Abstract
Parallel studies of primary breast carcinomas and corresponding distant metastases samples reveal considerable differences. Our aim was to highlight this issue from another perspective and provide further data based on 98 patient samples: 69 primary breast carcinoma and 85 distant metastases from bone, central nervous system (CNS) and lung (56 paired). Two independent series of immunohistochemical reactions with different antibodies for estrogen receptor (ER), progesterone receptor (PgR) and human epidermal growth factor receptor 2 (Her2), along with HER2 fluroscence in situ hybridization (FISH) were performed on tissue microarrays to classify breast carcinoma and distant metastases samples into Luminal A, Luminal B-proliferating, Luminal B-HER2+, HER2+ and triple negative (TNBC) surrogate breast cancer groups. Correlation and agreement between the two assessments of ER and PgR were fair-to-moderate, and almost perfect for HER2 and Ki67. There was 40% discordance concerning immunophenotype between breast carcinomas and distant metastases. Most common metastatic site of ER+ breast carcinoma was the skeletal system (59.2%), whereas that of TNBCs was the CNS (58.8%) and lungs (23.5%). Distant metastases in bones were mostly luminal (54.3%), in the CNS, Luminal B (53.2%), and in the lung, TNBC (37.5%). The change of drugable properties of primary breast cancers in the respective bone and CNS metastases suggests that characterization of the metastasis is necessary for appropriate treatment planning.
Keywords: breast cancer, metastasis, bone, central nervous system, lung, immunoprofiling
Introduction
Metastatic breast cancer is an incurable disease, and the optimal treatment for patients with progressive breast cancer is not yet established. Tailored therapies for primary breast carcinomas are available but those that could bring a breakthrough in metastatic disease are still awaited. Metastases from breast cancer for a long time were regarded as identical to the primary tumor, and this was also confirmed from a genetic aspect (Weigelt et al. 2003). However, for more than 30 years, there have been sporadic reports published that demonstrate the possibility of marked changes in the metastatic tumors when compared to the primaries (Holdaway and Bowditch 1983; Jakesz et al. 1985; Osborne 1985; Spataro et al. 1992; Li et al. 1994; Simon et al. 2001). Also, completely contradictory data were published: on the one hand, it was regarded to be rare for metastatic tumors to present with a change in its characteristics (Klinga et al. 1982; Andersen and Poulsen 1988), whereas others quite often found that metastatic tumors are different from the primary (Lower et al. 2005; Guarneri et al. 2008; Simmons et al. 2009; Aitken et al. 2010; Idirisinghe et al. 2010; Cummings et al. 2013). Early molecular genetic studies uncovered some possible mechanisms to explain the metastasis process and, from those studies, it is theoretically a logical conclusion that metastases could differ from the primary tumor (Vecchi et al. 2007; Ding et al. 2010). These findings were followed by recent reports demonstrating a change in the receptor status between primary breast cancer and distant metastases (Hoefnagel et al. 2012, Lindström et al. 2012).
If there is a high enough chance that a metastasis will differ from that of the primary tumor, patient management algorithms should be changed: following identification of a metastasis, every effort should be made to sample the metastasis and re-evaluate its basic predictive markers such as the status of estrogen and progesterone receptors (ER and PgR), and human epidermal growth factor receptor 2 (HER2). It is fortunate that these three predictive markers can be evaluated equally on cytology smears: ER and PgR with immunocytochemistry; HER2 with in situ hybridization (ISH). These tests are optimal in the intial instances (Fritzsche et al. 2010; Wilking et al. 2010). Thus, even in cases of bone metastases, or remote, non-resectable lung or liver metastases, or even central nervous system metastases, a re-evaluation of these markers seem feasible.
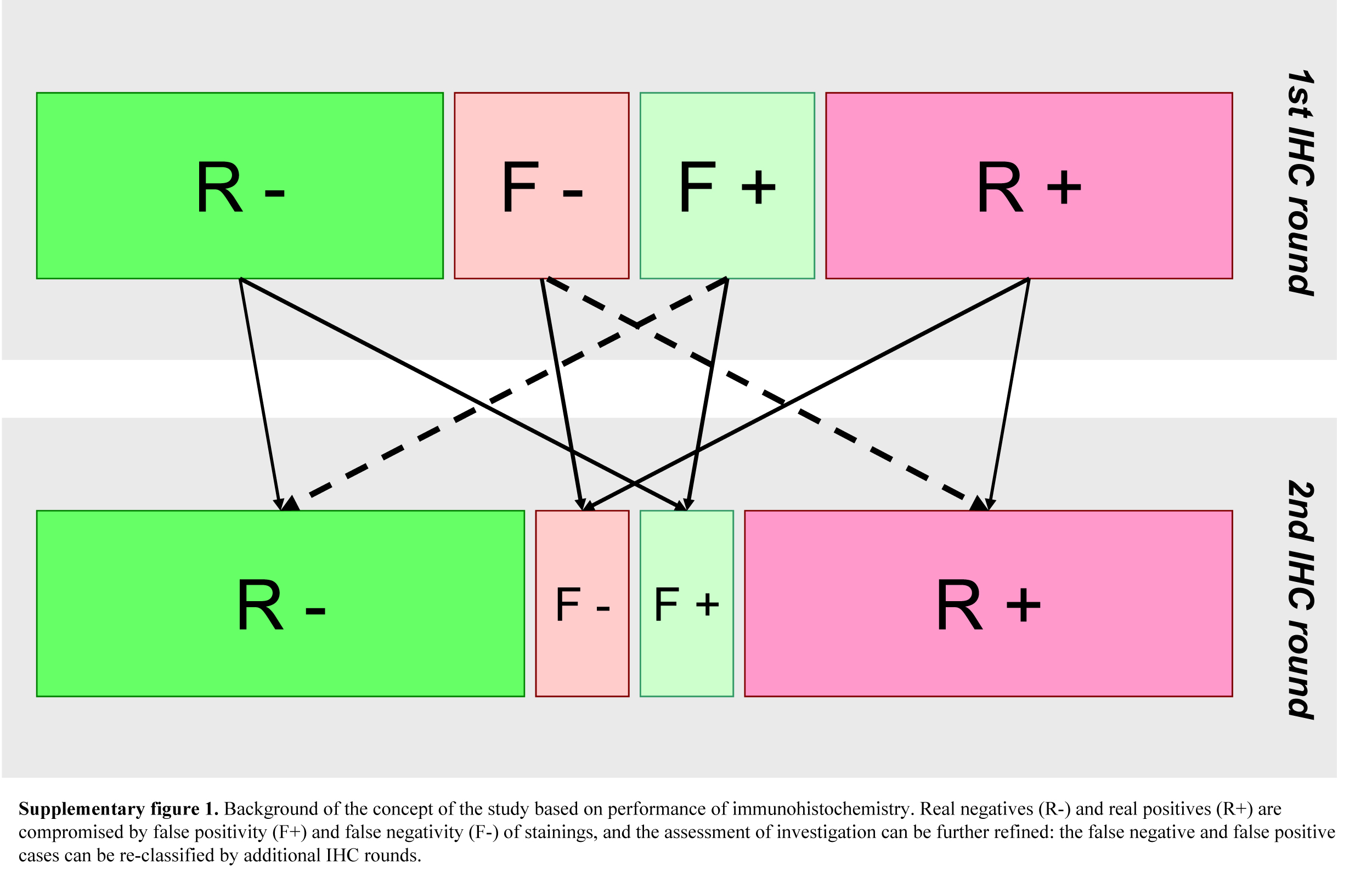
More recent studies have evaluated the immunophenotypes of primary breast carcinoma and their corresponding distant metastases (Hoefnagel et al. 2012; Lindström et al. 2012; Cummings et al. 2013); however, a publication also pointed out the effect of repeated laboratory evaluation in this setting (Pusztai et al. 2010). The recently published ASCO guideline highlights the need for repeating ER, PgR, HER2 testing in metastatic lesions (Van Poznak et al. 2015). Our aims with this study were to elucidate the occurrence rate of immunophenotypic changes using repeated ER, PgR, HER2 and Ki67 assessment in tumor samples, and additionally, in surgically biopsied or resectable distant metastases as compared to the primary breast carcinomas in our paired tumor samples (Supplemental Fig. 1).
Material & Methods
Ninety-eight women with metastatic breast cancer were enrolled in our study. Fifty-six primary breast carcinomas and paired distant metastastic breast carcinoma samples were collected at our Departments following permission of the Institutional Ethical Committee of the Semmelweis University, in concordance with the 1975 Helsinki Declaration (IKEB, #185/2007). The representative samples of 69 primary breast carcinomas and 85 metastases from three special institutions were collected: bone metastases were biopsied or operated at the Department of Orthopedics of the Semmelweis University, lung metastases were resected at the Department of Thoracic Surgery at the Korányi National Institute for Tuberculosis and Pulmonology or at the Department of Surgery of the Buda MAV Hospital, and central nervous system (CNS) metastases were operated at the National Institute of Neurosurgery. Metastatic tumors with available paraffin-embedded samples diagnosed between 2004 and 2007 were included in the study. Fifteen of the primary tumors were treated at the surgical departments of the Semmelweis University or the Buda MAV Hospital. A further 54 primary carcinoma samples were collected from the pathology departments of the main hospitals throughout the country (Supplemental Fig. 2). These institutions are participating in an external quality control system for ER, PgR, Ki67 and Her2 testing. Our samples were all neutrally buffered, formalin fixed and paraffin embedded. Bone samples were decalcified using EDTA solution, thus theoretically preserving the antigenicity of tissue proteins. All tissues in a certain department underwent the same protocol, and were treated chemically in an equal way (a tissue with positive staining from a given department could be used as positive control). Tissue microarrays were constructed using a simple manual device (Histopathology Ltd.; Pécs, Hungary). Two 2-mm cores (two from primary tumors and two from metastases each) were removed from the donor paraffin blocks after revision of the H&E slides by two investigators (JK, AMS). The regions for punching were selected based on the morphologically representative and most cellular areas of the tumor. The cores were then arranged in the recipient tissue microarray (TMA) blocks such that each patient’s primary tumor and metastasis samples were placed next to each other. Cores from the additional primary breast cancers and “orphan” metastases were subsequently placed in further TMA blocks (these cases were grouped by the site of the metastasis, resulting in TMA-bone, TMA-CNS, TMA-lung groups).
