microbial seascape and potentially impact AO productivity.
Acknowledgments
We thank the Canadian Coast Guard crew of the Canadian research icebreaker CCGS Amundsen. We are grateful to AM Comeau and E Medrinal for laboratory work, and to MP Simmons and SW Kembel for comments. This study was conducted as part of the MALINA Scientific Program led by MB and funded by the French National Research Agency (ANR), INSU-CNRS (Institut National des Sciences de l'Univers – Centre National de la Recherche Scientifique), CNES (Centre National d'Etudes Spatiales) and ESA (European Space Agency). Computing support was provided by CLUMEQ/Compute Canada. Additional funding was provided by the Natural Science and Engineering Council (NSERC) of Canada to CL, and Fond Québécois de Recherches Nature et Technologies (FQRNT) for Québec Océan, and is a contribution to ArcticNet. AF, AMa and AMo received scholarships from the Canadian Excellence Research Chair (CERC) in remote sensing of Canada's new Arctic frontier, and JC was supported by FQRNT and the EnviroNorth (CREATE program from NSERC).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Ardyna M, Gosselin M, Michel C, Poulin M, Tremblay J-É. (2011). Environmental forcing of phytoplankton community structure and function in the Canadian High Arctic: contrasting oligotrophic and eutrophic regions. Mar Ecol Prog Ser 442: 37–57. [Google Scholar]
- Arrigo KR, Perovich DK, Pickart RS, Brown ZW, van Dijken GL, Lowry KE et al. (2012). Massive phytoplankton blooms under Arctic sea ice. Science 336: 1408–1408. [DOI] [PubMed] [Google Scholar]
- Balzano S, Gourvil P, Siano R, Chanoine M, Marie D, Lessard S et al. (2012. a). Diversity of cultured photosynthetic flagellates in the northeast Pacific and Arctic Oceans in summer. Biogeosciences 11: 4553–4571. [Google Scholar]
- Balzano S, Marie D, Gourvil P, Vaulot D. (2012. b). Composition of the summer photosynthetic pico and nanoplankton communities in the Beaufort Sea assessed by T-RFLP and sequences of the 18S rRNA gene from flow cytometry sorted samples. ISME J 6: 1480–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SA, Stamatakis A. (2011). Aligning short reads to reference alignments and trees. Bioinformatics 27: 2068–2075. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. (2010. a). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010. b). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmack E, McLaughlin F. (2011). Towards recognition of physical and geochemical change in Subarctic and Arctic Seas. Prog Oceanogr 90: 90–104. [Google Scholar]
- Carmack EC, Macdonald RW, Jasper S. (2004). Phytoplankton productivity on the Canadian Shelf of the Beaufort Sea. Mar Ecol Prog Ser 277: 37–50. [Google Scholar]
- Carmack EC, Macdonald RW, Papadakis JE. (1989). Water mass structure and boundaries in the Mackenzie shelf estuary. J Geophys Res 94: 18043. [Google Scholar]
- Cermeno P, Dutkiewicz S, Harris RP, Follows M, Schofield O, Falkowski PG. (2008). The role of nutricline depth in regulating the ocean carbon cycle. Proc Natl Acad Sci USA 105: 20344–20349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell PD, Whitney LP, Haddock TL, Menden-Deuer S, Roy EG, Wells ML et al. (2013). Thalassiosira spp. community composition shifts in response to chemical and physical forcing in the northeast Pacific Ocean. Front Microbiol 4: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase JM. (2003). Community assembly: when should history matter? Oecologia 136: 489–498. [DOI] [PubMed] [Google Scholar]
- Chase JM, Leibold MA. (2002). Spatial scale dictates the productivity–biodiversity relationship. Nature 416: 427–430. [DOI] [PubMed] [Google Scholar]
- Chase JM, Myers JA. (2011). Disentangling the importance of ecological niches from stochastic processes across scales. Philos Trans R Soc Ser B 366: 2351–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bittinger K, Charlson ES, Hoffmann C, Lewis J, Wu GD et al. (2012). Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 28: 2106–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke KR, Ainsworth M. (1993). A method of linking multivariate community structure to environmental variables. Mar Ecol Prog Ser 92: 205–205. [Google Scholar]
