Abstract
Lysobactin, also known as katanosin B, is a potent antibiotic with in vivo efficacy against Staphylococcus aureus and Streptococcus pneumoniae. It was previously shown to inhibit peptidoglycan (PG) biosynthesis, but its molecular mechanism of action has not been established. Using enzyme inhibition assays, we show that lysobactin forms 1:1 complexes with Lipid I, Lipid II, and , substrates in the PG and wall teichoic acid (WTA) biosynthetic pathways. Therefore, lysobactin, like ramoplanin and teixobactin, recognizes the reducing end of lipid-linked cell wall precursors. We show that despite its ability to bind precursors from different pathways, lysobactin’s cellular mechanism of killing is due exclusively to Lipid II binding, which causes septal defects and catastrophic cell envelope damage.
The prevalence of antibiotic-resistant bacterial infections has increased dramatically over the past two decades and now poses a serious threat to public health. Today there is no antibiotic in clinical use to which resistance has not developed, and common infections that were once easily treated can result in permanent injury and even death. There is a pressing need to develop antibiotics that have novel mechanisms of action.
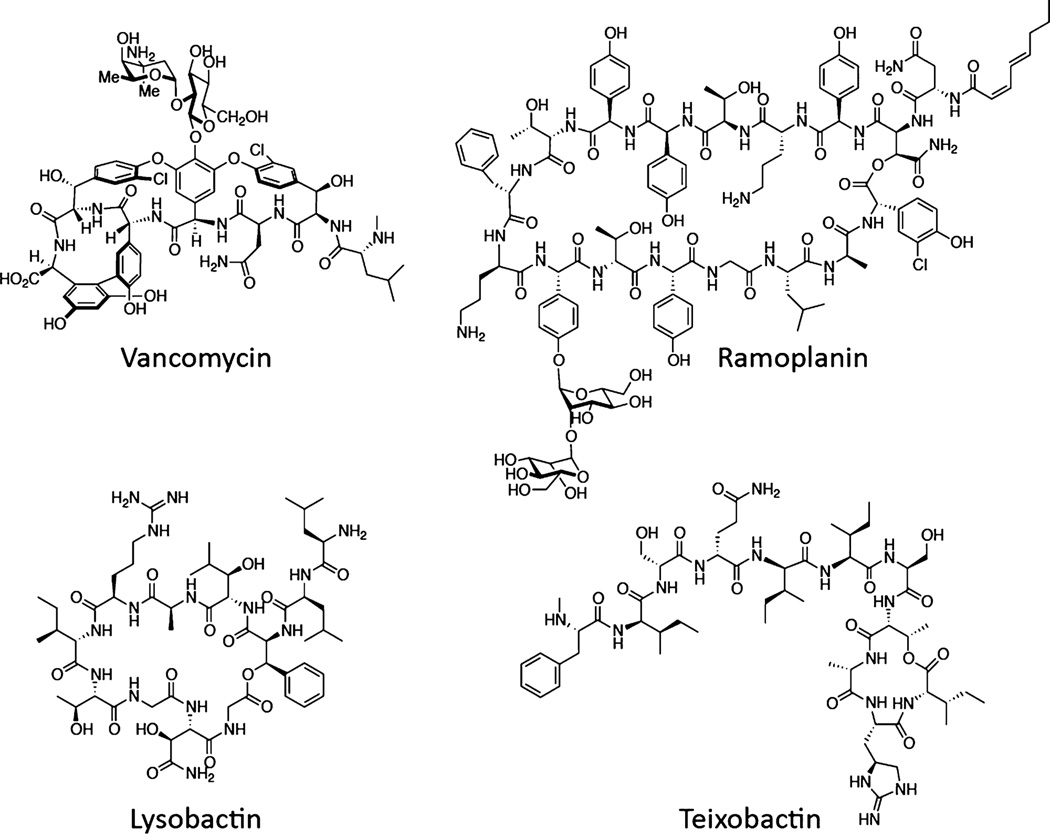
Among clinically used antibiotics, vancomycin (Figure 1) has had a remarkably long lifespan. Introduced in 1956, it is still used to treat methicillin-resistant Staphylococcus aureus (MRSA) and multidrug-resistant streptococcal infections, but clinical failure due to vancomycin resistance is increasingly common.1 Like other glycopeptide antibiotics, vancomycin inhibits biosynthesis of the bacterial cell wall by binding to a d-Ala-dAla found in peptidoglycan (PG) precursors.2 This mechanism of action, in which a substrate rather than a biosynthetic enzyme is the target, is difficult to overcome. Multiple genetic changes that remodel the cell envelope are required for even moderate resistance.3 Although vancomycin and related glycopeptides are the only clinically used substrate binders, they are not unique in having a substrate-binding mechanism. Other nonribosomal peptide synthetase (NRPS)-derived natural products that bind cell wall precursors include ramoplanin and the recently discovered teixobactin.4,5 The latter has garnered considerable attention not only because it represents a new structural class but also because it was shown to bind cell wall precursors from multiple biosynthetic pathways.5 In the course of our efforts to identify potent antimicrobial natural products from novel and known producing organisms, we found extracts of Lysobacter ezymogenes6 to be highly potent against Gram-positive bacteria. Isolation of the active compound revealed production of lysobactin, an NRPS-derived natural product that we here show has similar recognition properties as teixobactin, binding wall teichoic acid (WTA) as well as PG biosynthetic precursors. Lysobactin- and ramoplanin-induced cell death is due to inhibition of only PG biosynthesis.
