Abstract
The emerging association of assisted reproductive technologies with adverse perinatal outcomes has prompted the in-depth examination of clinical and laboratory protocols and procedures and their possible effects on epigenetic regulatory mechanism(s). The application of various approaches to study epigenetic regulation to problems in reproductive medicine has the potential to identify relative risk indicators for particular conditions, diagnostic biomarkers of disease state, and prognostic indicators of outcome. Moreover, when applied genome-wide, these techniques are likely to find novel pathways of disease pathogenesis and identify new targets for intervention. The analysis of DNA methylation, histone modifications, transcription factors, enhancer binding and other chromatin proteins, DNase-hypersensitivity and, micro- and other noncoding RNAs all provide overlapping and often complementary snapshots of chromatin structure and resultant “gene activity.” In terms of clinical application, the predictive power and utility of epigenetic information will depend on the power of individual techniques to discriminate normal levels of interindividual variation from variation linked to a disease state. At present, quantitative analysis of DNA methylation at multiple loci seems likely to hold the greatest promise for achieving the level of precision, reproducibility, and throughput demanded in a clinical setting.
Epigenetic changes occurring in response to assisted reproductive technologies may be linked to serious but rare disorders (e.g., Angelman syndrome) as well as more common conditions (e.g., low birth weight).
Since the first live-birth resulting from in vitro fertilization (IVF) in 1978, the combination of improved success rates and increased demand for treatment has led to a dramatic rise in the number of infants born as a result of this technology. In the United States alone, more than 130,000 assisted reproductive technology (ART) cycles were initiated in 2013, resulting in more than 42,000 live births and 55,000 infants (www.sartcorsonline.com). Worldwide, greater than three million children have been born with the assistance of in vitro reproductive technologies. (ESHRE 2006; Van Voorhis 2007).
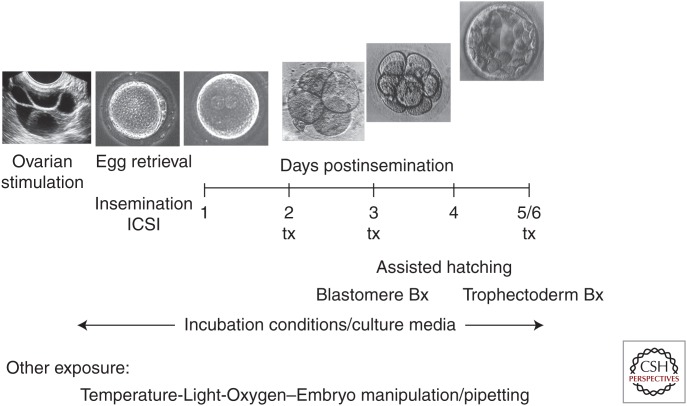
In recent years, there have been increasing concerns regarding the potential health impact of infants conceived with the assistance of IVF. Although twin and higher order multiple pregnancies are the main contributors to the morbidities associated with IVF conceptions, studies have indicated that even singleton pregnancies are at increased risk for a number of adverse perinatal outcomes such as preterm birth, low birth weight, congenital anomalies, preeclampsia, placental abruption, perinatal mortality, and several other pregnancy related complications (Wang et al. 2000; Ericson and Kallen 2001; Hansen et al. 2002; Schieve et al. 2002, 2004; Jackson et al. 2004; Shevell et al. 2005; Chung et al. 2006; Van Voorhis 2006; Kalra and Molinaro 2008; Kalra et al. 2011, 2012; Henningsen 2014). The contributing factors underlying these associations are not known, but the observations, particularly in singleton pregnancies, suggest a relationship of adverse outcomes to some aspects of IVF. There is ongoing debate whether an underlying inherent increased risk in subfertile couples is central to the pathogenesis of these adverse outcomes (Schieve 2004). Alternatively, these adverse outcomes may be related to the protocols used to hyperstimulate patients to maximize the number of oocytes retrieved (Sato et al. 2007; Market-Velker et al. 2010) and/or to the laboratory procedures used for oocyte retrieval, fertilization, and growth of the embryos (Fig. 1). Mechanical manipulation of gametes and embryos and exposure to temperature and oxygen variation (or even light) during a very critical time of development may potentially have profound and lasting effects.
Figure 1.
Schematic representation of the different exposures during the process of clinical human in vitro fertilization (IVF). From gonadotropin exposure during controlled ovarian stimulation (depicted as an ultrasound image) to culture and transfer (tx) at various stages of early development up to the blastocyst stage, the gametes and embryos are subjected to multiple chemical and physical exposures not encountered during unassisted conception.
Therefore, it is likely that multiple factors contribute to the observed adverse outcomes and their pathophysiologic cellular mechanism(s) may differ. Given that most of the clinical observations can be attributed to a “placenta process” (Shevell et al. 2005; Sun et al. 2009), data from the mouse model and our own published and preliminary observations in the human, lead us to hypothesize that some of the observed adverse outcomes may be the result of alterations in trophoblast function. This results in perturbations in the processes of implantation and placentation (Fowden et al. 2006). These processes depend on both embryonic factors regulating the adhesive and invasive properties of trophoblasts and maternal uterine and immunologic factors that play permissive and regulatory roles modulating trophoblast invasion (Norwitz et al. 2001). Most importantly, oxygen tension has emerged as a central regulator of trophoblast differentiation (Genbacev et al. 1996, 1997; Maltepe and Simon 1998; Red-Horse et al. 2004; Jiang and Mendelson 2005; Robins et al. 2007).
Given the now well accepted association of adult disease(s) to peri-conceptional and intrauterine environmental influences (Barker et al. 1993, 2002, 2005; Gluckman et al. 2008; Woo and Patti 2008; Simmons 2009), it is critical to methodically and comprehensibly evaluate the cellular and molecular effects following the in vitro preimplantation growth of human embryos. Elucidation of the specific contributions of treatment to the perinatal morbidity of children conceived with this technology is of both scientific and public health importance. Moreover, a comparison of results obtained from placentas and newborns derived after in vitro versus in vivo conceptions can not only provide information as to the pathogenesis of perinatal adverse events associated with ART, but can also suggest avenues for basic research that focus on understanding the regulatory mechanisms involved in preimplantation human embryonic development and placentation.
Here, we present background and summary information on some of the evolving technologies used to query epigenetic marks. We try to provide an assessment of their suitability as diagnostic or prognostic tools in a clinical setting. Although many of the technologies give deeply textured information on the relationship between individual epigenetic marks, or combinations of marks, and gene activity, not all are likely to provide the robust level of reproducibility and high-throughput amenable to routine clinical laboratory testing. The demands placed on the technology are, in part, a byproduct of the overriding issue in many clinical tests: Given the great diversity of “normal” human phenotypes, how does one distinguish an individual with an “abnormal” phenotype? Both clinicians and researchers should be aware of the strengths, weaknesses, and pitfalls of such technologies, as they are applied, in this case, to the evaluation of epigenetic changes during the clinical application of ARTs.