Immunohistochemical (IHC) reactions were performed with an automated immunostaining system (Ventana Benchmark XT, Roche Diagnostics; Mannheim, Germany) according to the manufacturer’s instructions in both rounds. Four years elapsed between the two investigations. In the first round, the antibodies used for ER, PgR, Ki67 and Her2 IHC were as follows: Novocastra NCL-ER-6F11, Novocastra NCL-PGR-312, DAKO M7240, Novocastra NCL-CB11. In the second round, the antibodies used for ER, PgR, Ki67 and Her2 IHC were as follows: Ventana Confirm antiEST receptor SP1, Ventana Confirm antiPROG receptor 1E2, DAKO M7240, Ventana-4B5 and Ventana Confirm antiHer2/neu 4B5. Ki67 was assessed with the same antibody in both rounds, thus, served as a control.
IHC reactions were evaluated by two investigators independently (JK, AMS). The status of ER and PgR was assessed by using the Allred scoring system (Allred et al. 1998), Ki67 positivity was measured as the ratio of the positive tumor cell nuclei in the sample, and Her2 immunohistochemistry was evaluated according to the standard protocol valid at the time of the 2nd round (i.e., positive by IHC only if more than 30% of tumor cells showed strong, complete membrane reaction). ER- and PgR-expressing tumors with lower than or equal to 14% Ki67-positive expression were considered as luminal A (LumA); for ER- or PgR- and Her2-positive tumors, LumBH (luminal B Her2 positive); and for ER- and PgR-expressing tumors with more than 14% Ki67 index, LumBP (luminal B proliferating) (Guiu et al. 2012). Triple-negative breast cancers (TNBC) were ER-, PgR-, Her2-negative tumors with no HER2 amplification. Her2-expressing and ER- and PgR-negative tumors were considered as part of the Her2 subgroup.
Fluorescence in situ hybridization (FISH) was performed on each TMA for the evaluation of HER2 gene status. FISH was performed using HER2/CE17 probes with the Poseidon kit (KBI-10735, Kreatech Diagnostics; Amsterdam, The Netherlands). The slides were evaluated using a Leica DM RXA microscope (Leica Microsystems GmbH; Wetzlar, Germany) supported by Leica CW4000 FISH software (Leica Microsystems Imaging Solutions Ltd.; Cambridge, UK). FISH results were evaluated according to the protocol valid at the time (Wolff et al. 2006): non-amplified if the HER2/CE17 ratio was less than 1.8, equivocal if the HER2/C17 ratio was between 1.8 and 2.2, and amplified if the HER2/CE17 ratio was over 2.2 (group Her2). When discordant with Her2 IHC, the FISH result was considered as adequate. One case showing HER2 polysomy was considered as Her2 positive due to the 3+ Her2 IHC reaction (100% of tumor cells).
Due to use of the TMA method, the very same tumor areas were compared between the runs in various approaches (Supplemental Fig. 3). All reactions were scored in a relatively short timeframe by one co-author to decrease deviation in inter- and intra-rater agreement.
Clinically relevant discrepancies only were subjected to statistical analysis. The scores of ER and PgR evaluated using the Allred method were dichotomized into <3 vs ≥3 groups (Allred et al. 1998). Her2 staining was assessed on a 0–3 scale supplemented by FISH analysis, and was split into two groups of negative vs positive (amplified) cases, and Ki67 staining, assessed on a 0–100% scale, was grouped into ≤14% vs >14% cohorts (Guiu et al. 2012). There were cases in all four reactions which were evaluable by the first round and not by the second and vice versa in all assessments.
To reach a final consensus profile based on the two staining rounds and FISH, the following approach was applied. When any of the tumor regions upon either staining round expressed ER or PgR with an Allred score of 3 or higher, those were accepted as hormone receptor-positive. Regarding Her2, the strongest staining and eventually HER2 FISH result was accepted. For Ki67, the highest ratio of stained cells among the cores was accepted to the final labeling index for a given tumor.
The data were recorded in a Microsoft Excel table (Microsoft Corp.; Redmond, WA). Statistical analysis using SPSS 15.0 Family Pack software was performed (SPSS Inc., Chicago, IL). A Chi-square test was used for statistical analysis of non-parametric variables. Spearman’s rank correlation (R) was used to assess the agreement of scores in the cross-experimental setting and Cohen’s Kappa test (κ) was used to evaluate the assigned categories derived from scores. All statistical tests were two sided. ‘p’ values of less than 0.05 were considered to be significant.
Results
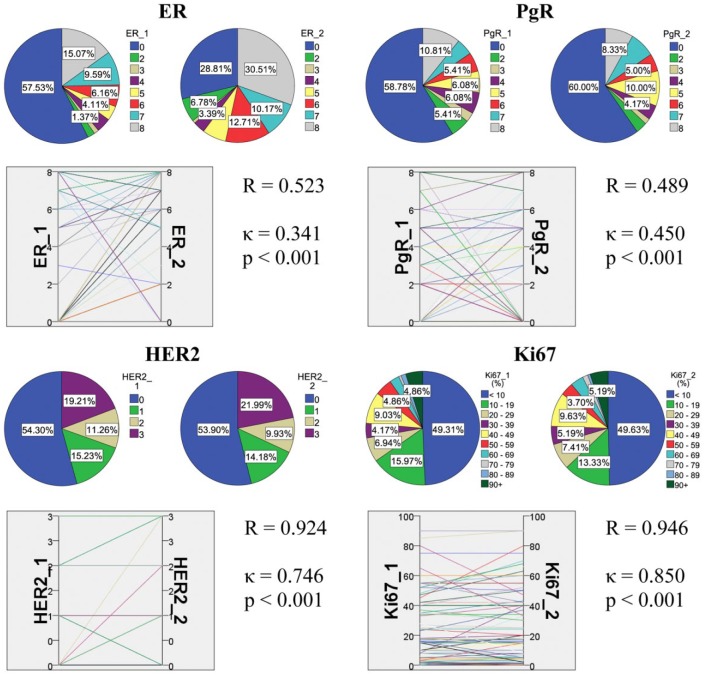
First, we have undertaken a technical analysis to gain an overall impression of the detection consistency or inconsistency of ER, PgR, Her2 and Ki67 expressions. In the first assessment, all samples, regardless of location, were included and underwent both rounds of IHC staining and evaluation. Reliable data of the expression in both runs were available for 112 tumors for ER, 112 for PgR, 141 for HER2 and 134 for Ki67. In line with this, a Spearman correlation showed moderate agreement between the two rounds of ER and PgR staining in the tumors, whereas the Kappa value showed fair agreement between the assigned categories for ER and moderate agreement for PgR. The Her2 and Ki67 expressions performed substantially better (Fig. 1).
Figure 1.
Summary of the results in all tumor samples regardless of location. The correlation and agreement between ER and PgR Allred scores, HER2 scores and Ki67 labeling index (LI) investigated by repeated immunoprofiling are shown. In each block, pie charts show the distribution of cases according to rounds 1 and 2, and parallel graph displays their relation supported by statistics.
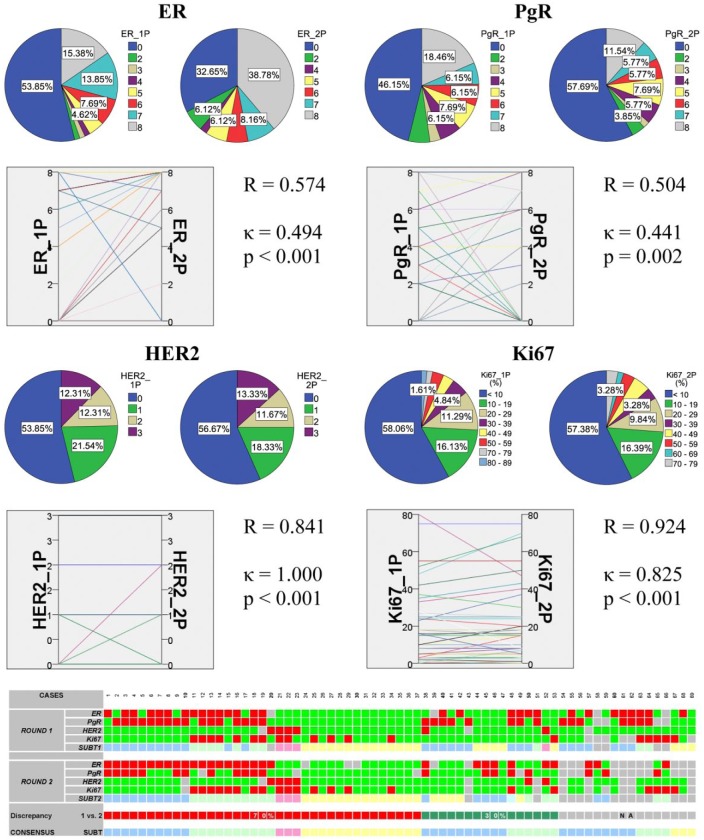
Forty-three primary breast carcinomas were available for both rounds of ER, 48 for PgR, 60 for both Her2 and Ki67 staining and analysis, respectively (Fig. 2). Again, the correlation was moderate for ER and PgR, while almost perfect for Ki67 and Her2 supplemented by FISH. The agreement (as assessed with a Kappa test) was also moderate for the hormone receptors and almost perfect for Her2 and Ki67. Sixty-nine primary breast tumors were immunoprofiled based on either or both reactions, among them 53 had two assignments based on the results of the two separate IHC rounds, and 30% of the tumors were discrepant (Fig. 2). Finally, a consensus profile was established for the primary tumors representing the combined analysis (Table 1).
Figure 2.