Figure 1.
Structures of four NRPS-derived natural products that target cell wall biosynthesis by binding substrates.
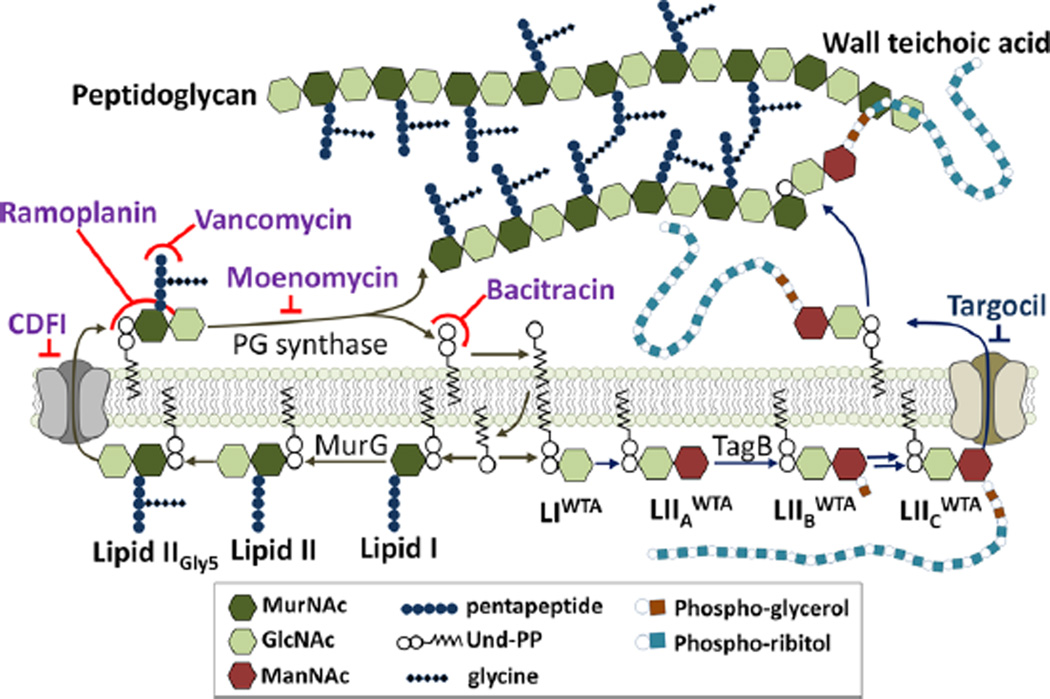
The bacterial cell wall in S. aureus is composed of thick layers of PG further modified with covalently bound WTA.7 The PG layers are essential for survival because they stabilize the cell membrane against high turgor pressure, thereby preventing osmotic lysis. As shown in Figure 2, the PG precursor Lipid II (LipidIIGly5) is synthesized inside the cell on an undecaprenyl phosphate (Und-P) “carrier lipid” and then flipped outside, where it is polymerized and cross-linked to make mature PG.8 Polymerization releases undecaprenyl pyrophosphate (Und-PP), which is dephosphorylated and recycled into the cell so that more Lipid II can be produced.9 The WTA biosynthetic pathway also involves intracellular assembly of a precursor on the Und-P carrier.7 After translocation to the surface of the cell, this precursor is attached to the C6 hydroxyl of residues in PG through a phosphodiester bond, liberating the carrier lipid.7 Vancomycin inhibits PG biosynthesis by binding to a d-Ala-d-Ala found at the terminus of the stem peptide of Lipid II, while ramoplanin and teixobactin bind to a region of Lipid II that includes the pyrophosphate and the first sugar but not the stem peptide.2b,4b,5 Teixobactin was also reported to bind a lipid-linked WTA precursor; therefore, it was proposed that teixobactin kills by inhibiting both the PG and WTA biosynthetic pathways.5
Figure 2.