EPIGENETIC REGULATION
Epigenetic regulation is thought to play a role in mediating the effects of many different exposures to long-term health and disease. However, linking an exposure and an outcome can be very difficult. Reproduction is an excellent model to study the relationship between an exposure and an outcome, primarily because there is access to tissue at birth (embryonic and extra-embryonic) and because there may be “early” consequences of exposure from as early as the period of preimplantation development to later in pregnancy, such as is observed in preeclampsia or intrauterine growth restriction (IUGR). Studies have shown that early exposure to high nutrient intake and rapid weight gain in infancy are associated with later metabolic risks in adolescence and adulthood (Fabricius-Bjerre et al. 2011). Exposures during pregnancy leading to IUGR have been linked to long-term undesirable outcomes in the offspring such as obesity, metabolic syndrome, and a predisposition toward long-term morbidity from type 2 diabetes and cardiovascular disease. Growing evidence suggests that the liver may represent one of the candidate organs targeted by programming, undergoing structural, functional, and epigenetic changes following exposure to an unfavorable intrauterine environment (Cianfarani et al. 2012). Low-birth weight and babies delivered preterm may represent populations at risk of long-term metabolic disorders and may be valuable cohorts to study the role of epigenetic programming.
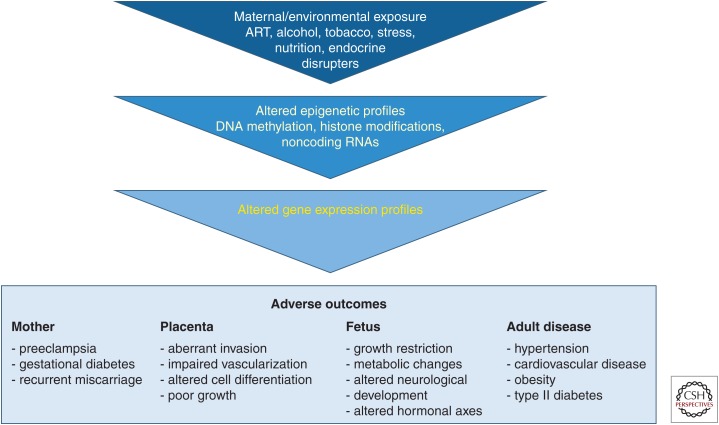
Our own interest in exploiting emerging technologies to study epigenetic regulation to answer questions in reproductive biology stems directly from the possibility that epigenetic structures can be altered by environmental factors. Since the pioneering work of Barker et al. (2002) on the fetal origins of adult disease, we have understood that what occurs in utero can have long-term consequences on health and disease. Multiple studies have found an association of the peri-conceptional, peri-implantation, or in utero environments and DNA methylation, and several are summarized in Table 1 (modified from data in Hogg et al. 2012). This programming, at the cellular and molecular levels, is still poorly understood, specifically as it relates to the role of epigenetic alterations to the development of adult disease (Fig. 2; modified from data in Hogg et al. 2012).
Table 1.
Associations between DNA methylation and different human in utero exposures
| Exposure type | Exposure in utero | Effect on DNA methylation | Tissue | Reference |
|---|---|---|---|---|
| Dietary/nutritional | Dutch famine | Altered DNA methylation at several imprinted genes, some sex-specific effects | Peripheral blood | Tobi et al. 2009 |
| Folate and vitamin B12 | Decreased global DNA methylation (LUMA) associated with maternal B12 status | Cord blood | McKay et al. 2012 | |
| Endocrine disrupting chemicals | Traffic air pollution | Lower global DNA methylation (methylamp) | Cord blood | Herbstman et al. 2012 |
| Antiepileptic drugs (valproic acid) | Reduced global DNA methylation (Illumina HumanMethylation27 array) and site-specific changes | Cord blood and placenta | Smith et al. 2012 |
Modified from data in Hogg et al. (2012).
Figure 2.
Many of the problems of reproductive health can be linked to problems with placental morphology and function. (Created from modified data in Hogg et al. 2012.)
Many different environmental effects, such as exposure to ethanol or altered maternal diet, have been linked to adverse perinatal outcomes that are believed to be regulated, in part, by epigenetic changes in the placenta. For example, ethanol exposure in an animal model led to changes in DNA methylation in the placenta, with DNA methylation in the embryo unaffected (Haycock 2009). Animal models have also linked maternal diabetes or a high-fat diet to epigenetic alterations in the placenta and offspring that can then lead to longer-term metabolic effects (Simmons 2009, 2013; Gatford and Simmons 2013). However, the majority of these studies focus on a small number of imprinted genes (Table 1). It is likely that all of these exposures, as well as ART, have more broad epigenetic consequences and it is critical to develop epigenetic technologies to broadly study these consequences.
Epigenomic Plasticity
Epigenetics is the study of differences in somatically heritable states of gene expression that are not the result of differences in DNA sequence. Epigenetic alterations may also allow changes in gene expression to become transgenerational. It is largely through epigenetic means that organisms achieve phenotypic plasticity, as well as the capacity for a single genotype to result in different cellular phenotypes that may also be tailored to the environment. The first studies of epigenetic variation within populations suggest high levels of phenotypically relevant variation, with the patterns of epigenetic regulation varying between individuals and genome regions as well as with environment (Johnson and Tricker 2010).
With respect to how one might measure the potential effects of ART on the epigenome and the effects of the epigenome on outcomes, there are three classes of epigenetic molecules that might be able to make these distinctions: (1) DNA methylation, (2) modifications of histones and other chromosomal proteins, and (3) noncoding RNAs, including miRNAs and long-noncoding RNAs. With special relevance to environmental effects on the epigenome, all three epigenetic regulatory mechanisms are not likely to be equally capable of distinguishing the epigenetic differences between individuals that may be of clinical interest with regard to procedures used in assisted reproduction. The scope of the problem lies in the observed level of interindividual variation, the expected effect-size of the laboratory exposure or clinical protocol or procedure and the precision and throughput with which the epigenetic measurements can be made. These considerations make DNA methylation the most likely candidate to be a biomarker of assisted reproduction exposures. DNA is a highly stable molecule. Levels of interindividual variation in global or site-specific methylation do vary but are constrained (i.e., methylation at any one site can vary, as a fraction of molecules measured, between zero and one) and there are high-precision, highly reproducible techniques available with the capacity for high-throughput (Eads et al. 2000; Weber et al. 2005; Khulan et al. 2006; Irizarry et al. 2008; Zuo et al. 2009; Bibikova et al. 2011; Jelinek et al. 2012). These techniques have the ability to distinguish differences in population means of the expected small magnitude in samples of moderate size. In other words, interindividual variation is low enough and precision of the DNA methylation measurement is high enough that it is likely that it can be used to distinguish the effects of the intervention (i.e., ART procedures or clinical protocols) on the epigenotype, even if those effects are expected to be small in magnitude. With current technologies, the same cannot be said for histone modifications or even for gene expression arrays interrogating long noncoding or miRNAs.
DNA Methylation
Epigenetics play an important role in fetal development. Following fertilization, areas of the genome that are not protected by imprinting undergo both active and passive demethylation of DNA. DNA methylation patterns then begin to differentiate by developmental stage and by tissue, with ∼20% of CpG sites showing tissue-specific methylation patterns. Different cell types, individuals, and disease states develop unique epigenomes. Because fetal development is a period of extensive cellular replication and growth, environmentally induced epigenetic changes may result in stable patterns of modified gene expression and phenotypic differences among exposed individuals (Menon et al. 2012). Such exposure-specific changes could represent epigenetic “signatures” of particular exposures and be used for both forensic and diagnostic purposes.