Summary of the results in all primary breast carcinoma samples. The correlation and agreement between ER and PgR Allred scores, HER2 scores and Ki67 LI investigated by repeated immunoprofiling are shown. In each block, pie charts show the distribution of cases according to rounds 1 and 2, and the parallel graph displays their relation supported by statistics. The bottom panel shows all 69 cases displayed on heatmap. For rounds 1 and 2 and consensus: bright green, ER score <3, PgR score <3, HER2 negative, Ki67 index ≤14%; red, ER score ≥3, PgR score ≥3, HER2 positive/amplified, Ki67 index >14%; light blue, luminal A; light green, luminal B (all); pink, HER2+; light yellow, TNBC. The “Discrepancy” line shows whether rounds 1 and 2 resulted in similar results: red, no change due to consistency; green, inconsistent results; gray, no subtype assigned in any of the rounds.
Table 1.
Consensus Immunoprofile of Primary Breast Carcinomas and Metastases of the Current Study.
| Consensus Subtypes of Primary Breast Carcinomas |
Total | |||||||
|---|---|---|---|---|---|---|---|---|
| Lum. A |
Lum. B |
Lum. B |
HER2+ | TNBC | NA | |||
| (HER2+) | (Ki67 high) | |||||||
| PRIMARY metastatic to | Bone | 17 (58.62%) | 1 (50.00%) | 11 (61.11%) | 1 (33.33%) | 3 (17.65%) | 2 (6.90%) | 35 (35.71%) |
| CNS | 11 (37.93%) | 1 (50.00%) | 6 (33.33%) | 1 (33.33%) | 10 (58.82%) | 18 (62.07%) | 47 (47.96%) | |
| Lung | 1 (3.45%) | 0 (0) | 1 (5.56%) | 1 (33.33%) | 4 (23.53%) | 9 (31.03%) | 16 (16.33%) | |
| Total | 29 (29.59%) | 2 (2.04%) | 18 (18.37%) | 3 (3.06%) | 17 (17.35%) | 29 (29.59%) | 98 (100%) | |
| Consensus Subtypes of Distant Metastases |
Total | |||||||
| Lum. A |
Lum. B |
Lum. B |
HER2+ | TNBC | NA | |||
| (HER2+) | (Ki67 high) | |||||||
| METASTASIS in | Bone | 19 (54.29%) | 1 (2.86%) | 2 (5.71%) | 3 (8.57%) | 6 (17.14%) | 4 (11.43%) | 35 (35.71%) |
| CNS | 5 (10.64%) | 16 (34.04%) | 9 (19.15%) | 3 (6.38%) | 6 (12.77%) | 8 (17.02%) | 47 (47.96%) | |
| Lung | 4 (25.00%) | 2 (12.50%) | 2 (12.50%) | 1 (6.25%) | 6 (37.50%) | 1 (6.25%) | 16 (16.33%) | |
| Total | 28 (28.57%) | 19 (19.38%) | 13 (13.26%) | 7 (7.14%) | 18 (18.36%) | 13 (13.26%) | 98 (100%) | |
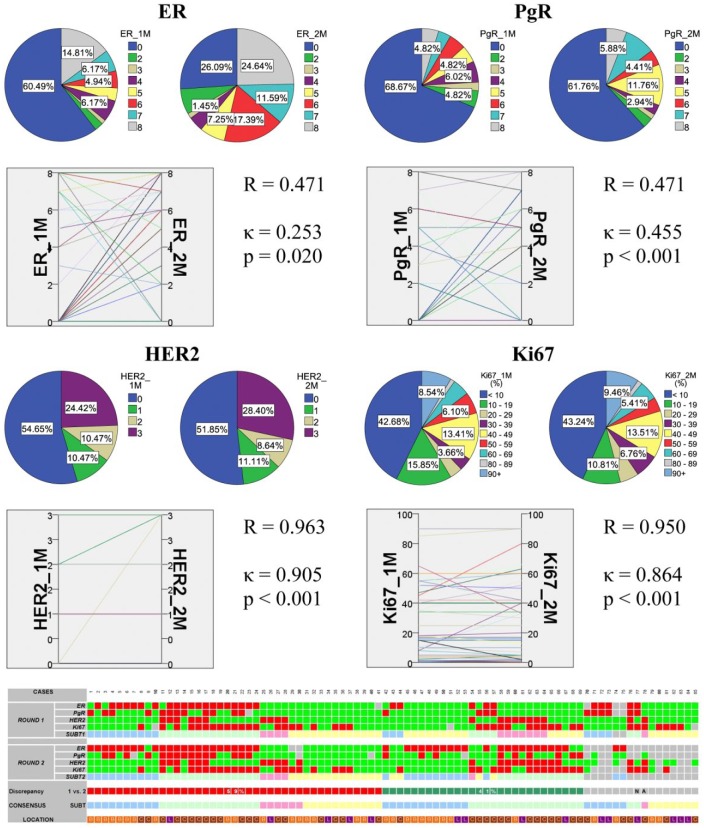
Regarding the 85 distant metastases, similar observation was noted: moderate correlation for ER and PgR, while almost perfect correlation for Her2 and Ki67. Agreement was only fair for ER, moderate for PgR, and still almost perfect for Her2 and Ki67 (Fig. 3). Eighty-five metastases were profiled, and 69 had two assigned categories based on the two rounds of immunohistochemistry. The discrepancy between them was 41%. Again a consensus profile was established based on the results of the two runs (Table 1).
Figure 3.
Summary of the results in all breast cancer metastases. The correlation and agreement between ER and PgR Allred scores, HER2 scores and Ki67 LI investigated by repeated immunoprofiling are shown. In each block, pie charts show the distribution of cases according to rounds 1 and 2, and parallel graph displays their relation supported by statistics. The bottom panel shows all 85 cases displayed as a heatmap. For rounds 1 and 2 and consensus: bright green, ER score <3, PgR score <3, HER2 negative, Ki67 index ≤14%; red, ER score ≥3, PgR score ≥3, HER2 positive/amplified, Ki67 index >14%; light blue, luminal A; light green, luminal B (all); pink, HER2+; light yellow, TNBC. The “Discrepancy” line shows whether rounds 1 and 2 resulted in similar results: red, no change due to consistency; green, inconsistent results; gray, no subtype assigned in any of the rounds. Consensus subtype is established with combination of assignments based on both rounds. The line location shows where the actual sample is located: orange, bone (B); brown, CNS (C); purple, lung (L).
Thereafter, we investigated the metastases also by their location. In the case of bone metastases, the correlation of ER and PgR expression—as defined in the two separate IHC runs—was fair, and almost perfect for Her2 and Ki67. Bone metastases showed slight agreement regarding ER expression, moderate agreement of PgR, and almost perfect agreement for Her2 and Ki67 (Supplemental Fig. 4). Thirty-one bone metastases were successfully profiled, among them 27 were investigated twice resulting in an overall 27% discrepancy between the two subtype assignments.
Between the two IHC runs, CNS metastases displayed a moderate correlation for both hormone receptors, and near perfect correlation for Her2 and Ki67 expressions. The agreement for the hormone receptors was fair, and almost perfect for Her2 expression and Ki67 labeling index (Supplemental Fig. 5). Thirty-nine CNS metastases were available for immunoprofiling, among them 33 were investigated with repeated IHC with an overall discrepancy ratio of 43%.
Although between the two separate runs of IHC reactions lung metastases showed substantial correlation for ER and Ki67 and perfect correlation for PgR and Her2, the agreement for ER was only slight, fair for Ki67, and perfect for PgR and Her2 (Supplemental Fig. 6). Fifteen lung metastases were profiled, of which nine could be thoroughly investigated by the two rounds of immunoprofiling. The overall discrepancy of the latter was 44%.
Next, we assessed the biological and clinical dimension of the investigation. The primary tumors displayed preferences of metastatic sites according to their immunophenotype: luminal tumors metastasized to bone (29/49, 59.2%), whereas triple negative tumors to the CNS (10/17, 58.8%) and to the lung (4/17, 23.5%). The metastases displayed the following characteristics: mostly luminal A in the skeletal system (17/29, 54.3%), luminal B in the brain (25/47, 53.2%) and TNBCs (6/16, 37.5%) in the lung (Table 1).
Finally, we analyzed the alteration of the phenotype of primary breast cancers as compared with that of the subsequent metastases. For this, we used the consensus immunoprofiles that were based on two IHC rounds supplemented by FISH for HER2 status. Out of the paired 56 cases, 29 were metastatic to bone, 21 to CNS and 6 to the lung (Fig. 4). Altogether, discrepancy between the primary and the metastatic tumors was detected in 41% of the cases (κ=0.166).
Figure 4.
Summary of the results in paired primary and metastatic breast cancer samples. The heatmap displays the results of ER, PgR, HER2 and Ki67 IHC investigated by repeated immunoprofiling in paired primary and metastatic breast cancer samples. The panel shows all 56 cases displayed with both IHC rounds, their resulting subtypes, the discrepancies of rounds 1 and 2, the consensus subtypes in both primaries and metastases, and the alteration of immunoprofile between the primary tumors and the distant metastases. For rounds 1 and 2 and consensus lines: bright green, ER score <3, PgR score <3, HER2 negative, Ki67 index ≤14%; red, ER score ≥3, PgR score ≥3, HER2 positive/amplified, Ki67 index >14%; light blue, luminal A; light green, luminal B (all); pink, HER2+; light yellow, TNBC. The “Discrepancy” lines show whether rounds 1 and 2 resulted in similar results. The “Alteration” line shows change in subtype in metastasis: red, no change due to consistency; green, inconsistent results; gray, no subtype assigned in any of the rounds. Consensus subtype is established with a combination of assignments based on both rounds in primaries and metastases alike. The line location shows where the actual sample is located: orange, bone (B); brown, CNS (C); purple, lung (L).