Schematic of pathways for biosynthesis of lipid-linked PG and WTA precursors from the common intermediate Und-P. Compounds targeting PG and WTA biosynthesis are shown in purple and blue, respectively.
Lysobactin, also known as katanosin B, is produced by several genera of Gram-negative gliding bacteria found in soil. First reported in 1987, it was shown to inhibit PG biosynthesis and found to have outstanding in vitro activity against MRSA and vancomycin-resistant Enterococcus (VRE) as well as efficacy against systemic staphylococcal and streptococcal infections in mice.10 Although it was speculated to act as a substrate binder, experimental evidence to establish this mechanism of action has not been reported.2 In 2007, two groups independently described the total synthesis of lysobactin, and in 2011 the gene cluster was identified and characterized.11 To enable assessment of analogues for possible development, we further characterized lysobactin’s activity and determined its mechanism of action.
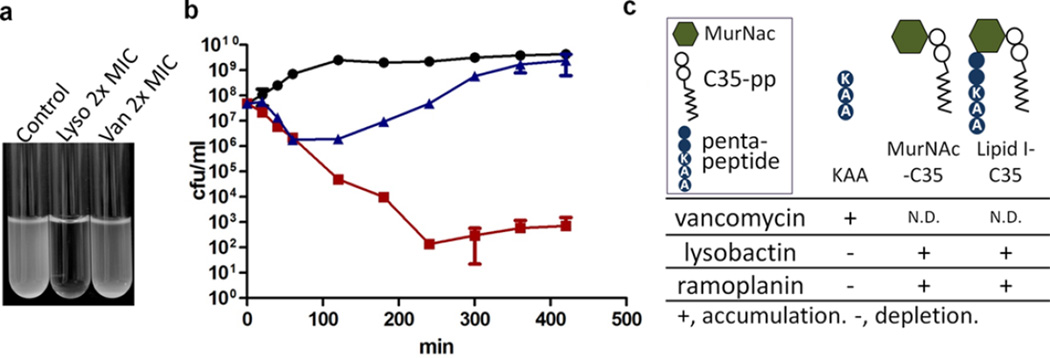
We found that lysobactin is rapidly bactericidal against S. aureus and also has significant activity against mycobacteria (Figures 3 and S2). The colony forming units (CFUs) of a growing S. aureus culture treated with lysobactin at 1.5 µg/mL (twice the minimum inhibitory concentration (MIC)) dropped more than five logs in 4 h, with lysis indicated by complete clearing of the culture tube within the same time period (Figure 3a,b). Although many antibiotics are not active against nongrowing cells, lysobactin was still bactericidal against stationary-phase cultures, although the rate of killing was lower (Figure S1b). As reported, ramoplanin was also bactericidal at 2× MIC (Figure S1a), but cultures treated with vancomycin at 2× MIC recovered after the CFUs dropped briefly.4e Vancomycin is known to have greatly reduced efficacy at high inoculum densities, explaining these results.12
Figure 3.
Lysobactin causes rapid lysis of S. aureus. (a) Cultures treated with lysobactin or vancomycin at 2× MIC. (b) Kill curves for S. aureus treated with no antibiotic (black circles), vancomycin (blue triangles), or lysobactin (red squares) at 2× MIC. (c) Addition of exogenous sugar–pyrophophate–lipids (5 µM) rescued S. aureus from killing by lysobactin (see Figure S3).
