Abstract
Friedreich's ataxia (FRDA) is an autosomal recessive neurological disease caused by expansions of guanine-adenine-adenine (GAA) repeats in intron 1 of the frataxin (FXN) gene. The expansion results in significantly decreased frataxin expression. We report that human FRDA cells can be corrected by zinc finger nuclease-mediated excision of the expanded GAA repeats. Editing of a single expanded GAA allele created heterozygous, FRDA carrier-like cells and significantly increased frataxin expression. This correction persisted during reprogramming of zinc finger nuclease-edited fibroblasts to induced pluripotent stem cells and subsequent differentiation into neurons. The expression of FRDA biomarkers was normalized in corrected patient cells and disease-associated phenotypes, such as decreases in aconitase activity and intracellular ATP levels, were reversed in zinc finger nuclease corrected neuronal cells. Genetically and phenotypically corrected patient cells represent not only a preferred disease-relevant model system to study pathogenic mechanisms, but also a critical step towards development of cell replacement therapy.
Introduction
Friedreich's ataxia (FRDA, OMIM229300) is the most common autosomal recessive ataxia, occurring in ~1–2/50,000 individuals, with a carrier frequency ranging from 1 : 50 to 1 : 100 individuals.1,2 Characteristic symptoms of FRDA include discoordination, slurred speech, peripheral neuropathy, and cardiomyopathy.3 At this time there is no effective treatment for FRDA. The disease is caused by hyperexpansion of the guanine-adenine-adenine (GAA) repeats located in the first intron of the FXN gene.2 The GAA repeat tract is polymorphic, with normal alleles not exceeding 30 repeats, and disease-causing alleles expanding up to 2,000 GAA repeats.2 FRDA patients homozygous for GAA expansion have low frataxin mRNA and protein levels when compared with heterozygous carriers and healthy controls. Importantly, heterozygous carriers of one expanded GAA allele are asymptomatic, indicating that correction of one allele is potentially curative. In various model systems and in patients' autopsy samples, the expanded GAA repeats induce chromatin changes leading to the epigenetic silencing of the FXN gene.4,5,6,7,8 This heterochromatinization of the FXN locus induced by repeat expansion can be recapitulated in model systems by inserting long GAA tracts outside of their natural sequence context into reporter genes.5,9,10 Although silencing triggers and the exact molecular mechanism of FXN silencing remain unknown,4 it is very likely that excision of the expanded GAA from FXN intron 1 would lead to the reactivation of frataxin expression.
GAA repeat-induced transcriptional inhibition is a major target for therapy (reviewed in refs. 11,12,13). The success of the strategies aimed to reactivate FXN expression depends predominately on the efficacy of potential inducers and their toxicity. Treatment with compounds stimulating FXN expression will require lifetime administration of an appropriate drug and could potentially be associated with developing resistance to the treatment. In addition, in the later stages of the disease, after cell death becomes apparent and damage irreversible, cell replacement therapy may provide the only potential therapeutic avenue. Therefore, it is essential to develop and test approaches for cellular therapy and regenerative medicine aimed to eliminate the source of transcriptional inhibition, the expanded GAA tract. The ability to derive induced pluripotent stem cells (iPSCs) from terminally differentiated patient cells allows us to obtain genetically matched cell types from different tissues. Correcting harmful mutations, such as repeat expansions, could allow for generation of patient-specific genetically corrected cells to be used for cell-specific disease models, regenerative medicine, tailored therapeutics, modeling of physiological developmental processes, and drug discovery.14,15,16,17,18,19
Concepts of induced, targeted repeat contractions were explored for myotonic dystrophy type 1 and fragile X syndrome using compounds with the potential to interfere with DNA conformations and/or with DNA repair pathways.20 However, the lack of specificity of these approaches may result in significant changes in cellular metabolism and consequently lead to a broad spectrum of side effects. Repeat-specific DNA cleavage was also demonstrated to induce repeat contractions.21,22 Another possible approach is gene targeting by homologous recombination to replace expanded repeats with a shorter tract. This approach was recently used to correct Huntington's disease iPSCs18; however, its application to hyperexpanded repeat sequences spanning several thousands of base pairs has not been demonstrated.
Recent advances in genome editing technologies, especially the development of engineered nucleases, including zinc finger nucleases (ZFNs), transcription activator-like effector nucleases, and clustered regularly interspaced short palindromic repeats-associated Cas9 nucleases, enable precise targeting of genome defects including deletions, insertions, and even individual point mutations.23 The combined binding specificity and cleavage accuracy make these engineered nucleases highly effective molecular tools that can be used to correct a plethora of genetic defects. In this study, we took a ZFN-mediated approach to excise the expanded GAA repeat region from the FXN gene in FRDA patient cells and restored expression of frataxin. The expanded tandem repeat sequences were edited via simultaneous cleavage by two ZFNs separated by several thousands of base pairs. Excision of the expanded GAAs from FXN resulted in a significant increase of frataxin expression in FRDA patient-derived lymphoblasts, fibroblasts, and iPSCs, along with a correction of the FRDA phenotype in iPSC-derived neuronal cells.
Results
Design of ZFNs that target sequences flanking the GAA tract
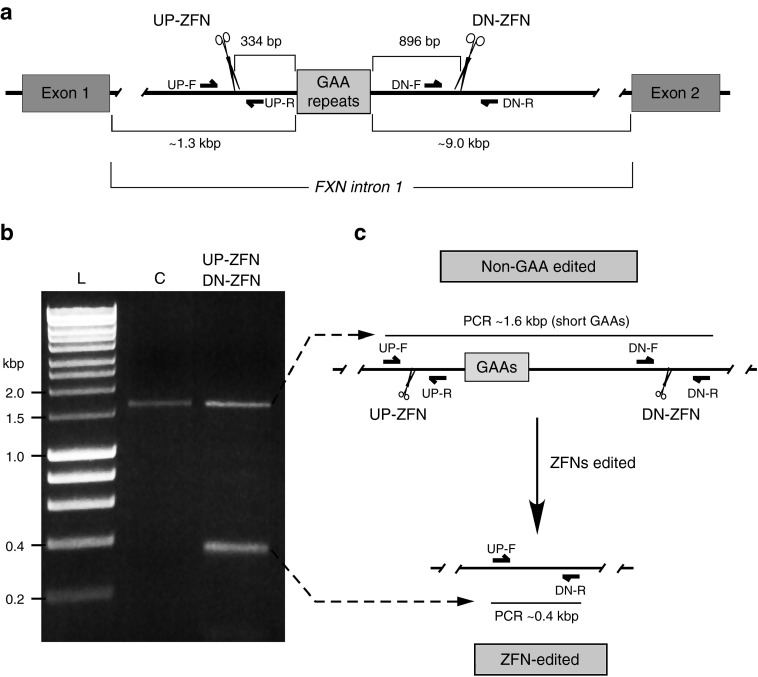
Intron 1 of the FXN gene spans ~10.4 kbp with the GAA repeats located proximal to exon 1, 1339 bp from the exon 1/intron 1 junction (Figure 1a). A very high abundance of interspersed repeat elements' upstream and downstream of the repeats24 precluded the design of specific ZFNs in the sequences immediately flanking the GAAs. Therefore, the ZFN cleavage site upstream of the GAA tract (UP-ZFN) is located 334 bp from the first GAA repeat and the ZFN cleavage sequence downstream of the repeats (DN-ZFN) is located 896 bp from the end of the GAA tract (Figure 1a and Supplementary Figure S1). To determine the feasibility of our approach, we conducted proof-of-principle ZFN analyses in K562 cells, a cell line amenable to genome editing.25 In vitro transcribed RNAs encoding the ZFNs (ZFN mRNA) were transfected into K562 cells, and the cleavage efficiency, as measured using the CEL I assay,26 was determined to be 4.3–11.7% for UP-ZFN and 20.6–26.8% for DN-ZFN (Supplementary Figure S2). Simultaneous double cleavage by UP-ZFN and DN-ZFN resulted in the excision of 1,230 bp + 3n bp (n designates number of repeats). After transfection of K562 cells harboring a nonexpanded GAA tract, both ZFN-edited (shorter) and non-GAA edited (longer) alleles were readily detected by PCR in the total population as early as 48 hours post-transfection (Figure 1b,c). The term “non-GAA edited” designates cells that were transfected by ZFNs; however, no sequence editing of the GAA region was detected DNA sequencing analyses of the UP- and DN-ZFN cleavage sites. Localization of UP- and DN-ZFN recognition sites as well as PCR primers used in the cleavage analyses are depicted in Supplementary Figure S1.