Discussion
More recent data show that metastatic tumors can indeed change their geno- or phenotype, for which the mechanism is not fully understood yet (Holdaway and Bowditch 1983; Hahnel and Twaddle 1985; Vecchi et al. 2007; Liedtke et al. 2009; Hoefnagel et al. 2012; Lindström et al. 2012). Tumor heterogeneity, clonal selection, early distant seeding of cancer cells and parallel evolution or the metastatic cascade model, and, therapy induced clonal evolution have all been suggested recently as biological explanation of discrepant metastases. A very recent review by Desmedt et al. (Cancer and Metastasis Review, 2016, accepted) provides insight into the present state of knowledge of the field (Kuukasjarvi et al. 1997; Ding et al. 2010; Cummings et al. 2013; Meric-Bernstam et al. 2014; Moelans et al. 2014; Brastianos et al. 2015). In this study, we evaluated the immunophenotype and HER2 FISH data of 56 paired primary tumors and distant metastases occurring in the skeletal system, CNS and respiratory system. The study was designed to derive more accurate results about this biological question based on more solid technical basis.
An obvious explanation for the longstanding failure of the successful treatment of metastatic breast cancer might be the longstanding dogma that metastatic tumors are identical to their primary carcinoma (Paget 1889). Our study provides further support to the accumulating evidence that breast cancer metastasis may substantially differ from the primary tumor, and that different types of breast cancer show preferential sites for distant relapse, as suggested earlier by other investigators (Smid et al. 2008). More recently, molecular genetic studies uncovered some of the features at the genome level that explain this particular characteristic of metastatic breast cancer. For example, a 17-gene signature was determined to be characteristic to breast cancers that developed brain metastasis during progression (Bos et al. 2009). A comprehensive review by the Steeg group summarized emerging knowledge that shed some light on the molecular background of brain metastatic breast cancer (Lin et al. 2013). Expression of adrenomedullin (Siclari et al. 2014) and RANK (Blake et al. 2014) by breast cancer cells were claimed to be characteristic to bone metastatic breast cancer. More recently, the key role of molecular mechanisms underlying cellular dormancy in typically bone metastatic cancers like breast- and prostate cancer was summarized in a review article (Quayle et al. 2015). In vivo mice experiments using prostate- and breast cancer cell lines suggest that characteristics of the “niche” are equally important in the formation of osteolytic bone metastases (Wang et al. 2015).
It was argued that ER changes may be the result of technical failures; e.g., differences in tissue fixation, antigen retrieval and antibodies, which might be an explanation of the discordant findings. In one study, it was shown that ER protein, mRNA and DNA levels differed when comparing primary tumors and lymph node metastases (Zheng et al. 2001). Furthermore, if we accept an 80% to 90% estimated overall accuracy of results of repeat biopsies (Bernards and Weinberg 2002; Miller et al. 2007), our analysis, simulating a repeated biopsy assessment, with still 41% discordance between primary and metastatic tumors supports that the discrepancy cannot be merely based on methodological errors (Pusztai et al. 2010).
We have shown that breast cancer upon progression to the skeletal system undergoes significant phenotypic changes. The majority of these tumors are ER positive, but a considerable number of the metastases loose their hormone receptors and HER2 expression alike and become triple-negative. This cohort showed difference in 27% of the cases between primary- and bone metastatic samples whereas those tumors metastasizing to the brain displayed a 43% discordance; this emphasizes that chemical processing (decalcification) of bone metastases alone cannot be responsible for most of the changes in hormone receptor expression in tumor cells. In a preclinical study, Hilton et al. (2011) described the feasibility and utility of bone marrow aspiration and trephine biopsies in breast cancer patients with skeletal metastases. They have found discrepancies of ER and PgR expression between the primary tumor and the bone or bone marrow metastasis in 38.5% and 42.3% of the cases, respectively. More recently, a study using breast cancer bone metastasis biopsy material confirmed common changes in ER and PgR receptor status and occasional changes in Her2 status (Aurilio et al. 2013).
Lung metastases were least similar to their primaries as compared with the other two groups (discordance rate 44% vs 43% for CNS; vs 27% for skeletal). It is important to note, however, that this group also had the lowest number of paired samples; even though, the presence of TNBC cases both in primary- and metastatic tumor samples could be noted.
In the case of CNS progression, a gain in Her2 expression and/or amplification accompanied by increased proliferative activity could be detected. Although, there are cases where HER2 gene amplification is lost and hormone receptors are expressed in the metastases, proliferation rate unequivocally increases even in these cases. In a recent study of paired primary breast cancers and respective brain metastases, it was shown that predictive IHC markers, especially hormone receptor expression, change in a substantial proportion of cases (Shen et al. 2015).
Altogether, three primary tumors and five metastases showed high Her2 expression or HER2 gene amplification without hormone receptor expression in the first round. Probably the improved detection of ER and PgR in the second round classified more tumors to the Luminal B-HER2 subtype, thereby shrinking the pure HER2 group.
Using similar material to ours (i.e., FFPE blocks of paired primary, locally recurrent and metastatic breast cancer cases), Meric-Bernstam et al. (2014) performed detailed molecular genetic studies, focusing on putative, targetable gene alterations, by identifying copy number changes and mutations in their samples. Although discrepancies at the gene level were remarkable, the authors concluded that primaries and local/distant relapses bear mostly similar genomic features. Nevertheless, the authors conclude that repeat biopsy of the metastatic tumor is advisable because clinically relevant, targetable genetic alterations may be revealed (Meric-Bernstam et al. 2014).
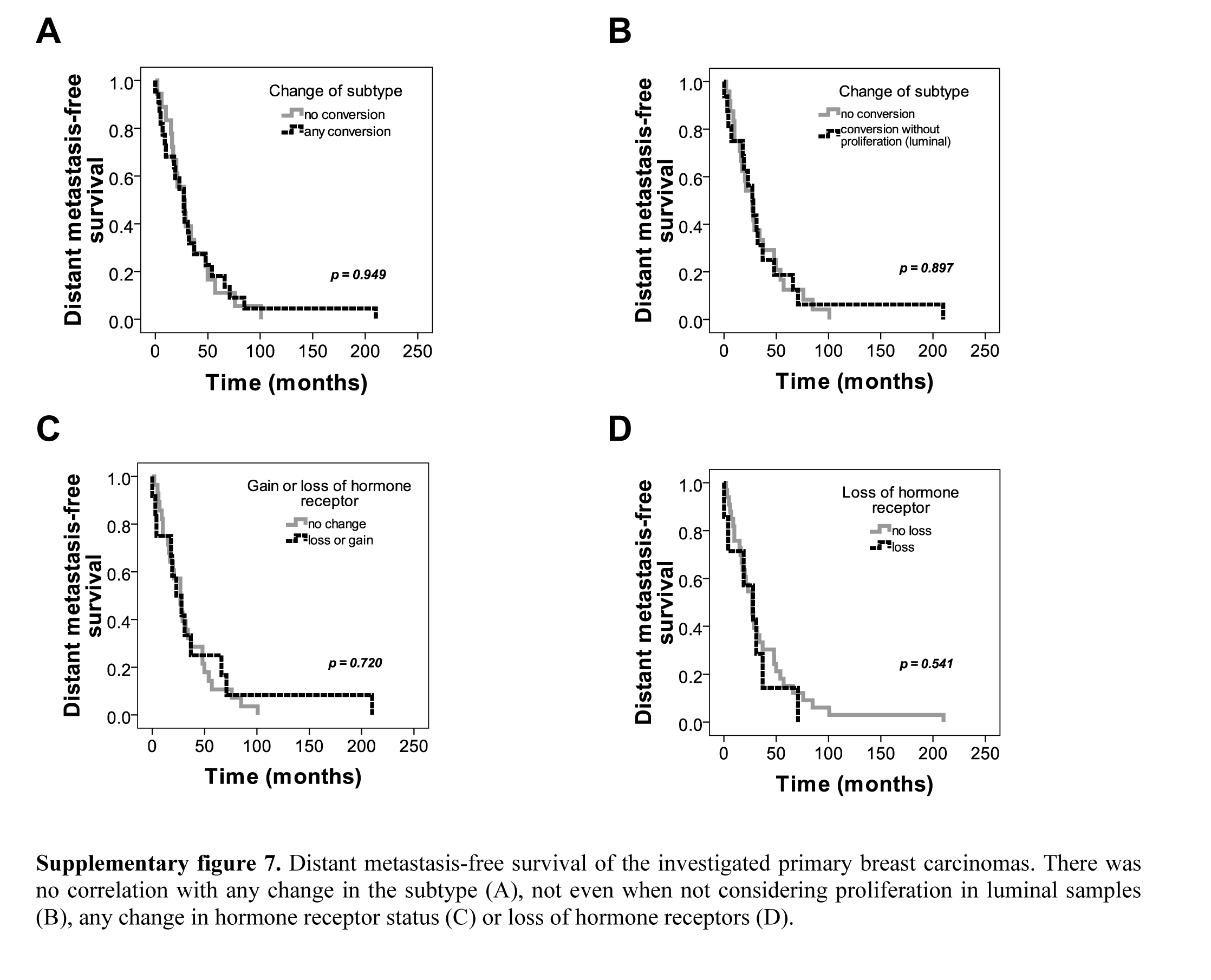
The distribution of subtypes in the distant locations is similar to the findings of others (Aversa et al. 2013; Cummings et al. 2013). Regarding prognostic relevance of changes in IHC predictive marker expression, the impact of hormone receptor conversion for overall survival was shown by Hoefnagel et al. (2012), with a conversion from positive to negative status having an independent, negative impact (Hoefnagel et al. 2012). In a Chinese patient cohort, it was also reported that overall survival may be worse for those breast cancer patients whose metastatic disease changes in terms of ER, PgR or Her2 statuses (Yang et al. 2014).
Surgical resection or biopsy of metastases from breast cancer is not always technically feasible; therefore, our patient cohort presented herein may not be fully representative of the whole population of advanced-stage breast cancer patients. However, Rothé et al. (2014) in a recent study suggest that even plasma circulating DNA could be used for molecular-based characterization of progressing breast cancer. Using next-generation sequencing, changes in clinically relevant tumor features were revealed, and it became evident that such a minimally invasive intervention (venous puncture) may be effective for the characterization of metastatic tumor cells (Rothe et al. 2014).
