Abstract
The tumor suppressor gene TP53 is the most frequently mutated gene in human cancer; this gene is subject to inactivation by mutation or deletion in >50% of sporadic cancers. Genes that encode proteins that regulate p53 function, such as MDM2, MDM4, and CDKN2A (p14ARF) are also frequently altered in tumors, and it is generally believed that the p53 pathway is likely to be inactivated by mutation in close to 100% of human tumors. Unlike most other cancer-relevant signaling pathways, some of the genes in the p53 pathway contain functionally significant single nucleotide polymorphisms (SNPs) that alter the amplitude of signaling by this protein. These variants, thus, have the potential to impact cancer risk, progression, and the efficacy of radiation and chemotherapy. In addition, the p53 pathway plays a role in other biological processes, including metabolism and reproductive fitness, so these variants have the potential to modify other diseases as well. Here we have chosen five polymorphisms in three genes in the p53 pathway for review, two in TP53, two in MDM2, and one in MDM4. These five variants were selected based on the quality and reproducibility of functional data associated with them, as well as the convincingness of epidemiological data in support of their association with disease. We also highlight two other polymorphisms that may affect p53 signaling, but for which functional or association data are still forthcoming (KITLG and ANRIL). Finally, we touch on three questions regarding genetic modifiers of the p53 pathway: Why did these variants arise? Were they under selection pressure? And, is there compelling evidence to support genotyping these variants to better predict disease risk and prognosis?
Some genes in the p53 pathway (e.g., MDM2, MDM4, and TP53) contain single nucleotide polymorphisms that alter signaling by the p53 protein. These may impact cancer risk, progression, and treatment.
With just over three billion base pairs in the human genome, there are an estimated 20,000–25,000 genes. Current estimates indicate that there may be single nucleotide polymorphism (SNP) approximately every 1000 base pairs in the human genome. Therefore, there are an estimated three million SNPs in the human population, and these have the potential to contribute to genetic variation, disease susceptibility, and the course of disease. In the past decade, there has been growing interest in performing genome-wide association studies (GWAS) to link various SNPs to human disease, often in the absence of any functional data on the variants. Despite the investment of considerable resources, to date only a modest number of GWAS analyses have revealed compelling associations between genetic polymorphisms and cancer, and even fewer have provided a functional basis for that association. Additionally, only rarely has information from GWAS association analyses made it into clinical awareness or impacted the course of treatment. However, as summarized herein, the data at present suggest that there may be compelling evidence to support genotyping of combined p53 pathway variants to better predict disease risk and prognosis.
The TP53 gene is unusual in a number of respects. First, there are a larger than normal number of SNPs in TP53, with 85 polymorphisms identified within 19 kilobases of DNA (Petitjean et al. 2007). Second, unlike the overwhelming majority of tumor suppressor genes and oncogenes, the TP53 gene is distinguished by the presence of three nonsynonymous polymorphisms that alter the amino acid sequence of the protein. Indeed, for two of these coding region SNPs, there are solid data that the amino acid change alters p53 function. Additionally, the p53 pathway is multifaceted, and although this protein has an obvious cancer-critical role in tumor suppression, this protein also regulates metabolism, redox state, and reproduction, among other functions (Vousden and Prives 2009). Therefore, some SNPs in the p53 pathway are predicted or reported to have relevance to immune, reproductive, and metabolic function. In this review, we focus on five SNPs in the p53 pathway; for more comprehensive treatment of other polymorphic loci in the p53 pathway, readers are referred to several excellent reviews (Pietsch et al. 2006; Whibley et al. 2009; Grochola et al. 2010; Denisov et al. 2012). Of the five variants discussed, two are coding region variants in TP53, two occur in the promoter of MDM2, and one is a polymorphism in the 3′ untranslated region of MDM4. There are some interesting differences in the frequencies of these SNPs in different populations; genotype frequencies for each polymorphism in different ethnic populations are provided in Table 1.
Table 1.
The allele frequencies of TP53 pathway variants
| p53 P72R, rsl042522 | |||||
| C | G | C | G | ||
| CEU CHB JPT YRI |
0.233 0.489 0.409 0.669 |
0.767 0.511 0.591 0.331 |
African Americans (AA) Caucasian |
0.59 0.26 |
0.41 0.74 |
|
p53 P47S, rsl800371 | |||||
| G | A | G | A | ||
| CEU CHB JPT YRI |
1 1 1 0.942 |
0 0 0 0.058 |
African Americans (AA) Caucasian |
0.975 1 |
0.025 0 |
|
MDM2 (SNP309), rs2279744 | |||||
| G | T | G | T | ||
| CEU CHB + JPT YRI |
0.3 0.642 0.085 |
0.7 0.358 0.915 |
African Americans (AA) Caucasian |
0.1 0.43 |
0.9 0.57 |
|
MDM2 (SNP285), rs117039649 | |||||
| G | C | G | C | ||
| CEU CHB JPT YRI |
0.97 1 1 1 |
0.03 0 0 0 |
African Americans (AA) Caucasian |
1 0.94 |
0 0.06 |
|
MDM4 3′ UTR (SNP 34091), rs4245739 | |||||
| C | A | C | A | ||
| CEU CHB JPT YRI |
0.31 0.05 0.06 0.19 |
0.69 0.95 0.94 0.81 |
African Americans (AA) Caucasian |
0.27 0.277 |
0.73 0.723 |
The combined data were collected from the 1000 Genomes database (www.1000genomes.org, www.1000genomes.org) as well as SNPedia (www.SNPedia.com, www.SNPedia.com).
CEU (Utah residents with Northern and Western European ancestry), CHB (Han Chinese in Beijing, China), JPT (Japanese in Tokyo, Japan), and YRI (Yoruba in Ibadan, Nigeria) populations. The frequency of each allele in African Americans (AA) and Caucasian Americans is shown on the right. The risk allele is shown in bold.