Figure 1.
ZFN targeting of intronic GAA repeats in the FXN gene. (a) Diagram of the FXN gene ZFN editing strategy. Exact location of the UP- and DN-ZFNs is shown relative to the proximal and distal end of the GAA repeat region. Approximate locations of the PCR primers (UP-F/R and DN-F/R) used to amplify ZFN targeted region are indicated. Exact locations and sequences of the primers and ZFNs are shown in Supplementary Figure S1. (b) Simultaneous cleavage by UP- and DN-ZFNs. K562 cells were cotransfected with UP/DN-ZFN mRNAs. In K562 cells harboring the short GAA tract, editing of intron 1 of the FXN gene leads to the release of ~1.2 kbp fragment. Deletion can be detected by PCR using UP-F and DN-R primers. Amplification of the non-GAA edited allele results in a 1.6-kbp fragment (lane C) while amplification of ZFN-edited DNA results in a 0.4-kbp fragment (lane UP-ZFN, DN-ZFN). L—Hyperladder I (BioLine). (c) Schematic illustrating ZFN-mediated editing of the FXN locus.
Nucleases were transfected as in vitro transcribed RNAs in order to decrease the possibility of off-target effects by reducing the level and persistence of ZFN expression in cells. In addition, the CompZr ZFNs (Sigma-Aldrich, St Louis, MO) used in our study contain FokI cleavage domains (termed ELD and KKR) that act as obligate heterodimers (Supplementary Figure S2). This approach has been shown to significantly reduce off-target cleavage.27 To further determine potential toxic effects of the UP/DN-ZFN pairs, we gauged the functional toxicity of UP- and DN-ZFN transfection and expression in FRDA fibroblasts using γH2AX immunostaining as a marker of DNA damage. The number of γH2AX foci was assessed in untransfected cells, mock transfected fibroblasts, cells treated with hydrogen peroxide, as well as cells transfected with GFP encoding plasmid, and GFP mRNA. A plasmid expressing the DFFB nuclease, which induces DNA fragmentation during apoptosis,28 was also used as a positive control. Transfection of the UP/DN-ZFN mRNAs did not lead to widespread double-strand DNA break formation typical for nonspecific DNA damage inducers. Instead, expression of the ZFN mRNAs resulted in an increased number of cells containing fewer than 5 γH2AX foci per nucleus (Supplementary Figure S3). Similarly, transfection with UP/DN-ZFN mRNAs did not result in significant toxicity as demonstrated by an XTT viability assay when compared with untransfected or mock-transfected fibroblasts (Supplementary Figure S3).
We also sought to determine specific off-target changes induced by the ZFNs. The initial, stringent CompZr ZFN design requires a minimum difference of two mismatches between the intended target site and any other locus in the genome per each arm of the nuclease (four mismatches per ZFN). In addition, we conducted in silico analyses of potential off-targets using three different computer algorithms: the ZFN-Site,29 Prognos,30 and Sigma–Aldrich algorithm (Supplementary Materials and Methods). No potential off-targets with less than six mismatches per ZFN were identified using any of the algorithms. The Sigma–Aldrich algorithm identified 22 potential off-target cleavage sites harboring six mismatches. Only 10 of the 22 loci were located in the vicinity of a gene or an annotated transcript (Supplementary Figure S4). We analyzed these 10 potential off-target locations in fibroblast cells using the CEL I assay (Supplementary Figure S5; Supplementary Materials and Methods). No bona fide off-targets of the UP/DN-ZFNs were identified among the 10 in silico predicted sites (Supplementary Figure S5).
Intronic sequences surrounding the GAA repeat tract do not affect expression of the FXN gene in K562 cells
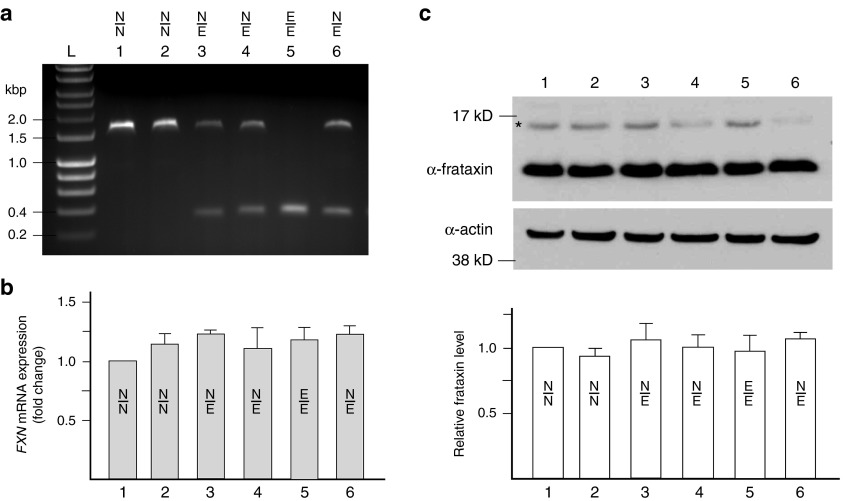
To determine whether excision of the short, nonpathogenic GAA repeats together with 1230 bp of flanking sequences of intron 1 affect expression of the FXN gene, we established 128 single-cell derived clonal lines from K562 cells transfected with UP/DN-ZFNs. K562 cells can be efficiently edited by ZFNs and were previously used to define regulatory elements of FXN expression.31 Six representative clones were genotyped using PCR and amplification of the 1.6 kbp band indicated the presence of the non-GAA edited allele (Figure 2a, lanes 1, 2, 3, 4, and 6), whereas the presence of a ~400 bp band demonstrated excision of the short GAA tract together with flanking sequences (Figure 2a, lanes 3–6). Among 128 clones analyzed, eight clones (6.3%) were found to be heterozygous for the corrected allele (Figure 2a clones 3, 4, and 6). In two cases (1.6%), we detected simultaneous editing of both FXN alleles (Figure 2a, clone 5).
Figure 2.
ZFN-mediated excision of the GAA repeat region in K562 cells does not affect frataxin expression. (a) Analysis of six representative clones derived from K562 cells cotransfected with UP/DN-ZFN mRNAs; N/N designates non-GAA edited clone (~1.6 kbp PCR product), N/E designates heterozygous clones with one edited and one non-GAA edited allele (~1.6 and 0.4 kbp PCR products), and E/E indicates clones with both alleles edited by ZFNs (0.4 kbp PCR product). (b) Results of qRT-PCR analysis of FXN expression. Heterozygous (samples 3, 4, and 6) or homozygous (sample 5) excision of the 1.2-kbp fragment of intron 1 of the FXN gene does not affect expression of FXN mRNA when compared with non-GAA edited cells (samples 1 and 2) or untransfected parental K562 cells (data not shown). Term “non-GAA edited” indicates cells that were nucleofected with UP- and DN-ZFN mRNAs and clonally expanded, but no sequence editing of the GAA region occurred as determined by DNA sequencing analyses of ZFN cleavage regions. Unless otherwise indicated, the non-GAA edited cells were used as controls in all experiments. (c) Editing of the FXN gene does not affect frataxin protein levels in K562 cells as determined by western blot. The asterisk denotes the precursor form of frataxin. Quantitation of the western blot (mature frataxin) is shown below. The results are reported as mean ± SD of three or more experiments. See also Supplementary Figure S6a.
