Abstract
One approach to understanding the process of speciation is to characterize the genetic architecture of post-zygotic isolation. As gene regulation requires interactions between loci, negative epistatic interactions between divergent regulatory elements might underlie hybrid incompatibilities and contribute to reproductive isolation. Here, we take advantage of a cross between house mouse subspecies, where hybrid dysfunction is largely unidirectional, to test several key predictions about regulatory divergence and reproductive isolation. Regulatory divergence between Mus musculus musculus and M. m. domesticus was characterized by studying allele-specific expression in fertile hybrid males using mRNA-sequencing of whole testes. We found extensive regulatory divergence between M. m. musculus and M. m. domesticus, largely attributable to cis-regulatory changes. When both cis and trans changes occurred, they were observed in opposition much more often than expected under a neutral model, providing strong evidence of widespread compensatory evolution. We also found evidence for lineage-specific positive selection on a subset of genes related to transcriptional regulation. Comparisons of fertile and sterile hybrid males identified a set of genes that were uniquely misexpressed in sterile individuals. Lastly, we discovered a nonrandom association between these genes and genes showing evidence of compensatory evolution, consistent with the idea that regulatory interactions might contribute to Dobzhansky-Muller incompatibilities and be important in speciation.
Forty years ago, King and Wilson argued that differences between chimpanzees and humans could not be explained by changes in protein sequences alone (King and Wilson 1975). Since then, there has been a lively debate about the relative importance of changes in gene regulation versus changes in gene structure in adaptive evolution (e.g., Hoekstra and Coyne 2007; Carroll 2008), and some recent studies have revealed a major role for regulatory changes in adaptation (e.g., Jones et al. 2012).
The role of gene regulation in speciation has received less attention. This is somewhat surprising since gene regulation requires interactions between loci, and disrupted interactions between loci in hybrids (Dobzhansky-Muller incompatibilities) are thought to underlie many examples of post-zygotic reproductive isolation. At the transcriptional level, gene expression is a consequence of the interaction of cis-regulatory elements and trans-acting factors. Cis-regulatory regions are stretches of noncoding DNA that bind trans-acting factors to regulate mRNA abundance. Thus, negative epistatic interactions between cis- and trans-regulatory elements in hybrids might be important in reproductive isolation (Landry et al. 2005; Tulchinsky et al. 2014).
One powerful way to identify cis and trans changes is to compare expression differences between species with expression differences between alleles in inter-specific hybrids (Fig. 1; Cowles et al. 2002; Wittkopp et al. 2004). This approach has now been used in a number of crosses in flies, yeast, mice, and plants (Table 1). These studies have led to an emerging understanding of regulatory divergence within and between species as well as some understanding of the causes of misexpression in hybrids.
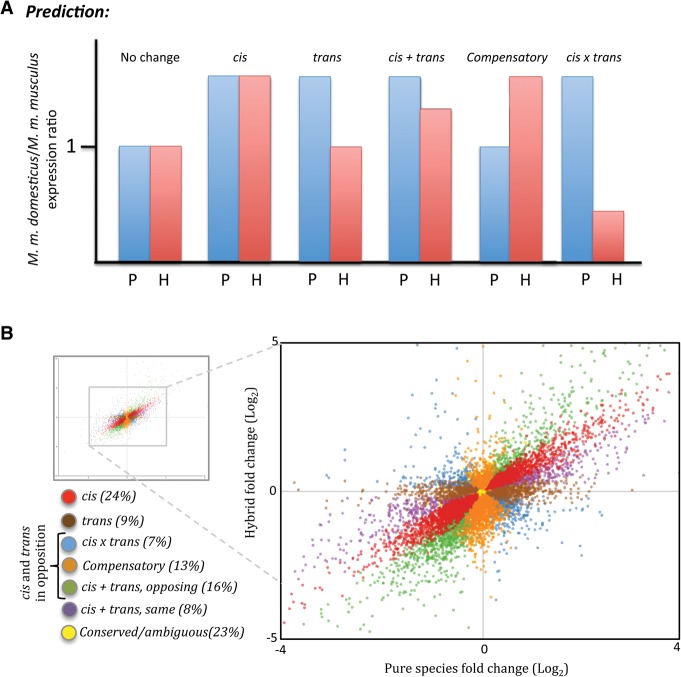
Figure 1.
(A) Categories of regulatory divergence between M. m. musculus and M. m. domesticus inferred from gene expression levels in pure subspecies and hybrids, where P and H are the ratio of reads mapping to M. m. domesticus versus M. m. musculus in the pure species and hybrids, respectively. (B) The relative distribution of regulatory categories in this data set. Each point represents one gene. Points represent log2 fold changes between reads mapping to each allele in the hybrid (M. m. domesticus/M. m. musculus) and the reads mapping to each subspecies (M. m. domesticus/M. m. musculus). Genes are color-coded based on their inferred regulatory category.
Table 1.