TP53: PRO72ARG (RS1042522)
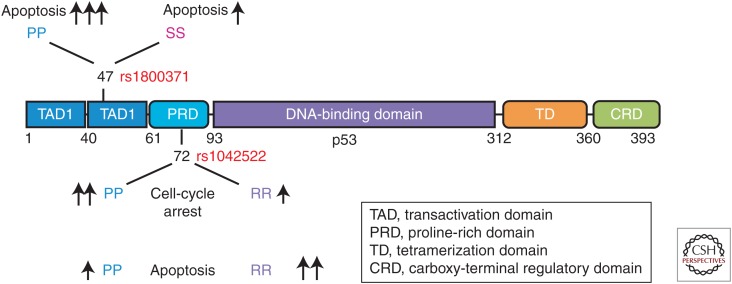
A common polymorphism at codon 72 of p53 (rs1042522, C>G) is one of the most widely studied variations in TP53 (Fig. 1). The different alleles in codon 72 of p53 encode either proline (P72) or arginine (R72), and this SNP gives rise to variants of distinct electrophoretic mobility, suggestive of a structural change in the protein (Harris et al. 1986; Matlashewski et al. 1987). The codon 72 SNP is located in exon 4, in a segment of TP53 that is referred to as the proline-rich domain (PRD) of p53, which is fundamental for apoptosis induction in response to stress (Walker and Levine 1996; Sakamuro et al. 1997; Roth et al. 2000; Toledo et al. 2007). Data from several laboratories indicate that the codon 72 SNP modifies the function of p53. Three groups have reported that in response to DNA damage, the P72 variant of p53 shows a greater ability to induce growth arrest and senescence, and to transactivate the cell-cycle regulator CDKN1A (p21/waf1) compared to R72 (Bonafe et al. 2002, 2004; Pim and Banks 2004; Azzam et al. 2011; Frank et al. 2011). Chromatin immunoprecipitation analyses indicated that the increased transactivation of CDKN1A was because of enhanced binding of the P72 variant to the p53 response elements in this gene (Frank et al. 2011). This increased ability of P72 to induce growth arrest and senescence is believed to underlie the association of P72 with longevity, as seen in two independent studies (Gaspari et al. 2003; Bojesen and Nordestgaard 2008).
Figure 1.
The functional domains of p53, and the localization of coding region SNPs. The SNPs rs1800371 (P47S) and rs1042522 (P72R) are localized in the transactivation domain 2 (TAD2) and proline-rich domain (PRD), respectively. The arrows depict the levels of apoptosis and cell-cycle arrest induced by each variant.
The R72 variant of p53 has been shown to possess superior ability to induce programmed cell death. This is due both to increased ability of this protein variant to traffic to the mitochondria and engage BCL2 family members, as well as to increased ability of R72 to transactivate a subset of proapoptotic p53 target genes (Dumont et al. 2003; Bonafe et al. 2004; Pim and Banks 2004; Sullivan et al. 2004; Jeong et al. 2010; Frank et al. 2011; Kung et al. 2015). Several microarray analyses have now been performed on matched cells that differ in their codon 72 status. Each of these studies has reported consistent and reproducible differences in gene expression between P72 and R72 variants. The Murphy laboratory created humanized p53 knockin mice containing the P72 and R72 alleles, and analyzed the transcriptional profiles of sibling littermate mice whose thymocytes were isolated after γ radiation. This group found that of >500 genes induced in a p53-dependent manner, there were less than two dozen that were differentially regulated by the codon 72 variants, with a cutoff for differences of 1.5-fold or greater. Interestingly, most of these genes were preferentially transactivated by the P72 variant, and they were also coordinately regulated by p53 and NF-κB. This group went on to show that increased binding of the P72 variant to the p65 subunit of NF-κB explains the increased transactivation of this subset of genes (Frank et al. 2011). The Buckhaults group recently performed RNA Seq on the human colon carcinoma line RKO, which is normally heterozygous P72/R72, but for which they generated clones in which one allele or the other was knocked out by somatic cell recombination (Weige et al. 2014). This group identified 22 genes that were significantly differentially regulated by the P72 and R72 variants in response to etoposide (p < 0.01). Unlike the data obtained by the Murphy group in mouse thymocytes, the majority of these genes were transactivated to a greater extent by the R72 variant; it is possible that this reflected differences in cell type and the genotoxic stresses used. The Buckhaults group identified the gene encoding the β-subunit of PRDM1, a transcriptional repressor, as one that was up-regulated to significantly higher levels in two different R72 clones, compared to P72. Interestingly, this group also identified this gene as one whose silencing conferred enhanced survival following treatment with nutlin-3a. PRDM1 is, thus, a gene that underlies part of the enhanced apoptotic potential of the R72 variant. More recently, the Murphy group performed microarray analysis of normal human fibroblasts that were homozygous P72 or R72. This group identified the SUMO-ligase TRIML2 as an R72-enhanced transcript. Interestingly, this group also identified TRIML2 as a p53-binding protein, and interaction with TRIML2 was found to enhance SUMOylation of p53 with SUMO-2, allowing this protein to have enhanced ability to transactivate a subset of p53 target genes like PIDD, PIG3 (TP53I3), and PIG6 (PRODH) (Kung et al. 2015). The combined data suggest that P72 and R72 variants possess altered transcriptional and apoptotic potential owing to multiple mechanisms: altered ability to bind to p53 target promoters, as well as altered ability to bind to transcriptional regulators.
Despite strong data indicating that the codon 72 polymorphism affects the growth arrest versus cell death decision in cells, persuasive data linking these variants to cancer risk, prognosis, or the efficacy of therapy are lacking. One could argue that the inconclusive nature of the association studies is attributable to a variety of reasons, such as the failure to account for the presence of p53 mutations and inaccuracy of genotyping (the current genotyping primers for this “G” to “C” change actually sequence the reverse strand, so the G allele is actually the P72 variant, and often gets miscoded as R72). However, it must be noted that GWAS have failed to identify the codon 72 SNP as a risk factor in cancer. Interestingly, given p53’s role in metabolism, GWAS have positively linked the codon 72 SNP to body-mass index (BMI)/obesity (p < 0.005), and to log 10 fibrinogen levels (p < 0.006) (Parry et al. 2004; Kengne et al. 2013). Both BMI and fibrinogen levels are strongly associated with metabolic disorders, such as coronary heart disease and type-II diabetes, thus raising the possibility that codon 72 influences the role of p53 in metabolism. In support of this, several studies have linked the R72 allele of TP53 with the risk of developing type-II diabetes (Gaulton et al. 2008; Burgdorf et al. 2011; Bonfigli et al. 2013). Further analysis of the impact of codon 72 variants on metabolic disease is thus warranted.
Before discounting a role for the codon 72 polymorphism and cancer, mention should be made of a growing literature supporting an impact of codon 72 on the ability of mutant p53 protein to perform its “gain-of-function” activities. Mutations in the DNA-binding domain of p53 cause the protein to accumulate, often in a misfolded, denatured conformation. The mutant protein then binds to other proteins in the cell, often inactivating or altering their function, and hence behaving as a gain-of-function modifier. As two examples, mutant forms of p53 can bind and inactivate the p53 family member p73 (Marin et al. 2000) or bind and stabilize the sterol regulatory transcription factor SREBP1/2 (Freed-Pastor et al. 2012), thus leading to new properties of the cancer cell. Kaelin and colleagues were the first to report that certain tumor-derived mutant forms of p53, particularly those known to have a “denatured” conformation, were more apt to bind and inactivate p73 when they encoded R72. In a small sample set of squamous cell cancers from individuals heterozygous for codon 72, this group also reported that the overwhelming majority of tumors lost the P72 allele and retained the mutated R72 protein (Marin et al. 2000). Crook and colleagues confirmed these results in an independent study, and showed that individuals with mutations in the R72 allele of p53 had markedly poorer survival for head and neck cancer, compared to P72 (Bergamaschi et al. 2003). Moreover, several other groups have shown that in heterozygous P72/R72 individuals, there is much more frequent loss of the P72 allele and retention of the mutant R72 allele in cancers of the breast, head and neck, urinary tract, and colon (Tada et al. 2001; Bonafe et al. 2003; Schneider-Stock et al. 2004; Siddique et al. 2005). Although these data offer strong evidence for a role of the codon 72 polymorphism of p53 in cancer, they suggest that this association should be analyzed only in tumors that have mutant p53.