Next, we evaluated whether excision of the short GAA tract changed the steady state levels of FXN mRNA and frataxin. Quantitative RT-PCR of the mature FXN mRNA (primers located in exons 3 and 4) and western blot analyses of cell lines heterozygous or homozygous for editing of the FXN locus revealed no differences in FXN gene expression compared to control cell lines (Figure 2b,c). K562 cells transfected with UP/DN-ZFNs but non-GAA edited, as determined by DNA sequencing of ZFNs target sites, served as control. These results demonstrate that the short GAA repeats and their flanking sequences, including the previously identified E-box,7 can be removed from intron 1 without unintended effects on FXN expression in K562 cells. Importantly, all other currently known cis regulatory elements of FXN transcription are located outside of the ZFN cleavage region.31,32
In a control experiment, we induced a deletion of ~3 kbp spanning a large fragment of intron 1 and exon 1 of the FXN gene together with a fragment of the 5′UTR and all transcription start sites, and we detected a near 50% decrease in FXN mRNA and protein production in K562 cells (data not shown). This result demonstrated that in the event of ZFN editing of the GAA repeat region, removal of critical expression regulators would be manifested by decreased FXN expression and not compensated by the second allele.
Double-strand DNA breaks induced by ZFN cleavage are predominantly repaired by nonhomologous end joining. This mutation-prone pathway frequently involves a resection of the DNA ends at the site of the break resulting in removal of additional DNA sequence beyond the ZFN cleavage site. To determine the exact DNA sequence of the ZFN-edited alleles, we cloned the targeted FXN region from four independent lines. Sequencing revealed deletions extending up to 94 bp beyond the ZFN-induced breaks (Supplementary Figure S6a).
Heterochromatin surrounding the expanded GAA repeats does not interfere with ZFN cleavage
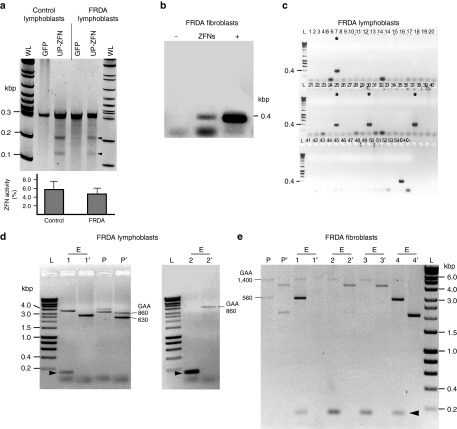
Numerous studies have demonstrated formation of a heterochromatin-like environment in the vicinity of expanded GAAs.4,8,33 Furthermore, it has been shown that accessibility of chromatin may decrease efficiency or even inhibit ZFN cleavage.34 In order to determine ZFN cleavage efficiency in FRDA cells, we transfected the UP-ZFN (located closer to the GAAs than DN-ZFN) into GM15850 FRDA lymphoblasts (Coriell Institute for Medical Research, Camden, NJ), which contain 630/860 GAA repeats and a well-characterized heterochromatin signature at the FXN locus.4,35 In parallel, we transfected the UP-ZFN into GM15851 control cells containing short GAA tracts and determined the efficiency of ZFN cleavage in both cell lines using the CEL I assay. The heterochromatin environment had no significant effect on UP-ZFN cleavage (Figure 3a). These data indicated that regardless of heterochromatin-like changes in the region upstream of the expanded GAA repeats, the flanking sequences are accessible to the ZFN and can be efficiently cleaved.
Figure 3.
Genetic correction of expanded GAA repeats in FRDA lymphoblasts and fibroblasts. (a) CEL I analysis to determine the efficiency of UP-ZFN cleavage in the heterochromatin region in cells harboring expanded GAAs; FRDA lymphoblasts (GM15850), control lymphoblasts (GM15851); arrowheads indicate expected cleavage products. Quantitative analysis of UP-ZFN activity in FRDA and control lymphoblasts is shown below. WL designates Wide Range DNA marker (Sigma), L—Hyperladder I. (b) Detection of the ZFN-edited alleles in the population of FRDA68 fibroblasts 48 hours post-transfection with UP- and DN-ZFNs (ZFN lane). K562 clone 5 (Figure 2a) was used as a positive control (lane +), whereas a PCR reaction with no template served as a negative control (lane –). (c) Detection of ZFN-edited clones from FRDA lymphoblasts by PCR with UP-F/R primers. Asterisks indicate positive, ZFN-edited clones; C+ positive control (K562 clone 5, Figure 2a), C-negative PCR control. (d) Analysis of the GAA repeat region in two ZFN-edited (E1 and E2) and non-GAA edited clone (P) derived from FRDA lymphoblasts using two PCR primer sets: ZFN-Ext F/R (lanes E1, E2, P) and ZFN-Int (lanes E1′, E2′, and P′) which produce amplicons that differ in the length of the sequences flanking the GAA repeats. The location of all primers relative to the GAA repeats and ZFN cleavage sites is depicted in Supplementary Figure S1. (e) Analysis of the GAA repeat region in identified ZFN-edited and non-GAA edited clones derived from FRDA68 fibroblasts. Clones P, E1, E2, E3, and E4 were analyzed using ZFN-Ext primers; clones P′, E1′, E2′, E3′, and E4′ were amplified with ZFN-Int primers. The short, ~0.2 kbp DNA fragment resulting from ZFN-mediated genetic correction of one of the alleles is indicated by an arrowhead. In some cases, because of the inherent difficulties of amplifying extremely long tracts of GAAs, a long exposure of the gel is required to visualize GAA amplification products (lane E1′). See also Supplementary Figure S6b,c.
Excision of the expanded GAAs in FRDA lymphoblasts and fibroblasts
To excise the expanded GAA tract and establish corrected FRDA cell lines, we cotransfected UP-ZFN and DN-ZFN mRNAs into FRDA cell lines GM15850 lymphoblasts and FRDA68 fibroblasts (560/1400 GAA repeats). A fraction of transfected cells was seeded on 96-well plates at an average density of ~0.5 cells per well 48–72 hours postnucleofection, and DNA from the remaining cells was analyzed by PCR with UP-F and DN-R primers to detect the ZFN-mediated excision. A 0.4-kbp product corresponding to the corrected allele was identified in the population of transfected cells (Figure 3b) as well as in single-cell derived clones (Figure 3c), indicating successful editing of the expanded GAAs.
Thirty (7 in GM15850 line and 23 in FRDA68 line) successfully edited, single-cell derived clones were identified among 649 analyzed (305 from GM15850 and 344 FRDA68). The large difference between the editing efficiency of lymphoblasts when compared with fibroblasts (2.3 versus 6.7%) is likely to be a consequence of approximately threefold lower efficiency of transfection of lymphoblasts when compared with fibroblasts.
In order to distinguish between homozygous and heterozygous editing events, we conducted PCR with primers specifically designed to amplify pathologically expanded GAA tracts. Analysis of the GAA repeat region was conducted using two PCR primer sets which amplify products that differ in the length of the sequences flanking the GAA repeats. PCR amplification using primers located outside of the UP-ZFN and DN-ZFN target sites (ZFN-External-F/R primers; Supplementary Figure S1) yields products that represent both the non-GAA edited allele (~3–5 kbp depending on number of GAAs) and ZFN-edited allele (~0.2 kbp) (Figure 3d,e). Because of the inherent difficulties in amplifying extremely long tracts of GAAs, the short 0.2-kbp product can outcompete the long GAA repeat-containing fragment during PCR (lane E2, Figure 3d). Therefore, a second set of nested primers (ZFN-Internal-F/R primers; Supplementary Figure S1), which hybridize inside the DNA region excised by ZFNs, was used to selectively amplify only the non-GAA edited allele (Figure 3d,e, lanes indicated by apostrophe). Amplification using primers spanning the longer flanking region revealed a short 0.2-kbp product specific for the edited alleles (indicated by arrowhead in Figure 3d,e) and a second, GAA repeat containing allele. Unlike in the K562 cells, only heterozygous ZFN corrections were identified. In some cases, clonal selection of the edited cells was incomplete and the DNA band from the corrected allele can be detected on the gel together with both expanded alleles (Figure 3e, lane E1), indicating that the population consists of edited cells and non-GAA edited, parental cells.