We are aware of the weaknesses of our study: the fact that many of our samples were collected from numerous hospitals did not allow us to assess detailed clinico-pathological data of the primary tumors. Therefore, for most of the primary breast carcinoma cases, we have no data regarding the mode of discovery (screen detected vs symptomatic), type of surgery (mastectomy vs breast conservation), pTNM stage, or other basic prognostic factors. Similarly, no data regarding postoperative oncological treatment were available. At the time our patients’ primary tumors were operated, for ER-positive tumors, adjuvant endocrine therapy was already standard. At that time, trastuzumab was not yet available as an adjuvant standard treatment for patients with HER2-positive breast cancer. Nevertheless, we are convinced that, even without these data, a relevant finding from our study is that pathologists should be careful while selecting ER and PgR antibodies for diagnostic purposes. It may be also of interest for oncologists who, by requesting re-evaluation of the patient’s primary breast cancer’s sample at the time when distant metastasis occurs, may get at least a partial explanation for the failure of the treatment he or she recommended postoperatively. While acknowledging that subtyping breast cancer into clinically meaningful classes based on IHC panels has its inherent limitations, it still provides the data necessary for adjuvant treatment decisions in the vast majority of cases. Therefore, in the everyday practice, biopsy and accurate characterization of the metastasis may open the prospect for appropriate, targeted therapy in cases of progressive breast cancer.