Studies that have identified regulatory divergence due to changes in cis and trans between species
Lacking in these studies is a direct association with reproductive isolation through a hybrid sterility or inviability phenotype. House mice (Mus musculus) provide a good opportunity for making links between hybrid sterility phenotypes, misexpression in hybrids, and regulatory divergence between lineages. House mice consist of three main subspecies that diverged recently and are isolated to varying degrees by hybrid male sterility. Over the past four decades, house mice have been developed as a model system for the study of mammalian hybrid sterility (e.g., Forejt and Iványi 1974; Forejt 1985, 1996; Oka et al. 2004, 2007, 2010, 2014; Britton-Davidian et al. 2005; Good et al. 2008a, 2010; Mihola et al. 2009; Bhattacharyya et al. 2013, 2014). Genes underlying hybrid sterility are polymorphic between different laboratory strains and in natural populations (Forejt and Iványi 1974; Good et al. 2008a; Vyskočilová et al. 2009; Bhattacharyya et al. 2014). Importantly, crosses between a wild-derived inbred line of M. m. musculus (PWK/PhJ) and a wild-derived inbred line of M. m. domesticus (LEWES/EiJ) result in infertile hybrid males in one direction and fertile hybrid males in the reciprocal direction. Infertile hybrid males in this cross have significantly reduced testis weight and sperm count compared to pure subspecies (Good et al. 2008a). For simplicity, hereafter we refer to these hybrid males with lowered fertility as “sterile” though sterility is not complete in all individuals. By comparing sterile and fertile hybrid males, it is possible to disentangle misexpression that is associated with sterility from misexpression that is simply a consequence of hybridization.
In a previous study using genome-wide microarray data, hybrid male sterility in this cross was associated with widespread overexpression of the M. m. musculus X Chromosome during spermatogenesis and misexpression at a number of autosomal genes (Good et al. 2010). This work suggested that differences in gene regulation might be important in reproductive isolation. More recently, Turner et al. (2014) mapped sterility quantitative trait loci (QTL) and expression QTL (eQTL) in an F2 cross using different strains of M. m. musculus and M. m. domesticus. They identified a large role for trans-eQTL as well as a number of complex regulatory network interactions related to sterility (Turner et al. 2014). However, the mapping approach was not designed to identify allele-specific expression patterns in F1s and did not address the relative importance of cis and trans changes to regulatory divergence between these subspecies.
Here, we compare expression differences between house mouse subspecies with expression patterns in sterile and fertile F1 hybrids. This allows us to address a number of related issues. First, we describe the proportion of changes between subspecies that are due to changes in cis, trans, or both. Second, when both kinds of changes occur, they may occur in the same direction or in the opposite direction. If gene expression is largely under stabilizing selection, as experimental work suggests (Denver et al. 2005; Lemos et al. 2005; Gilad et al. 2006), cis and trans- variants that act in opposite directions may be more common than expected by chance. We test this prediction. Third, the identification of cis-eQTL allows us to ask whether differences in expression are driven by positive selection (Bullard et al. 2010; Fraser et al. 2010, 2011) and, if so, to identify classes of genes that are under selection. Fourth, we identify misexpression (i.e., changes >1.25-fold on a log2 scale between the hybrid and both parents) in sterile and fertile hybrids. Comparing sterile and fertile hybrids allows us to identify those genes that are misexpressed only in sterile mice and thereby associate misexpression with hybrid sterility. While this approach does not distinguish between the specific genes causing sterility from those that are misexpressed as a downstream consequence of causative genes, it does identify a set of candidate genes for reproductive isolation and it makes specific testable predictions. In particular, we test the hypothesis that these candidate genes are disproportionately governed by compensatory evolution, as expected if regulatory interactions contribute to Dobzhansky-Muller incompatibilities.
Results
Extensive cis-regulatory divergence between M. m. musculus and M. m. domesticus
To characterize the contribution of cis- and trans-acting variants to divergence between M. m. musculus and M. m. domesticus, we compared expression differences in whole testis between subspecies with allele-specific expression in their fertile hybrid using three replicates per genotype (Fig. 1A). Since hybrids inherit alleles from both parents that meet in the same trans-acting environment, differences in expression between parents that are also seen between alleles in hybrids can be inferred to be the result of one or more cis-regulatory variants (Cowles et al. 2002). Alternatively, when a gene is differentially expressed between subspecies but not between alleles in the hybrid, we can infer divergence in one or more trans variants (Wittkopp et al. 2004).
Only reads that could be assigned preferentially to either M. m. musculus or M. m. domesticus were retained for analysis (see Supplemental Table S1 for read counts). This allowed us to measure allele-specific expression in hybrids by comparing the relative number of reads mapping to the genome of each subspecies. After excluding genes with low read counts from the analysis, 9851 autosomal genes could be tested for regulatory divergence (see Supplemental Methods). Of genes that could be tested, ∼24% (2349 genes) showed evidence of divergence due to one or more variants acting in cis alone, 9% (883 genes) showed evidence of divergence due to one more variants acting in trans alone, and 44% (4349 genes) showed evidence of divergence in both cis and trans (Fig. 1B).