Removal of the expanded GAA repeats together with predicted flanking sequences was confirmed by DNA sequencing of selected clones. Resection of the DNA ends spanning up to 55 bp as a result of nonhomologous end joining repair of double-strand DNA breaks was also observed (Supplementary Figure S6b,c).
Correction of FRDA cells alleviates the molecular consequences of GAA expansion
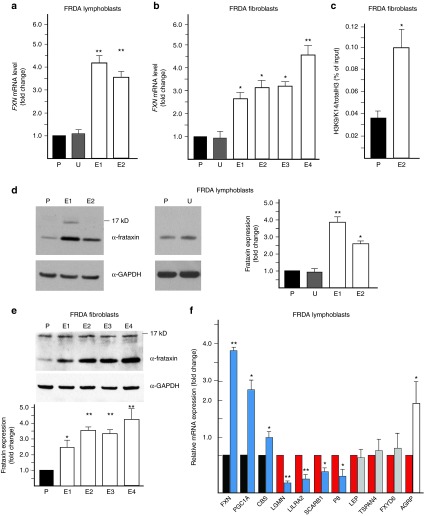
To assess whether heterozygous correction of the FRDA cells affects FXN expression, we measured levels of FXN mRNA and protein in the ZFN-edited cells lines. Importantly, excision of the expanded GAA repeats in both GM15850 lymphoblasts and FRDA68 fibroblasts increased expression of FXN mRNA (Figure 4a,b) and protein (Figure 4d,e) ~2.5–4.5-fold when compared with the non-GAA edited parental cells. GM15850 and FRDA68 cells transfected with UP/DN-ZFNs, but non-GAA edited, served as controls.
Figure 4.
Excision of the GAA tract increases frataxin expression and corrects the molecular phenotype of FRDA cells. (a) Determination of FXN mRNA expression using qRT-PCR in the ZFN-edited FRDA lymphoblast clones (E1 and E2) relative to the non-GAA edited control (P) and untransfected control (U). (b) Determination of FXN mRNA expression using qRT-PCR in the ZFN-edited FRDA fibroblast clones (E1–E4) relative to the non-GAA edited control (P) and untransfected control (U). (c) Chromatin immunoprecipitation with an antibody specific to acetylated lysines 9 and 14 on histone H3 (H3K9K14ac) in ZFN-corrected FRDA fibroblast clone E4 and non-GAA edited FRDA fibroblasts (P). (d) Analysis of frataxin expression in ZFN-edited FRDA lymphoblast clones E1 and E2 as determined by western blot. P designates non-GAA edited FRDA lymphoblasts; U designates untransfected FRDA lymphoblasts. Quantitative analysis of frataxin expression is shown in the graph. (e) Analysis of frataxin expression by western blot in the ZFN-corrected FRDA fibroblast clones (E1–E4) relative to the non-GAA edited control (P). Quantitation of the western blot is shown below. (f) The effect of ZFN correction on the FRDA signature of lymphoblast cells. A set of FRDA expression biomarkers37 was analyzed by qRT-PCR. The expression of the selected mRNAs was normalized to the non-GAA edited FRDA lymphoblasts. The mRNAs known to be underexpressed in FRDA lymphocytes are indicated as black bars while the overexpressed are shown as red bars.37 Editing of the expanded FXN allele reverses the changes related to frataxin deficiency in six biomarkers (blue bars), does not affect the expression of three mRNAs (gray bars), and has an inverse effect on expression of a single mRNA (white bar). The designations * and ** indicate statistically significant differences with P-values <0.05 and <0.01, respectively. The error bars represent SD of three or more experiments.
To determine the potential role of the E-box motif on FXN expression in FRDA fibroblasts, we generated a FRDA68 fibroblast cell line that retained the E-box motif but lacked one of the expanded GAAs. Importantly, expression analyses demonstrated similar FXN mRNA levels in cell lines lacking the expanded GAA tract irrespective of the presence of the E-box motif (data not shown).
Although consistently increased in each of the edited clones, we detected variability in the level of FXN expression between the corrected clones (Figure 4a,b,d,e). A strong correlation exists between the GAA repeat number and FXN expression36; therefore, the magnitude of increase of FXN expression depends on the number of GAAs in the remaining noncorrected allele. Hence, editing the shorter repeat expansion (e.g., 560 GAAs in FRDA68 fibroblasts) resulted in a smaller increase of FXN expression when compared with the excision of the longer GAA tract (e.g., 1400 GAAs in FRDA68; compare lanes E2 and E3 with E4 in Figure 3e with quantitation in Figure 4b,e).
As the expansion of the GAA repeats in FRDA is associated with epigenetic silencing, predominantly with changes in H3K9ac/me in the vicinity of the GAA repeats,4,7,35 the removal of expanded repeats even from a single allele should result in partial correction of this epigenetic alteration. To test this, we used chromatin immunoprecipitation to assess histone modifications in the region upstream of the UP-ZFN cleavage site. A significant increase of H3K9ac and K14ac was detected in corrected fibroblast cells (clone E4), which express the highest levels of frataxin, relative to the non-GAA edited controls (Figure 4c), indicating rescue of the epigenetic defect induced by expanded GAAs.
Next, we tested whether higher frataxin expression in the corrected lymphoblasts influenced the recently identified lymphocyte-specific FRDA biomarker signature.37 Using two edited clones, we determined that expression of 6 out of the 10 gene expression biomarkers was rescued by removal of the GAA expansion (Figure 4f). No significant changes in mRNA expression were observed for three biomarkers while the expression of only one biomarker was inversely affected by FXN correction. Overall, ZFN-mediated editing of the GAA repeat region had a similar effect on expression of FRDA biomarkers to the treatment of FRDA lymphocytes by histone deacetylase inhibitor 106, which transiently increases FXN expression.37 Thus, correction of the GAA expansion not only increased frataxin expression in FRDA cells, but also improved the molecular phenotype of the disease.