DNA methylation is a very important epigenetic mark that has suspected regulatory roles in a broad range of biological processes and diseases. DNA methylation plays a crucial role in embryonic development, maintenance of pluripotency, X-chromosome inactivation and genomic imprinting through regulation of transcription, chromatin structure, and chromosomal stability (Robertson 2005).
In humans, DNA methylation occurs predominantly at cytosine bases in the form of 5-methylcytosines (mCs) at methyl cytosine-guanine dinucleotides (mCGs). hydroxymethylcytosines (hmCs), formylmethylcytosines (fmCs), carboxymethylctyosines (cmCs), and possibly other, yet unknown, forms also occur at much lower frequencies. These bases comprise the human DNA methylome and interindividual differences in the methylome may identify genomic regions involved in cell differentiation and disease. Experimental and bioinformatic methods are now available for applying genome-wide DNA methylation mapping to a broad range of phenotypes, diseases, and biological questions (Bock 2012). However, there is a growing need for establishing powerful and efficient tools for integrative data analysis to filter biologically relevant findings from the growing mass of epigenome data.
Gene-specific methods of measuring DNA methylation include sodium bisulfite conversion of unmethylated cytosine to uracil, followed by methylation-specific (MS) polymerase chain reaction (PCR), pyrosequencing or mass spectrometry-based analyses. Bisulfite conversion is needed for many downstream applications to distinguish methylated DNA from nonmethylated DNA. Sodium bisulfite oxidatively deaminates cytosine to uracil, however, 5-methylcytosine is resistant to this conversion. Although bisulfite treatment creates easily assayed, qualitative differences in sequence from differences in epigenotype, the method involves a harsh chemical treatment resulting in high fragmentation and low-yield of DNA posttreatment. Of necessity, the technique also reduces sequence complexity (converting all unmethylated cytosines to uracil/thymine), which becomes a challenge for primer design for individual genes, as well as downstream mapping from “reads” back to the genome in sequencing-based whole-genome approaches. Incomplete bisulfite conversion can also result in false positive methylation results. There are several methylation-sensitive restriction endonucleases that have the ability to cleave DNA at specific sequences, which is dependent upon the DNA methylation state of the sequence. Combined bisulfate restriction analysis (COBRA) combines bisulfite treatment with sequence-specific restriction endonucleases for locus-specific analysis of DNA methylation. MS PCR allows for highly sensitive detection of locus-specific DNA methylation using PCR amplification of bisulfite-converted DNA (Herman et al. 1998). It is sensitive, quick, and inexpensive. However, there are problems with specificity (PCR design is difficult because the reduced complexity of converted DNA minimizes primer specificity); it is not quantitative, only one or two CpG sites are interrogated at a time and there is no bisulfite conversion control. Pyrosequencing is site-specific, quantitative, and high-throughput, however, it only covers short reads (∼100 bp) and the assay design is difficult in CG dense areas. Mass spectrometry-based methods allow for relatively inexpensive long reads. However, site-specific effects may be lost if CpG sites are close together or fall on small fragments.
Global restriction enzyme-based methods of measuring DNA methylation focus on repetitive elements such as long interspersed element 1 (LINE1), an autonomous retrotransposon comprising ∼17% of the total genome, and Alu, a SINE family (short interspersed element) which is a nonautonomous retrotransposon comprising ∼11% of the genome. There are quick and quantitative screens for these regions, collectively. However, the assay design for these screens can be challenging (which region/CpGs to include, problems with variability), and sample quality and tissue of origin can affect the success of the assay (Lander et al. 2001). The restriction enzyme based luminometric methylation assay (LUMA) determines methylation at all CCGG sites (HpaII cleaves unmethylated CCGG; MspI cleaves both methylated and unmethylated CCGG). Although this method is advantageous because one does not need to know the exact target sequence, it does not give specific information on which genes are affected (Karimi et al. 2006).
DNA sequencing has become more affordable and capable of high-throughput approaches. Whole-genome sequencing of bisulfite-converted or MeDIP-precipitated DNA to detect genome-wide DNA methylation profiles is being more routinely used. With the rapid development of new methods for epigenomic analysis, there is an acute need for a systematic assessment of available technologies (Laird 2010; Stolzenberg et al. 2011).
Genome-wide analysis of DNA methylation can also be performed using the DREAM (digital restriction enzyme analysis of methylation) assay. Genomic DNA is sequentially digested using SmaI and XmaI enzymes and the methylation-specific signatures created at the ends of each cleavage product are analyzed using next generation sequencing. DREAM analyses ∼150K unique CpG sites, 39K of which are in CpG islands and 30K are at transcription start sites of 13K RefSeq genes. Compared with bisulfate pyrosequencing, DREAM is cost effective, quantitative, and reproducible (Jelinek et al. 2012).
HELP (HpaII tiny fragment enrichment by ligation-mediated PCR) is another whole-genome methylation analysis technique. This assay is quantitative (compared methylation-sensitive HpaII sites with the methylation-insensitiveisoschizomer MspI sites) and supported by open source bioinformatic tools for analysis. One version of the HELP assay generated representations of sizes 200−2000 bp, allowing the testing of roughly 800K loci throughout the human genome (Khulan et al. 2006; Oda et al. 2009). The MspI representation allows simultaneous copy number variability testing (HELP-CNV) and as little as 10 ng of starting DNA can be used for the assay (nanoHELP). Another protocol uses HELP with massively parallel sequencing (MPS) rather than microarrays (HELP-seq). This technique is similar to methyl-seq and allows for more sensitive detection of hypomethylated loci. A large number of polymorphisms in HpaII sites (>3%) are also detected. These are a significant source of variability for microarray-based studies. A HELP-tagging assay (combining the power of the MspI normalization of HELP with the increased comprehensiveness of the methyl-sensitive cut counting, MSCC, assay) is based on Illumina sequencing and is highly quantitative (Suzuki et al. 2010).
Epigenome-wide analyses (arrays or parallel sequencing) of DNA methylation include platforms such as Illumina’s BeadChips for human DNA (27K CpG island-heavy versus 450K with better whole-genome coverage), promoter arrays (NimbleGen, Agilent), which are biased but available for multiple species, and tiling arrays (Affymetrix, NimbleGen) for human DNA. These latter arrays are unbiased and have more than 41 million probes. These protocols may include antibody precipitation of methylated DNA or MBP before array hybridization. Parallel sequencing or deep sequencing methods include BS-Seq, MeDIP-Seq, and MethylPlex-Seq. Methylated DNA immunoprecipitation (MeDIP) uses an antibody that specifically recognizes methylated DNA, and the immunoprecipitated methylated DNA sequences can be identified by PCR, Next-Gen sequencing or microarray hybridization approaches. These methods are global, with respect to genome coverage; however, a reference sequence is needed for the BS-Seq method, an antibody is required for the MeDIP method and in all cases and results must be validated (Pomraning et al. 2009).