We conclude that metastases of breast carcinomas change their properties often enough to prompt the formulation of a diagnostic algorithm of metastatic breast cancer patients: sampling of the metastasis should be performed whenever feasible before further treatment is decided (Macfarlane et al. 2012; Van Poznak et al. 2015). Also, ideally, the primary tumor should be re-assessed at the same time the metastasis is sampled. This would be especially important in CNS metastases, where Her2 positivity may be gained in cases of ER-positive/Her2-negative primaries.
This study also shows that technical reasons (e.g., choice of hormone receptor antibodies) might exist behind different immunoprofiles of primary breast carcinomas and their metastasis. Therefore, the careful selection of antibodies used for diagnostic purpose is mandatory.
In our study, in line with that of Metzger-Filho et al. (2013), we demonstrate that there is a certain preference of metastatic sites for luminal and triple-negative carcinomas, being the skeletal system and brain/lung, respectively.
Supplementary Material
Supplementary Material
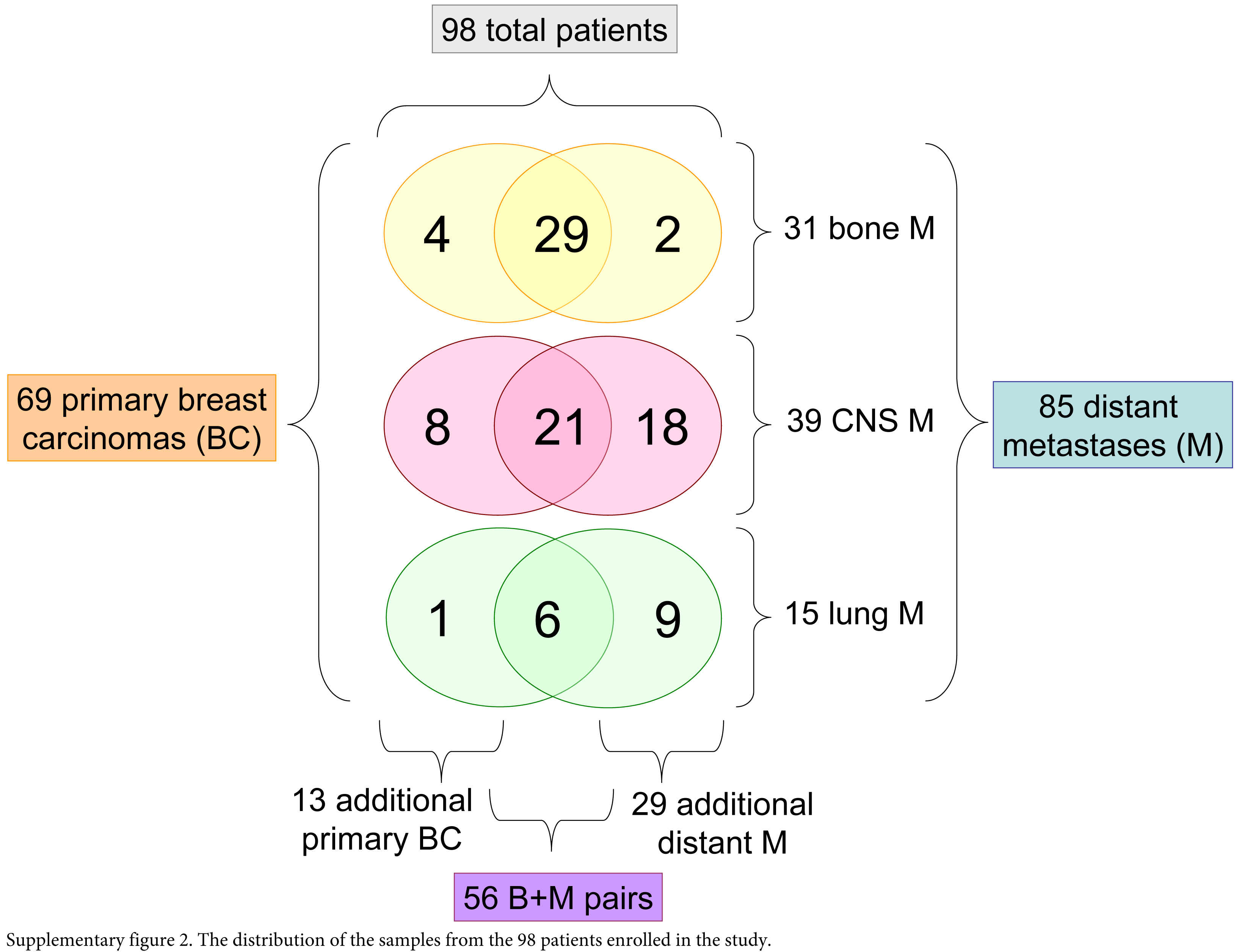
Supplementary Material
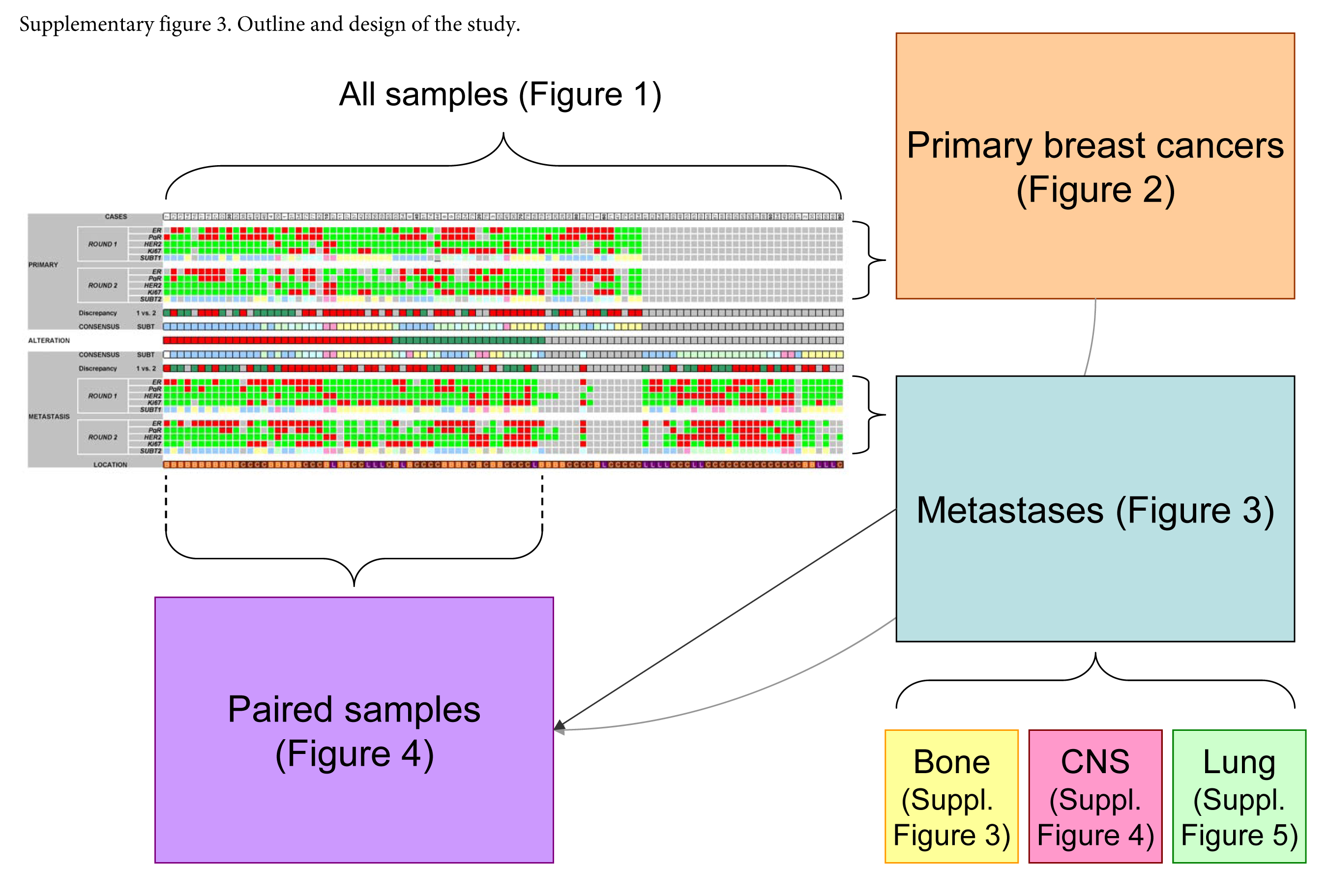
Supplementary Material
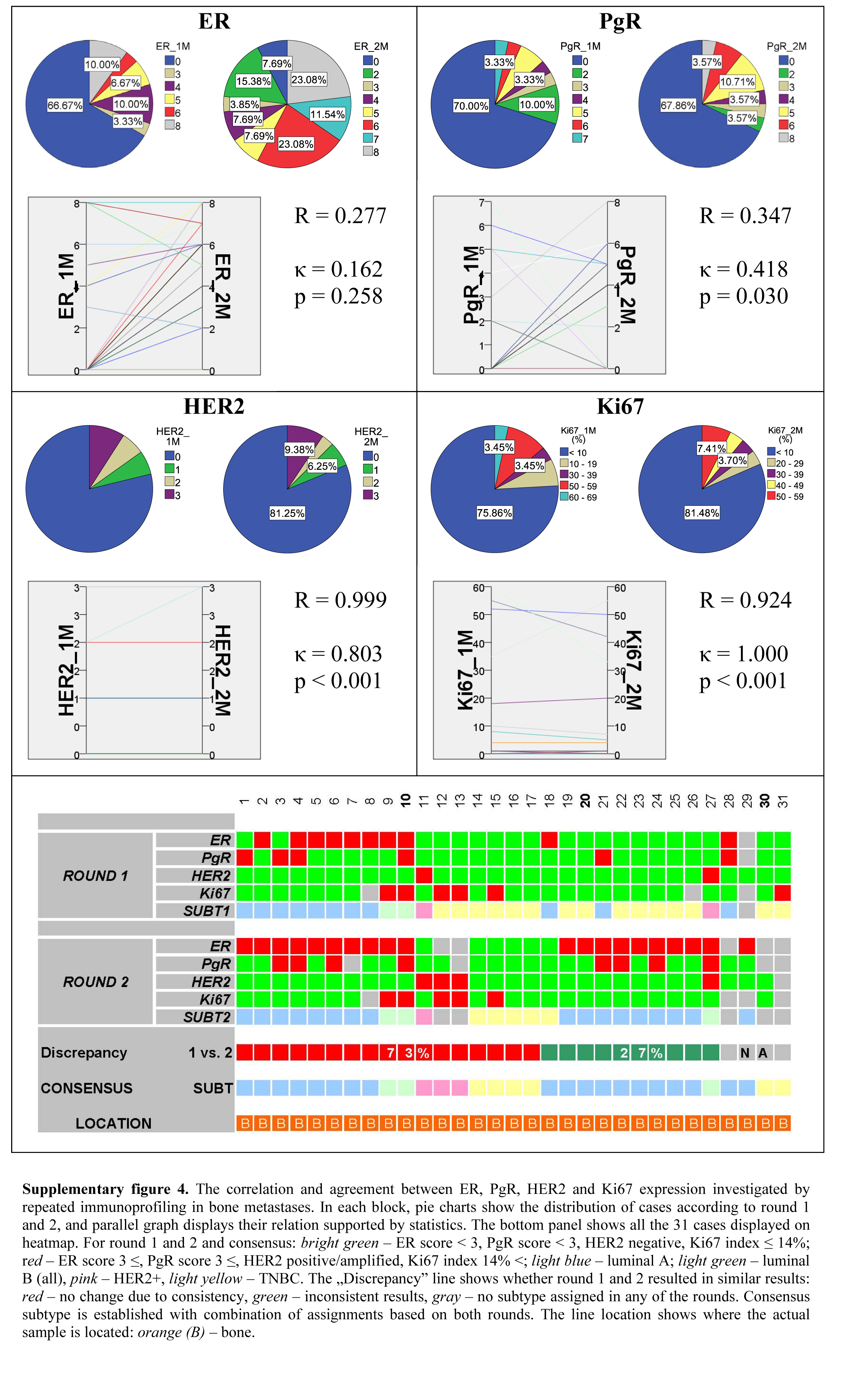
Supplementary Material
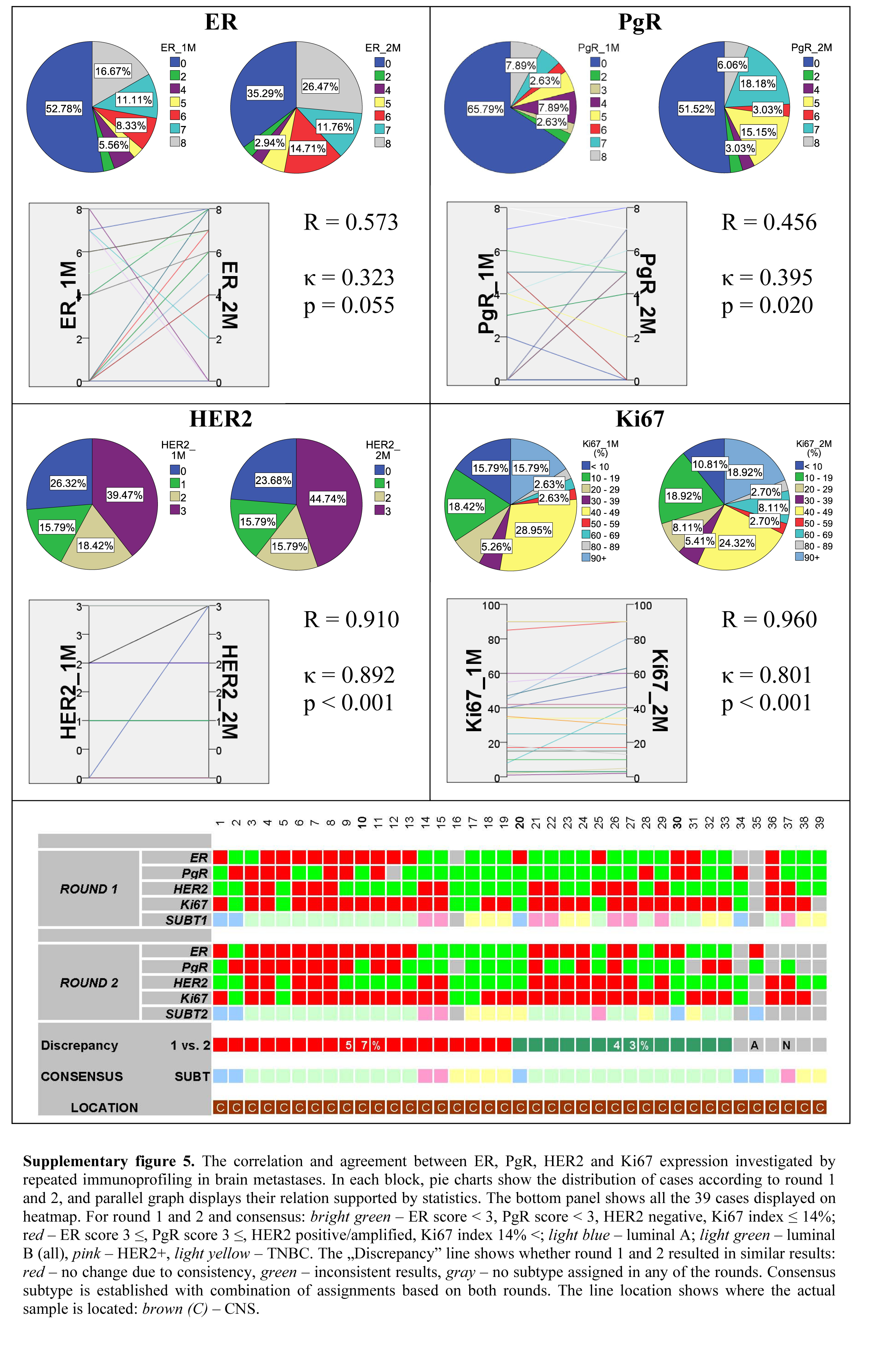
Supplementary Material
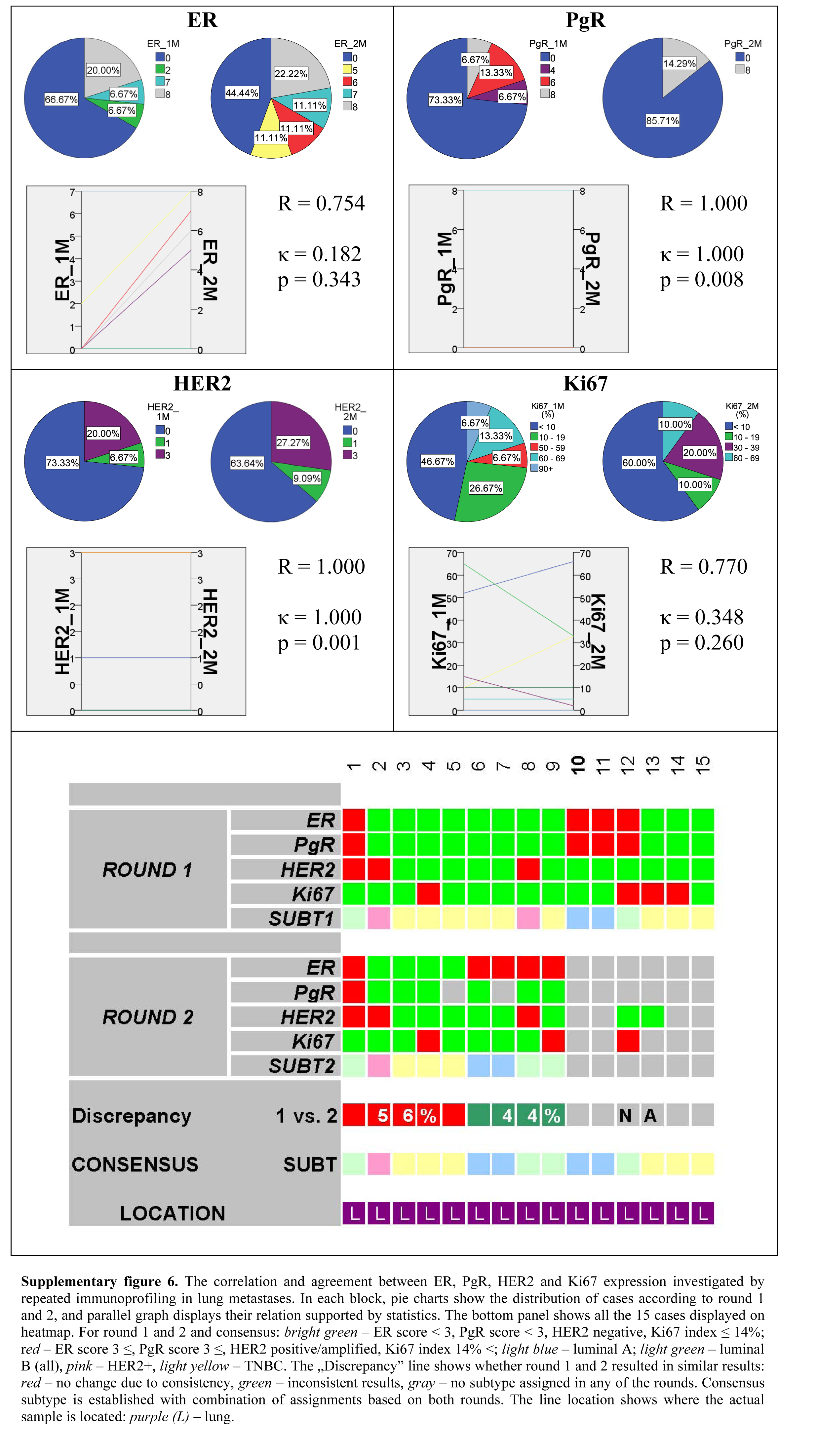
Supplementary Material
Acknowledgments
We are grateful for all colleagues who let us have the various samples for the study: Meggyesházi N, Kas J, Harsányi L, Baranyai Zs, Soltész I, Bálint KE. We thank Erzsébet Azumah, Csilla Jaczó, Magdolna Pekár and Erika Samodai for their tremendous technical assistance. We also express our special thanks to József Tímár and Lajos Pusztai for sharing their comments.