Generation and characterization of ZFN-corrected FRDA iPSCs and neuronal cells
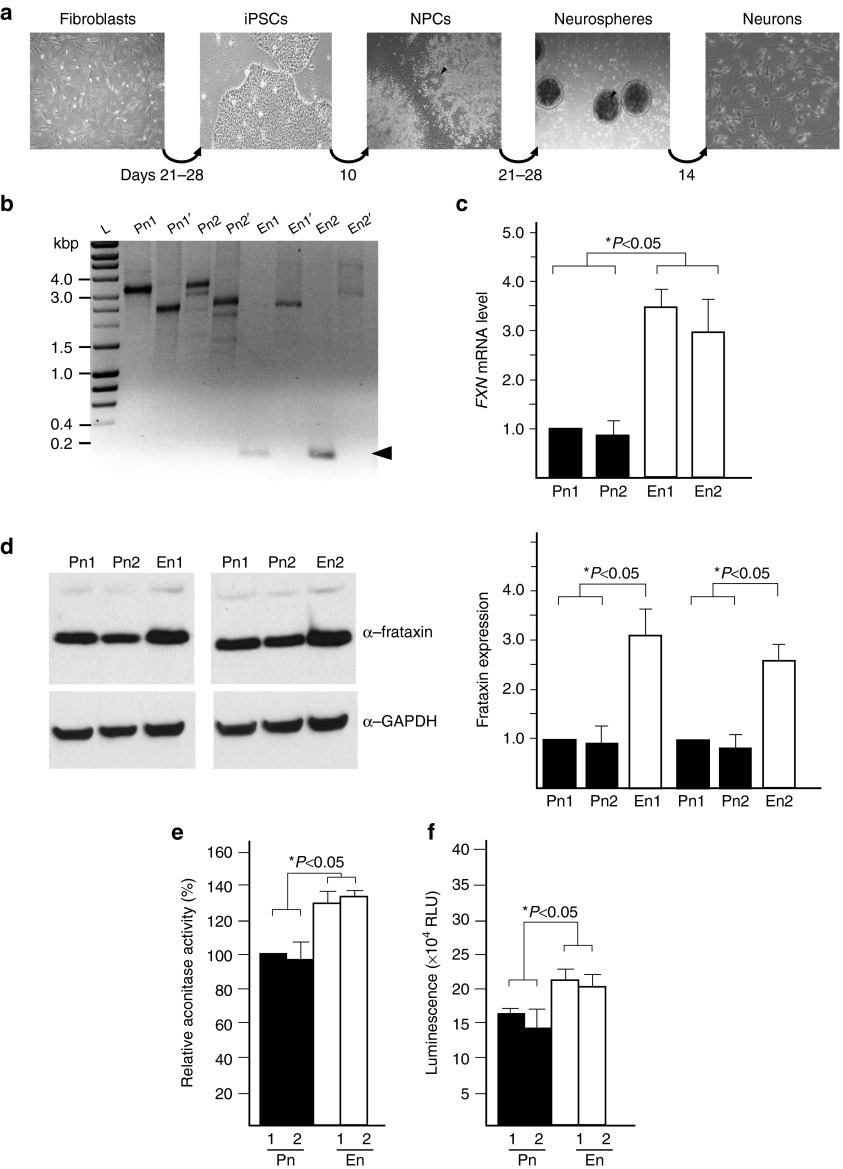
Both the ZFN-edited FRDA fibroblasts and lymphoblasts demonstrated a correction of the molecular FRDA phenotype as shown by increased FXN expression and rescue of several FRDA biomarkers. In order to determine whether excision of the expanded GAA tract and increased FXN expression will translate to an improved physiological phenotype in cells directly relevant to the pathogenesis of FRDA, we reprogrammed ZFN-corrected and non-GAA edited fibroblast lines to iPSCs and subsequently differentiated them to neuronal cells (Figure 5a).
Figure 5.
Genetic correction of the GAA expansion ameliorates phenotypic defects in iPSC-derived neuronal cells. (a) Schematic illustrating the protocol used to obtain neurons from FRDA fibroblast cells. The arrowheads designate neural rosettes. (b) Analysis of the size of the GAA repeat tract in iPSC-derived neuronal cells using GAA-Int and GAA-Ext PCR primers as described in the legend to Figure 3. The multiple bands visible on the agarose gel result from instability of the GAA repeats during the processes of iPSC reprogramming and differentiation to neuronal cells.6,53 Pn1 and Pn2 represent neuronal cells obtained from non-GAA edited iPSCs; En1 and En2 represent neuronal cells obtained from ZFN-edited iPSCs. The ~0.2 kbp DNA fragment resulting from ZFN-mediated genetic correction is indicated by an arrowhead. (c) FXN mRNA expression was determined using qRT-PCR in the corrected neurons (En1 and En2) and noncorrected neurons (Pn1 and Pn2). The data was normalized to the expression of Pn1. (d) Western blot analysis of frataxin levels in Pn1, Pn2, En1, and En2 neurons. (e) Analysis of the aconitase activity in the Pn1, Pn2, En1, and En2 neurons. (f) Intracellular levels of ATP were measured in En1 and En2 neurons and Pn1 and Pn2 neurons. The error bars represent SD of three or more experiments. See also Supplementary Figures S7 and S8.
We employed retroviral transduction of Oct3/4, Sox2, Klf4, and c-Myc transcription factors to derive four iPSC lines: two ZFN-corrected (iPS_Ei1, iPS_Ei2) and two parental lines (iPS_Pi1, iPS_Pi2). All lines were characterized in detail for the expression of pluripotency markers and karyotype status (Supplementary Figure S7a–d). In addition, the pluripotency status of all iPSC lines was evaluated using a qRT-PCR ScoreCard panel of 93 markers of pluripotency, mesodermal, ectodermal, and endodermal differentiation (Supplementary Figure S7e). Finally, the capacity of the iPSC cell lines to differentiate into all three germ layers was assessed by immunostaining embryoid bodies with Tuj1, Sox17, and ASMA specific antibodies (Supplementary Figure S8a).
Next, we differentiated the non-GAA edited, parental, and edited iPSC lines into neuronal cells (Figure 5b, Pn1, Pn2 and En1, En2, respectively). Tuj1-positive cells exhibited neuronal morphology and expressed high levels of several markers of mature neuronal cells (Supplementary Figure S8b). Importantly, GAA-corrected neurons expressed approximately threefold higher level of frataxin mRNA and protein when compared with non-GAA edited neurons (Figure 5c,d). As expected, based on the results of previous studies,6,38 no overt phenotypic differences in proliferation or morphology were observed between parental and ZFN-corrected iPSCs or neuronal cells.
Correction of GAA expansion increases aconitase activity and ATP levels in neuronal cells
FRDA patient tissues and cell lines exhibit impaired activities of several Fe–S containing enzymes, such as aconitase and mitochondrial respiratory chain complexes,39,40 as well as lower ATP levels that can be rescued by increased expression of frataxin.41 To determine whether the increase of frataxin expression in the edited cells affects these pathophysiological hallmarks of FRDA cells, we measured aconitase activity and the level of ATP in the corrected and non-GAA edited neurons.
Neuronal cells demonstrated ~30% increase in aconitase activity in two corrected lines when compared with the parental, non-GAA edited neurons (Figure 5e). Similarly, the total cellular ATP content of the ZFN-corrected neurons was ~25% higher than in the non-GAA edited neurons, indicating improved cell viability of the cells expressing a higher level of frataxin (Figure 5f). Overall, these data demonstrate that excision of one copy of the expanded GAAs in FRDA cells and subsequent generation of neuronal cells results in correction of the molecular phenotype of FRDA.
Discussion
Recent advances in reprogramming of somatic cells into pluripotent cells, their differentiation to various cell types, and especially the progress made in genome editing using sequence specific nucleases have introduced opportunities to develop new disease models and to test novel approaches of regenerative therapy. FRDA is an ideal target for such therapeutic strategies for the following reasons: (i) GAA repeat expansion is the most predominant genetic defect leading to FRDA, (ii) all patients carry expanded repeats on at least one allele, and (iii) the repeats are located in the first intron of the gene, the FXN coding sequence is unchanged and can be translated into functional frataxin. Furthermore, data from asymptomatic carriers indicate that correction of a single allele in patients' cells would be curative.2
In this study, we demonstrated that simultaneous cleavage using two ZFNs, with recognition sites located upstream and downstream of the expanded GAA repeat tract, corrects the genetic defect of FRDA cells. Editing of the intronic sequence has no effect on expression of the FXN gene in K562 cells harboring short repeats, but significantly increases frataxin mRNA and protein expression in FRDA patient cells. Furthermore, creating cell lines heterozygous for the GAA expansion (an equivalent of asymptomatic carrier cells) via correction of one expanded allele reverses the FRDA molecular signature in FRDA lymphoblast cells and epigenetic defects in FRDA fibroblasts. Most significantly, this targeted editing strategy results in increased aconitase activity and ATP levels in the corrected FRDA iPSC-derived neurons when compared with their non-GAA edited counterparts.