To determine whether lysobactin could be a substrate binder, we added exogenous cell wall precursors to S. aureus treated with lysobactin. Whereas the stem peptide mimic Lys-d-Ala-d-Ala antagonized the effects of vancomycin, it had no effect on the MIC of lysobactin, as previously reported.13 In contrast, synthetic Lipid I14 and an analogue lacking the stem peptide protected S. aureus from killing by lysobactin. These results suggested that lysobactin does indeed act via a substrate-binding mechanism (Figure 3c and S3).
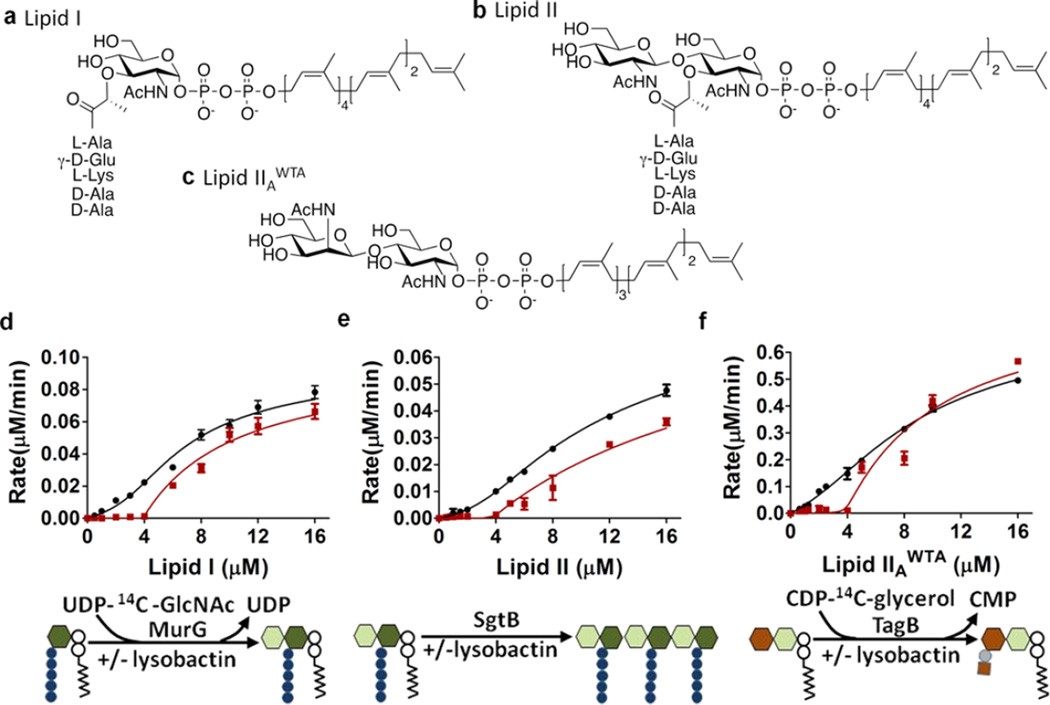
To confirm a substrate-binding mechanism and characterize lysobactin’s recognition preferences, we monitored the reaction rate as a function of substrate concentration for three enzymes that use cell wall precursors, MurG, SgtB, and TagB. MurG catalyzes the formation of Lipid II from Lipid I; SgtB catalyzes the polymerization of the PG precursor Lipid II; TagB catalyzes the transfer of phosphoglycerol to a lipid-linked WTA disaccharide intermediate, (Figure 2).14–16 Substrate binders produce a characteristic enzyme inhibition curve in which the reaction rate is negligible at low substrate concentrations because there is no free substrate but jumps as soon as substrate becomes available.4 The inhibitor:substrate ratio at which reaction is first observed provides the stoichiometry of the complex. Lysobactin inhibited all three enzymes and the velocity versus substrate concentration curves were characteristic of a substrate-binding mode of inhibition (Figures 4 and S5). Product was observed once the substrate concentration exceeded the concentration of lysobactin. Hence, lysobactin forms a 1:1 complex with cell wall precursors and inhibits all three enzymes comparably in vitro. However, micromolar concentrations of substrates were used in these experiments, and any difference in binding affinity may be obscured.
Figure 4.