For any epigenome-wide analyses, study design considerations include sample requirements (amount and quality; i.e., some restriction endonuclease digests require >2 μg DNA, Illumina Bead Arrays can still be effective with degraded DNA, whereas affinity methods are more tolerable of DNA impurity but require larger amounts), sample throughput (high-throughput 96 or 384 sample assays are low in labor but high in reagent costs), genome coverage and resolution (restriction endonuclease technologies are limited to the number/distribution of recognition sites and some technologies work better for smaller genomes), accuracy and reproducibility (fragment lengths affect hybridization, sequencing, and reproducibility/false positives and negatives, incomplete bisulfite conversion and validation), and finally bioinformatic and data storage requirements (Laird 2010). DNA methylation analyses, in terms of cost, resolution, genome coverage, and accuracy were previously reviewed by Harris et al. (2010) and Bock (2012).
ASSISTED REPRODUCTIVE TECHNOLOGIES (ART)
Major epigenetic events that occur during germ cell development and preimplantation stages of embryo development precisely coincide with when clinical and laboratory procedures related to ART are being done. The combination of epidemiological data indicating that children conceived through ART are at an increased risk for a number of undesirable outcomes and the demonstration that model organisms subjected to similar manipulations exhibit epigenetic alterations at specific loci (Mann et al. 2004; Rivera et al. 2008) suggest an association between the two.
ART may affect proper establishment and/or maintenance of epigenetic marks. The most provocative observations have been the combination of demonstrated effects of culture media on DNA methylation and imprinted gene expression in model systems (Mann et al. 2004; Rivera et al. 2008) and the suggestion that two very rare diseases resulting from alterations at imprinted genes (such as Beckwith–Wiedemann and Angelman syndromes) (DeBaun et al. 2003; Maher et al. 2003; Odom and Segars 2010; Lazaraviciute et al. 2014) have been observed at greater frequency among children conceived in vitro. These data are suggestive because the timing of epigenetic reprogramming and the timing of clinical protocols used in assisted reproduction overlap to a great degree.
Given that the timing of interventions used in assisted reproduction coincide with the period when genome-wide changes in DNA methylation are thought to take place, it is only a short logical extension to compare DNA methylation levels between children conceived in vitro and children conceived in vivo to determine whether differences observed in model organisms are also observed in human conceptions. Such comparisons have been performed in multiple laboratories, initially targeting small numbers of imprinted genes (reviewed by Batcheller et al. 2011; systematic review and meta-analysis by Lazaraviciute et al. 2014; Nelissen et al. 2014; Whitelaw et al. 2014; Melamed et al. 2015), under the assumption that imprinted genes might be more susceptible to effects of epigenetic disruption (Table 2), and in a modest numbers of patients.
Table 2.
Studies comparing epigenetic differences between children conceived in vivo or using ART
| Tissue | Methodology | Sample size | Genes analyzed | Difference observed | Reference |
|---|---|---|---|---|---|
| Cord blood and placenta | GoldenGate array and RT-PCR | 10 ART, 13 in vivo | 736 genes | Yes | Katari et al. 2009 |
| Placenta | Bisulfite PCR | 78 ART, 38 in vivo | H19, GTL2, PEG1, KCNQ1OT1, ZAC, PEG3, SNRPN, XIST | Yes | Kobayashi et al. 2009 |
| Cord blood, peripheral blood, amnion/chorion | Bisulfite PCR | 77 ICSI, 35 IVF, 73 in vivo | KvDMR1, H19, SNRPN, MEST, GRB10, MEG3, IG-DMR, GNAS, NESP55, GNAS, NESPas, GNAS XL α-s, GNAS Exon1A | No | Tierling et al. 2010 |
| Cord blood and placenta | Methylation specific-PCR and RT-PCR | 45 ART, 56 in vivo | IGF2/H19 and IGF2R DMR, X-inactivation | Yes | Turan et al. 2010 |
| Placenta | RT-PCR | 65 ART, 924 in vivo | IGF2, H19, KCNQ1OT1, CDKN1C | Yes | Katagiri et al. 2010 |
| Placenta (CVS) | Bisulfite PCR | 42 ART, 29 in vivo | H19, MEG3, LIT1, MEST, NESP55, PEG3, SNRPN, NANOG, APC | Yes | Zechner et al. 2010 |
| Cord Blood | Bisulfite PCR, cloning and sequencing | 29 IVF and 30 in vivo conceived twin pairs | KvDMR1, PEG1, H19/IGF2 DMR | No | Li et al. 2011 |
| Cord blood and peripheral blood | Bisulfite sequencing | 30 IVF, 30 ICSI and 60 in vivo | PEG3, L3MBTL, PHILDA2 | No | Feng et al. 2011 |
| Placenta | Pyrosequencing | 35 IVF, 35 in vivo | H19, MEST | Yes | Nelissen et al. 2013 |
| Chorionic villi and muscle samples | Pyrosequencing, bisulfite sequencing PCR | 75 ART stillbirth, 73 ART multifetal reduction, 90 natural conception stillbirths, 82 natural conception induced abortion | H19, LIT1, SNRPN | Yes | Zheng et al. 2013 |
Modified from data in Batcheller et al. (2011).
However, it is also critical to expand our knowledge beyond imprinted genes and CpG islands. DNA methylation on the edges of CpG islands, so-called “island shores,” may play a role in affecting gene expression or affect chromatin structure and secondarily gene expression (Irizarry et al. 2009). It is therefore increasingly necessary to examine global DNA methylation and not focus solely on imprinted genes and the known imprinting control regions. A recent study found that methylation of LINE1 was associated with birth weight and methylation of LINE1 and AluYb8 methylation also differed among infants exposed to tobacco and alcohol (Wilhelm-Benartzi et al. 2012).
MOVING FROM BIOMARKERS TO PHYSIOLOGY AND FROM ASSOCIATIONS TO CAUSE/EFFECT: WHAT WILL IT TAKE?