Footnotes
Author Contributions: Conception and design: JK, AMS; financial support: JK, MS, AMS; sample acquisition: LVL, EV, ET, JF, ZH, MS, AMS; data acquisition: LVL, ET, JF, ZH, MS, AMS; immunohistochemical work-up: AMT, BS, AMS; FISH evaluation: AMS, BS; analysis and data interpretation: JK, AMS, BS, BG; writing of manuscript: JK, AMS, BS, BG.
Competing Interests: The authors declared no potential competing interests with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Legacy of Szőllős András Péterné, the MKOT-Roche-2008, the MKOT-GSK-2010, the TÁMOP 4.2.1.B-09/1/KMR-2010-0001 and the TÁMOP 4.2.2/B-10/1-2010-0013 grant. The research project was supported by the European Union and Hungary, co-financed by the European Social Fund under the identification number TÁMOP 4.2.4.A/2-11-1-2012-0001 implemented within the framework of the “National Excellence Program - Development and operation of support system for domestic students and scientists’ convergence program”. This paper was also supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences, MKOT 2014-2016, SE-OTKA 2014-2015, KTIA_NAP_13-2014-0021, and OTKA-K-116151 grants.
Research Material Access Statement: Digital slides are available on request: cac@korb2.sote.hu
References
- Aitken SJ, Thomas JS, Langdon SP, Harrison DJ, Faratian D. (2010). Quantitative analysis of changes in ER, PR and HER2 expression in primary breast cancer and paired nodal metastases. Ann Oncol 21:1254-1261. [DOI] [PubMed] [Google Scholar]
- Allred D, Harvey J, Berardo M, Clark G. (1998). Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol 11:155-168. [PubMed] [Google Scholar]
- Andersen J, Poulsen HS. (1988). Relationship between estrogen receptor status in the primary tumor and its regional and distant metastases. An immunohistochemical study in human breast cancer. Acta Oncol 27:761-765. [DOI] [PubMed] [Google Scholar]
- Aurilio G, Monfardini L, Rizzo S, Sciandivasci A, Preda L, Bagnardi V, Disalvatore D, Pruneri G, Munzone E, Della Vigna P, Renne G, Bellomi M, Curigliano G, Goldhirsch A, Nole F. (2013). Discordant hormone receptor and human epidermal growth factor receptor 2 status in bone metastases compared to primary breast cancer. Acta Oncol 52:1649-1656. [DOI] [PubMed] [Google Scholar]
- Aversa C, Geuna E, Martinello R, Milani A, Rossi V, Redana S, Valabrega G, Aglietta M, Montemurro F. (2013). Metastatic breast cancer subtypes and central nervous system metastases. J Clin Oncol 31:e11581. [DOI] [PubMed] [Google Scholar]
- Bernards R, Weinberg RA. (2002). Metastasis genes: A progression puzzle. Nature 418:823-823. [DOI] [PubMed] [Google Scholar]
- Blake ML, Tometsko M, Miller R, Jones JC, Dougall WC. (2014). RANK expression on breast cancer cells promotes skeletal metastasis. Clin Exp Metastasis 31:233-245. [DOI] [PubMed] [Google Scholar]
- Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massague J. (2009). Genes that mediate breast cancer metastasis to the brain. Nature 459:1005-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brastianos PK, Carter SL, Santagata S, Cahill DP, Taylor-Weiner A, Jones RT, Van Allen EM, Lawrence MS, Horowitz PM, Cibulskis K, Ligon KL, Tabernero J, Seoane J, Martinez-Saez E, Curry WT, Dunn IF, Paek SH, Park SH, McKenna A, Chevalier A, Rosenberg M, Barker FG, 2nd, Gill CM, Van Hummelen P, Thorner AR, Johnson BE, Hoang MP, Choueiri TK, Signoretti S, Sougnez C, Rabin MS, Lin NU, Winer EP, Stemmer-Rachamimov A, Meyerson M, Garraway L, Gabriel S, Lander ES, Beroukhim R, Batchelor TT, Baselga J, Louis DN, Getz G, Hahn WC. (2015). Genomic Characterization of Brain Metastases Reveals Branched Evolution and Potential Therapeutic Targets. Cancer Discov 5:1164-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings MC, Simpson PT, Reid LE, Jayanthan J, Skerman J, Song S, McCart Reed AE, Kutasovic JR, Morey AL, Marquart L, O’Rourke P, Lakhani SR. (2013). Metastatic progression of breast cancer: insights from 50 years of autopsies. J Pathol 232:23-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, Abbott RM, Hoog J, Dooling DJ, Koboldt DC, Schmidt H, Kalicki J, Zhang Q, Chen L, Lin L, Wendl MC, McMichael JF, Magrini VJ, Cook L, McGrath SD, Vickery TL, Appelbaum E, Deschryver K, Davies S, Guintoli T, Lin L, Crowder R, Tao Y, Snider JE, Smith SM, Dukes AF, Sanderson GE, Pohl CS, Delehaunty KD, Fronick CC, Pape KA, Reed JS, Robinson JS, Hodges JS, Schierding W, Dees ND, Shen D, Locke DP, Wiechert ME, Eldred JM, Peck JB, Oberkfell BJ, Lolofie JT, Du F, Hawkins AE, O’Laughlin MD, Bernard KE, Cunningham M, Elliott G, Mason MD, Thompson DM, Jr., Ivanovich JL, Goodfellow PJ, Perou CM, Weinstock GM, Aft R, Watson M, Ley TJ, Wilson RK, Mardis ER. (2010). Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464:999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsche FR, Bode PK, Moch H, Kristiansen G, Varga Z, Bode B. (2010). Determination of the Her-2/neu gene amplification status in cytologic breast cancer specimens using automated silver-enhanced in-situ hybridization (SISH). Am J Surg Pathol 34:1180-1185. [DOI] [PubMed] [Google Scholar]
- Guarneri V, Giovannelli S, Ficarra G, Bettelli S, Maiorana A, Piacentini F, Barbieri E, Dieci MV, D’Amico R, Jovic G, Conte P. (2008). Comparison of HER-2 and hormone receptor expression in primary breast cancers and asynchronous paired metastases: impact on patient management. Oncologist 13:838-844. [DOI] [PubMed] [Google Scholar]
- Guiu S, Michiels S, André F, Cortes J, Denkert C, Di Leo A, Hennessy BT, Sorlie T, Sotiriou C, Turner N, Van de Vijver M, Viale G, Loi S, Reis-Filho JS. (2012). Molecular subclasses of breast cancer: how do we define them? The IMPAKT 2012 Working Group Statement. Annals of Oncology 23:2997-3006. [DOI] [PubMed] [Google Scholar]
- Hahnel R, Twaddle E. (1985). The relationship between estrogen receptors in primary and secondary breast carcinomas and in sequential primary breast carcinomas. Breast Cancer Res Treat 5:155-163. [DOI] [PubMed] [Google Scholar]
- Hilton JF, Amir E, Hopkins S, Nabavi M, DiPrimio G, Sheikh A, Done SJ, Gianfelice D, Kanji F, Dent S, Barth D, Bouganim N, Al-Najjar A, Clemons M. (2011). Acquisition of metastatic tissue from patients with bone metastases from breast cancer. Breast Cancer Res Treat 129:761-765. [DOI] [PubMed] [Google Scholar]
- Hoefnagel LDC, Moelans CB, Meijer SL, van Slooten H-J, Wesseling P, Wesseling J, Westenend PJ, Bart J, Seldenrijk CA, Nagtegaal ID, Oudejans J, van der Valk P, van Gils CH, van der Wall E, van Diest PJ. (2012). Prognostic value of estrogen receptor α and progesterone receptor conversion in distant breast cancer metastases. Cancer 118:4929-4935. [DOI] [PubMed] [Google Scholar]
- Holdaway IM, Bowditch JV. (1983). Variation in receptor status between primary and metastatic breast cancer. Cancer 52:479-485. [DOI] [PubMed] [Google Scholar]
- Idirisinghe PK, Thike AA, Cheok PY, Tse GM, Lui PC, Fook-Chong S, Wong NS, Tan PH. (2010). Hormone receptor and c-ERBB2 status in distant metastatic and locally recurrent breast cancer. Pathologic correlations and clinical significance. Am J Clin Pathol 133:416-429. [DOI] [PubMed] [Google Scholar]
- Jakesz R, Dittrich C, Hanusch J, Kolb R, Lenzhofer R, Moser K, Rainer H, Reiner G, Schemper M, Spona J. (1985). Simultaneous and sequential determinations of steroid hormone receptors in human breast cancer. Influence of intervening therapy. Ann Surg 201:305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinga K, Kaufmann M, Runnebaum B, Kubli F. (1982). Distribution of estrogen and progesterone receptors on primary tumor and lymph nodes in individual patients with breast cancer. Oncology 39:337-339. [DOI] [PubMed] [Google Scholar]
- Kuukasjarvi T, Karhu R, Tanner M, Kahkonen M, Schaffer A, Nupponen N, Pennanen S, Kallioniemi A, Kallioniemi OP, Isola J. (1997). Genetic heterogeneity and clonal evolution underlying development of asynchronous metastasis in human breast cancer. Cancer Res 57:1597-1604. [PubMed] [Google Scholar]