TP53: PRO47SER (RS1800371)
In 1993, Felley-Bosco and Harris uncovered a rare germ-line polymorphism in codon 47 of the p53 gene that replaces the wild-type proline residue (CCG) with a serine (TCG) (Fig. 1). At the time, functional assays for p53 were limiting, and the group performed only two assays for p53 function: colony suppression and transient transcription assays using a reporter gene. Using these assays, these investigators were unable to detect functional differences between WT p53 and the S47 variant. Interestingly, this group did note an unusual ethnic bias of the variant, and reported the T allele frequency to be 4%–5%, but only in African-American populations and not Caucasians (Felley-Bosco et al. 1993). This ethnic bias has been seen in larger sample sizes, and present data indicate that the S47 variant is present at a low allele frequency in African Americans (∼1.5%–2%) and Africans (5%–6%), but has not been detected in >8600 Caucasian Americans (www.1000 Genomes .org).
Several years after the discovery of the S47 allele, seminal work by the Fornace and Taya groups showed that serine 46 phosphorylation of p53 was a critical modification for p53-mediated apoptosis (Bulavin et al. 1999; Oda et al. 2000). The Murphy group first noted that the S47 variant replaced the proline necessary for phosphorylation of serine 46 by proline-directed kinases, and this group used in vitro kinase assays to show that the S47 variant could not be phosphorylated on serine 46 by purified p38MAPK. Further, they showed that inducible cell lines containing the S47 variant were impaired for apoptosis, relative to WT p53 (Li et al. 2005). However, these studies relied on inducible versions of WT p53 and S47 in engineered cell lines, as opposed to endogenous p53. Further, at the time, large cohorts of DNA samples from African Americans were not available, so the impact of the S47 variant on cancer risk was not assessed.
In 2015, the Murphy group generated a mouse model for the S47 polymorphism, and surprisingly found that mice expressing the S47 variant, in either homozygous or heterozygous form, were markedly susceptible to cancer. Reminiscent of mouse models for Li–Fraumeni syndrome generated by Lozano and Jacks (Lang et al. 2004; Olive et al. 2004), the S47 mouse did not develop the prevalent T-cell lymphoma and sarcoma seen in homozygous p53 knockout mice, but rather developed more epithelial tumor types, including intestinal and pancreatic adenocarcinoma. Interestingly, gene expression analysis of human and murine cells containing WT p53 and the S47 variant revealed that the majority of p53 target genes were transactivated identically. In fact, only three p53 target genes differed following cisplatin treatment: one of these is implicated in apoptosis (PMAIP1 or NOXA) and two are implicated in metabolism (SCO2 and GLS2). This group found that the S47 variant was associated with markedly impaired apoptosis in response to several genotoxic stresses but particularly cisplatin, and further that the heterozygous S47/WT cells showed markedly impaired apoptosis, similar to the homozygous S47/S47 variant. This finding prompted these researchers to analyze breast cancer samples from African-American women, and a statistically significant association between S47 and premenopausal breast cancer in African-American women was observed (Jennis et al. 2015). These findings suggest that the S47 variant may play a role in the increased risk breast and other cancers in African Americans.
MDM2: SNP309T>G (RS2279744) AND SNP285G>C (RS117039649)
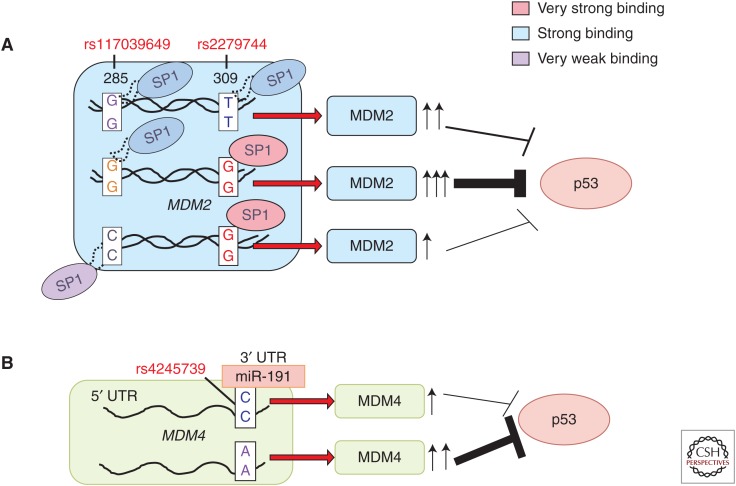
Among all proteins in biology and cancer whose function is to regulate the p53 tumor suppressor protein, MDM2 seems by far to be the most critical. As evidence of this fact, the genetic knockout of MDM2 leads to early embryonic lethality, but this lethality is rescued by concomitant knockout of p53 (Montes de Oca Luna et al. 1995). In an analysis of the DNA damage response in >50 human lymphocyte cell lines, the Levine group first noticed a defined variability in p53 response, suggestive of a genetic component influencing p53’s response to DNA damage (Harris et al. 2005). Analysis of the sequence of the MDM2 gene revealed the presence of a SNP located within the second (P2) promoter of MDM2 that tracked with the p53 apoptotic response. This SNP, a T to G change at position 309 of intron 1, was found to create an Sp1 transcription factor-binding site, leading to an average of an eightfold increased expression of MDM2, along with an attenuation of p53 signaling in cells stressed with DNA-damaging agents (Fig. 2) (Bond et al. 2004). Notably, in families with Li–Fraumeni syndrome (germline mutations in p53), the presence of the G allele of SNP309 was associated with an average age of tumor development that was 9–12 years earlier, depending on tumor type. Moreover, Li–Fraumeni patients with two SNP309 G alleles were more frequently diagnosed with multiple primary tumors (Bougeard et al. 2006).
Figure 2.