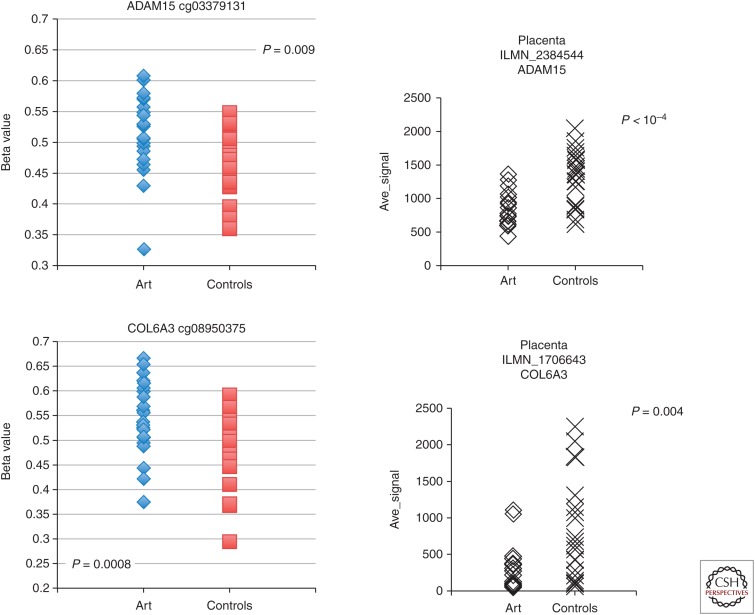
In multiple studies, associations have been observed between DNA methylation levels at specific sites and conception in vitro (Table 2); however, some inconsistencies have been observed. Many of the differences between studies can be viewed as the most likely outcome of replication studies involving small numbers of individuals when the level of interindividual variation is large and the expected difference between groups is small. As shown from the data presented in Figure 3, there is significant interindividual variation in both methylation and transcript levels. Nevertheless, it is clear that many of the small differences in mean methylation level between groups are also matched by differences in transcript level (Fig. 3), suggesting that the ART-associated differences in DNA methylation may play an important role. Many such differences are difficult to show because the level of interindividual difference in transcript level is even larger than the interindividual difference in methylation levels (Katari et al. 2009). In total, the studies comparing DNA methylation between children conceived in vitro and children conceived in vivo point to subtle differences at many genes. In agreement with data derived from experiments in the mouse, extraembryonic tissues, in general, exhibit more and larger DNA methylation differences than embryonic tissues (Mann et al. 2004; Rivera et al. 2008; de Waal et al. 2014).
Figure 3.
Interindividual variation in gene-specific DNA methylation and transcription. Interindividual variation in DNA methylation (left) at two CpG sites in genes that also show significant differences in transcript level (right) between ART and control placentas. Methylation was measured using Illumina’s Infinium 27K methylation array and transcript level was measured using Illimina’s Human HT-12 Expression BeadChip in placenta from 24 children conceived in vitro (ART) and 24 children conceived in vivo (controls).
CONCLUDING REMARKS
Infertile couples are increasingly turning to ART to treat their infertility. Of growing concern is that ART-conceived pregnancies and children are at increased risk for specific loss-of-imprinting disorders, as well as congenital malformations, IUGR, and preeclampsia. Published reports from our own and other laboratories show that following hormonal stimulation along with the in vitro manipulation of human gametes and/or embryos, extra-embryonic and fetal tissues exhibit particular susceptibility to both methylation and specific gene expression differences.
Epigenetic changes occurring in response to ART manipulations have been linked with adverse outcomes in the offspring. Although initially ART was associated with diseases known to be caused by epigenetic alterations, such as Angelman syndrome and Beckwith–Wiedemann syndrome (Odom and Segars 2010), it is not these rare outcomes that are most concerning. The developmental origins of adult disease hypothesis has gained strength as a powerful explanation for risk of some of the most prevalent and morbid human conditions. It is likely that less severe epigenetic alterations are responsible for other, more common, adverse outcomes associated with IVF such as low birth weight, preeclampsia, and preterm labor. The full impact of impaired fetal growth may not be completely evident until years after birth when later developmental and reproductive milestones can be affected. Many of these outcomes are associated with long-term health problems such as hypertension, diabetes, and other adverse metabolic effects.
Studying the effects of ART on the epigenome may help to predict long-term consequences of our early interventions. It is critical to investigate our interventions to determine whether they are the cause of these epigenetic differences. It is particularly important to investigate the role of each individual intervention used in ART (i.e., superovulation, ICSI, extended culture, etc.) on epigenetic outcome and to adapt current ART protocols to minimize risk to the pregnancy, the neonate and the long-term health of the offspring. Finally, it is also critical to use this information to further our understanding of epigenetic programming.
Advancement in the field of epigenetics may help us understand many of the outcomes following ART treatments and processes that decrease IVF success rates, such as poor/abnormal embryo development, failed implantation, pregnancy loss, and developmental anomalies. We know that chromosomally normal embryos do not implant 100% of the time and changes in DNA methylation may also play a role in lower “egg quality” associated with age (Yue et al. 2012). Abnormal maintenance of methylation may further play a role in early pregnancy loss (Yin et al. 2012; Zheng et al. 2013). Therefore, understanding epigenetic marks that predict adverse disease events may help us with developing biomarkers that could aid in early diagnosis and treatment of conditions that could lead to improved quality of life or even prevention of disease. Furthermore, we know very little about the extended consequences of any ART-associated epigenetic differences. Clinical ART has only been in existence for less than four decades so there may be yet undiscovered consequences in the millions of children conceived by IVF.
Finally, epidemiologic data supporting a relationship between ARTs, low birth weight and rare conditions involving imprinted genes also makes it plausible that additional exposures around the time of conception may impact fetal growth. Growing evidence supports the concept that developmental programming may occur from a window that spans peri-conception through postnatal life. Nearly half of all pregnancies conceived in the United States are unplanned (3.1 million). Such pregnancies are associated with an increased risk of delayed or lack of prenatal care, fetal exposure to harmful chemicals such as endocrine disrupters, smoking and alcohol, and low birth weight. These associations highlight the fact that maternal exposures both preconception and in utero are critical for programming fetal development and when these exposures are unfavorable perinatal morbidity increases.
It should be noted that these issues go beyond the effects of clinical and laboratory protocols involved in assisted reproductive technologies. Environmental and nutritional exposures differ among populations of women according to socioeconomic status, race, and age. Epigenetic differences may not only be secondary to exposure (ART or other in utero exposure) but may also be dependent on the maternal genotype. A recent study found that polymorphisms in genes involved in folate absorption and metabolism may affect DNA methylation patterns (McKay et al. 2012). Abnormal methylation of paternal DNA is also important to study because DNA methylation may be altered in the sperm of men with infertility. One study has shown that in some infertile men perturbations in methylation may contribute to abnormal embryo development (Aston et al. 2012). The possibility that some aspect(s) of infertility per se contributes to altered phenotype and/or epigenotype is open for consideration and further investigation (Song et al. 2015).
Ultimately, we need additional epidemiological research incorporating epigenetic markers of environmental exposure, taking a life-course approach with regards to assessing outcomes. Compelling results from such studies could help to broaden the scope of maternal care to one that places more emphasis on preconception surveillance and treatment. Modifications to maternal health, both before and at the earliest stages of gestation, could impact upon fetal development and reduce adult disease disk. To aid us in achieving this, techniques for measuring DNA methylation are very useful in quantifying epigenetic effects of environment and nutrition, correlating developmental epigenetic variation with phenotypes, understanding epigenetics of cancer and chronic diseases, measuring the effects of drugs on DNA methylation or deriving new biological insights into mammalian genomes (Jelinek et al. 2012). To conclude, it is necessary to study not only different tissues, but different time points during development and adulthood. Although there are conflicting hypotheses, epigenetic marks may be static and tissue-specific differences in DNA methylation may change during different times of development (Novakovic et al. 2011; Yuen et al. 2011).
ACKNOWLEDGMENTS
The authors thank Dr. Nahid Turan for her help in the preparation of this manuscript. This work is supported by the National Institutes of Health P50 HD068157 (C.C., C.S., S.B.), R01 HD048730 (C.S., C.C.), and the K12 Reproductive Scientists Development Program (RSDP) to M.M.