In vitro, lysobactin inhibits three cell wall biosynthetic enzymes by binding to their substrates. (a–c) Chemical structures of the substrates used for the in vitro enzyme assays. (d–f) Plots of reaction rate as a function of substrate concentration for (d) MurG, (e) SgtB, and (f) TagB in the absence (black) and presence (red) of lysobactin (4 µM). See Figure 2 for the symbol key.
Ramoplanin was previously reported to bind Lipid I and Lipid II with a stoichiometry of 2:1, but it was never tested for binding to WTA precursors. We found that ramoplanin inhibits TagB by binding as a dimer to (Figure S6).4b We conclude that the ability to recognize cell wall precursors from different biosynthetic pathways may be a common feature of substrate binders that recognize the sugar–pyrophophosphate–lipid portion of cell wall intermediates.17 As teixobactin has also been reported to bind cell wall precursors with a 2:1 stoichiometry, lysobactin is distinctive among these NRPS-derived antibiotics for its 1:1 stoichiometry.
While in vitro studies on substrate binders provide useful information on recognition preferences and stoichiometry, they do not establish the cellular targets responsible for their antibiotic activity. Identifying these targets requires in vivo assays. We recently introduced a method to detect changes in Lipid II pool levels after treatment of bacterial cultures with antibiotics, and below we demonstrate the utility of this assay for establishing the cellular mechanism of lysobactin.18
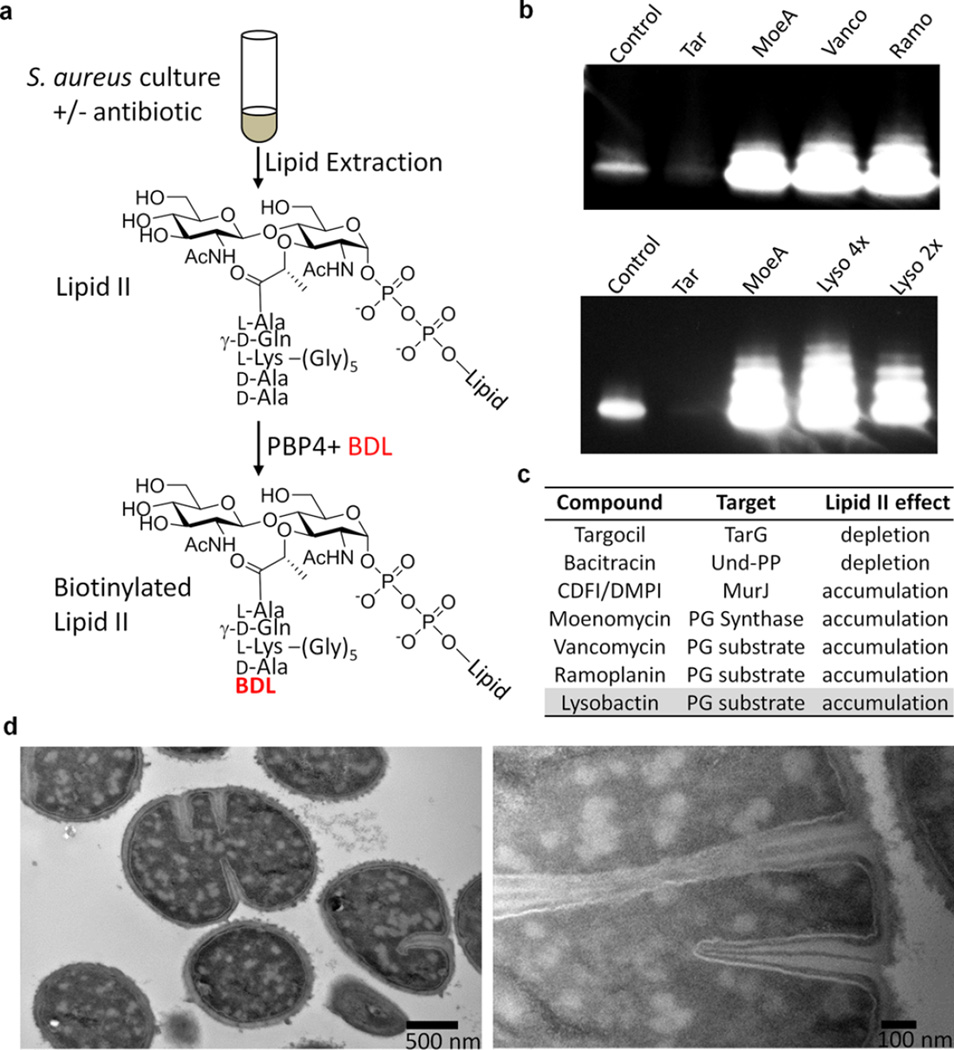
We first examined how Lipid II pool levels change upon treatment of S. aureus with several antibiotics having known mechanisms of action. Following a 10 min incubation in the presence of antibiotic, total cellular lipids were extracted from 2 mL cultures of S. aureus, and Lipid II was labeled with biotin via PBP4-mediated exchange of the terminal d-Ala with biotin–d-Lys (BDL) (Figure 5a). Moenomycin and vancomycin, which prevent Lipid II utilization by PG synthases, resulted in accumulation of Lipid II, as shown by the appearance of several intense chemiluminescent bands on a Western blot (Figure 5b). Treatment with lysostaphin, which cleaves pentaglycine cross-links, resulted in collapse to a single Lipid II band (Figure S7a), showing that the higher bands resulted from PBP4-mediated cross-linking of Lipid II during biotin labeling. CDFI and DMPI, reported as S. aureus MurJ inhibitors, also resulted in Lipid II accumulation (Figure S7b), consistent with the proposed mechanism of action.19 Treatment with ramoplanin unexpectedly resulted in apparent depletion of Lipid II (Figure S8a). However, ramoplanin 2:1 complexes are known to form stable fibrils, and we speculated that these fibrils were resistant to disassembly and therefore to Lipid II extraction (Figure S8b).4c,e It is worth noting that Lipid II