(A) The impact of SNP309 and SNP285 on MDM2 levels. The 309 G variant creates a stronger binding site for the transcription factor Sp1, leading to increased MDM2 transcription, as compared to the T variant. The SNP285 C allele creates a weaker binding site for Sp1. The combined SNP285 G and SNP309 G alleles are predicted to lead to the greatest level of MDM2 transcription, and the greatest effect on the p53 pathway. (B) The impact of SNP34091 on MDM4 levels. The C allele in SNP34091 creates a binding site for microRNA miR-191 in the 3′ UTR, leading to decreased mRNA and translation of MDM4. The A allele produces increased MDM4, and thus is the risk allele, owing to dampening of the p53 pathway.
Shortly after the study by the Levine group, numerous investigations were published supporting a role for SNP309 in cancer risk, progression, and age of onset. As one example, the Mdm2 SNP309 T allele was found associated with earlier onset prostate cancer (p = 0.004), higher Gleason scores (p = 0.004), and higher stages in men undergoing a radical prostatectomy (p = 0.011) (Sun et al. 2010). Several other studies confirmed a significant association between SNP309 variants and cancer risk (Boersma et al. 2006; Bond et al. 2006; Phillips et al. 2010; Ma et al. 2013; Qin et al. 2013). However, a small number of studies failed to find evidence for an association between SNP309 and cancer (Schmidt et al. 2007; Zhuo et al. 2012). At least two reasons emerged for these early conflicting results. First, the Levine group discovered that the Sp1-binding site created by the SNP309 G allele cooperated with estrogen response elements in the promoter of MDM2; this led to the finding of hormone- and gender-specific associations between SNP309 and cancer (Bond et al. 2006). In fact, many of the most compelling associations between SNP309 and cancer risk have been for cancer in premenopausal women. Second, while studying SNP309 status in Norwegian breast cancer patients, the Lonning group discovered a second MDM2 polymorphism (rs117039649; SNP285G>C) located 24 base pairs upstream of SNP309, which eliminated an Sp1-binding site (Knappskog et al. 2011). In two separate studies, this group found that SNP285C reduced the risk for female malignancies (breast, ovarian, and endometrial cancer) (Knappskog et al. 2012; Gansmo et al. 2015). This group hypothesized that SNP285C may counteract the impact of SNP309G in the small percentage of western Caucasians in which these SNPs co-occur (Knappskog and Lonning 2011). This hypothesis remains to be tested.
The most compelling role for SNP309 and cancer risk came, interestingly enough, from a mouse model. In such inbred models, potentially confounding genetic modifiers of risk like SNP285C and other alleles as well as lifestyle, nutrition, and other factors are eliminated. As such, analyses in mouse models can provide a more unbiased assessment of risk association. Post and Lozano generated a humanized version of MDM2 in which mouse intron 1 was replaced by human intron 1, containing either the T or G alleles of SNP309. As in human, SNP309 G/G mice possessed between twofold and fourfold increased levels of MDM2 mRNA in multiple tissues, along with reduced p53 response following DNA damage. Notably, SNP309 G/G mice showed markedly higher incidence of spontaneous cancer, thereby implicating the G allele as a direct player in cancer initiation. Further, the Lozano group showed that, like the human studies on Li–Fraumeni syndrome, G/G mice had a markedly earlier age of onset for cancer (p = 0.0005). Additionally, more SNP309 G/G mice developed multiple primary tumors, again reminiscent of the data in humans (Post et al. 2010).
MDM4 SNP34091 (RS4245739)
The MDM4 oncoprotein cooperates with MDM2 to negatively regulate p53, and amplification of MDM4 is evident in a subset of human cancers. The current consensus is that MDM4 binds and inactivates p53, whereas MDM2 binds, inactivates, and targets p53 for degradation (Toledo et al. 2007). An A>C change exists in the 3′ UTR of MDM4 (rs4245739) (Fig. 2). This A>C change creates a microRNA-binding site for mIR-191, which is a microRNA that is commonly highly expressed in many tissues and tumors. The A allele of this polymorphism is resistant to mIR-191-mediated regulation, leading to higher levels of MDM4 protein (Wynendaele et al. 2010; Stegeman et al. 2015). As such, the A allele is considered the risk allele, and it is predicted to lead to increased MDM4 levels and presumably to reduced p53 function. The group of Bartel reported that the A allele is significantly more common in high-grade ovarian carcinomas compared to low-grade carcinomas; interestingly, as in the case with the MDM2 SNP309, there was a hormonal component, and tumors without expression of the estrogen receptor showed a more marked association (Wynendaele et al. 2010).
Unlike other p53 pathway SNPs, the MDM4 SNP34091 was identified in GWAS as an allele linked to susceptibility to cancer. Initial studies linked this polymorphism to prostate cancer. The significance of this association was very strong, p = 2.1 × 10−11, although the per allele odds ratio was modest (1.10; 95% CI: 1.05–1.14) (Eeles et al. 2013). It should be noted that SNP34091 is linked with several other MDM4 SNPs (Atwal et al. 2009), including at least one other SNP, rs1380576 (r2 = 0.89), which has been linked to prostate cancer aggressiveness (p = 0.047) (Sun et al. 2010). Therefore, it remains unclear which of these SNPs is the underlying cause for the association. For breast cancer, SNP34091 was identified by GWAS as one of four loci associated with estrogen receptor (ER)–negative (but not ER-positive) breast cancer (p = 2 × 10−12) (Garcia-Closas et al. 2013). A subsequent independent study confirmed the association between SNP34091 with breast cancer, again with the AA genotype as the risk genotype, with a possible additive effect with the codon 72 Pro/Pro allele (Liu et al. 2013). Therefore, there are compelling data to implicate SNP34091 in cancer risk. However, functional data linking this polymorphism to the amplitude of p53 signaling are lacking, and it remains unclear whether the cancer associations of this SNP are because of the impact on the p53 pathway.
OTHER POLYMORPHIC VARIANTS IN THE p53 PATHWAY
With the number of functionally significant polymorphisms in the TP53, MDM2, and MDM4 genes that alter the amplitude of signaling in this pathway, it becomes interesting to note the p53 pathway member that is conspicuously absent from this list, namely p14ARF, encoded by an alternate reading frame from the CDKN2A gene on chromosome 9p21. GWAS and epidemiological studies make no mention of polymorphisms in the CDKN2A gene that impact p14ARF function or cancer risk. However, closely linked to the CDKN2A locus, and transcribed in the opposite direction, is a long-noncoding RNA called ANRIL (antisense noncoding RNA in the INK4 locus, also called CDKN2B-AS). Long noncoding RNAs are well known to mediate gene expression by a variety of mechanisms, and often these RNAs regulate the genes that are neighboring to them. ANRIL has been shown to specifically bind to two polycomb proteins, CBX7 (PRC1) and SUZ12 (PRC2), and by virtue of this interaction, ANRIL affects the transcription of the CDKN2A locus (Yap et al. 2010; Aguilo et al. 2011; Kotake et al. 2011). Antisense silencing of ANRIL leads to increased expression of the CDKN2A locus, and to a shortening of the life span of normal human fibroblasts (Yap et al. 2010). Notably, numerous GWAS have linked polymorphisms in the ANRIL locus to the level of expression of this long noncoding RNA, and to cancer risk (for review, see Congrains et al. 2013). It is tempting to speculate that the impact of these SNPs on cancer risk is related to their ability to regulate p14ARF level and the p53 pathway, but this remains to be determined.