- Comeau AM, Li WKW, Tremblay J-É, Carmack EC, Lovejoy C. (2011). Arctic Ocean microbial community structure before and after the 2007 record sea ice minimum. PLoS One 6: e27492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvelier ML, Ortiz A, Kim E, Moehlig H, Richardson DE, Heidelberg JF et al. (2008). Widespread distribution of a unique marine protistan lineage. Environ Microbiol 10: 1621–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Zang R, Letcher SG, Liu S, He F. (2011). Disturbance regime changes the trait distribution, phylogenetic structure and community assembly of tropical rain forests. Oikos 121: 1263–1270. [Google Scholar]
- Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fitter AH. (2009). Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J 4: 337–345. [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhauer N, Scheu S, Jousset A. (2012). Bacterial diversity stabilizes community productivity. PLoS One 7: e34517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forest A, Babin M, Stemmann L, Picheral M, Sampei M, Fortier L et al. (2013). Ecosystem function and particle flux dynamics across the Mackenzie Shelf (Beaufort Sea, Arctic Ocean): an integrative analysis of spatial variability and biophysical forcings. Biogeosciences 10: 2833–2866. [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28: 3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goslee SC, Urban DL. (2007). The ecodist package for dissimilarity-based analysis of ecological data. J Stat Softw 22: 1–19. [Google Scholar]
- Gotelli NJ, Entsminger GL. (2003). Swap algorithms in null model analysis. Ecology 84: 532–535. [Google Scholar]
- Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JBH. (2012). Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol 10: 497–506. [DOI] [PubMed] [Google Scholar]
- Harding T, Jungblut AD, Lovejoy C, Vincent WF. (2011). Microbes in high Arctic snow and implications for the cold biosphere. Appl Environ Microb 77: 3234–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell FE Jr. (2008). Hmisc: Harrell Miscellaneous. R package version 3.12-2. Availabe at: http://cran.r-project.org/package=Hmisc (accessed 2 October 2014).
- Helmus MR, Keller WB, Paterson MJ, Yan ND, Cannon CH, Rusak JA. (2010). Communities contain closely related species during ecosystem disturbance. Ecol Lett 13: 162–174. [DOI] [PubMed] [Google Scholar]
- Hill VJ, Matrai PA, Olson E, Suttles S, Steele M, Codispoti LA et al. (2013). Synthesis of integrated primary production in the Arctic Ocean: II. In situ and remotely sensed estimates. Prog Oceanogr 110: 107–125. [Google Scholar]
- Horner-Devine MC, Leibold MA, Smith VH, Bohannan BJM. (2003). Bacterial diversity patterns along a gradient of primary productivity. Ecol Lett 6: 613–622. [Google Scholar]
- Horner-Devine MC, Bohannan BJM. (2006). Phylogenetic clustering and overdispersion in bacterial communities. Ecology 87: 100–108. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Mayer L, Coakley B, Dowdeswell JA, Forbes S, Fridman B et al. (2012). The international bathymetric chart of the Arctic Ocean (IBCAO) version 3.0. Geophys Res Lett 39: L12609. [Google Scholar]
- Katoh K, Frith MC. (2012). Adding unaligned sequences into an existing alignment using MAFFT and LAST. Bioinformatics 28: 3144–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Creevey CJ, Pentony MM, Naughton TJ, Mclnerney JO. (2006). Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 6: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kembel SW. (2009). Disentangling niche and neutral influences on community assembly: assessing the performance of community phylogenetic structure tests. Ecol Lett 12: 949–960. [DOI] [PubMed] [Google Scholar]
- Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD et al. (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26: 1463–1464. [DOI] [PubMed] [Google Scholar]
- Klausmeier CA, Litchman E. (2001). Algal games: the vertical distribution of phytoplankton in poorly mixed water columns. Limnol Oceanogr 46: 1998–2007. [Google Scholar]
- Leibold MA, Economo EP, Peres-Neto P. (2010). Metacommunity phylogenetics: separating the roles of environmental filters and historical biogeography. Ecol Lett 13: 1290–1299. [DOI] [PubMed] [Google Scholar]
- Leibold MA, Holyoak M, Mouquet N, Amarasekare P, Chase JM, Hoopes MF et al. (2004). The metacommunity concept: a framework for multi-scale community ecology. Ecol Lett 7: 601–613. [Google Scholar]
- Li WKW, McLaughlin FA, Lovejoy C, Carmack EC. (2009). Smallest algae thrive as the Arctic Ocean freshens. Science 326: 539. [DOI] [PubMed] [Google Scholar]
- Lovejoy C. (2012). Polar marine microbiology. In: Miller RV, Whyte LG, (eds) Polar Microbiology: Life in a Deep Freeze. ASM Press: Washington, DC, pp 1–17. [Google Scholar]
- Lovejoy C, Potvin M. (2011). Microbial eukaryotic distribution in a dynamic Beaufort Sea and the Arctic Ocean. J Plankton Res 33: 431–444. [Google Scholar]
- Lovejoy C, Vincent WF, Bonilla S, Roy S, Martineau M-J, Terrado R et al. (2007). Distribution, phylogeny, and growth of cold-adapted picoprasinophytes in Arctic Seas. J Phycol 43: 78–89. [Google Scholar]
- Lozupone C, Knight R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microb 71: 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald RW, Carmack EC, McLaughlin FA, Iseki K, Macdonald DM, O'Brien MC. (1989). Composition and modification of water masses in the Mackenzie shelf estuary. J Geophys Res 94: 18057. [Google Scholar]
- Martin J, Tremblay JÉ, Gagnon J, Tremblay G, Lapoussière A, Jose C et al. (2010). Prevalence, structure and properties of subsurface chlorophyll maxima in Canadian Arctic waters. Mar Ecol Prog Ser 412: 69–84. [Google Scholar]
- Martin J, Tremblay JÉ, Price NM. (2012). Nutritive and photosynthetic ecology of subsurface chlorophyll maxima in Canadian Arctic waters. Biogeosciences 9: 5353–5371. [Google Scholar]
- Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL et al. (2006). Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4: 102–112. [DOI] [PubMed] [Google Scholar]
- Martiny JBH, Eisen JA, Penn K, Allison SD, Horner-Devine MC. (2011). Drivers of bacterial beta-diversity depend on spatial scale. Proc Natl Acad Sci USA 108: 7850–7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka A, Bricaud A, Benner R, Para J, Sempéré R, Prieur L et al. (2012). Tracing the transport of colored dissolved organic matter in water masses of the Southern Beaufort Sea: relationship with hydrographic characteristics. Biogeosciences 9: 925–940. [Google Scholar]
- Mayfield MM, Levine JM. (2010). Opposing effects of competitive exclusion on the phylogenetic structure of communities. Ecol Lett 13: 1085–1093. [DOI] [PubMed] [Google Scholar]
- McKie-Krisberg ZM, Sanders RW. (2014). Phagotrophy by the picoeukaryotic green alga Micromonas: implications for Arctic Oceans. ISME J 8: 1953–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin FA, Carmack EC. (2010). Deepening of the nutricline and chlorophyll maximum in the Canada Basin interior, 2003–2009. Geophys Res Lett 37: article L24602. [Google Scholar]
- Monier A, Terrado R, Thaler M, Comeau AM, Medrinal E, Lovejoy C. (2013). Upper Arctic Ocean water masses harbor distinct communities of heterotrophic flagellates. Biogeosciences 10: 4273–4286. [Google Scholar]
- Nelson CE, Carlson CA, Ewart CS, Halewood ER. (2013). Community differentiation and population enrichment of Sargasso Sea bacterioplankton in the euphotic zone of a mesoscale mode-water eddy. Environ Microbiol 16: 871–887. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB et al. (2007). Vegan: Community Ecology Package. R package version 2.0-8. Available at: http://cran.r-project.org/package=vegan (accessed 2 October 2014).