Here, we demonstrated that in spite of heterochromatin formation in the direct vicinity of the expanded GAA tracts, sequences surrounding the repeats could be efficiently cleaved by our ZFNs. Perhaps chromatin remodeling associated with cell-cycle and replication renders repeat-proximal sequences temporarily accessible to the ZFNs. In addition, as much as 20–30% of FXN mRNA can be detected in patient cells relative to unaffected control cells,36 rather indicating a partially repressive chromatin status within the FXN locus.
Given that intronic sequences harbor regulatory elements controlling gene expression, a potential caveat of this work was the excision of a significant portion of FXN intron 1 (1.2 kbp). The vast majority of identified cis elements controlling FXN expression were discovered using plasmid reporters carrying sequences in the vicinity of exon 1, more than 1 kbp upstream of the UP-ZFN cleavage site.31,32 However, analyses conducted using a luciferase reporter in mouse C2C12 cells indicated ~50% drop in transcriptional activity after deletion of an E-box motif located between the GAA tract and the UP-ZFN cleavage site.7 Our data showed that excision of this previously identified E-box motif did not influence FXN expression in K562 cells harboring short GAAs. Cell-type specificity as well as the endogenous chromatin environment, as opposed to episomal reporters, may contribute to the observed differences in the putative regulatory role of the E-box motif and further emphasizes the importance of retaining the natural chromosomal context in studies defining transcription control elements.
In cases of diseases caused by insufficient expression of a particular gene, the most logical intervention strategy is the supplementation of the deficient protein. This approach has been successfully employed to correct the cardiac phenotype in an FRDA mouse model (Mck-Cre-FxnL3/L-), which lacks frataxin expression in cardiac and skeletal muscle.42 However, sustained expression of exogenous frataxin may potentially induce toxic effects, especially if the protein is expressed at higher than physiological levels. In addition, molecular consequences of the GAA repeat expansion that are not directly linked to frataxin deficiency, but rather to the expanded repeats themselves, have been reported.43 These phenotypic changes would not be remedied by ectopic expression of the FXN gene. Moreover, chemical reactivation of transcription of the FXN gene with HDAC inhibitors or other chromatin targeting drugs can partially rescue frataxin deficiency; however, the long-term consequences of elevated levels of transcripts containing expanded GAAs are unknown. Increased expression of the FXN pre-mRNA could potentially lead to the accumulation of toxic RNAs or synthesis of toxic proteins via a repeat-associated non-ATG translation mechanism.44 Increased transcription through the expanded GAAs could also result in augmented somatic expansions of the repeat sequence.45 All of these potential side effects can be circumvented by excision of the pathologic repeat sequences using the specific genome editing approach presented in this work.
Neurons derived from FRDA iPSCs do not demonstrate a strong phenotype. Decreased mitochondrial membrane potential and changes in spontaneous and evoked action potentials were recently reported in iPSC-derived FRDA neurons when compared with unaffected controls.38 However, the small number of samples studied as well as inherent variability between patient and control cells can additionally obscure the relatively mild phenotype of the FRDA neurons. The establishment of non-GAA edited FRDA patient cells and their ZFN-corrected heterozygous counterparts allowed us to detect not only a significant increase in FXN expression and correction of the FRDA biomarker signature, but also increased aconitase activity and ATP levels. Up to a 40% difference in aconitase activity has been reported between neuronal cells derived from a FRDA transgenic and control mouse.46 Considering that ZFN-edited neuronal cells are equivalent to the heterozygous FRDA mutation carriers, a ~30% increase is in agreement with a partial rescue of frataxin deficiency. It is likely that correction of the GAA expansion of both alleles would be necessary for a complete reversal of the mitochondrial phenotype. As FRDA affects a specific subset of neuronal cells, primarily large sensory and dentate nuclei neurons,1 efficient differentiation of patient-derived iPSCs into these cell types may be required to uncover the robust functional phenotype of the disease. Moreover, FRDA is a slowly progressing disease; thus extended culturing conditions or an additional stimulus may be required to accentuate the phenotype.
FRDA is a multisystem disorder and frataxin deficiency affects several organs.1 Besides the nervous system, silencing of FXN expression leads to dysfunction of pancreatic beta cells resulting in glucose intolerance and frequently diabetes.1 More importantly, frataxin deficiency results in cardiomyopathy, the major cause of death in FRDA patients.47 Therefore, correction of iPSCs opens the possibility of a regenerative intervention that may counteract the debilitating consequences of frataxin insufficiency in tissues other than the nervous system. Severity and progression of FRDA correlate with the number of GAA repeats and FXN expression,36 indicating that even a small increase of frataxin levels will have a beneficial effect. It is also likely that partial replacement of the mutated cells with corrected cells will be sufficient to improve functions of affected organ/tissues and postpone disease progression.
Frataxin expression is variable among FRDA patients and among unaffected individuals, with mRNA levels spanning a fourfold range or twofold range, respectively (Li et al., unpublished data). Considering these differences in frataxin expression, the impact of an allelic correction in FRDA cells will vary and may depend on which allele is edited. For example, the extent of phenotype improvement will likely be different when correcting an allele containing long GAAs in FRDA cells expressing relatively high levels of frataxin when compared with the correction of the shorter of the two expanded alleles in FRDA cells with very low frataxin expression.
Furthermore, at least 12 other human diseases are caused by expansion of repeat sequences located in noncoding regions of the particular causal genes, including fragile X syndrome, myotonic dystrophies type 1 and 2, and amyotrophic lateral sclerosis.48 Results of our work provide a proof-of-principle for similar therapeutic strategies aimed towards other expansion disorders. In addition, an important aspect of our study was creating new, disease-specific in vitro models. ZFN-corrected and non-GAA edited FRDA cell lines may serve as closely matched pairs resembling control-patient counterparts in studies of disease mechanism or as a platform for drug screening and discovery.16 Although the use of ZFNs requires challenging analysis of the individual clones, recent improvements in the development of more robust editing strategies supported by rigorous safety studies may allow for much more efficient correction of repeat expansions, and perhaps even in vivo use of the editing nucleases without a need for ex vivo culturing, expansion, and reprogramming of patient cells.49 This emphasizes the importance of proof-of-concept in vitro studies to develop tools and standards that can subsequently translate to in vivo therapeutic approaches. The development and validation of highly specific engineered nucleases may also advance other approaches aimed to reactivate FXN expression, such us precise, locus specific delivery of gene expression activators, and DNA or chromatin modifying enzymes.50
Materials and Methods
Excision of the GAA repeat region using custom ZFNs. Editing of intron 1 of the FXN gene was conducted in K562, GM15850, and FRDA68 cells using custom UP-ZFN and DN-ZFN ZFNs as described in the Results section. Cell line specific nucleofection protocols were used to deliver ZFN mRNAs into the cells. The detailed ZFN editing protocol including determination of ZFN off-targets and toxicity are described in Supplementary Materials and Methods section.
Reprogramming of fibroblasts to iPS cells. Human iPS cells were obtained from non-GAA edited and ZFN-edited fibroblasts using retroviral transduction of Oct3/4, Sox2, Klf-4, and c-Myc transcription factors as described earlier.6,51 Characterization of the iPSC lines is described in the Supplementary Materials and Methods section.