Footnotes
Editors: Diana W. Bianchi and Errol R. Norwitz
Additional Perspectives on Molecular Approaches to Reproductive and Newborn Medicine available at www.perspectivesinmedicine.org
REFERENCES
- Aston KI, Punj V, Liu L, Carrell DT. 2012. Genome-wide sperm deoxyribonucleic acid methylation is altered in some men with abnormal chromatin packaging or poor in vitro fertilization embryogenesis. Fertil Steril 97(2): 285–292. [DOI] [PubMed] [Google Scholar]
- Barker DJ, Hales CN, Fall CH, Osmond C, Phipps K, Clark PM. 1993. Type 2 (non-insulin-dependent) diabetes mellitus, hypertension and hyperlipidaemia (syndrome X): Relation to reduced fetal growth. Diabetologia 36: 62–67. [DOI] [PubMed] [Google Scholar]
- Barker DJ, Eriksson JG, Forsén T, Osmond C. 2002. Fetal origins of adult disease: Strength of effects and biological basis. Int J Epidemiol 31: 1235–1239. [DOI] [PubMed] [Google Scholar]
- Barker DJ, Osmond C, Forsen JJ, Kajantie E, Eriksson JG. 2005. Trajectories of growth among children who have coronary events as adults. N Engl J Med 353: 1802–1809. [DOI] [PubMed] [Google Scholar]
- Batcheller A, Cardozo E, Maguire M, DeCherney AH, Segars JH. 2011. Are there subtle genome-wide epigenetic alterations in normal offspring conceived by assisted reproductive technologies? Fertil Steril 96: 1306–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, et al. 2011. High density DNA methylation array with single CpG site resolution. Genomics 98: 288–295. [DOI] [PubMed] [Google Scholar]
- Bock C. 2012. Analysing and interpreting DNA methylation data. Nat Rev Genet 13: 705–719. [DOI] [PubMed] [Google Scholar]
- Chung K, Coutifaris C, Chalian R, Lin K, Ratcliffe SJ, Castelbaum AJ, Freedman MF, Barnhart KT. 2006. Factors influencing adverse perinatal outcomes in pregnancies achieved through use of assisted reproductive technology. Fertil Steril 86: 1634–1641. [DOI] [PubMed] [Google Scholar]
- Cianfarani S, Agostoni C, Bedogni G, Berni Canani R, Brambilla P, Nobili V, Pietrobelli A. 2012. Effect of intrauterine growth retardation on liver and long-term metabolic risk. Int J Obes (Lond) 36: 1270–1277. [DOI] [PubMed] [Google Scholar]
- DeBaun MR, Niemitz EL, Feinberg AP. 2003. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LITI and H19. Am J Hum Genet 72: 156–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Waal E, Mak W, Calhoun S, Stein P, Ord T, Krapp C, Coutifaris C, Schultz RM, Bartolomei MS. 2014. In vitro culture increases the frequency of stochastic epigenetic errors at imprinted genes in placental tissues from mouse concepti produced through assisted reproductive technologies. Biol Reprod 90: 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Dandenberg PV, Laird PW. 2000. MethyLight: A high-throughput assay to measure DNA methylation. Nucleic Acids Res 28: E32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson A, Kallen B. 2001. Congenital malformations in infants born after IVF: A population-based study. Hum Reprod 16: 504–509. [DOI] [PubMed] [Google Scholar]
- ESHRE (European Society of Human Reproduction and Embryology). 2006. World report on assisted reproduction technology. www.eshre.eu [Google Scholar]
- Fabricius-Bjerre S, Jensen RB, Færch K, Larsen T, Mølgaard C, Michaelsen KF, Vaag A, Greisen G. 2011. Impact of birth weight and early infant weight gain on insulin resistance and associated cardiovascular risk factors in adolescence. PLoS ONE 6: e20595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C, Tian S, Zhang Y, He J, Zhu XM, Zhang D, Sheng JZ, Huang HF. 2011. General imprinting status is stable in assisted reproduction conceived offspring. Fertil Steril 96: 1417–1423. [DOI] [PubMed] [Google Scholar]
- Fowden AL, Sibley C, Reik W, Constancia M. 2006. Imprinted genes, placental development and fetal growth. Horm Res 65: 50–58. [DOI] [PubMed] [Google Scholar]
- Gatford KL, Simmons RA. 2013. Prenatal programming of insulin secretion in intrauterine growth restriction. Clin Obstet Gynecol 56: 520–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genbacev O, Joslin R, Damsky CH, Pllliotti BM, Fisher SJ. 1996. Hypoxia alters early early gestational human cytotrophoblast differentiation/invasion in vitro and models the placental defects that occur in preeclampsia. J Clin Invest 97: 540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genbacev O, Zhou U, Ludlow JW, Fisher SJ. 1997. Regulation of human placental development by oxygen tension. Science 277: 1669–1672. [DOI] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA, Cooper C, Thornburg KL. 2008. Effect in in utero and early-life conditions on adult health and disease. N Engl J Med 359: 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Kurinczuk JJ, Bower C, Webb S. 2002. The risk of major birth defects after intracytoplasmic sperm injection and in-vitro fertilization. N Engl J Med 346: 725–730. [DOI] [PubMed] [Google Scholar]
- Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y. 2010. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic odifications. Nat Biotechnol 28: 1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycock PC, Ramsay M. 2009. Exposure of mouse embryos to ethanol during preimplantation development: Effect on DNA methylation in the H19 imprinting control region. Biol Reprod 81(4): 618–627. [DOI] [PubMed] [Google Scholar]
- Henningsen AK, Pinborg A. 2014. Birth and perinatal outcomes and complications for babies conceived following ART. Semin Fetal Neonatal Med 19: 234–238. [DOI] [PubMed] [Google Scholar]
- Herbstman JB, Tang D, Zhu D, Qu L, Sjödin A, Li Z, Camann D, Perera FP. 2012. Prenatal exposure to polycyclic aromatic hydrocarbons, benzo[a]pyrene-DNA adducts, and genomic DNA methylation in cord blood. Environ Health Perspect 120: 733–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JPJ, Markowitz S, Willson JKV, Hamilton SR, Kinzler KW. 1998. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci 95: 6870–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg K, Price EM, Hanna CW, Robinson WP. 2012. Prenatal and perinatal environmental influences on the human fetal and placental epigenome. Clin Pharmacol Ther 92: 716–726. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Carvalho B, Wu H, Brandenburg SA, Jeddeloh JA, Wen B, Feinberg AP. 2008. Comprehensive high-throughput arrays for relative methylation (CHARM). Genome Res 18: 780–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M. 2009. Genome-wide methylation analysis of human colon cancer reveals similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41: 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R, Gibson KA, Wu YW, Croughan MS. 2004. Perinatal outcomes in singletons following in vitro fertilization: A meta-analysis. Obstet Gynecol 130: 551–563. [DOI] [PubMed] [Google Scholar]