p53 binds to a consensus site composed of RRRCWWGYYY (n) RRRCWWGYYY, where R is purine, Y is pyrimidine, W is A or T, and (n) is a spacer of 0–13 base pairs. Recently, several groups have identified SNPs in the response elements of p53 target genes that alter the ability of p53 to bind and transactivate these genes (Tomso et al. 2005; Menendez et al. 2006; Bandele et al. 2011; Wang et al. 2014). Notably, a SNP in the p53 response element of the gene-encoding KIT ligand (KITLG) disrupts the G residue that exists in the core CWWG motif, and dramatically decreases the ability of p53 to bind and transactivate the promoter (Zeron-Medina et al. 2013). This SNP is notable because it resides in a region that has been highlighted by GWAS studies for association with cancer risk. For example, this SNP, rs4590952 (G/A), is in linkage disequilibrium with three other SNPs that are associated with risk for developing testicular cancer (Kanetsky et al. 2009; Rapley et al. 2009; Turnbull et al. 2010). Although the existent data are suggestive, at present, the broad relevance of this locus to cancer and the particular importance of KITLG remains to be determined.
CONCLUDING REMARKS
A number of intriguing questions remain regarding p53 pathway variants. Why did these variants arise? Were they selected for in response to environmental or other stresses? And, is it worthwhile to collect genotype data for these variants to predict cancer risk? Unfortunately, the answers for some of these questions are not yet clear. With regard to environmental selection, initial data from the analysis of the allele frequencies of the P72 and R72 variants in human populations showed a linear distribution of these alleles that stratifies with latitude and winter temperature (Beckman et al. 1994; Shi et al. 2009). High exposure to UV light is correlated with the presence of the P72 allele in dark-skinned populations near the equator, and p53-induced melanin pigmentation in these individuals was suggested to provide sunburn resistance to such populations (Hirshfield et al. 2010; Jeong et al. 2010). In contrast, the R72 allele was found to be more prevalent in light-skinned Caucasian populations that have migrated north where sunlight exposure is low (McGregor et al. 2002). Another study found evidence for an association between codon 72 alleles and winter temperature that was more significant than the association with UV exposure, suggesting a role for codon 72 variants in metabolism (Shi et al. 2009). However, that these alleles may be under natural selection is controversial, and random evolutionary sweeps may explain their ethnic bias. The Onel group examined the relationship between codon 72 allele frequencies and environmental variables in 971 unrelated individuals from 52 unique populations worldwide, using Bayesian and Spearman rank correlation methods; these researchers found no compelling evidence for selective pressures related to the geographic distribution of these variants (Sucheston et al. 2011).
Some data suggest an impact for codon 72 alleles on fecundity. Levine and co-workers found a notable influence of the codon 72 polymorphism on reproduction. They discovered that the R72 variant shows twofold higher transcriptional activity toward the LIF gene, which encodes a cytokine that is required for optimal implantation and reproduction (Kang et al. 2009; Feng et al. 2011). In support of this association, this group reported that the Pro72 allele was increased in women who have recurrent implantation failure and who undergo in vitro fertilization (IVF) (Kang et al. 2009). These latter data suggest that the R72 variant may have been selected for its role in fecundity.
A remaining question is whether these alleles in the p53 pathway should be collectively genotyped to provide better predictors for cancer risk. Whereas the genotype of any one SNP in the p53 pathway may have a modest impact on cancer risk and progression, it seems clear that the presence of multiple risk alleles in an individual has the potential to confer significant cancer risk to that person. One vision might be of a “p53 pathway chip” on which oligonucleotide arrays that distinguish between different SNPs in several genes in the p53 pathway are tiled, and used for hybridization to determine one’s overall p53 pathway activity. Indeed, such a chip could be expanded to include SNPs in other cancer-relevant genes, such as drug metabolizing enzymes and DNA repair proteins, to offer a snapshot assessment of combined cancer risk. Before this reality can be realized, however, more GWAS-type analyses wherein cancer-risk SNPs are assessed in combination need to be performed to establish the formulas for cancer risk from the cooperative or antagonistic interaction between two different SNPs. This seems to point to the future of “personalized” cancer risk and therapy efforts.
ACKNOWLEDGMENTS
The authors thank Donna George, Julie Leu, and Che-Pei Kung for critical reading of the manuscript. This work is supported by funds from NCI CA102184 (Murphy).