extraction into organic solvent is also prevented by complexation with teixobactin.5 We were able to extract Lipid II into the organic phase after increasing the pH above 10, which likely disrupts the ramoplanin–Lipid II complexes. We then found that cells treated with ramoplanin had in fact accumulated large amounts of Lipid II (Figures 5b and S8a).18 Bacitracin, which binds to Und-PP, prevents recycling of the carrier lipid, resulting in depletion of Lipid II (Figure S7c). Finally, targocil, a WTA flippase (TarGH) inhibitor, also resulted in depletion of Lipid II (Figure 5b), in this case because the carrier lipid accumulates in WTA intermediates and thus is not available to make PG precursor (see Figure 2).
Figure 5.
Lysobactin sequesters Lipid II, triggering catastrophic cell envelope damage. (a) Schematic of the method used to detect Lipid II after extraction from cells. (b) Western blots of Lipid II isolated from cells after treatment with the indicated antibiotics and then labeled with biotin–d-Lys (BDL) using S. aureus PBP4. (c) Table summarizing how inhibition of or binding to various targets in the PG and WTA pathways affects pool levels of Lipid II. (d) TEM images showing the septal defects and distinctive cytoplasmic changes in S. aureus resulting from lysobactin treatment (1.5 µg/mL).
We tested lysobactin and found that it resulted in a dramatic accumulation of Lipid II in S. aureus. Therefore, we have concluded that although both lysobactin and ramoplanin bind to WTA precursors in vitro, the ability to do so does not contribute substantially to the cellular mechanism of action for either one because Lipid II is not depleted. Indeed, our results conclusively show that Lipid II is the cellular target of both compounds.
We finally examined the terminal phenotypes of S. aureus cells treated with lysobactin and ramoplanin using transmission electron microscopy (TEM). Treated at 2× MIC, both compounds caused major septal defects as well as distinctive cytoplasmic changes. It has been suggested that PG substrates recruit components of the division machinery, and the septal defects observed are consistent with mislocalization of cell division proteins in the absence of free Lipid II (Figures 5d and S9).20
In conclusion, we have shown that lysobactin is a substrate binder that binds the reducing end of lipid-linked cell wall precursors. Although lysobactin, like ramoplanin and teixobactin, recognizes cell wall precursors from different biosynthetic pathways, we have shown that its cellular mechanism of action is due to its ability to bind the PG precursor Lipid II. One in nine MurNAc units of PG is modified with WTA, suggesting that the precursor flux is substantially higher through the PG pathway.21 Because most of the cell wall precursors present on the cell surface are PG precursors, they comprise the major target of lysobactin.