- Peres-Neto PR, Legendre P. (2010). Estimating and controlling for spatial structure in the study of ecological communities. Global Ecol Biogeog 19: 174–184. [Google Scholar]
- Peterson BJ, Holmes RM, McClelland JW, Vörösmarty CJ, Lammers RB, Shiklomanov AI et al. (2002). Increasing river discharge to the Arctic Ocean. Science 298: 2171–2173. [DOI] [PubMed] [Google Scholar]
- Pontarp M, Sjöstedt J, Lundberg P. (2013). Experimentally, induced habitat filtering in marine bacterial communities. Mar Ecol Prog Ser 477: 77–86. [Google Scholar]
- Post E, Bhatt US, Bitz CM, Brodie JF, Fulton TL, Hebblewhite M et al. (2013). Ecological consequences of sea-ice decline. Science 341: 519–524. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. (2010). FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainville L, Lee CM, Woodgate RA. (2011). Impact of wind-driven mixing in the Arctic Ocean. Oceanography 24: 136–145. [Google Scholar]
- Ricklefs RE. (2007). History and diversity: explorations at the intersection of ecology and evolution. Am Nat 170: S56–S70. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screen JA, Simmonds I. (2010). The central role of diminishing sea ice in recent Arctic temperature amplification. Nature 464: 1334–1337. [DOI] [PubMed] [Google Scholar]
- Seenivasan R, Sausen N, Medlin LK, Melkonian M. (2013). Picomonas judraskeda gen. et sp. nov.: the first identified member of the Picozoa phylum nov., a widespread group of picoeukaryotes, formerly known as ‘Picobiliphytes'. PLoS One 8: e59565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr EB, Sherr BF, Wheeler PA, Thompson K. (2003). Temporal and spatial variation in stocks of autotrophic and heterotrophic microbes in the upper water column of the central Arctic Ocean. Deep-Sea Res Part I 50: 557–571. [Google Scholar]
- Smith VH. (2007). Microbial diversity–productivity relationships in aquatic ecosystems. FEMS Microbiol Ecol 62: 181–186. [DOI] [PubMed] [Google Scholar]
- Soininen J, Korhonen JJ, Karhu J, Vetterli A. (2011). Disentangling the spatial patterns in community composition of prokaryotic and eukaryotic lake plankton. Limnol Oceanogr 56: 508–520. [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Stegen JC, Lin X, Konopka AE, Fredrickson JK. (2012). Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J 6: 1653–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay J-É, Gagnon J. (2009). The effects of irradiance and nutrient supply on the productivity of Arctic waters: a perspective on climate change. In: Nihoul JCJ, Kostianoy AG, (eds) NATO Science for Peace and Security Series C: Environmental Security. Springer: Dordrecht, Netherlands, pp 73–93. [Google Scholar]
- Waide RB, Willig MR, Steiner CF, Mittelbach G, Gough L, Dodson SI et al. (1999). The relationship between productivity and species richness. Annu Rev Ecol Syst 30: 257–300. [Google Scholar]
- Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. (2002). Phylogenies and community ecology. Annu Rev Ecol Syst 33: 475–505. [Google Scholar]
- Wheeler PA, Gosselin M, Sherr E, Thibaultc D, Kirchman DL, Benner R et al. (1996). Active cycling of organic carbon in the central Arctic Ocean. Nature 380: 697–699. [Google Scholar]
- White JR, Nagarajan N, Pop M. (2009). Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol 5: e1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams WJ, Carmack EC, Shimada K, Melling H, Aagaard K, Macdonald RW et al. (2006). Joint effects of wind and ice motion in forcing upwelling in Mackenzie Trough, Beaufort Sea. Cont Shelf Res 26: 2352–2366. [Google Scholar]
- Yoon HS, Price DC, Stepanauskas R, Rajah VD, Sieracki ME, Wilson WH et al. (2011). Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science 332: 714–717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.