Footnotes
Editors: Guillermina Lozano and Arnold J. Levine
Additional Perspectives on The p53 Protein available at www.perspectivesinmedicine.org
REFERENCES
- Aguilo F, Zhou MM, Walsh MJ. 2011. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res 71: 5365–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwal GS, Kirchhoff T, Bond EE, Montagna M, Menin C, Bertorelle R, Scaini MC, Bartel F, Bohnke A, Pempe C, et al. 2009. Altered tumor formation and evolutionary selection of genetic variants in the human MDM4 oncogene. Proc Natl Acad Sci 106: 10236–10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam GA, Frank AK, Hollstein M, Murphy ME. 2011. Tissue-specific apoptotic effects of the p53 codon 72 polymorphism in a mouse model. Cell Cycle 10: 1352–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandele OJ, Wang X, Campbell MR, Pittman GS, Bell DA. 2011. Human single-nucleotide polymorphisms alter p53 sequence-specific binding at gene regulatory elements. Nucleic Acids Res 39: 178–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman G, Birgander R, Sjalander A, Saha N, Holmberg PA, Kivela A, Beckman L. 1994. Is p53 polymorphism maintained by natural selection? Hum Hered 44: 266–270. [DOI] [PubMed] [Google Scholar]
- Bergamaschi D, Gasco M, Hiller L, Sullivan A, Syed N, Trigiante G, Yulug I, Merlano M, Numico G, Comino A, et al. 2003. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell 3: 387–402. [DOI] [PubMed] [Google Scholar]
- Boersma BJ, Howe TM, Goodman JE, Yfantis HG, Lee DH, Chanock SJ, Ambs S. 2006. Association of breast cancer outcome with status of p53 and MDM2 SNP309. J Natl Cancer Inst 98: 911–919. [DOI] [PubMed] [Google Scholar]
- Bojesen SE, Nordestgaard BG. 2008. The common germline Arg72Pro polymorphism of p53 and increased longevity in humans. Cell Cycle 7: 158–163. [DOI] [PubMed] [Google Scholar]
- Bonafe M, Salvioli S, Barbi C, Mishto M, Trapassi C, Gemelli C, Storci G, Olivieri F, Monti D, Franceschi C. 2002. p53 codon 72 genotype affects apoptosis by cytosine arabinoside in blood leukocytes. Biochem Biophys Res Commun 299: 539–541. [DOI] [PubMed] [Google Scholar]
- Bonafe M, Ceccarelli C, Farabegoli F, Santini D, Taffurelli M, Barbi C, Marzi E, Trapassi C, Storci G, Olivieri F, et al. 2003. Retention of the p53 codon 72 arginine allele is associated with a reduction of disease-free and overall survival in arginine/proline heterozygous breast cancer patients. Clin Cancer Res 9: 4860–4864. [PubMed] [Google Scholar]
- Bonafe M, Salvioli S, Barbi C, Trapassi C, Tocco F, Storci G, Invidia L, Vannini I, Rossi M, Marzi E, et al. 2004. The different apoptotic potential of the p53 codon 72 alleles increases with age and modulates in vivo ischaemia-induced cell death. Cell Death Differ 11: 962–973. [DOI] [PubMed] [Google Scholar]
- Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, et al. 2004. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119: 591–602. [DOI] [PubMed] [Google Scholar]
- Bond GL, Hirshfield KM, Kirchhoff T, Alexe G, Bond EE, Robins H, Bartel F, Taubert H, Wuerl P, Hait W, et al. 2006. MDM2 SNP309 accelerates tumor formation in a gender-specific and hormone-dependent manner. Cancer Res 66: 5104–5110. [DOI] [PubMed] [Google Scholar]
- Bonfigli AR, Sirolla C, Testa R, Cucchi M, Spazzafumo L, Salvioli S, Ceriello A, Olivieri F, Festa R, Procopio AD, et al. 2013. The p53 codon 72 (Arg72Pro) polymorphism is associated with the degree of insulin resistance in type 2 diabetic subjects: A cross-sectional study. Acta Diabetol 50: 429–436. [DOI] [PubMed] [Google Scholar]
- Bougeard G, Baert-Desurmont S, Tournier I, Vasseur S, Martin C, Brugieres L, Chompret A, Bressac-de Paillerets B, Stoppa-Lyonnet D, Bonaiti-Pellie C, et al. 2006. Impact of the MDM2 SNP309 and p53 Arg72Pro polymorphism on age of tumour onset in Li–Fraumeni syndrome. J Med Genet 43: 531–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulavin DV, Saito S, Hollander MC, Sakaguchi K, Anderson CW, Appella E, Fornace AJ Jr. 1999. Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J 18: 6845–6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgdorf KS, Grarup N, Justesen JM, Harder MN, Witte DR, Jorgensen T, Sandbaek A, Lauritzen T, Madsbad S, Hansen T, et al. 2011. Studies of the association of Arg72Pro of tumor suppressor protein p53 with type 2 diabetes in a combined analysis of 55,521 Europeans. PLoS ONE 6: e15813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congrains A, Kamide K, Ohishi M, Rakugi H. 2013. ANRIL: Molecular mechanisms and implications in human health. Int J Mol Sci 14: 1278–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont P, Leu JI, Della Pietra AC III, George DL, Murphy M. 2003. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet 33: 357–365. [DOI] [PubMed] [Google Scholar]
- Eeles RA, Olama AA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, Ghoussaini M, Luccarini C, Dennis J, Jugurnauth-Little S, et al. 2013. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet 45: 385–391, 391e381–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov EV, Cherdyntseva NV, Litviakov NV, Malinovskaya EA, Babyshkina NN, Belyavskaya VA, Voevoda MI. 2012. TP53 gene polymorphisms in cancer risk: The modulating effect of ageing, ethnicity and TP53 somatic abnormalities. In Tumor suppressor genes (ed. Cheng DY). InTech, Rijeka, Croatia. [Google Scholar]
- Felley-Bosco E, Weston A, Cawley HM, Bennett WP, Harris CC. 1993. Functional studies of a germ-line polymorphism at codon 47 within the p53 gene. Am J Hum Genet 53: 752–759. [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Zhang C, Kang HJ, Sun Y, Wang H, Naqvi A, Frank AK, Rosenwaks Z, Murphy ME, Levine AJ, et al. 2011. Regulation of female reproduction by p53 and its family members. FASEB J 25: 2245–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank AK, Leu JI, Zhou Y, Devarajan K, Nedelko T, Klein-Szanto A, Hollstein M, Murphy ME. 2011. The codon 72 polymorphism of p53 regulates interaction with NF-κB and transactivation of genes involved in immunity and inflammation. Mol Cell Biol 31: 1201–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed-Pastor WA, Mizuno H, Zhao X, Langerod A, Moon SH, Rodriguez-Barrueco R, Barsotti A, Chicas A, Li W, Polotskaia A, et al. 2012. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 148: 244–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gansmo LB, Knappskog S, Romundstad P, Hveem K, Vatten L, Lonning PE. 2015. Influence of MDM2 SNP309 and SNP285 status on the risk of cancer in the breast, prostate, lung and colon. Int J Cancer 137: 96–103. [DOI] [PubMed] [Google Scholar]
- Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, Orr N, Rhie SK, Riboli E, Feigelson HS, et al. 2013. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet 45: 392–398, 398e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspari L, Pedotti P, Bonafe M, Franceschi C, Marinelli D, Mari D, Garte S, Taioli E. 2003. Metabolic gene polymorphisms and p53 mutations in healthy centenarians and younger controls. Biomarkers 8: 522–528. [DOI] [PubMed] [Google Scholar]
- Gaulton KJ, Willer CJ, Li Y, Scott LJ, Conneely KN, Jackson AU, Duren WL, Chines PS, Narisu N, Bonnycastle LL, et al. 2008. Comprehensive association study of type 2 diabetes and related quantitative traits with 222 candidate genes. Diabetes 57: 3136–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grochola LF, Zeron-Medina J, Meriaux S, Bond GL. 2010. Single-nucleotide polymorphisms in the p53 signaling pathway. Cold Spring Harb Perspect Biol 2: a001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris N, Brill E, Shohat O, Prokocimer M, Wolf D, Arai N, Rotter V. 1986. Molecular basis for heterogeneity of the human p53 protein. Mol Cell Biol 6: 4650–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SL, Gil G, Robins H, Hu W, Hirshfield K, Bond E, Bond G, Levine AJ. 2005. Detection of functional single-nucleotide polymorphisms that affect apoptosis. Proc Natl Acad Sci 102: 16297–16302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirshfield KM, Rebbeck TR, Levine AJ. 2010. Germline mutations and polymorphisms in the origins of cancers in women. J Oncol 2010: 297671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennis M, Kung CP, Basu S, Budina-Kolomets A, Leu JIJ, Khaku S, Cai KQ, Campbell MR, Porter DK, Wang X, et al. 2015. An African-specific polymorphic variant in the TP53 gene impairs tumor suppressor function. Drexel University, Philadelphia, PA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong BS, Hu W, Belyi V, Rabadan R, Levine AJ. 2010. Differential levels of transcription of p53-regulated genes by the arginine/proline polymorphism: p53 with arginine at codon 72 favors apoptosis. FASEB J 24: 1347–1353. [DOI] [PubMed] [Google Scholar]
- Kanetsky PA, Mitra N, Vardhanabhuti S, Li M, Vaughn DJ, Letrero R, Ciosek SL, Doody DR, Smith LM, Weaver J, et al. 2009. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat Genet 41: 811–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Feng Z, Sun Y, Atwal G, Murphy ME, Rebbeck TR, Rosenwaks Z, Levine AJ, Hu W. 2009. Single-nucleotide polymorphisms in the p53 pathway regulate fertility in humans. Proc Natl Acad Sci 106: 9761–9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kengne AP, Czernichow S, Stamatakis E, Hamer M, Batty GD. 2013. Fibrinogen and future cardiovascular disease in people with diabetes: Aetiological associations and risk prediction using individual participant data from nine community-based prospective cohort studies. Diab Vasc Dis Res 10: 143–151. [DOI] [PubMed] [Google Scholar]
- Knappskog S, Lonning PE. 2011. MDM2 promoter SNP285 and SNP309; phylogeny and impact on cancer risk. Oncotarget 2: 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappskog S, Bjornslett M, Myklebust LM, Huijts PE, Vreeswijk MP, Edvardsen H, Guo Y, Zhang X, Yang M, Ylisaukko-Oja SK, et al. 2011. The MDM2 promoter SNP285C/309G haplotype diminishes Sp1 transcription factor binding and reduces risk for breast and ovarian cancer in Caucasians. Cancer Cell 19: 273–282. [DOI] [PubMed] [Google Scholar]
- Knappskog S, Trovik J, Marcickiewicz J, Tingulstad S, Staff AC, Romundstad P, Hveem K, Vatten L, Salvesen HB, Lonning PE. 2012. SNP285C modulates oestrogen receptor/Sp1 binding to the MDM2 promoter and reduces the risk of endometrial but not prostatic cancer. Eur J Cancer 48: 1988–1996. [DOI] [PubMed] [Google Scholar]
- Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. 2011. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 30: 1956–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung CP, Khaku S, Jennis M, Zhou Y, Murphy ME. 2015. Identification of TRIML2, a novel p53 target, that enhances p53 SUMOylation and regulates the transactivation of proapoptotic genes. Mol Cancer Res 13: 250–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, et al. 2004. Gain of function of a p53 hot spot mutation in a mouse model of Li–Fraumeni syndrome. Cell 119: 861–872. [DOI] [PubMed] [Google Scholar]
- Li X, Dumont P, Della Pietra A, Shetler C, Murphy ME. 2005. The codon 47 polymorphism in p53 is functionally significant. J Biol Chem 280: 24245–24251. [DOI] [PubMed] [Google Scholar]
- Liu J, Tang X, Li M, Lu C, Shi J, Zhou L, Yuan Q, Yang M. 2013. Functional MDM4 rs4245739 genetic variant, alone and in combination with P53 Arg72Pro polymorphism, contributes to breast cancer susceptibility. Breast Cancer Res Treat 140: 151–157. [DOI] [PubMed] [Google Scholar]
- Ma YY, Guan TP, Yao HB, Yu S, Chen LG, Xia YJ, He XJ, Wang HJ, Jiang XT, Tao HQ. 2013. The MDM2 309T>G polymorphism and ovarian cancer risk: A meta-analysis of 1534 cases and 2211 controls. PLoS ONE 8: e55019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin MC, Jost CA, Brooks LA, Irwin MS, O’Nions J, Tidy JA, James N, McGregor JM, Harwood CA, Yulug IG, et al. 2000. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet 25: 47–54. [DOI] [PubMed] [Google Scholar]
- Matlashewski GJ, Tuck S, Pim D, Lamb P, Schneider J, Crawford LV. 1987. Primary structure polymorphism at amino acid residue 72 of human p53. Mol Cell Biol 7: 961–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor JM, Harwood CA, Brooks L, Fisher SA, Kelly DA, O’Nions J, Young AR, Surentheran T, Breuer J, Millard TP, et al. 2002. Relationship between p53 codon 72 polymorphism and susceptibility to sunburn and skin cancer. J Invest Dermatol 119: 84–90. [DOI] [PubMed] [Google Scholar]
- Menendez D, Krysiak O, Inga A, Krysiak B, Resnick MA, Schonfelder G. 2006. A SNP in the flt-1 promoter integrates the VEGF system into the p53 transcriptional network. Proc Natl Acad Sci 103: 1406–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378: 203–206. [DOI] [PubMed] [Google Scholar]
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, et al. 2000. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102: 849–862. [DOI] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. 2004. Mutant p53 gain of function in two mouse models of Li–Fraumeni syndrome. Cell 119: 847–860. [DOI] [PubMed] [Google Scholar]
- Parry DJ, Grant PJ, Scott DJA. 2004. Atherothrombotic risk factor clustering in healthy male relatives of male patients with intermittent claudication. J Vasc Surg 40: 891–898. [DOI] [PubMed] [Google Scholar]
- Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. 2007. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum Mutat 28: 622–629. [DOI] [PubMed] [Google Scholar]
- Phillips CL, Gerbing R, Alonzo T, Perentesis JP, Harley IT, Meshinchi S, Bhatla D, Radloff G, Davies SM. 2010. MDM2 polymorphism increases susceptibility to childhood acute myeloid leukemia: A report from the Children’s Oncology Group. Pediatr Blood Cancer 55: 248–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietsch EC, Humbey O, Murphy ME. 2006. Polymorphisms in the p53 pathway. Oncogene 25: 1602–1611. [DOI] [PubMed] [Google Scholar]