Unlike ramoplanin and teixobactin, lysobactin binds Lipid II with a stoichiometry of 1:1. Because lysobactin binding does not prevent Lipid II extraction at neutral pH, we speculate that the complexes do not form higher-order aggregates as they do in the case of ramoplanin. Ramoplanin’s membrane disruption effects, which have to date prevented its successful clinical development, may be related to its propensity to oligomerize on cell surfaces even when Lipid II is not present. We note in closing that lysobactin, unlike ramoplanin, is not hemolytic against human red blood cells at concentrations well above its MIC (40×) (Figure S10). In view of its potent activity against a broad spectrum of important pathogens, lysobactin may be a promising candidate for further development.
Supplementary Material
Acknowledgments
This research was supported by NIH Grants AI09914, GM076710, GM066174, and U19 AI109764 and a Singapore A*STAR NSS (Ph.D.) scholarship to Y.Q. We thank Terry Roemer (Merck) for sharing DMPI and CDFI and Mohlopheni Marakalala for the MIC assay on Mtb.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.5b11807.
Experimental methods and supplementary figures (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Johnson AP, Uttley AH, Woodford N, George RC. Clin. Microbiol. Rev. 1990;3:280–291. doi: 10.1128/cmr.3.3.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Clardy J, Fischbach MA, Walsh CT. Nat. Biotechnol. 2006;24:1541–1550. doi: 10.1038/nbt1266. [DOI] [PubMed] [Google Scholar]; (b) Breukink E, de Kruijff B. Nat. Rev. Drug Discovery. 2006;5:321–332. doi: 10.1038/nrd2004. [DOI] [PubMed] [Google Scholar]
- 3.(a) Walsh CT, Fisher SL, Park IS, Prahalad M, Wu Z. Chem. Biol. 1996;3:21–28. doi: 10.1016/s1074-5521(96)90079-4. [DOI] [PubMed] [Google Scholar]; (b) Courvalin P. Clin. Infect. Dis. 2006;42:S25–S34. doi: 10.1086/491711. [DOI] [PubMed] [Google Scholar]; (c) Howden BP, Davies JK, Johnson PD, Stinear TP, Grayson ML. Clin. Microbiol. Rev. 2010;23:99–139. doi: 10.1128/CMR.00042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Fang X, Tiyanont K, Zhang Y, Wanner J, Boger D, Walker S. Mol. BioSyst. 2006;2:69–76. doi: 10.1039/b515328j. [DOI] [PubMed] [Google Scholar]; (b) Hu Y, Helm JS, Chen L, Ye XY, Walker S. J. Am. Chem. Soc. 2003;125:8736–8737. doi: 10.1021/ja035217i. [DOI] [PubMed] [Google Scholar]; (c) Lo MC, Helm JS, Sarngadharan G, Pelczer I, Walker S. J. Am. Chem. Soc. 2001;123:8640–8641. doi: 10.1021/ja011080p. [DOI] [PubMed] [Google Scholar]; (d) Oman TJ, Lupoli TJ, Wang TS, Kahne D, Walker S, van der Donk WA. J. Am. Chem. Soc. 2011;133:17544–17547. doi: 10.1021/ja206281k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Walker S, Chen L, Hu Y, Rew Y, Shin D, Boger DL. Chem. Rev. 2005;105:449–476. doi: 10.1021/cr030106n. [DOI] [PubMed] [Google Scholar]
- 5.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schaberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K. Nature. 2015;517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen P, Cook FD. Int. J. Syst. Bacteriol. 1978;28:367– 393. [Google Scholar]
- 7.Brown S, Santa Maria J, Walker S. Annu. Rev. Microbiol. 2013;67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Higashi Y, Strominger JL, Sweeley CC. Proc. Natl. Acad. Sci. U. S. A. 1967;57:1878–1884. doi: 10.1073/pnas.57.6.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Higashi Y, Strominger JL, Sweeley CC. J. Biol. Chem. 1970;245:3697–3702. [PubMed] [Google Scholar]