Neuronal differentiation of iPSCs. Neuronal differentiation was conducted using AggreWell 800 plates (Stem Cell Technologies, Vancouver, British Columbia; cat. 27865) according to the manufacturer's protocol. The iPSCs were cultured for ~5 days followed by accutase treatment to obtain single cell suspensions. Approximately 3 × 106 cells were plated per each well of the AggreWell plate in STEMdiff medium supplemented with 10 μmol/l ROCK inhibitor Y27632. After 24 hours, two-thirds of the medium was replaced with fresh STEMdiff medium without Y27632. Aggregates were cultured for 5 days with daily media changes, then harvested, and plated onto poly-L-ornithine (Sigma, cat. P4957) and laminin-coated plates (Life Technologies, Carlsbad, CA; cat. 23017-015). After 5–7 days, distinct neural rosettes were separated from progenitor cells using STEMdiff Neural Rosette Selection Reagent (Stem Cell Technologies, cat. 05832), transferred to low attachment plates, and cultured in Neurobasal A medium (Invitrogen, Carlsbad, CA; cat. 10888-022) supplemented with N-2 Supplement (Invitrogen, cat. 17502048), B27 Supplement (Invitrogen, cat. 17504), insulin–transferin–selenium-A supplement (Invitrogen, cat. 51300), 20 ng/ml of basic fibroblast growth factor (bFGF) and 20 ng/ml of epidermal growth factor (EGF) (R&D Systems, Minneapolis, MN; cat. 236-EG). Neurospheres were cultured as described in ref. 52 and terminal neuronal differentiation was conducted as described in ref. 38. In brief, neurospheres were treated with accutase to obtain a single-cell suspension and plated on poly-D-lysine (Sigma, cat. P0899) and laminin-coated plates in Neurobasal A media without bFGF and EGF. To induce neuronal differentiation, 100 ng/ml of recombinant human neurotrophin 3 (Peprotech, Rocky Hill, NJ; cat. 450-03) and 100 ng/ml recombinant human brain-derived neurotrophic factor (Peprotech, cat. 450-02) were added to the medium. Unless otherwise indicated, cells were cultured for 2–4 weeks and half of the media volume was replaced with a fresh media every 2–3 days.
Determination of cellular ATP levels and aconitase activity. Neuronal cells were plated on poly-D-lysine and laminin-coated white 96-well plates at a density of 1–5 × 104 cells per well and cultured for 3–4 weeks in neurobasal A medium supplemented with N-2, B27, and ITS as described earlier. Cells were counted prior to the analyses and the total intracellular level of ATP was measured using a Luminescence ATP Detection Assay Kit (Abcam, Cambridge, MA; cat. 113849) and Synergy2 plate reader (BioTek, Winooski, VT).
Aconitase activity was determined in neuronal cell lysates as described in46 using the Aconitase Assay Kit (Cayman Chemical Company, Ann Arbor, MI; cat. 705502). Reactions were incubated at 37°C for 15 minutes, followed by absorbance measurements every minute for 25 minutes at 340 nm to determine the reaction rate and to calculate aconitase activity. Ten micrograms of neuronal cell extract was used to normalize the aconitase activity to citrate synthase activity using a Citrate Synthase Assay Kit (Sigma, cat. CS0720) according to the manufacturer's instructions.
Statistical analyses. Statistical analyses were conducted using GraphPad Prism 6. Statistical significance was determined by performing paired two-tailed Student's t-test and P < 0.05 was considered significant.
SUPPLEMENTARY MATERIAL Figure S1. Sequence of an FXN intron 1 fragment based on NCBI reference sequence: NG_008845.2. Figure S2. Characterization of UP-and DN-ZFNs. Figure S3. Toxicity of UP- and DN-ZFNs. Figure S4. In silico analysis of the potential off-target sites for UP- and DN-ZFNs using Sigma–Aldrich algorithm. Figure S5. CEL I analysis of in silico identified potential off-target sites. Figure S6. DNA sequence analyses of the cell lines edited by UP-ZFN and DN-ZFN. Figure S7. Characterization of FRDA patient-derived iPS cells (clones Pi1 and Pi2) and ZFN-edited FRDA patient-derived iPS cells (clones Ei1 and Ei2). Figure S8. Differentiation potential of the ZFN-edited and non-GAA edited iPSC lines. Supplementary Materials and Methods
Acknowledgments
These studies were supported by NIH 7R01NS081366 from NINDS to M.N., a grant from the Muscular Dystrophy Association (MDA0789 to M.N.), Friedreich's Ataxia Research Alliance (FARA to M.N.), and scholarship from UAB Civitan Emerging Scholar Award to Y.L. The authors wish to thank Shondra Miller (University of Washington, MI) for help with ZFNs and Elizabeth McIvor, Claudia Reyes for technical help. The authors do not declare any conflict of interest. Y.L., U.P., A.B., N.R., J.S.B., and M.N. conducted experiments. D.L. provided FRDA patient biopsy material. J.S.B., S.Y.R.D., and M.N. designed experiments. J.S.B. and M.N. wrote the paper. All authors read the manuscript, contributed comments and suggestions, and approved the final version of the manuscript.
Supplementary Material
References
- Koeppen, AH (2011). Friedreich's ataxia: pathology, pathogenesis, and molecular genetics. J Neurol Sci 303: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campuzano, V, Montermini, L, Moltò, MD, Pianese, L, Cossée, M, Cavalcanti, F et al. (1996). Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271: 1423–1427. [DOI] [PubMed] [Google Scholar]
- Delatycki, MB and Corben, LA (2012). Clinical features of Friedreich ataxia. J Child Neurol 27: 1133–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, D, Jenssen, K, Burnett, R, Soragni, E, Perlman, SL and Gottesfeld, JM (2006). Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat Chem Biol 2: 551–558. [DOI] [PubMed] [Google Scholar]
- Soragni, E, Herman, D, Dent, SY, Gottesfeld, JM, Wells, RD and Napierala, M (2008). Long intronic GAA*TTC repeats induce epigenetic changes and reporter gene silencing in a molecular model of Friedreich ataxia. Nucleic Acids Res 36: 6056–6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku, S, Soragni, E, Campau, E, Thomas, EA, Altun, G, Laurent, LC et al. (2010). Friedreich's ataxia induced pluripotent stem cells model intergenerational GAA⋅TTC triplet repeat instability. Cell Stem Cell 7: 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, E, Mahishi, L, Entezam, A, Kumari, D and Usdin, K (2007). Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res 35: 3383–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mahdawi, S, Pinto, RM, Ismail, O, Varshney, D, Lymperi, S, Sandi, C et al. (2008). The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum Mol Genet 17: 735–746. [DOI] [PubMed] [Google Scholar]
- Grant, L, Sun, J, Xu, H, Subramony, SH, Chaires, JB and Hebert, MD (2006). Rational selection of small molecules that increase transcription through the GAA repeats found in Friedreich's ataxia. FEBS Lett 580: 5399–5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda, CJ, Santos, MM, Ohshima, K, Smith, J, Li, L, Bunting, M et al. (2002). Frataxin knockin mouse. FEBS Lett 512: 291–297. [DOI] [PubMed] [Google Scholar]
- Gottesfeld, JM (2007). Small molecules affecting transcription in Friedreich ataxia. Pharmacol Ther 116: 236–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert, MD and Whittom, AA (2007). Gene-based approaches toward Friedreich ataxia therapeutics. Cell Mol Life Sci 64: 3034–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandi, C, Sandi, M, Anjomani Virmouni, S, Al-Mahdawi, S and Pook, MA (2014). Epigenetic-based therapies for Friedreich ataxia. Front Genet 5: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, K, Tanabe, K, Ohnuki, M, Narita, M, Ichisaka, T, Tomoda, K et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872. [DOI] [PubMed] [Google Scholar]
- Yamanaka, S and Blau, HM (2010). Nuclear reprogramming to a pluripotent state by three approaches. Nature 465: 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, H, Nagata, N, Kurokawa, H and Yamanaka, S (2014). iPS cells: a game changer for future medicine. EMBO J 33: 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J, Vodyanik, MA, Smuga-Otto, K, Antosiewicz-Bourget, J, Frane, JL, Tian, S et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920. [DOI] [PubMed] [Google Scholar]