- Pim D, Banks L. 2004. p53 polymorphic variants at codon 72 exert different effects on cell cycle progression. Int J Cancer 108: 196–199. [DOI] [PubMed] [Google Scholar]
- Post SM, Quintas-Cardama A, Pant V, Iwakuma T, Hamir A, Jackson JG, Maccio DR, Bond GL, Johnson DG, Levine AJ, et al. 2010. A high-frequency regulatory polymorphism in the p53 pathway accelerates tumor development. Cancer Cell 18: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Peng Q, Tang W, Lao X, Chen Z, Lai H, Deng Y, Mo C, Sui J, Wu J, et al. 2013. An updated meta-analysis on the association of MDM2 SNP309 polymorphism with colorectal cancer risk. PLoS ONE 8: e76031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapley EA, Turnbull C, Al Olama AA, Dermitzakis ET, Linger R, Huddart RA, Renwick A, Hughes D, Hines S, Seal S, et al. 2009. A genome-wide association study of testicular germ cell tumor. Nat Genet 41: 807–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J, Koch P, Contente A, Dobbelstein M. 2000. Tumor-derived mutations within the DNA-binding domain of p53 that phenotypically resemble the deletion of the proline-rich domain. Oncogene 19: 1834–1842. [DOI] [PubMed] [Google Scholar]
- Sakamuro D, Sabbatini P, White E, Prendergast GC. 1997. The polyproline region of p53 is required to activate apoptosis but not growth arrest. Oncogene 15: 887–898. [DOI] [PubMed] [Google Scholar]
- Schmidt MK, Reincke S, Broeks A, Braaf LM, Hogervorst FB, Tollenaar RA, Johnson N, Fletcher O, Peto J, Tommiska J, et al. 2007. Do MDM2 SNP309 and TP53 R72P interact in breast cancer susceptibility? A large pooled series from the breast cancer association consortium. Cancer Res 67: 9584–9590. [DOI] [PubMed] [Google Scholar]
- Schneider-Stock R, Boltze C, Peters B, Szibor R, Landt O, Meyer F, Roessner A. 2004. Selective loss of codon 72 proline p53 and frequent mutational inactivation of the retained arginine allele in colorectal cancer. Neoplasia 6: 529–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Tan SJ, Zhong H, Hu W, Levine A, Xiao CJ, Peng Y, Qi XB, Shou WH, Ma RL, et al. 2009. Winter temperature and UV are tightly linked to genetic changes in the p53 tumor suppressor pathway in Eastern Asia. Am J Hum Genet 84: 534–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique MM, Balram C, Fiszer-Maliszewska L, Aggarwal A, Tan A, Tan P, Soo KC, Sabapathy K. 2005. Evidence for selective expression of the p53 codon 72 polymorphs: Implications in cancer development. Cancer Epidemiol Biomarkers Prev 14: 2245–2252. [DOI] [PubMed] [Google Scholar]
- Stegeman S, Moya L, Selth LA, Spurdle AB, Clements JA, Batra J. 2015. A genetic variant of MDM4 influences regulation by multiple microRNAs in prostate cancer. Endocr Relat Cancer 22: 265–276. [DOI] [PubMed] [Google Scholar]
- Sucheston L, Witonsky DB, Hastings D, Yildiz O, Clark VJ, Di Rienzo A, Onel K. 2011. Natural selection and functional genetic variation in the p53 pathway. Hum Mol Genet 20: 1502–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan A, Syed N, Gasco M, Bergamaschi D, Trigiante G, Attard M, Hiller L, Farrell PJ, Smith P, Lu X, et al. 2004. Polymorphism in wild-type p53 modulates response to chemotherapy in vitro and in vivo. Oncogene 23: 3328–3337. [DOI] [PubMed] [Google Scholar]
- Sun T, Lee GS, Oh WK, Pomerantz M, Yang M, Xie W, Freedman ML, Kantoff PW. 2010. Single-nucleotide polymorphisms in p53 pathway and aggressiveness of prostate cancer in a Caucasian population. Clin Cancer Res 16: 5244–5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada M, Furuuchi K, Kaneda M, Matsumoto J, Takahashi M, Hirai A, Mitsumoto Y, Iggo RD, Moriuchi T. 2001. Inactivate the remaining p53 allele or the alternate p73? Preferential selection of the Arg72 polymorphism in cancers with recessive p53 mutants but not transdominant mutants. Carcinogenesis 22: 515–517. [DOI] [PubMed] [Google Scholar]
- Toledo F, Lee CJ, Krummel KA, Rodewald LW, Liu CW, Wahl GM. 2007. Mouse mutants reveal that putative protein interaction sites in the p53 proline-rich domain are dispensable for tumor suppression. Mol Cell Biol 27: 1425–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomso DJ, Inga A, Menendez D, Pittman GS, Campbell MR, Storici F, Bell DA, Resnick MA. 2005. Functionally distinct polymorphic sequences in the human genome that are targets for p53 transactivation. Proc Natl Acad Sci 102: 6431–6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull C, Rapley EA, Seal S, Pernet D, Renwick A, Hughes D, Ricketts M, Linger R, Nsengimana J, Deloukas P, et al. 2010. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat Genet 42: 604–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Prives C. 2009. Blinded by the light: The growing complexity of p53. Cell 137: 413–431. [DOI] [PubMed] [Google Scholar]
- Walker KK, Levine AJ. 1996. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc Natl Acad Sci 93: 15335–15340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Pittman GS, Bandele OJ, Bischof JJ, Liu G, Brothers JF II, Spira A, Bell DA. 2014. Linking polymorphic p53 response elements with gene expression in airway epithelial cells of smokers and cancer risk. Hum Genet 133: 1467–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weige CC, Birtwistle MR, Mallick H, Yi N, Berrong Z, Cloessner E, Duff K, Tidwell J, Clendenning M, Wilkerson B, et al. 2014. Transcriptomes and shRNA suppressors in a TP53 allele-specific model of early-onset colon cancer in African Americans. Mol Cancer Res 12: 1029–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whibley C, Pharoah PD, Hollstein M. 2009. p53 polymorphisms: Cancer implications. Nat Rev Cancer 9: 95–107. [DOI] [PubMed] [Google Scholar]
- Wynendaele J, Bohnke A, Leucci E, Nielsen SJ, Lambertz I, Hammer S, Sbrzesny N, Kubitza D, Wolf A, Gradhand E, et al. 2010. An illegitimate microRNA target site within the 3′ UTR of MDM4 affects ovarian cancer progression and chemosensitivity. Cancer Res 70: 9641–9649. [DOI] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. 2010. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 38: 662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeron-Medina J, Wang X, Repapi E, Campbell MR, Su D, Castro-Giner F, Davies B, Peterse EF, Sacilotto N, Walker GJ, et al. 2013. A polymorphic p53 response element in KIT ligand influences cancer risk and has undergone natural selection. Cell 155: 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo W, Zhang L, Zhu B, Ling J, Chen Z. 2012. Association of MDM2 SNP309 variation with lung cancer risk: Evidence from 7196 cases and 8456 controls. PLoS ONE 7: e41546. [DOI] [PMC free article] [PubMed] [Google Scholar]