- Li BD, Byskosh A, Molteni A, Duda RB. (1994). Estrogen and progesterone receptor concordance between primary and recurrent breast cancer. J Surg Oncol 57:71-77. [DOI] [PubMed] [Google Scholar]
- Liedtke C, Broglio K, Moulder S, Hsu L, Kau SW, Symmans WF, Albarracin C, Meric-Bernstam F, Woodward W, Theriault RL, Kiesel L, Hortobagyi GN, Pusztai L, Gonzalez-Angulo AM. (2009). Prognostic impact of discordance between triple-receptor measurements in primary and recurrent breast cancer. Ann Oncol 20:1953-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin NU, Amiri-Kordestani L, Palmieri D, Liewehr DJ, Steeg PS. (2013). CNS metastases in breast cancer: old challenge, new frontiers. Clin Cancer Res 19:6404-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström LS, Karlsson E, Wilking UM, Johansson U, Hartman J, Lidbrink EK, Hatschek T, Skoog L, Bergh J. (2012). Clinically Used Breast Cancer Markers Such As Estrogen Receptor, Progesterone Receptor, and Human Epidermal Growth Factor Receptor 2 Are Unstable Throughout Tumor Progression. Journal of Clinical Oncology 30:2601-2608. [DOI] [PubMed] [Google Scholar]
- Lower EE, Glass EL, Bradley DA, Blau R, Heffelfinger S. (2005). Impact of metastatic estrogen receptor and progesterone receptor status on survival. Breast Cancer Res Treat 90:65-70. [DOI] [PubMed] [Google Scholar]
- Macfarlane R, Seal M, Speers C, Woods R, Masoudi H, Aparicio S, Chia SK. (2012). Molecular Alterations Between the Primary Breast Cancer and the Subsequent Locoregional/Metastatic Tumor. The Oncologist 17:172-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric-Bernstam F, Frampton GM, Ferrer-Lozano J, Yelensky R, Perez-Fidalgo JA, Wang Y, Palmer GA, Ross JS, Miller VA, Su X, Eroles P, Barrera JA, Burgues O, Lluch AM, Zheng X, Sahin A, Stephens PJ, Mills GB, Cronin MT, Gonzalez-Angulo AM. (2014). Concordance of genomic alterations between primary and recurrent breast cancer. Mol Cancer Ther 13:1382-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger-Filho O, Sun Z, Viale G, Price KN, Crivellari D, Snyder RD, Gelber RD, Castiglione-Gertsch M, Coates AS, Goldhirsch A, Cardoso F. (2013). Patterns of Recurrence and Outcome According to Breast Cancer Subtypes in Lymph Node-Negative Disease: Results From International Breast Cancer Study Group Trials VIII and IX. J Clin Oncol 31:3083-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K, Ibrahim M, Barnett S, Jasani B. (2007). Technical aspects of predictive and prognostic markers in breast cancer: What UK NEQAS data shows. Curr Diagn Pathol 13:135-149. [Google Scholar]
- Moelans CB, van der Groep P, Hoefnagel LD, van de, Vijver MJ, Wesseling P, Wesseling J, van der Wall E, van Diest PJ. (2014). Genomic evolution from primary breast carcinoma to distant metastasis: Few copy number changes of breast cancer related genes. Cancer Lett 344:138-146. [DOI] [PubMed] [Google Scholar]
- Osborne CK. (1985). Heterogeneity in hormone receptor status in primary and metastatic breast cancer. Semin Oncol 12:317-326. [PubMed] [Google Scholar]
- Paget S. (1889). The distribution of secondary growths in cancer of the breast. LANCET 133:571-573. [PubMed] [Google Scholar]
- Pusztai L, Viale G, Kelly CM, Hudis CA. (2010). Estrogen and HER-2 Receptor Discordance Between Primary Breast Cancer and Metastasis. Oncologist 15:1164-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quayle L, Ottewell PD, Holen I. (2015). Bone Metastasis: Molecular Mechanisms Implicated in Tumour Cell Dormancy in Breast and Prostate Cancer. Curr Cancer Drug Targets 15:469-480. [DOI] [PubMed] [Google Scholar]
- Rothe F, Laes JF, Lambrechts D, Smeets D, Vincent D, Maetens M, Fumagalli D, Michiels S, Drisis S, Moerman C, Detiffe JP, Larsimont D, Awada A, Piccart M, Sotiriou C, Ignatiadis M. (2014). Plasma circulating tumor DNA as an alternative to metastatic biopsies for mutational analysis in breast cancer. Ann Oncol 25:1959-1965. [DOI] [PubMed] [Google Scholar]
- Shen Q, Sahin AA, Hess KR, Suki D, Aldape KD, Sawaya R, Ibrahim NK. (2015). Breast cancer with brain metastases: clinicopathologic features, survival, and paired biomarker analysis. Oncologist 20:466-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siclari VA, Mohammad KS, Tompkins DR, Davis H, McKenna CR, Peng X, Wessner LL, Niewolna M, Guise TA, Suvannasankha A, Chirgwin JM. (2014). Tumor-expressed adrenomedullin accelerates breast cancer bone metastasis. Breast Cancer Res 16:458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons C, Miller N, Geddie W, Gianfelice D, Oldfield M, Dranitsaris G, Clemons MJ. (2009). Does confirmatory tumor biopsy alter the management of breast cancer patients with distant metastases? Ann Oncol 20:1499-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Nocito A, Hubscher T, Bucher C, Torhorst J, Schraml P, Bubendorf L, Mihatsch MM, Moch H, Wilber K, Schotzau A, Kononen J, Sauter G. (2001). Patterns of her-2/neu amplification and overexpression in primary and metastatic breast cancer. J Natl Cancer Inst 93:1141-1146. [DOI] [PubMed] [Google Scholar]
- Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JG, Foekens JA, Martens JW. (2008). Subtypes of breast cancer show preferential site of relapse. Cancer Res 68:3108-3114. [DOI] [PubMed] [Google Scholar]
- Spataro V, Price K, Goldhirsch A, Cavalli F, Simoncini E, Castiglione M, Rudenstam CM, Collins J, Lindtner J, Gelber RD. (1992). Sequential estrogen receptor determinations from primary breast cancer and at relapse: prognostic and therapeutic relevance. The International Breast Cancer Study Group (formerly Ludwig Group). Ann Oncol 3:733-740. [DOI] [PubMed] [Google Scholar]
- Van Poznak C, Somerfield MR, Bast RC, Cristofanilli M, Goetz MP, Gonzalez-Angulo AM, Hicks DG, Hill EG, Liu MC, Lucas W, Mayer IA, Mennel RG, Symmans WF, Hayes DF, Harris LN. (2015). Use of Biomarkers to Guide Decisions on Systemic Therapy for Women With Metastatic Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol 33:2695-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Reeves KJ, Brown HK, Fowles AC, Docherty FE, Ottewell PD, Croucher PI, Holen I, Eaton CL. (2015). The frequency of osteolytic bone metastasis is determined by conditions of the soil, not the number of seeds; evidence from in vivo models of breast and prostate cancer. J Exp Clin Cancer Res 34:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchi M, Confalonieri S, Nuciforo P, Vigano MA, Capra M, Bianchi M, Nicosia D, Bianchi F, Galimberti V, Viale G, Palermo G, Riccardi A, Campanini R, Daidone MG, Pierotti MA, Pece S, Di Fiore PP. (2007). Breast cancer metastases are molecularly distinct from their primary tumors. Oncogene 27:2148-2158. [DOI] [PubMed] [Google Scholar]
- Weigelt B, Glas AM, Wessels LF, Witteveen AT, Peterse JL, van’t Veer LJ. (2003). Gene expression profiles of primary breast tumors maintained in distant metastases. Proc Natl Acad Sci U S A 100:15901-15905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilking U, Karlsson E, Skoog L, Hatschek T, Lidbrink E, Elmberger G, Johansson H, Lindstrom L, Bergh J. (2011). HER2 status in a population-derived breast cancer cohort: discordances during tumor progression. Breast Cancer Res Treat 125:553-561. [DOI] [PubMed] [Google Scholar]
- Wolff AC, Hammond MEH, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM, Hayes DF. (2006). American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations for Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer. J Clin Oncol 25:118-145. [DOI] [PubMed] [Google Scholar]
- Yang YF, Liao YY, Yang M, Peng NF, Xie SR, Xie YF. (2014). Discordances in ER, PR and HER2 receptors between primary and recurrent/metastatic lesions and their impact on survival in breast cancer patients. Med Oncol 31:214. [DOI] [PubMed] [Google Scholar]
- Zheng WQ, Lu J, Zheng JM, Hu FX, Ni CR. (2001). Variation of ER status between primary and metastatic breast cancer and relationship to p53 expression*. Steroids 66:905-910. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials