Abstract
Although previous studies have documented a bottleneck in the transmission of mtDNA genomes from mothers to offspring, several aspects remain unclear, including the size and nature of the bottleneck. Here, we analyze the dynamics of mtDNA heteroplasmy transmission in the Genomes of the Netherlands (GoNL) data, which consists of complete mtDNA genome sequences from 228 trios, eight dizygotic (DZ) twin quartets, and 10 monozygotic (MZ) twin quartets. Using a minor allele frequency (MAF) threshold of 2%, we identified 189 heteroplasmies in the trio mothers, of which 59% were transmitted to offspring, and 159 heteroplasmies in the trio offspring, of which 70% were inherited from the mothers. MZ twin pairs exhibited greater similarity in MAF at heteroplasmic sites than DZ twin pairs, suggesting that the heteroplasmy MAF in the oocyte is the major determinant of the heteroplasmy MAF in the offspring. We used a likelihood method to estimate the effective number of mtDNA genomes transmitted to offspring under different bottleneck models; a variable bottleneck size model provided the best fit to the data, with an estimated mean of nine individual mtDNA genomes transmitted. We also found evidence for negative selection during transmission against novel heteroplasmies (in which the minor allele has never been observed in polymorphism data). These novel heteroplasmies are enhanced for tRNA and rRNA genes, and mutations associated with mtDNA diseases frequently occur in these genes. Our results thus suggest that the female germ line is able to recognize and select against deleterious heteroplasmies.
Heteroplasmy (intra-individual variation) in mitochondrial DNA (mtDNA) plays an important role in mtDNA-related diseases and has also been implicated in aging and cancer (Greaves et al. 2012; Wallace 2012; Chinnery and Hudson 2013; Lombès et al. 2014). Most mtDNA mutations that cause diseases due to defects in mitochondrial function exist as heteroplasmies and only cause disease symptoms when the frequency of the mutant allele exceeds a particular threshold (Wallace and Chalkia 2013). Below this threshold, individuals are asymptomatic, presumably because there are sufficient functional mitochondria for normal metabolism. Changes in the frequency of pathogenic mutations during the transmission of heteroplasmies from mothers to offspring can thus play an important role in the disease risk of the offspring. However, most of our knowledge concerning the dynamics of heteroplasmy transmission comes from studies of pathogenic mutations (Monnot et al. 2011; Shen et al. 2012; de Laat et al. 2013; Wallace and Chalkia 2013), which in blood have been shown to decrease over time and hence may not accurately reflect the overall level of such pathogenic mutations within an individual (Poulton and Morten 1993; ‘t Hart et al. 1996; Rahman et al. 2001; Rajasimha et al. 2008). Mouse models have also been utilized (Cree et al. 2008; Fan et al. 2008; Freyer et al. 2012; Ross et al. 2013), but to date, there have been only a few studies of normal patterns of heteroplasmy transmission in humans (Sekiguchi et al. 2003; Goto et al. 2011; Sondheimer et al. 2011; Guo et al. 2013; Rebolledo-Jaramillo et al. 2014), including studies of oocytes and placenta (Marchington et al. 1997, 2002; Jacobs et al. 2007), and several questions remain.
For example, although it is clear that a bottleneck occurs during the transmission of mtDNA genomes from mothers to offspring, the size of the bottleneck remains a contentious issue. Previous estimates of the effective number of transmitted mtDNA genomes range widely, from eight to 200 (Brown et al. 2001; Guo et al. 2013; Rebolledo-Jaramillo et al. 2014). However, all previous studies have assumed a constant size for the bottleneck across individuals; the effect of allowing the bottleneck size to vary among individuals has not been investigated. Moreover, it has been suggested that mtDNA genomes may not behave as independent entities but instead are organized into discrete units called “nucleoids,” each of which contains 5–10 mtDNA genomes (Jacobs et al. 2000; Cao et al. 2007; Khrapko 2008), although recently it has been suggested that the number may be much smaller, on the order of one mtDNA genome per nucleoid (Kukat et al. 2011). Each nucleoid is thought to be homoplasmic for mtDNA genome sequences; thus, mtDNA heteroplasmy at the cellular level would reflect nucleoids that are homoplasmic for different sequence variants. Nucleoid structures within cells have been studied microscopically and biochemically (Bogenhagen 2012), and nucleoid-based models have been found to provide a better fit to the segregation of heteroplasmic mtDNA genomes in cell lines than do simple bottleneck models in some studies (Cao et al. 2007; Khrapko 2008), but not in others (Cree et al. 2008). However, to date, nucleoid-based models have not been investigated in the transmission of mtDNA heteroplasmy from mothers to offspring.
Another issue is the degree to which negative (or purifying) selection may act on deleterious variants during the transmission of mtDNA heteroplasmy. There are conflicting results and views as to whether changes in the frequency of a heteroplasmic mutation from mother to offspring are governed solely by genetic drift, or whether there is an additional role for negative (purifying) selection (Jenuth et al. 1997; Durham et al. 2006; Stewart et al. 2008a,b; Wonnapinij et al. 2008; Wallace and Chalkia 2013; Rebolledo-Jaramillo et al. 2014). Negative selection during heteroplasmy transmission, as evidenced by a decrease in the frequency of presumably deleterious heteroplasmic variants in offspring compared to mothers, must operate on the female germ line and/or early in development after fertilization, and hence differs from negative selection operating on homoplasmic variants that reduce viability or fertility (Holt et al. 2014). The opportunities for, and extent of, such negative selection during heteroplasmy transmission in humans remain largely unknown.
Here, we utilize the Genomes of the Netherlands (GoNL) project (Boomsma et al. 2014; Genome of the Netherlands Consortium 2014), consisting of whole-genome sequence data from blood samples from 250 families, to carry out the largest study to date (to our knowledge) of the dynamics of heteroplasmy transmission across the entire mtDNA genome. We utilize the data on changes in minor allele frequency (MAF) from mothers to offspring at heteroplasmic sites to compare different models for the inheritance of mtDNA genomes, and we analyze the data for evidence of negative selection during heteroplasmy transmission.
Results
Heteroplasmy characteristics
After quality filtering, there were data from 246 families representing 228 trios, eight DZ twin quartets, and 10 MZ twin quartets (total of 254 independent transmissions of mtDNA from mother to offspring, assuming independent transmissions of mtDNA to the DZ twins), sequenced to an average mtDNA coverage of approximately 1200× (Supplemental Table S1; Supplemental Fig. S1). We first used strict criteria, requiring all heteroplasmies in each individual to pass the quality control measures (including a minimum minor allele frequency [MAF] of 2%) to identify heteroplasmies. These quality control measures also include criteria for detecting contamination and the potential influence of nuclear inserts of mtDNA (NUMTs), as discussed in detail elsewhere (Li et al. 2012; Li and Stoneking 2012). We then called additional heteroplasmies in mothers or offspring if the same two alleles were detected in both (regardless of frequency) and if the heteroplasmy at that position in either the mother or the offspring had passed the quality control measures. With these criteria, we identified 189 heteroplasmies at 163 positions in the 228 trio mothers and 159 heteroplasmies at 137 positions in the 228 trio offspring (Table 1; Supplemental Table S1); the heteroplasmy data for the twin quartets are considered separately (Supplemental Tables S2, S3). A list of all of the heteroplasmies identified and associated MAF can be found in Supplemental Data Set S1. We also analyzed a subset of these heteroplasmies (Supplemental Tables S4, S5) with a different detection method, namely droplet digital PCR (ddPCR), and found a very high and significant correlation (Pearson's correlation = 0.998, P < 0.00001) between MAF estimated from sequencing versus MAF estimated from ddPCR (Supplemental Fig. S2). Even for low frequency heteroplasmies (MAF < 0.05), the correlation is still quite high (Pearson's correlation = 0.890, P < 0.00001). Thus, the heteroplasmies identified by our criteria in the sequencing data are reproducible by a different method.
Table 1.
Summary of the heteroplasmy data for trios
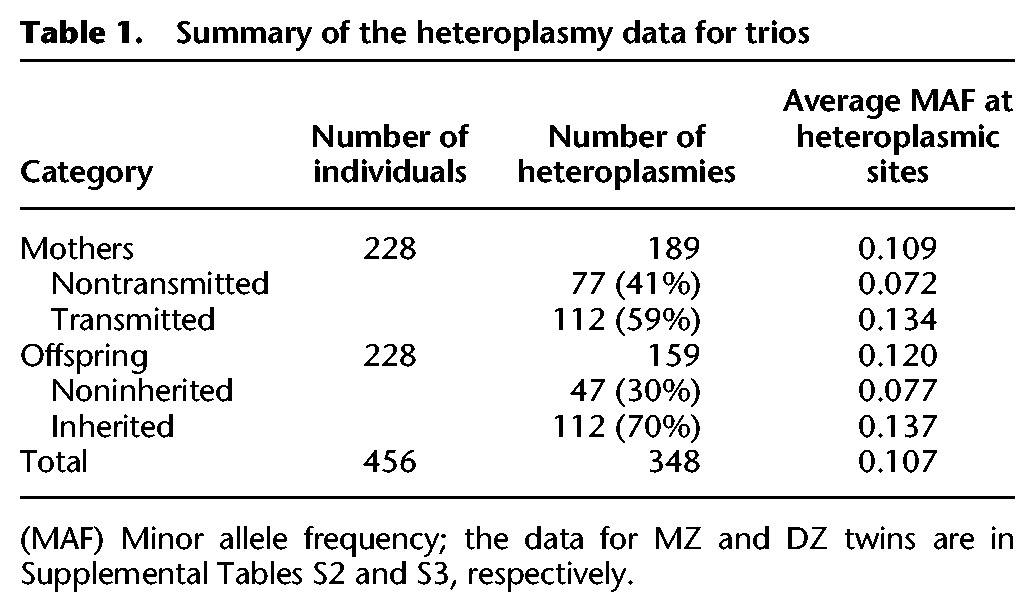
We first compared the number of heteroplasmies in the parents and offspring, using only those heteroplasmies that had passed all quality control measures so as not to introduce a bias when comparing mothers or offspring to fathers. There were significantly fewer heteroplasmies in the offspring than in the parents (P = 0.0179, Mann-Whitney U test), which presumably reflects the age-related accumulation of heteroplasmies (Liu et al. 1998; Sondheimer et al. 2011; Williams et al. 2013). Indeed, there is a small but significant correlation between age and number of heteroplasmies in the parents (Spearman's rho = 0.139, P = 0.000236). The number of heteroplasmies identified in the offspring is significantly correlated with the number of heteroplasmies in mothers (rho = 0.328, P = 3 × 10−7), but not with that in fathers nor with parental age at conception. The number of heteroplasmies per individual (using only unrelated individuals, e.g., the parents) does not differ significantly from a Poisson distribution (χ2 test, P = 0.63) (Supplemental Fig. S3), indicating that there is no evidence to indicate that heteroplasmies occur preferentially in some individuals; the mean number of heteroplasmies per individual was 0.7 (range 0–5). Finally, the MAF distribution across heteroplasmic sites did not differ significantly between mothers and fathers (P = 0.10, Mann-Whitney U test).
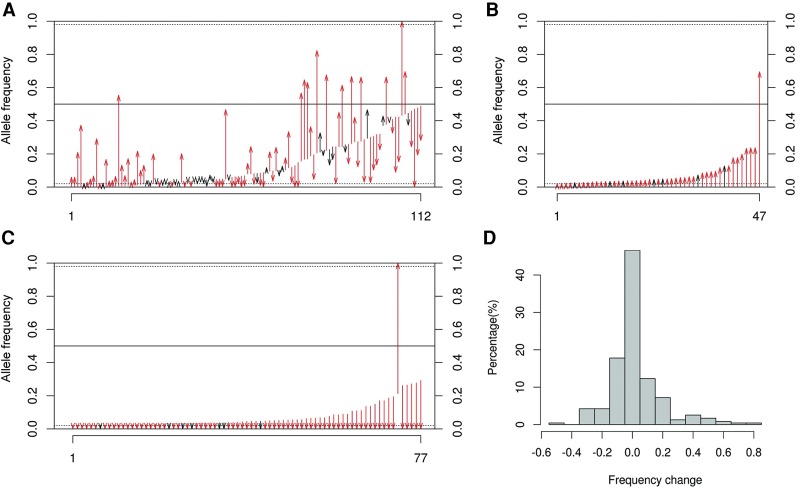
The average shift in heteroplasmy MAF between mothers and offspring was 0.108 (regardless of direction), with a maximum of 0.787 (Fig. 1). Although the MAF at all heteroplasmic sites did not differ significantly between mothers and offspring (P = 0.08, Mann-Whitney U test) (Table 1), the transmitted heteroplasmies in mothers had a significantly higher MAF than the nontransmitted heteroplasmies (P = 5 × 10−15) (Table 1). Similarly, the inherited heteroplasmies in offspring had a significantly higher MAF than the noninherited (i.e., de novo) heteroplasmies (P = 1 × 10−8) (Table 1), suggesting that heteroplasmies with higher MAF are more likely to be transmitted to the next generation and also have a higher mutant allele frequency than de novo mutations (Table 1). Differences in coverage could potentially explain these results; however, we did not find any significant coverage differences between transmitted versus nontransmitted heteroplasmies in mothers or between inherited versus noninherited heteroplasmies in offspring (Supplemental Fig. S4).
Figure 1.
Minor allele frequency (MAF) changes between mothers and offspring. Red arrows indicate cases in which the MAF changed significantly between mother and offspring (P < 0.00001, Fisher's exact test). (A) MAF change for heteroplasmies detectable in both mothers and offspring. (B) MAF change for heteroplasmies detected in offspring but not in mothers. (C) MAF change for heteroplasmies detected in mothers but not in offspring. (D) Summary of the MAF changes observed for all heteroplasmies.
We observe a higher concordance (i.e., smaller difference in MAF) in transmitted heteroplasmies in MZ than in DZ twins. For the 10 sets of MZ twins, the MAF differs significantly between the members of a twin pair at just one of seven heteroplasmic sites (Supplemental Table S2), whereas for the eight sets of DZ twins, the MAF differs significantly at six of nine heteroplasmic sites (Supplemental Table S3). MZ twins are therefore more concordant for MAF than are DZ twins, although this result is of borderline significance (P = 0.06, Fisher's exact test).
The major alleles at 18 positions differed between mothers and offspring (Supplemental Table S6). The biggest change was observed at np 15152, where the A allele increased from a frequency of 21.3% in the mother to fixation in the offspring. Interestingly, the observed G > A mutation at this position is a nonsynonymous mutation, resulting in a change from Gly to Ser at amino acid position 136 in the MT-CYB gene. This amino acid change is predicted to have a high risk of a functional effect (Reva et al. 2011) and has only been reported previously in the Acadian population (Secher et al. 2014) and in a patient with type 2 diabetes mellitus (Ohkubo et al. 2000). Only one of the shifts in the major allele from mother to offspring involved a putative de novo mutation (i.e., the mutant allele could not be detected in the mother): the mutant allele at np 8405, which had a frequency of 69.5% in the offspring (Supplemental Table S6). All the other putative de novo mutations had a frequency of <30% in the offspring (Fig. 1C). We emphasize that all of these putative de novo mutations could still reflect heteroplasmies that were actually transmitted from the mothers but were present in the mothers at too low a frequency for our methods to detect. Therefore, these results should be interpreted as providing an upper bound for the incidence and frequency shifts of de novo mutations.
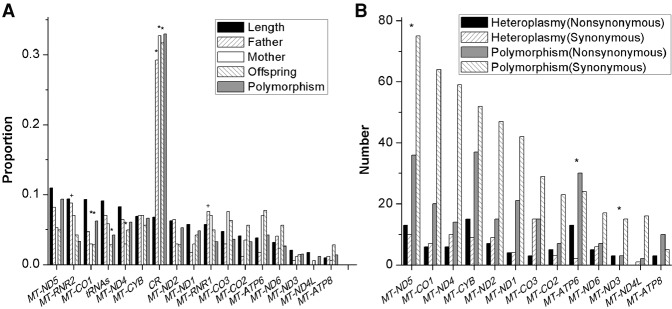
The distribution of heteroplasmies across different regions of the mtDNA genome did not differ among fathers, mothers, and offspring, but overall heteroplasmies were not distributed at random across the mtDNA genome (Fig. 2). Instead, heteroplasmies occurred preferentially in the noncoding control region (CR), as observed previously (He et al. 2010; Li et al. 2010; Rebolledo-Jaramillo et al. 2014; Ye et al. 2014), whereas MT-CO1, MT-ND4, and tRNAs had fewer heteroplasmies than expected based on gene length (Fig. 2). The heteroplasmy incidence is significantly correlated with the estimated mutation rate for each site (Pearson's rho = 0.421, P = 7 × 10−14), as observed previously (Li et al. 2010), which is in accordance with expectations under neutrality. However, compared to polymorphisms observed among the consensus mtDNA genome sequences from the same individuals, heteroplasmic mutations are enriched for nonsynonymous (NS) mutations relative to synonymous (SS) mutations (NS:SS ratio is 1.09 for heteroplasmies and 0.46 for polymorphisms, P = 2 × 10−6, Fisher's exact test; the details for each gene are shown in Fig. 2). Moreover, the NS:SS ratio decreases with increasing MAF (Supplemental Fig. S5), and the proportion of mutations that are predicted to have a high or medium risk of a functional effect is significantly higher in low frequency (MAF < 5%) than in high frequency (MAF > 5%) heteroplasmies (73% versus 31%, P = 0.0017, Fisher's exact test) (Supplemental Fig. S6).
Figure 2.
Distribution of heteroplasmies in different mtDNA gene regions. (A) Overall distribution of heteroplasmies. Black bars represent the expected heteroplasmy frequency for each gene region based purely on length. Uphill-striped, white, and downhill-striped bars represent the observed proportion of heteroplasmies identified in fathers, mothers, and offspring, respectively. Gray bars represent the polymorphism frequency for each gene region (inferred from PhyloTree Build 15 [van Oven and Kayser 2009]). Asterisks indicate significant differences (P < 0.01) between the observed and expected proportion of heteroplasmy based on gene region length, and the plus signs indicate significant differences between the observed and expected proportion of heteroplasmy based on polymorphism frequency. There were no significant differences observed between fathers and mothers, fathers and offspring, or mothers and offspring in the distribution of heteroplasmies across gene regions. (B) Number of nonsynonymous (NS) and synonymous (SS) heteroplasmies and polymorphisms observed in different genes. Asterisks indicate that the NS:SS ratio for heteroplasmies is significantly greater than the NS:SS ratio for polymorphisms in the respective gene (P < 0.05).
Negative selection
Overall, there is no evidence for nonrandom shift in the MAF (either increasing or decreasing) from mother to offspring (P = 0.903, Wilcoxon signed-rank test). In addition, as shown in Figure 3A, the average MAF was lower for NS heteroplasmies than for SS or noncoding (NC) heteroplasmies in both mothers and offspring (MAF = 0.174 for NS heteroplasmies versus 0.275 for SS-NC heteroplasmies in the mothers; MAF = 0.169 for NS heteroplasmies versus 0.249 for SS-NC heteroplasmies in the offspring). However, these differences in MAF between NS and SS-NC heteroplasmies are not significant in either the mothers (P = 0.127, Mann-Whitney U test) or the offspring (P = 0.317, Mann-Whitney U test).
Figure 3.
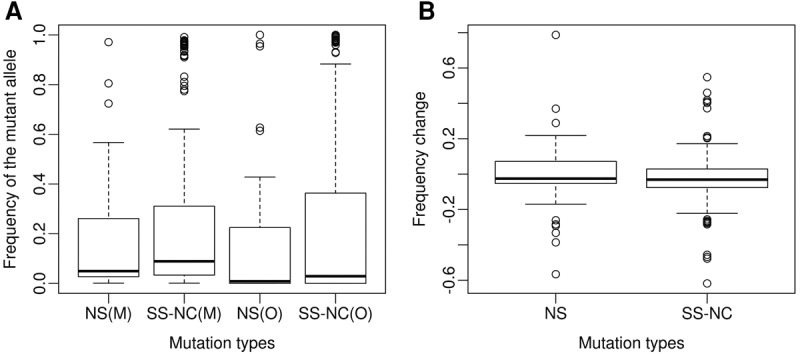
Frequency of the mutant allele (relative to the revised Cambridge Reference Sequence [rCRS]) and frequency change of the mutant allele for different mutation types. (A) Frequency of the mutant allele for: NS mutations in the mother [NS(M)]; SS and NC mutations in the mother [SS-NC(M)]; NS mutations in the offspring [NS(O)]; and SS and NC mutations in the offspring [SS-NC(O)]. (B) Distribution of the frequency change of NS and SS-NC mutations during transmission from mothers to offspring. The same results were obtained when the analyses were done for the minor allele at each position rather than the non-rCRS allele.
NS heteroplasmic mutations were further categorized, based on the likelihood of having a functional impact, into high risk, medium risk, low risk, and neutral (Reva et al. 2011). Although high risk mutations had a lower frequency and were more likely to be eliminated in the next generation (Fig. 4), the difference between the high risk and other categories was not significant (P = 0.26). Overall, these results do not provide compelling evidence for negative selection against NS mutations during transmission from mothers to offspring.
Figure 4.
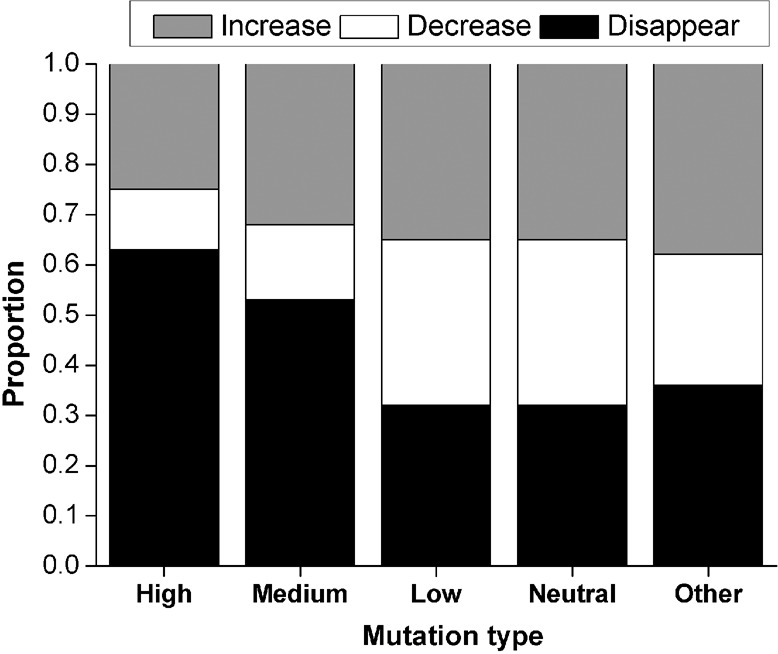
Frequency change during transmission for different types of NS mutations. NS mutations were categorized in terms of likely functional impact on the protein as high risk, medium risk, low risk, or neutral (Reva et al. 2011). Other: SS and NC mutations.
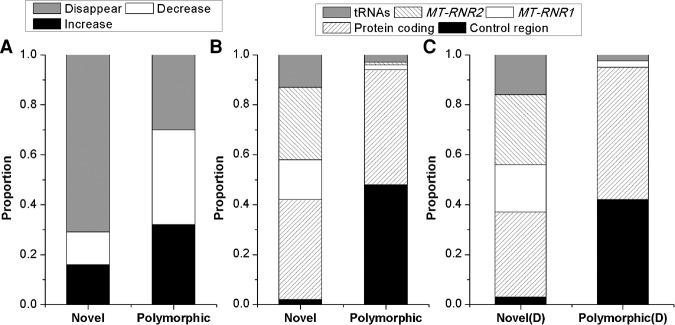
However, we do find significant differences in the transmission of polymorphic heteroplasmies (heteroplasmies for which both the major and minor alleles have been previously observed as polymorphic variants in comparisons among mtDNA sequences from worldwide individuals) (van Oven and Kayser 2009) versus novel heteroplasmies (heteroplasmies for which the alleles have not been reported previously as polymorphic variants). Novel heteroplasmies had a significantly lower MAF than polymorphic heteroplasmies in both mothers (novel MAF: 0.074; polymorphic MAF: 0.310; P = 0.0001, Mann-Whitney U test) and offspring (novel MAF: 0.056; polymorphic MAF: 0.286; P = 4 × 10−6). Moreover, the MAF decreased in frequency from mothers to offspring more often for novel heteroplasmies than for polymorphic heteroplasmies (84.4% of novel heteroplasmies versus 68.0% of polymorphic heteroplasmies; P = 0.049) (Fig. 5A). Consequently, a significantly larger proportion of novel heteroplasmies were eliminated in the offspring (71.1% of novel heteroplasmies versus 27.9% of polymorphic heteroplasmies, P = 2.2 × 10−6) (Fig. 5A).
Figure 5.
Transmission characteristics of heteroplasmies for novel versus polymorphic heteroplasmies. (A) Proportion of minor alleles observed in mothers that disappear entirely, decrease in frequency, or increase in frequency in the offspring for novel versus polymorphic heteroplasmies. (B) Distribution across genic regions of all heteroplasmies involving novel versus polymorphic heteroplasmies. (C) Distribution across genic regions for novel heteroplasmies that disappeared in the offspring [Novel(D)] compared to polymorphic heteroplasmies that disappeared in the offspring [Polymorphic(D)]. The MT-RNR1 and MT-RNR2 genes and the tRNA genes are overrepresented in the novel heteroplasmies.
This difference in the disappearance of novel versus polymorphic heteroplasmies remains significant even when controlling for the lower average MAF of novel heteroplasmies (Supplemental Fig. S7). Interestingly, novel heteroplasmies are overrepresented in ribosomal RNA (rRNA) and transfer RNA (tRNA) genes (Fig. 5B,C): These genes account for 58% of the novel heteroplasmies versus just 5% of the polymorphic heteroplasmies (P = 5 × 10−13, Fisher's exact test). Moreover, 88% of the novel mutations in rRNA and tRNA genes decreased in frequency in the offspring, and 77% disappeared entirely, compared to 67% of polymorphic mutations in rRNA and tRNA genes decreasing in frequency in the offspring and 33% disappearing entirely. As discussed subsequently, disease-associated mtDNA mutations often occur in the rRNA/tRNA genes; these results thus suggest that negative selection is occurring against novel heteroplasmies in the rRNA and tRNA genes that are likely to be deleterious.
Bottleneck models
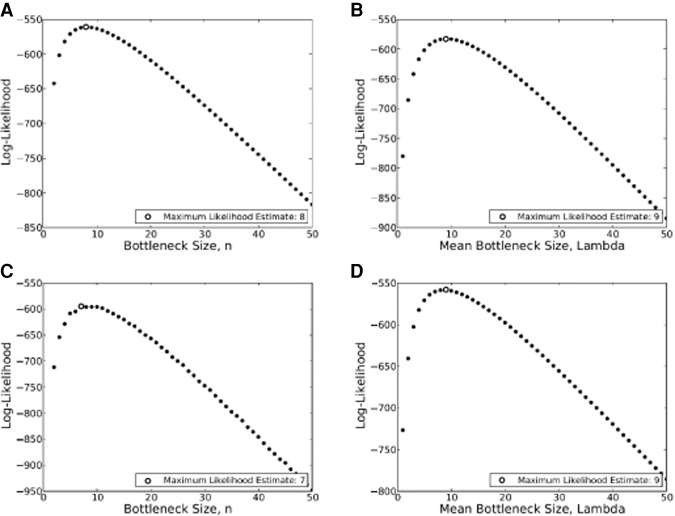
We used the distribution of the changes in MAF between mothers and offspring at heteroplasmic sites (Fig. 1D) to investigate the size and nature of the transmission bottleneck. We used one offspring and one heteroplasmic position per family so as to avoid any complications due to potential non-independence of heteroplasmies within families. The specific positions used are identified in Supplemental Data Set S1; similar results were obtained when different sets of positions were used. Previous studies of the bottleneck size have assumed a constant size for the bottleneck across individuals and treated each mtDNA genome as an independent unit (Guo et al. 2013; Rebolledo-Jaramillo et al. 2014); neither variable-size bottleneck models nor nucleoid models have been investigated. We therefore fit four models to the data: a constant-size bottleneck, a variable-size bottleneck, a constant-size bottleneck with nucleoids, and a variable-size bottleneck with nucleoids (see Methods). We took into account the observed coverage and associated sequencing error rate for each position in the analysis (see Methods). Under the constant-size bottleneck model, the maximum-likelihood estimate (MLE) of the number of transmitting mtDNA genomes was eight (Fig. 6A); and under the variable-size bottleneck model, the MLE was a mean of nine transmitted mtDNA genomes (Fig. 6B). For both nucleoid models, we assumed an average of 7.5 mtDNA genomes per nucleoid, in accordance with empirical data (Cao et al. 2007; Khrapko 2008; Bogenhagen 2012). The MLE was seven nucleoids for the constant-size bottleneck model (Fig. 6C); and for the variable-size bottleneck model, the MLE was a mean of nine transmitted nucleoids (Fig. 6D). The Akaike Information Criterion (AIC) for the various models (Supplemental Table S7) indicates that the variable-size bottleneck model with mtDNA as independent entities provides the best fit to the data (the smaller the AIC value, the smaller the loss in information when fitting the model).
Figure 6.
Likelihood curves for the observed data (changes in MAF from mothers to offspring) under different models for the transmission of mtDNA. (A) Likelihood curve for the constant-size bottleneck model with mtDNA genomes as segregating units. (B) Likelihood curve for the variable-size bottleneck model with mtDNA genomes as segregating units. (C) Likelihood curve for the constant-size nucleoid model. (D) Likelihood curve for the variable-size nucleoid model.
Discussion
This study of the transmission of human mtDNA heteroplasmy across the entire mtDNA genome is one of the largest to date and provides several important insights. However, before discussing these insights, it is necessary to consider the accuracy of the data. First, we note that since the GoNL data were originally aligned and mapped to the hg19 reference, a newer reference (GRChg38) has become available; remapping the data to GRCh38 in theory might change the results. However, the mtDNA reference sequence has not changed, so the only difference that could arise would be in the identification of NUMTs. We therefore did a BLAST search of the mtDNA genome against hg19 and GRCh38; there is a ∼3% increase in the number and total length of NUMTs in GRCh38 compared to hg19. Because additional steps are taken in the pipeline to remove potential NUMT sequences, as discussed in detail in Li et al. (2012), and the additional confirmation by ddPCR indicates that NUMTs are not being misidentified as heteroplasmies, it is highly unlikely that remapping reads to GRCh38 would result in any significant differences in the conclusions of our study.
Although next-generation sequencing platforms are extremely useful for detecting low-level heteroplasmy, rigorous criteria are needed to call heteroplasmies in order to avoid false positives due to sequencing errors (Bandelt and Salas 2012). Moreover, contamination with another sample can lead to the false appearance of heteroplasmy (Just et al. 2014). The pipeline we have implemented for calling heteroplasmies includes strict criteria for avoiding false positives due to sequencing errors, as well as for detecting contamination and avoiding the influence of NUMTs (Li et al. 2012; Li and Stoneking 2012). In addition, contamination was previously assessed independently in the GoNL genomic sequence data that we analyzed (Genome of the Netherlands Consortium 2014). Most importantly, a subset of the heteroplasmies identified by our pipeline from the Illumina sequence data were verified by an independent method (Supplemental Fig. S2; Supplemental Table S5); it is thus highly unlikely that the heteroplasmies we identified are sequencing errors, artifacts due to NUMTs, or contamination.
MZ versus DZ twins
We also find fewer heteroplasmies exhibiting significant differences in MAF in MZ twin pairs than in DZ twin pairs (Supplemental Tables S2, S3), which would not be expected if these reflected sequencing errors. This result suggests that for inherited heteroplasmies, the MAF in the offspring is largely influenced by the MAF in the oocyte at the time of fertilization rather than subsequent drift and/or selection during development. We note that a previous study did not report a higher concordance in heteroplasmy in MZ versus DZ twin pairs (Andrew et al. 2011); however, this study used a probe-based assay to interrogate a limited number of positions in the control region and hence only evaluated the presence/absence of heteroplasmy at a site, not the MAF. Another study, which utilized Sanger sequencing of PCR products of the control region, did find higher concordance in MAF for MZ than DZ twins (Bendall et al. 1996). Still, our observations are based on a relatively small number of twin pairs (10 MZ and eight DZ), and therefore need further evaluation in a larger data set.
Bottleneck models
We used the shifts in heteroplasmy MAF from mothers to offspring to estimate the size of the bottleneck that occurs during the transmission of mtDNA genomes. The size of the bottleneck was estimated under four models: a constant-size bottleneck model, in which each mtDNA genome is a segregating unit and the bottleneck size does not vary between individuals; a variable-size bottleneck model, in which each mtDNA genome is a segregating unit and the bottleneck size is allowed to vary between individuals; a constant-size nucleoid model, in which a nucleoid containing a variable number of homoplasmic mtDNA genomes is the segregating unit and the bottleneck size does not vary between individuals; and a variable-size nucleoid model, in which a nucleoid containing a variable number of homoplasmic mtDNA genomes is the segregating unit and the bottleneck size is allowed to vary between individuals. The best fitting model (as determined by AIC values) was a variable-size bottleneck, with an estimated mean of nine individual mtDNA genomes transmitted from mothers to offspring.
This number is smaller than a recent estimate of 30–35 mtDNA genomes transmitted, based on 39 mother-offspring pairs (Rebolledo-Jaramillo et al. 2014). Although this previous study assumed a constant-size bottleneck model, our estimate for a similar constant-size bottleneck model is also smaller, about eight mtDNA genomes transmitted (Fig. 6; Supplemental Table S7). The reason for this discrepancy is probably because we do not assume that the observed MAF in the offspring is identical to the MAF at transmission, as is usually assumed. Instead, we model the replication process from the bottleneck to the actual mtDNA population in the offspring, thereby allowing for genetic drift during the replication process (see Methods). Doing so allows for substantial changes in MAF during the replication process, but such changes will only be substantial if the bottleneck size is small. Incorporating drift in this way has two consequences: First, the same bottleneck model can be consistent with the few variants in the data set that have drastic changes in allele frequency and with the large set of variants in the data set that show a smaller change; second, small MAF in the offspring do not require very large bottleneck sizes. Consider that without a drift model, the smallest nonzero allele frequency possible is 1/n, where n is the bottleneck size. Hence, without modeling drift, all descendants with a very low MAF provide strong evidence for a large bottleneck size. However, by including drift, the final MAF in the offspring can be substantially smaller than the frequency at the bottleneck. As drift can only have substantial effects if the bottleneck size is small, this explains the estimate of a relatively small number of transmitted mtDNA genomes.
A variable-size bottleneck with each mtDNA genome as a segregating unit fit the data better than models involving nucleoids. However, this is not necessarily evidence against nucleoids, as we assumed an average of 7.5 mtDNA genomes per nucleoid, in accordance with some observations (Cao et al. 2007; Khrapko 2008; Bogenhagen 2012). If instead the number of mtDNA genomes per nucleoid is smaller, then the results based on nucleoids will approach the results based on mtDNA genomes as segregating units; in the limit, if each nucleoid contains exactly one mtDNA genome, as suggested by some studies (Kukat et al. 2011), then both models will give identical results. Our results therefore argue against the existence of nucleoids with several mtDNA genomes, but not necessarily against nucleoids with smaller numbers of mtDNA genomes. The most important conclusion is that the size of the bottleneck varies among individuals, whereas all previous attempts to model the size of the bottleneck have assumed that it is constant among individuals. Identifying the factors that influence this between-individual variation in bottleneck size would be of great interest and might have consequences for understanding the transmission of mtDNA-related diseases.
One limitation of our approach is that we are using the MAF observed in the mother's blood several years after conception as the estimate for the MAF in the egg at the time of conception. In the absence of data on heteroplasmy in human eggs, this limitation is unavoidable, although one way to improve the estimate would be to utilize heteroplasmy data from multiple tissues, as was recently done elsewhere (Rebolledo-Jaramillo et al. 2014). To further investigate this potential limitation, from a previous study of heteroplasmy variation across different tissues (Li et al. 2015), we calculated the correlation in MAF at heteroplasmic positions in blood and ovarian tissue from the same individual. There were 52 heteroplasmies with MAF > 0.02 detected in either blood or ovarian tissue (or both) in individuals with data from both tissues, and the MAF in blood exhibits a modest but nonetheless significant correlation with that in ovarian tissue (Pearson's correlation = 0.62, P < 0.0001). This would suggest that the MAF in blood is a reasonable proxy for the MAF in ovarian tissue, although data on heteroplasmy in human eggs would still be desirable.
A significant correlation between the mother's age at conception and the number of heteroplasmies detected in the offspring was reported previously (Rebolledo-Jaramillo et al. 2014). However, there is no such correlation in the GoNL data (Pearson's rho = −0.03, P = 0.65), although the range of mothers’ ages at conception is similar between the two studies (range = 18–44). The reason for this difference is unclear, and further studies are warranted.
Negative selection
Our results provide additional insights into the role of negative selection on heteroplasmic variants. We first examined nonsynonymous (NS) heteroplasmies in the coding region. As found previously (Li et al. 2010; Ye et al. 2014), NS heteroplasmies occur proportionally more often (relative to SS heteroplasmies) than do NS polymorphisms among individuals. Moreover, the ratio of NS:SS heteroplasmies is higher for low-frequency heteroplasmies (Supplemental Fig. S5), and low-frequency NS heteroplasmies are enriched for NS mutations that are likely to have a functional impact (Supplemental Fig. S6). These results are consistent with the threshold model for deleterious mtDNA mutations (Wallace and Chalkia 2013): Such mutations are tolerated at a low frequency within a cell because there are still sufficient nonmutant mtDNA genomes to carry out normal mitochondrial metabolism. However, above a certain frequency threshold, the deleterious mutations have a negative impact on mitochondrial function and so can never reach “fixation” within an individual. Thus, NS mutations that are likely to be deleterious are observed at heteroplasmic frequencies below the frequency threshold, but are never observed as polymorphisms (i.e., homoplasmic variants among individuals).
However, we do not see any compelling evidence for negative selection acting on NS heteroplasmies during transmission from mothers to offspring. The distribution of the average change in MAF from mothers to offspring does not differ for NS heteroplasmies versus SS-NC heteroplasmies (Fig. 3B). Although there is a tendency for NS heteroplasmies with a higher risk of a functional effect to be reduced in frequency or disappear entirely in offspring versus mothers (Fig. 4), the difference between high-risk versus other types of NS heteroplasmies is not significant. These results are in contrast to a previous study (Rebolledo-Jaramillo et al. 2014), which reported a significant decrease in the MAF for 11 NS heteroplasmies when comparing mothers to offspring. However, we have recomputed (based on a one-tailed sign test) the P-value associated with the decrease in MAF in this previous study and obtained a higher value (0.033) than that reported by the authors (9.54 × 10−7); with Bonferroni correction for multiple tests, this P-value is no longer significant. Moreover, in this previous study, the change in MAF for NS sites does not differ significantly from the MAF change for SS or NC sites, which we also found to be the case for the GoNL data. Overall, there does not seem to be any compelling evidence for selection in humans against NS heteroplasmies during transmission from mothers to offspring.
This conclusion is in contrast with previous studies based on the mouse mutator mtDNA model (Fan et al. 2008; Stewart et al. 2008a,b), which found evidence for purifying selection against NS mutations. It may be that the purifying selection in the mouse mutator model operates at a different level, as the NS mutations generated in the mouse mutator model may be starting at a much higher frequency than would be observed in the human family data, possibly already above the threshold at which an impact on mitochondrial metabolism could occur. Although the differences do not reach statistical significance, we do see a tendency for NS mutations with a higher risk of functional impact to be reduced in frequency in offspring. Further studies of potential purifying selection against NS mutations during heteroplasmy transmission in humans are warranted.
In contrast to the situation with NS heteroplasmies, we do see clear evidence for negative selection acting on heteroplasmies for novel mutations versus heteroplasmies for polymorphic mutations (Fig. 5). These novel heteroplasmies are enriched for mutations in rRNA and tRNA genes; intriguingly, disease-causing mtDNA mutations frequently occur in these genes (Taylor and Turnbull 2005; Tuppen et al. 2010; Wallace and Chalkia 2013). However, none of the observed rRNA/tRNA heteroplasmies are annotated as disease-associated on the MITOMAP website (www.mitomap.org). Based on criteria developed from conservation of mitochondrial tRNA sequences across species as well as potential for disrupting stem–loop pairing (Kondrashov 2005), three of six novel heteroplasmies and two of three polymorphic heteroplasmies are predicted to be deleterious. However, because ∼70% of tRNA mutations are predicted to be deleterious by these criteria, it is not clear how to interpret these numbers; further studies are needed to investigate the functional consequences of these novel heteroplasmies.
Overall, there is growing evidence for a role of negative selection in decreasing the frequency of potentially deleterious mutations during the transmission of mtDNA genomes from mothers to offspring (Fan et al. 2008; Stewart et al. 2008a). This negative selection could operate during the bottleneck that occurs during transmission of mtDNA from mothers to offspring, as this bottleneck might by chance raise the frequency of deleterious heteroplasmic mutations above the threshold at which impairment of mitochondrial function occurs (Freyer et al. 2012). Alternatively, there may be selective replication favoring nonmutant mtDNA during the amplification that occurs after the bottleneck, as has been recently found in Drosophila (Hill et al. 2014). Regardless of the underlying mechanism, it is important to recognize that negative selection during the transmission of deleterious heteroplasmies is not the same as negative selection against homoplasmic deleterious mtDNA mutations that reduce fertility or viability; the latter operates according to the well-known principles of Darwinian natural selection, whereas the former implies the existence of a mechanism in the female germ line for recognizing and preferentially discriminating against such deleterious heteroplasmies, perhaps via impaired mitochondrial function (Holt et al. 2014). Understanding how this putative mechanism operates could be of crucial importance in providing new therapeutic targets for reducing the transmission of disease-associated mtDNA mutations.
Methods
The characteristics of the study population and the production of the sequence data have been described in detail elsewhere (Boomsma et al. 2014; Genome of the Netherlands Consortium 2014); the sequence data can be accessed at the European Genome-Phenome Archive (study accession number EGAS00001000644). Briefly, genomic DNA was purified from blood samples from 769 individuals from across The Netherlands (consisting of 231 trios, 11 monozygotic [MZ] twin quartets, and eight dizygotic [DZ] twin quartets) and sequenced to an average genomic coverage of ∼14× on the Illumina HiSeq 2000 platform. The reads were aligned and mapped, and heteroplasmies called (with MAF > 0.02) as described in detail in the Supplemental Material (Processing mtDNA data). A subset of the inferred heteroplasmies were selected for independent verification via droplet digital PCR, which was performed as described previously (Li et al. 2015), with additional details in the Supplemental Material (Digital Droplet PCR). Potential contamination in the whole-genome sequence data was previously assessed by the GoNL Consortium (Genome of the Netherlands Consortium 2014); additional measures were taken to identify potential contamination as described in the Supplemental Material (Potential contamination). After removal of potential contaminants, there were 756 samples (228 trios, eight DZ twin quartets, and 10 MZ twin quartets) for further analysis.
The relative mutation rate for each heteroplasmic position was estimated as the number of mutations observed at that position in PhyloTree Build 15 (van Oven and Kayser 2009). Mutations noted in PhyloTree were defined as polymorphic mutations, whereas mutations that were not reported in PhyloTree were defined as novel mutations.
The size of the bottleneck during transmission of mtDNA genomes from mothers to offspring was estimated using a maximum-likelihood approach for four different models: a constant-size bottleneck model and a variable-size bottleneck model in which each mtDNA genome is the segregating unit; and a constant-size bottleneck model and a variable-size bottleneck model in which a nucleoid containing a variable number of identical mtDNA genomes (with mean = 7.5 genomes per nucleoid) is the segregating unit. The equations used for each model and further details concerning the modeling approach are in the Supplemental Material (Estimating the size of the bottleneck during mtDNA transmission and Modeling the replication process).
Genome of the Netherlands Consortium
Steering group
Cisca Wijmenga17,18 (principal investigator), Morris A. Swertz,17,18,19 P. Eline Slagboom,20 Gert-Jan B. van Ommen,21 Cornelia M. van Duijn,22 Dorret I. Boomsma,23 and Paul I.W. de Bakker24,25,26,27
Ethical, legal, and social issues
Jasper A. Bovenberg28
Cohort collection and sample management
P. Eline Slagboom,20 Anton J.M. de Craen,20 Marian Beekman,20 Albert Hofman,22 Dorret I. Boomsma,23 Gonneke Willemsen,23 Bruce Wolffenbuttel,29 and Mathieu Platteel17
Sequencing
Yuanping Du,30 Ruoyan Chen,30 Hongzhi Cao,30 Rui Cao,30 Yushen Sun,30 and Jeremy Sujie Cao30
Analysis group
Morris A. Swertz,17,18,19 (co-chair), Freerk van Dijk,17,18 Pieter B.T. Neerincx,17,18 Patrick Deelen,17,18 Martijn Dijkstra,17,18 George Byelas,17,18 Alexandros Kanterakis,17,18 Jan Bot,31 Kai Ye,20 Eric-Wubbo Lameijer,20 Martijn Vermaat,19,21,32 Jeroen F.J. Laros,19,21,32 Johan T. den Dunnen,21,32 Peter de Knijff,21 Lennart C. Karssen,22 Elisa M. van Leeuwen,22 Najaf Amin,22 Vyacheslav Koval,33 Fernando Rivadeneira,33 Karol Estrada,33 Jayne Y. Hehir-Kwa,34 Joep de Ligt,34 Abdel Abdellaoui,23 Jouke-Jan Hottenga,23 V. Mathijs Kattenberg,19,23 David van Enckevort,19 Hailiang Mei,19 Mark Santcroos,35 Barbera D.C. van Schaik,35 Robert E. Handsaker,27,36 Steven A. McCarroll,27,36 Evan E. Eichler,37 Arthur Ko,37 Peter Sudmant,37 Laurent C. Francioli,24 Wigard P. Kloosterman,24 Isaac J. Nijman,24 Victor Guryev,38 and Paul I.W. de Bakker24,25,26,27 (co-chair)
Supplementary Material
Acknowledgments
This study makes use of data generated by the Genome of the Netherlands (GoNL) Project. Samples were contributed by LifeLines, The Leiden Longevity Study, The Netherlands Twin Register, The Rotterdam Studies, and the Genetic Research in Isolated Populations Program. The sequencing was carried out in collaboration with the BGI. Funding for the GoNL was provided by the Netherlands Organization for Scientific Research under award number 184021007, dated July 9, 2009, and made available as a Rainbow Project of the Biobanking and Biomolecular Research Infrastructure Netherlands (BBMRI-NL). This research was supported by the National Science Foundation, Directorate for Biological Sciences (Graduate Research Fellowship Grant No. DGE 1256260 to R.R.); the Netherlands Bioinformatics Center (M.V. and J.F.J.L.); and the Max Planck Society (M.L., M.W., R.S., and M.S.). The Netherlands Twin Register acknowledges funding from the Netherlands Organization for Scientific Research (NWO) and MagW/ZonMW (grants 904-61-090, 985-10-002, 912-10-020, 904-61-193, 480-04-004, 463-06-001, 451-04-034, 400-05-717, Addiction-31160008, Middelgroot-911-09-032, Spinozapremie 56-464-14192); VU University's Institute for Health and Care Research (EMGO+) and Neuroscience Campus Amsterdam (NCA); and the European Research Council (ERC Advanced Award 230374).
Author contributions: M.S. and P.d.K. conceived the study; P.I.W.deB., J.A.B., C.M.v.D., G.J.B.v.O., P.E.S., M.A.S., C.W., D.I.B., and G.N.C. organized and oversaw the sampling and the production of the sequence data; M.L., R.R., M.V., J.F.J.L., and M.v.O. analyzed the data with input and supervision from M.K., S.Z., P.d.K., and M.S.; M.W. and R.S. performed the ddPCR experiments; M.L., R.R., P.d.K., and M.S. wrote the manuscript with input from the other authors; all authors read and approved the final manuscript.
Footnotes
Department of Genetics, University Medical Center Groningen and University of Groningen, Groningen 9700 RB, The Netherlands
Genomics Coordination Center, University Medical Center Groningen and University of Groningen, Groningen 9700 RB, The Netherlands
Netherlands Bioinformatics Centre, Nijmegen 6500 HB, The Netherlands
Section Molecular Epidemiology, Department of Medical Statistics and Bioinformatics, Leiden University Medical Center, Leiden 2300 RC, The Netherlands
Department of Human Genetics, Leiden University Medical Center, Leiden 2300 RC, The Netherlands
Department of Epidemiology, Erasmus Medical Center, Rotterdam 3000 CA, The Netherlands
Department of Biological Psychology, VU University, Amsterdam 1081 BT, The Netherlands
Department of Medical Genetics, University Medical Center Utrecht, Utrecht 3584 CG, The Netherlands
Department of Epidemiology, University Medical Center Utrecht, Utrecht 3584 CG, The Netherlands
Division of Genetics, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02116, USA
Broad Institute of Harvard and Massachusetts Institute of Technology, Cambridge, MA 02142, USA
Legal Pathways Institute for Health and Bio Law, Aerdenhout 2111, The Netherlands
Department of Endocrinology, University Medical Center Groningen, Groningen 9700 RB, The Netherlands
BGI, Shenzhen 518083, China
Leiden Institute of Advanced Computer Science, Leiden University, Leiden 2300 RC, The Netherlands
Center for Human and Clinical Genetics and Leiden Genome Technology Center, Leiden University, Leiden 2300 RC, The Netherlands
Department of Internal Medicine, Erasmus Medical Center, Rotterdam 3000 CA, The Netherlands
Department of Human Genetics, Radboud University Nijmegen Medical Centre, Nijmegen 6500 HB, The Netherlands
Bioinformatics Laboratory, Department of Clinical Epidemiology, Biostatistics and Bioinformatics, Amsterdam Medical Center, Amsterdam 1090 GE, The Netherlands
Department of Genetics, Harvard Medical School, Boston, MA 02115, USA
Department of Genome Sciences, University of Washington, Seattle, WA 98101, USA
Hubrecht Institute, Utrecht 3508 AD, The Netherlands
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.203216.115.
Contributor Information
Collaborators: Genome of the Netherlands Consortium, Manfred Kayser, Dorret I. Boomsma, Sebastian Zöllner, Peter de Knijff, and Mark Stoneking
References
- Andrew T, Calloway CD, Stuart S, Lee SH, Gill R, Clement G, Chowienczyk P, Spector TD, Valdes AM. 2011. A twin study of mitochondrial DNA polymorphisms shows that heteroplasmy at multiple sites is associated with mtDNA variant 16093 but not with zygosity. PLoS One 6: e22332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Salas A. 2012. Current next generation sequencing technology may not meet forensic standards. Forensic Sci Int Genet 6: 143–145. [DOI] [PubMed] [Google Scholar]
- Bendall KE, Macaulay VA, Baker JR, Sykes BC. 1996. Heteroplasmic point mutations in the human mtDNA control region. Am J Hum Genet 59: 1276–1287. [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen DF. 2012. Mitochondrial DNA nucleoid structure. Biochim Biophys Acta 1819: 914–920. [DOI] [PubMed] [Google Scholar]
- Boomsma DI, Wijmenga C, Slagboom EP, Swertz MA, Karssen LC, Abdellaoui A, Ye K, Guryev V, Vermaat M, van Dijk F, et al. 2014. The Genome of the Netherlands: design, and project goals. Eur J Hum Genet 22: 221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DT, Samuels DC, Michael EM, Turnbull DM, Chinnery PF. 2001. Random genetic drift determines the level of mutant mtDNA in human primary oocytes. Am J Hum Genet 68: 533–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Shitara H, Horii T, Nagao Y, Imai H, Abe K, Hara T, Hayashi J, Yonekawa H. 2007. The mitochondrial bottleneck occurs without reduction of mtDNA content in female mouse germ cells. Nat Genet 39: 386–390. [DOI] [PubMed] [Google Scholar]
- Chinnery PF, Hudson G. 2013. Mitochondrial genetics. Br Med Bull 106: 135–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cree LM, Samuels DC, de Sousa Lopes SC, Rajasimha HK, Wonnapinij P, Mann JR, Dahl HH, Chinnery PF. 2008. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat Genet 40: 249–254. [DOI] [PubMed] [Google Scholar]
- de Laat P, Koene S, Heuvel LP, Rodenburg RJ, Janssen MC, Smeitink JA. 2013. Inheritance of the m.3243A>G mutation. JIMD Rep 8: 47–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham SE, Samuels DC, Chinnery PF. 2006. Is selection required for the accumulation of somatic mitochondrial DNA mutations in post-mitotic cells? Neuromuscul Disord 16: 381–386. [DOI] [PubMed] [Google Scholar]
- Fan W, Waymire KG, Narula N, Li P, Rocher C, Coskun PE, Vannan MA, Narula J, Macgregor GR, Wallace DC. 2008. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science 319: 958–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyer C, Cree LM, Mourier A, Stewart JB, Koolmeister C, Milenkovic D, Wai T, Floros VI, Hagström E, Chatzidaki EE, et al. 2012. Variation in germline mtDNA heteroplasmy is determined prenatally but modified during subsequent transmission. Nat Genet 44: 1282–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genome of the Netherlands Consortium. 2014. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet 46: 818–825. [DOI] [PubMed] [Google Scholar]
- Goto H, Dickins B, Afgan E, Paul IM, Taylor J, Makova KD, Nekrutenko A. 2011. Dynamics of mitochondrial heteroplasmy in three families investigated via a repeatable re-sequencing study. Genome Biol 12: R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves LC, Reeve AK, Taylor RW, Turnbull DM. 2012. Mitochondrial DNA and disease. J Pathol 226: 274–286. [DOI] [PubMed] [Google Scholar]
- Guo Y, Li CI, Sheng Q, Winther JF, Cai Q, Boice JD, Shyr Y. 2013. Very low-level heteroplasmy mtDNA variations are inherited in humans. J Genet Genomics 40: 607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, Diaz LA Jr, Kinzler KW, Vogelstein B, Papadopoulos N. 2010. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature 464: 610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JH, Chen Z, Xu H. 2014. Selective propagation of functional mitochondrial DNA during oogenesis restricts the transmission of a deleterious mitochondrial variant. Nat Genet 46: 389–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt IJ, Speijer D, Kirkwood TB. 2014. The road to rack and ruin: selecting deleterious mitochondrial DNA variants. Philos Trans R Soc Lond B Biol Sci 369: 20130451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs HT, Lehtinen SK, Spelbrink JN. 2000. No sex please, we're mitochondria: a hypothesis on the somatic unit of inheritance of mammalian mtDNA. Bioessays 22: 564–572. [DOI] [PubMed] [Google Scholar]
- Jacobs L, Gerards M, Chinnery PF, Dumoulin J, de Coo I, Geraedts J, Smeets H. 2007. mtDNA point mutatioins are present at various levels of heteroplasmy in human oocytes. Mol Hum Reprod 13: 149–154. [DOI] [PubMed] [Google Scholar]