- Jelinek J, Liang S, Lu Y, He R, Ramagli LS, Shpall EJ, Estecio MR, Issa JP. 2012. Conserved DNA methylation patterns in healthy blood cells and extensive changes in leukemia measured by a new quantitative technique. Epigenetics 7: 1368–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B, Mendelson CR. 2005. O2 enhancement of human trophoblast differentiation and hCYP19 (Aromatase) gene expression are mediated by proteasomal degradation of USF1 and USF2. Molec Cell Biol 25: 8824–8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LJ, Tricker PJ. 2010. Epigenomic plasticity within populations: Its evolutionary significance and potential. Heredity 105: 113–121. [DOI] [PubMed] [Google Scholar]
- Kalra SK, Molinaro T. 2008. The association of in vitro fertilization and perinatal morbidity. Semin Reprod Med 26: 423–435. [DOI] [PubMed] [Google Scholar]
- Kalra SK, Ratcliffe SJ, Coutifaris C, Molinaro T, Barnhart KT. 2011. Ovarian stimulation and low birth weight in newborns conceived through in vitro fertilization. Obstet Gynecol 118: 863–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra SK, Ratcliffe SJ, Barnhart KT, Coutifaris C. 2012. Extended embryo culture and an increased risk of preterm delivery. Obstet Gynecol 120: 69–75. [DOI] [PubMed] [Google Scholar]
- Karimi M, Johansson S, Ekstrom TJ. 2006. Using LUMA: A luminometric-based assay for global DNA-methylation. Epigenetics 1: 45–48. [DOI] [PubMed] [Google Scholar]
- Katagiri Y, Aoki C, Tamaki-Ishihara Y, Fukuda Y, Kitamura M, Matsue Y, So A, Morita M. 2010. Effects of assisted reproduction technology on placental imprinted gene expression. Obstet Gynecol Int 2010: 437528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katari S, Turan N, Bibikova M, Erinle O, Chalian R, Foster M, Gaughan JP, Coutifaris C, Sapienza C. 2009. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum Mol Genet 18: 3769–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khulan B, Thompson RF, Ye K, Fazzari MJ, Suzuki M, Stasiek E, Figueroa ME, Glass JL, Chen Q, Montagna C. 2006. Comparative isoschizomer profiling of cytosine methylation: The HELP assay. Genome Res 16: 1046–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Hiura H, John RM, Sato A, Otsu E, Kobayashi N, Suzuki R, Suzuki F, Hayashi C, Utsunomiya T. 2009. DNA methylation errors at imprinted loci after assisted conception originate in the parental sperm. Eur J Hum Genet 17: 1582–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird PW. 2010. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet 11: 191–203. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. Erratum: Nature412: 565 and Nature411: 720. [DOI] [PubMed] [Google Scholar]
- Lazaraviciute G, Kauser M, Bhattacharya S, Haggarty P, Bhattacharya S. 2014. A systematic review and meta-analysis of DNA methylation levels and imprinting disorders in children conceived by IVF/ICSI compared with children conceived spontaneously. Hum Reprod Update 20: 840–852. [DOI] [PubMed] [Google Scholar]
- Li L, Wang L, Le F, Liu X, Yu P, Sheng J, Huang H, Jin F. 2011. Evaluation of DNA methylation status at differentially methylated regions in IVF- conceived newborn twins. Fertil Steril 95: 1975–1979. [DOI] [PubMed] [Google Scholar]
- Maher ER, Brueton LA, Bowdin SC. 2003. Beckwith-Wiedemann syndrome and assisted reproduction technology (ART). J Med Genetics 40: 62–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltepe E, Simon MC. 1998. Oxygen, genes, and development: An analysis of the role of hypoxic gene regulation during murine vascular development. J Molec Med 76: 391–401. [DOI] [PubMed] [Google Scholar]
- Mann MR, Lee SS, Doherty AS, Verona RI, Nolen LD, Schultz RM, Bartolomei MS. 2004. Selective loss of imprinting in the placenta following preimplantation development in culture. Development 131: 3727–3735. [DOI] [PubMed] [Google Scholar]
- Market-Velker BA, Zhang L, Magri LS, Bonvissuto AC, Mann MRW. 2010. Dual effects of superovulation: Loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum Molec Genet 19: 36–51. [DOI] [PubMed] [Google Scholar]
- McKay JA, Groom A, Potter C, Coneyworth LJ, Ford D, Mathers JC, Relton CL. 2012. Genetic and non-genetic influences during pregnancy on infant global and site specific DNA methylation: Role for folate gene variants and vitamin B12. PLoS ONE 7: e33290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed N, Choufani S, Wilkins-Haug LE, Koren G, Weksberg R. 2015. Comparison of genome-wide and gene-specific DNA methylation between ART and naturally conceived pregnancies. Epigenetics 10.4161/15592294.2014.988041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon R, Conneely KN, Smith AK. 2012. DNA methylation: An epigenetic risk factor in preterm birth. Reprod Sci 19: 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. 2009. Long non-coding RNAs: Insights into functions. Nat Rev Genet 10: 155–159. [DOI] [PubMed] [Google Scholar]
- Nelissen EC, Dumoulin JC, Daunay A, Evers JL, Tost J, Van Montfoort AP. 2013. Placentas from pregnancies conceived by IVF/ICSI have a reduced DNA methylation level at the H19 and MEST differentially methylated regions. Hum Reprod 28: 1117–1126. [DOI] [PubMed] [Google Scholar]
- Nelissen EC, Dumoulin JC, Busato F, Ponger L, Eijssen LM, Evers JL, Tost J, van Montfoort AP. 2014. Altered gene expression in human placentas after IVF/ICSI. Hum Reprod 29: 2821–2831. [DOI] [PubMed] [Google Scholar]
- Norwitz ER, Schust DJ, Fisher SJ. 2001. Implantation and the survival of early pregnancy. N Engl J Med 345: 1400–1408. [DOI] [PubMed] [Google Scholar]
- Novakovic B, Yuen RK, Gordon L, Penaherrera MS, Sharkey A, Moffett A, Craig JM, Robinson WP, Saffery R. 2011. Evidence for widespread changes in promoter methylation profile in human placenta in response to increasing gestational age and environmental/stochastic factors. BMC Genomics 12: 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda M, Glass JL, Thompson RF, Mo Y, Olivier EN, Figueroa ME, Selzer RR, Richmond TA, Zhang X, Dannenberg L. 2009. High-resolution genome-wide cytosine methylation profiling with simultaneous copy number analysis and optimization for limited cell numbers. Nucleic Acids Res 37: 3829–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom LN, Segars J. 2010. Imprinting disorders and assisted reproductive technology. Curr Opin Endocrinol Diabetes Obes 17: 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M, Ferrari R. 2012. Epigenetic analysis: ChIP-chip and ChIP-seq. Methods Mol Biol 802: 377–387. [DOI] [PubMed] [Google Scholar]
- Petronis A. 2010. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature 465: 721–727. [DOI] [PubMed] [Google Scholar]
- Pomraning KR, Smith KM, Freitag M. 2009. Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods 47: 142–150. [DOI] [PubMed] [Google Scholar]