- 9.Siewert G, Strominger JL. Proc. Natl. Acad. Sci. U. S. A. 1967;57:767–773. doi: 10.1073/pnas.57.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Bonner DP, O’Sullivan J, Tanaka SK, Clark JM, Whitney RR. J. Antibiot. 1988;41:1745–1751. doi: 10.7164/antibiotics.41.1745. [DOI] [PubMed] [Google Scholar]; (b) Kato T, Hinoo H, Terui Y, Kikuchi J, Shoji J. J. Antibiot. 1988;41:719–725. doi: 10.7164/antibiotics.41.719. [DOI] [PubMed] [Google Scholar]; (c) Shoji J, Hinoo H, Matsumoto K, Hattori T, Yoshida T, Matsuura S, Kondo E. J. Antibiot. 1988;41:713–718. doi: 10.7164/antibiotics.41.713. [DOI] [PubMed] [Google Scholar]
- 11.(a) Guzman-Martinez A, Lamer R, VanNieuwenhze MS. J. Am. Chem. Soc. 2007;129:6017–6021. doi: 10.1021/ja067648h. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hall EA, Kuru E, VanNieuwenhze MS. Org. Lett. 2012;14:2730–2733. doi: 10.1021/ol300926d. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Hou J, Robbel L, Marahiel MA. Chem. Biol. 2011;18:655–664. doi: 10.1016/j.chembiol.2011.02.012. [DOI] [PubMed] [Google Scholar]; (d) von Nussbaum F, Anlauf S, Benet-Buchholz J, Habich D, Kobberling J, Musza L, Telser J, Rubsamen-Waigmann H, Brunner NA. Angew. Chem. Int. Ed. 2007;46:2039–2042. doi: 10.1002/anie.200604232. [DOI] [PubMed] [Google Scholar]
- 12.LaPlante KL, Rybak MJ. Antimicrob. Agents Chemother. 2004;48:4665–4672. doi: 10.1128/AAC.48.12.4665-4672.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maki H, Miura K, Yamano Y. Antimicrob. Agents Chemother. 2001;45:1823–1827. doi: 10.1128/AAC.45.6.1823-1827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Ha S, Chang E, Lo MC, Men H, Park P, Ge M, Walker S. J. Am. Chem. Soc. 1999;121:8415–8426. [Google Scholar]; (b) Men H, Park P, Ge M, Walker S. J. Am. Chem. Soc. 1998;120:2484–2485. [Google Scholar]
- 15.Terrak M, Nguyen-Disteche M. J. Bacteriol. 2006;188:2528– 2532. doi: 10.1128/JB.188.7.2528-2532.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ginsberg C, Zhang YH, Yuan Y, Walker S. ACS Chem. Biol. 2006;1:25–28. doi: 10.1021/cb0500041. [DOI] [PubMed] [Google Scholar]
- 17.(a) Schneider T, Kruse T, Wimmer R, Wiedemann I, Sass V, Pag U, Jansen A, Nielsen AK, Mygind PH, Raventos DS, Neve S, Ravn B, Bonvin AM, De Maria L, Andersen AS, Gammelgaard LK, Sahl HG, Kristensen HH. Science. 2010;328:1168–1172. doi: 10.1126/science.1185723. [DOI] [PubMed] [Google Scholar]; (b) Varney KM, Bonvin AM, Pazgier M, Malin J, Yu W, Ateh E, Oashi T, Lu W, Huang J, Diepeveen-de Buin M, Bryant J, Breukink E, Mackerell AD, Jr, de Leeuw EP. PLoS Pathog. 2013;9:e1003732. doi: 10.1371/journal.ppat.1003732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiao Y, Lebar MD, Schirner K, Schaefer K, Tsukamoto H, Kahne D, Walker S. J. Am. Chem. Soc. 2014;136:14678–14681. doi: 10.1021/ja508147s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huber J, Donald RG, Lee SH, Jarantow LW, Salvatore MJ, Meng X, Painter R, Onishi RH, Occi J, Dorso K, Young K, Park YW, Skwish S, Szymonifka MJ, Waddell TS, Miesel L, Phillips JW, Roemer T. Chem. Biol. 2009;16:837–848. doi: 10.1016/j.chembiol.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 20.Pinho MG, Errington J. Mol. Microbiol. 2005;55:799–807. doi: 10.1111/j.1365-2958.2004.04420.x. [DOI] [PubMed] [Google Scholar]
- 21.Bera A, Biswas R, Herbert S, Kulauzovic E, Weidenmaier C, Peschel A, Gotz F. J. Bacteriol. 2007;189:280–283. doi: 10.1128/JB.01221-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.