- Jenuth JP, Peterson AC, Shoubridge EA. 1997. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat Genet 16: 93–95. [DOI] [PubMed] [Google Scholar]
- Just RS, Irwin JA, Parson W. 2014. Questioning the prevalence and reliability of human mitochondrial DNA heteroplasmy from massively parallel sequencing data. Proc Natl Acad Sci 111: E4546–E4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrapko K. 2008. Two ways to make an mtDNA bottleneck. Nat Genet 40: 134–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA. 2005. Prediction of pathogenic mutations in mitochondrially encoded human tRNAs. Hum Mol Genet 14: 2415–2419. [DOI] [PubMed] [Google Scholar]
- Kukat C, Wurm CA, Spåhr H, Falkenberg M, Larsson NG, Jakobs S. 2011. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc Natl Acad Sci 108: 13534–13539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Stoneking M. 2012. A new approach for detecting low-level mutations in next-generation sequence data. Genome Biol 13: R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Schönberg A, Schaefer M, Schroeder R, Nasidze I, Stoneking M. 2010. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am J Hum Genet 87: 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Schroeder R, Ko A, Stoneking M. 2012. Fidelity of capture-enrichment for mtDNA genome sequencing: influence of NUMTs. Nucleic Acids Res 40: e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Schroder R, Ni S, Madea B, Stoneking M. 2015. Extensive tissue-related and allele-related mtDNA heteroplasmy suggests positive selection for somatic mutations. Proc Natl Acad Sci 112: 2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu VW, Zhang C, Nagley P. 1998. Mutations in mitochondrial DNA accumulate differentially in three different human tissues during ageing. Nucleic Acids Res 26: 1268–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombès A, Auré K, Bellané-Chantelot C, Gilleron M, Jardel C. 2014. Unsolved issues related to human mitochondrial diseases. Biochimie 100: 171–176. [DOI] [PubMed] [Google Scholar]
- Marchington DR, Hartshorne GM, Barlow D, Poulton J. 1997. Homopolymeric tract heteroplasmy in mtDNA from tissues and single oocytes: support for a genetic bottleneck. Am J Hum Genet 60: 408–416. [PMC free article] [PubMed] [Google Scholar]
- Marchington DR, Scott Brown MS, Lamb VK, van Golde RJ, Kremer JA, Tuerlings JH, Mariman EC, Balen AH, Poulton J. 2002. No evidence for paternal mtDNA transmission to offspring or extra-embryonic tissues after ICSI. Mol Hum Reprod 8: 1046–1049. [DOI] [PubMed] [Google Scholar]
- Monnot S, Gigarel N, Samuels DC, Burlet P, Hesters L, Frydman N, Frydman R, Kerbrat V, Funalot B, Martinovic J, et al. 2011. Segregation of mtDNA throughout human embryofetal development: m.3243A>G as a model system. Hum Mutat 32: 116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo E, Aida K, Chen J, Hayashi JI, Isobe K, Tawata M, Onaya T. 2000. A patient with type 2 diabetes mellitus associated with mutations in calcium sensing receptor gene and mitochondrial DNA. Biochem Biophys Res Commun 278: 808–813. [DOI] [PubMed] [Google Scholar]
- Poulton J, Morten K. 1993. Noninvasive diagnosis of the MELAS syndrome from blood DNA. Ann Neurol 34: 116. [DOI] [PubMed] [Google Scholar]
- Rahman S, Poulton J, Marchington D, Suomalainen A. 2001. Decrease of 3243 A→G mtDNA mutation from blood in MELAS syndrome: a longitudinal study. Am J Hum Genet 68: 238–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasimha HK, Chinnery PF, Samuels DC. 2008. Selection against pathogenic mtDNA mutations in a stem cell population leads to the loss of the 3243→G mutation in blood. Am J Hum Genet 82: 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebolledo-Jaramillo B, Su MS, Stoler N, McElhoe JA, Dickins B, Blankenberg D, Korneliussen TS, Chiaromonte F, Nielsen R, Holland MM, et al. 2014. Maternal age effect and severe germ-line bottleneck in the inheritance of human mitochondrial DNA. Proc Natl Acad Sci 111: 15474–15479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva B, Antipin Y, Sander C. 2011. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res 39: e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JM, Stewart JB, Hagström E, Brené S, Mourier A, Coppotelli G, Freyer C, Lagouge M, Hoffer BJ, Olson L, et al. 2013. Germline mitochondrial DNA mutations aggravate ageing and can impair brain development. Nature 501: 412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secher B, Fregel R, Larruga JM, Cabrera VM, Endicott P, Pestano JJ, González AM. 2014. The history of the North African mitochondrial DNA haplogroup U6 gene flow into the African, Eurasian and American continents. BMC Evol Biol 14: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi K, Kasai K, Levin BC. 2003. Inter- and intragenerational transmission of a human mitochondrial DNA heteroplasmy among 13 maternally-related individuals and differences between and within tissues in two family members. Mitochondrion 2: 401–414. [DOI] [PubMed] [Google Scholar]
- Shen SS, Liu C, Xu ZY, Hu YH, Gao GF, Wang SY. 2012. Heteroplasmy levels of mtDNA1555A>G mutation is positively associated with diverse phenotypes and mutation transmission in a Chinese family. Biochem Biophys Res Commun 420: 907–912. [DOI] [PubMed] [Google Scholar]
- Sondheimer N, Glatz CE, Tirone JE, Deardorff MA, Krieger AM, Hakonarson H. 2011. Neutral mitochondrial heteroplasmy and the influence of aging. Hum Mol Genet 20: 1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JB, Freyer C, Elson JL, Larsson NG. 2008a. Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nat Rev Genet 9: 657–662. [DOI] [PubMed] [Google Scholar]
- Stewart JB, Freyer C, Elson JL, Wredenberg A, Cansu Z, Trifunovic A, Larsson NG. 2008b. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol 6: e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ‘t Hart LM, Jansen JJ, Lemkes HH, De Knijff P, Maassen JA. 1996. Heteroplasmy levels of a mitochondrial gene mutation associated with diabetes mellitus decrease in leucocyte DNA upon aging. Hum Mutat 7: 193–197. [DOI] [PubMed] [Google Scholar]
- Taylor RW, Turnbull DM. 2005. Mitochondrial DNA mutations in human disease. Nat Rev Genet 6: 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. 2010. Mitochondrial DNA mutations and human disease. Biochim Biophys Acta 1797: 113–128. [DOI] [PubMed] [Google Scholar]
- van Oven M, Kayser M. 2009. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat 30: E386–E394. [DOI] [PubMed] [Google Scholar]
- Wallace DC. 2012. Mitochondria and cancer. Nat Rev Cancer 12: 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Chalkia D. 2013. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb Perspect Med 3: a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SL, Mash DC, Züchner S, Moraes CT. 2013. Somatic mtDNA mutation spectra in the aging human putamen. PLoS Genet 9: e1003990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonnapinij P, Chinnery PF, Samuels DC. 2008. The distribution of mitochondrial DNA heteroplasmy due to random genetic drift. Am J Hum Genet 83: 582–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Lu J, Ma F, Keinan A, Gu Z. 2014. Extensive pathogenicity of mitochondrial heteroplasmy in healthy human individuals. Proc Natl Acad Sci 111: 10654–10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.