- An, MC, Zhang, N, Scott, G, Montoro, D, Wittkop, T, Mooney, S et al. (2012). Genetic correction of Huntington's disease phenotypes in induced pluripotent stem cells. Cell Stem Cell 11: 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G, McCain, ML, Yang, L, He, A, Pasqualini, FS, Agarwal, A et al. (2014). Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med 20: 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes-Pereira, M and Monckton, DG (2006). Chemical modifiers of unstable expanded simple sequence repeats: what goes up, could come down. Mutat Res 598: 15–34. [DOI] [PubMed] [Google Scholar]
- Mittelman, D, Moye, C, Morton, J, Sykoudis, K, Lin, Y, Carroll, D et al. (2009). Zinc-finger directed double-strand breaks within CAG repeat tracts promote repeat instability in human cells. Proc Natl Acad Sci USA 106: 9607–9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard, GF, Viterbo, D, Khanna, V, Mosbach, V, Castelain, L and Dujon, B (2014). Highly specific contractions of a single CAG/CTG trinucleotide repeat by TALEN in yeast. PLoS One 9: e95611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H and Kim, JS (2014). A guide to genome engineering with programmable nucleases. Nat Rev Genet 15: 321–334. [DOI] [PubMed] [Google Scholar]
- Clark, RM, Dalgliesh, GL, Endres, D, Gomez, M, Taylor, J and Bidichandani, SI (2004). Expansion of GAA triplet repeats in the human genome: unique origin of the FRDA mutation at the center of an Alu. Genomics 83: 373–383. [DOI] [PubMed] [Google Scholar]
- Chen, F, Pruett-Miller, SM, Huang, Y, Gjoka, M, Duda, K, Taunton, J et al. (2011). High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods 8: 753–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulinski, J, Besack, D, Oleykowski, CA, Godwin, AK and Yeung, AT (2000). CEL I enzymatic mutation detection assay. Biotechniques 29: 44–6, 48. [DOI] [PubMed] [Google Scholar]
- Miller, JC, Holmes, MC, Wang, J, Guschin, DY, Lee, YL, Rupniewski, I et al. (2007). An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol 25: 778–785. [DOI] [PubMed] [Google Scholar]
- Enari, M, Sakahira, H, Yokoyama, H, Okawa, K, Iwamatsu, A and Nagata, S (1998). A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature 391: 43–50. [DOI] [PubMed] [Google Scholar]
- Cradick, TJ, Ambrosini, G, Iseli, C, Bucher, P and McCaffrey, AP (2011). ZFN-site searches genomes for zinc finger nuclease target sites and off-target sites. BMC Bioinformatics 12: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine, EJ, Cradick, TJ, Zhao, CL, Lin, Y and Bao, G (2014). An online bioinformatics tool predicts zinc finger and TALE nuclease off-target cleavage. Nucleic Acids Res 42: e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, K, Singh, A, Crooks, DR, Dai, X, Cong, Z, Pan, L et al. (2010). Expression of human frataxin is regulated by transcription factors SRF and TFAP2. PLoS One 5: e12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puspasari, N, Rowley, SM, Gordon, L, Lockhart, PJ, Ioannou, PA, Delatycki, MB et al. (2011). Long range regulation of human FXN gene expression. PLoS One 6: e22001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveliev, A, Everett, C, Sharpe, T, Webster, Z and Festenstein, R (2003). DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature 422: 909–913. [DOI] [PubMed] [Google Scholar]
- van Rensburg, R, Beyer, I, Yao, XY, Wang, H, Denisenko, O, Li, ZY et al. (2013). Chromatin structure of two genomic sites for targeted transgene integration in induced pluripotent stem cells and hematopoietic stem cells. Gene Ther 20: 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, E, Napierala, M and Dent, SY (2011). Hyperexpansion of GAA repeats affects post-initiation steps of FXN transcription in Friedreich's ataxia. Nucleic Acids Res 39: 8366–8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filla, A, De Michele, G, Cavalcanti, F, Pianese, L, Monticelli, A, Campanella, G et al. (1996). The relationship between trinucleotide (GAA) repeat length and clinical features in Friedreich ataxia. Am J Hum Genet 59: 554–560. [PMC free article] [PubMed] [Google Scholar]
- Coppola, G, Burnett, R, Perlman, S, Versano, R, Gao, F, Plasterer, H et al. (2011). A gene expression phenotype in lymphocytes from Friedreich ataxia patients. Ann Neurol 70: 790–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hick, A, Wattenhofer-Donzé, M, Chintawar, S, Tropel, P, Simard, JP, Vaucamps, N et al. (2013). Neurons and cardiomyocytes derived from induced pluripotent stem cells as a model for mitochondrial defects in Friedreich's ataxia. Dis Model Mech 6: 608–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidari, MM, Houshmand, M, Hosseinkhani, S, Nafissi, S and Khatami, M (2009). Complex I and ATP content deficiency in lymphocytes from Friedreich's ataxia. Can J Neurol Sci 36: 26–31. [DOI] [PubMed] [Google Scholar]
- Lodi, R, Cooper, JM, Bradley, JL, Manners, D, Styles, P, Taylor, DJ et al. (1999). Deficit of in vivo mitochondrial ATP production in patients with Friedreich ataxia. Proc Natl Acad Sci USA 96: 11492–11495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow, M, Pfister, MF, Yee, AJ, Schubert, M, Michael, L, Zhang, CY et al. (2000). Frataxin activates mitochondrial energy conversion and oxidative phosphorylation. Proc Natl Acad Sci USA 97: 12239–12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdomini, M, Belbellaa, B, Monassier, L, Reutenauer, L, Messaddeq, N, Cartier, N et al. (2014). Prevention and reversal of severe mitochondrial cardiomyopathy by gene therapy in a mouse model of Friedreich's ataxia. Nat Med 20: 542–547. [DOI] [PubMed] [Google Scholar]
- Bayot, A, Reichman, S, Lebon, S, Csaba, Z, Aubry, L, Sterkers, G et al. (2013). Cis-silencing of PIP5K1B evidenced in Friedreich's ataxia patient cells results in cytoskeleton anomalies. Hum Mol Genet 22: 2894–2904. [DOI] [PubMed] [Google Scholar]
- Cleary, JD and Ranum, LP (2014). Repeat associated non-ATG (RAN) translation: new starts in microsatellite expansion disorders. Curr Opin Genet Dev 26: 6–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditch, S, Sammarco, MC, Banerjee, A and Grabczyk, E (2009). Progressive GAA.TTC repeat expansion in human cell lines. PLoS Genet 5: e1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandi, C, Sandi, M, Jassal, H, Ezzatizadeh, V, Anjomani-Virmouni, S, Al-Mahdawi, S et al. (2014). Generation and characterisation of Friedreich ataxia YG8R mouse fibroblast and neural stem cell models. PLoS One 9: e89488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne, RM and Wagner, GR (2012). Cardiomyopathy in Friedreich ataxia: clinical findings and research. J Child Neurol 27: 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak, U, McIvor, E, Dent, SY, Wells, RD and Napierala, M (2013). Expanded complexity of unstable repeat diseases. Biofactors 39: 164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, PD, Lander, ES and Zhang, F (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, AW, Wang, H, Yang, H, Shi, L, Katz, Y, Theunissen, TW et al. (2013). Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res 23: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak, U, Hirsch, C, Ku, S, Gottesfeld, JM, Dent, SYR and Napierala, M (2012). Selecting and isolating colonies of human induced pluripotent stem cells reprogrammed from adult fibroblasts. J Vis Exp 60: e3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dottori, M and Pera, MF (2008). Neural differentiation of human embryonic stem cells. Methods Mol Biol 438: 19–30. [DOI] [PubMed] [Google Scholar]
- Liu, J, Verma, PJ, Evans-Galea, MV, Delatycki, MB, Michalska, A, Leung, J et al. (2011). Generation of induced pluripotent stem cell lines from Friedreich ataxia patients. Stem Cell Rev 7: 703–713. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.