- Red-Horse K, Zhou Y, Genbacev O, Prakobphol A, Foulk R, McMaster M, Fisher SJ. 2004. Trophoblast differentiation during embryo implantation and formation of the maternal-fetal interface. J Clin Invest 114: 744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera RM, Stein P, Weaver JR, Mager J, Schultz RM, Bartolomei MS. 2008. Manipulations of mouse embryos prior to implantation result in aberrant expression of imprinted genes on day 9.5 of development. Hum Mol Genet 17: 1–14. [DOI] [PubMed] [Google Scholar]
- Robertson KD. 2005. DNA methylation and human disease. Nat Rev Genet 6: 597–610. [DOI] [PubMed] [Google Scholar]
- Robins JC, Heizer A, Hardiman A, Hubert M, Handwerger S. 2007. Oxygen tension directs the differentiation pathway of human cytotrophoblast cells. Placenta 28: 1141–1146. [DOI] [PubMed] [Google Scholar]
- Ross JP, Shaw JM, Molloy PL. 2013. Identification of differentially methylated regions using streptavidin bisulfite ligand methylation enrichment (SuBLiME), a new method to enrich for methylated DNA prior to deep bisulfite genomic sequencing. Epigenetics 8: 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Otsu E, Negishi H, Utsunomiya T, Arima T. 2007. Aberrant DNA methylation of imprinted loci in superovulated oocytes. Hum Reprod 22: 26–35. [DOI] [PubMed] [Google Scholar]
- Schieve LA, Meikle SF, Ferre C, Peterson HB, Jeng GT, Wilcox LS. 2002. Low and very low birth weight in infants conceived with use of assisted reproductive technology. N Engl J Med 346: 725–730. [DOI] [PubMed] [Google Scholar]
- Schieve LA, Ferre C, Peterson HB, Macaluso M, Reynolds MA, Wright VC. 2004. Perinatal outcome among singleton infants conceived through assisted reproductive technology in the United States. Obstet Gynecol 103: 1144–1153. [DOI] [PubMed] [Google Scholar]
- Shevell T, Malone FC, Vicaver J, Porter JF, Luthy DA, Cornstock CH, Hankins GD, Eddleman K, Dolan S, Dugoff L, et al. 2005. Assisted reproductive technology and pregnancy outcome. Obstet Gynecol 106: 1039–1045. [DOI] [PubMed] [Google Scholar]
- Simmons RA. 2009. Developmental origins of adult disease. Pediatr Clin North Am 56: 449–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons RA. 2013. Programming of DNA methylation in type 2 diabetes. Diabetologia 56: 947–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AK, Conneely KN, Newport DJ, Kilaru V, Schroeder JW, Pennell PB, Knight BT, Cubells JC, Stowe ZN, Brennan PA. 2012. Prenatal antiepileptic exposure associates with neonatal DNA methylation differences. Epigenetics 7: 458–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Ghosh J, Mainigi M, Turan N, Weinerman R, Truongcao M, Coutifaris C, Sapienza C. 2015. DNA methylation differences between in vitro- and in vivo conceived children are associated with ART procedures rather than infertility. Clin Epigenetics 10.1186/s13148-015-0071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolzenberg DS, Grant PA, Bekiranov S. 2011. Epigenetic methodologies for behavioral scientists. Horm Behav 59: 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LM, Walker M, Cao HL, Yang Q, Duan T, Kingdom J. 2009. Assisted reproductive technology and placenta-mediated adverse pregnancy outcomes. Obstet Gynecol 114: 818–824. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Jing Q, Lia D, Pascual M, McLellan A, Greally JM. 2010. Optimized design and data analysis of tag-based cytosine methylation assays. Genome Biol 11: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierling S, Souren NY, Gries J, Loporto C, Groth M, Lutsik P, Neitzel H, Utz-Billing I, Gillessen-Kaesbach G, Kentenich H. 2010. Assisted reproductive technologies do not enhance the variability of DNA methylation imprints in human. J Med Genet 47: 371–376. [DOI] [PubMed] [Google Scholar]
- Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, Slagboom PE, Heijmans BT. 2009. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet 18: 4046–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turan N, Katari S, Gerson LF, Chalian R, Foster MW, Gaughan JP, Coutifaris C, Sapienza C. 2010. Inter- and intra-individual variation in allele-specific DNA methylation and gene expression in children conceived using assisted reproductive technology. PLoS Genet 6: e1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Voorhis BJ. 2006. Outcomes from assisted reproductive technology. Obstet Gynecol 107: 183. [DOI] [PubMed] [Google Scholar]
- Van Voorhis BJ. 2007. In vitro fertilization. N Engl J Med 356: 379–386. [DOI] [PubMed] [Google Scholar]
- Wang YA, Sullivan EA, Black D, Dean J, Bryant J, Chapman M. 2000. Preterm birth and low birth weight after assisted reproductive technology-related pregnancy in Australia between 1996 and 2000. Fertil Steril 83: 1650–1658. [DOI] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schübeler D. 2005. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37: 853–862. [DOI] [PubMed] [Google Scholar]
- Whitelaw N, Bhattacharya S, Hoad G, Horgan GW, Hamilton M, Haggarty P. 2014. Epigenetic status in the offspring of spontaneous and assisted conception. Hum Reprod 29: 1452–1458. [DOI] [PubMed] [Google Scholar]
- Wilhelm-Benartzi CS, Andres Houseman E, Maccani MA, Poage GM, Koestler DC, Langevin SM, Gagne LA, Banister CE, Padbury JF, Marsit CJ. 2012. In utero exposures, infant growth, DNA methylation of repetitive elements and developmentally related genes in human placenta. Environ Health Perspect 120: 296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo M, Patti ME. 2008. Diabetes risks begins in utero. Cell Metab 8: 5. [DOI] [PubMed] [Google Scholar]
- Yin LJ, Zhang Y, Lv P, He W, Wu Y, Liu A, Ding G, Dong M, Qu F, Xu C, et al. 2012. Insufficient maintenance DNA methylation is associated with abnormal embryonic development. BMC Med 10: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue MX, Fu XW, Zhou GB, Hou YP, DU M, Wang L, Zhu SE. 2012. Abnormal DNA methylation in oocytes could be associated with a decrease in reproductive potential in old mice. J Assist Reprod Genet 29: 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen RK, Neumann SM, Fok AK, Peñaherrera MS, McFadden DE, Robinson WP, Kobor MS. 2011. Extensive epigenetic reprogramming in human somatic tissues between fetus and adult. Epigenetics Chromatin 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner U, Pliushch G, Schneider E, El Hajj N, Tresch A, Shufaro Y, Seidmann L, Coerdt W, Müller AM, Haaf T. 2010. Quantitative methylation analysis of developmentally important genes in human pregnancy losses after ART and spontaneous conception. Mol Hum Reprod 16: 704–713. [DOI] [PubMed] [Google Scholar]
- Zheng HY, Tang Y, Niu J, Li P, Ye DS, Chen X, Shi XY, Li L, Chen SL. 2013. Aberrant DNA methylation of imprinted loci in human spontaneous abortions after assisted reproduction techniques and natural conception. Hum Reprod 28: 265–273. [DOI] [PubMed] [Google Scholar]
- Zuo T, Tycko B, Liu T, Lin H, Huang T. 2009. Methods in DNA methylation profiling. Epigenomics 1: 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]