The median regulatory divergence between subspecies in trans alone (0.58 log2 fold change) was significantly lower than the median divergence in cis alone (0.65 log2 fold change; Wilcoxon rank-sum test, P = 0.00019). Genes with an upper-quartile log2 fold change between subspecies (|log2 fold change| > 0.96) were also enriched for variants acting in cis alone relative to those in trans alone (40% cis alone, 9% trans alone; Fisher's exact test, P = 0.0003).
Widespread compensatory evolution
Genes with evidence of divergence in cis and trans can be further subdivided into categories based on their contribution to expression differences between subspecies and their direction of action. Genes with evidence of divergence in both cis and trans were divided into three subgroups (see Supplemental Methods and Fig. 1; Landry et al. 2005; McManus et al. 2010): (1) cis × trans, where there was significant differential expression between subspecies, significant differential expression between alleles in the hybrid, and where the subspecies with higher expression contributed the lower expressed allele in the hybrid; (2) compensatory, where the subspecies did not show differences in expression, but alleles in hybrids were significantly different; and (3) cis + trans, where there was significant differential expression between subspecies, significant differential expression between alleles in the hybrid, and where the subspecies with the higher expression level contributed the higher expressed allele in the hybrid. We further subdivided genes in this last category, cis + trans, into cases where cis and trans variants act in the same direction and cases where these variants act in opposition (Supplemental Fig. S1). Of genes with evidence of both cis and trans divergence, the majority were categorized as cis + trans (24%, or 2392 genes); in the majority of these, cis and trans variants act in opposition (1626 genes) rather than in the same direction (766 genes) (Fig. 1B). Thirteen percent of genes were categorized as compensatory (1309 genes). A minority of genes showed evidence of cis × trans divergence (7%, 648 genes) (Supplemental Table S2).
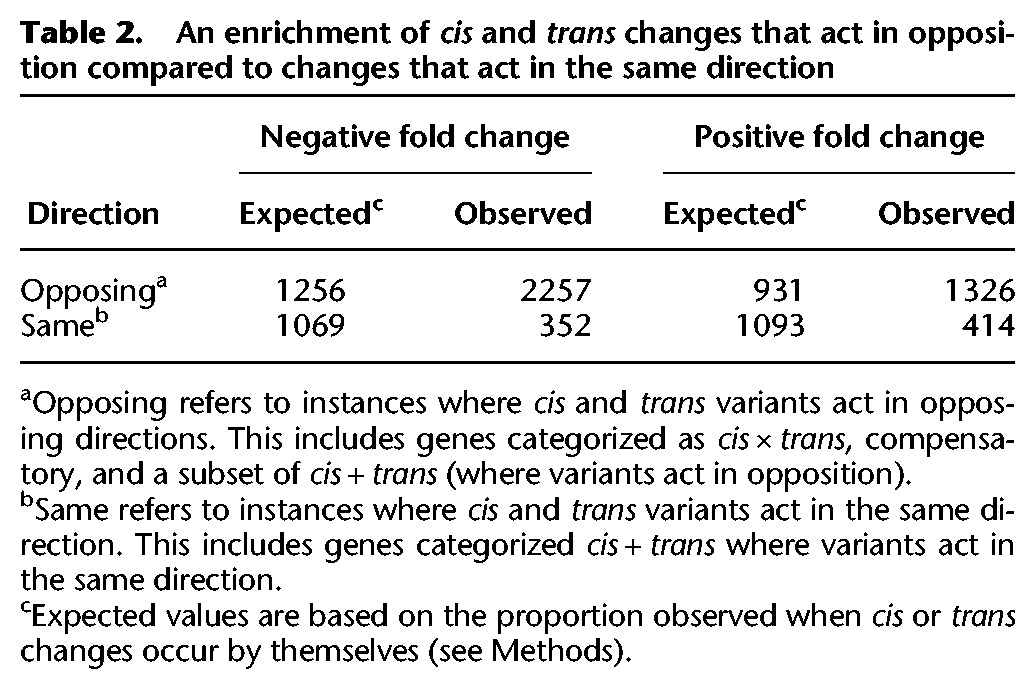
Under a neutral model, we expect an equal number of genes to show divergence due to cis and trans variants acting in opposition and cis and trans variants acting in the same direction. An excess of cis and trans changes acting to reinforce one another would be consistent with directional selection to alter expression level. Alternatively, an excess of cis and trans variants acting in opposition would be evidence for compensatory evolution and widespread stabilizing selection to maintain expression level. Genes categorized as cis × trans, compensatory, and a subset of cis + trans (where variants act in opposition) show evidence of cis and trans changes acting in opposite directions (Fig. 1B). In contrast, a subset of genes categorized as cis + trans show evidence of cis and trans changes that are acting in the same direction. By deriving neutral expectations from the number of independent cis and trans changes acting in the same and opposite directions, we tested for bias in directionality (see Methods). The proportion of cis and trans changes that act in opposition was extremely inflated compared to the neutral expectation (P < 0.0001) (Table 2), providing evidence for widespread compensatory evolution.
Table 2.
An enrichment of cis and trans changes that act in opposition compared to changes that act in the same direction
Adaptive evolution of cis-regulatory elements
Changes in cis variants are potentially targets for selection on gene expression level as cis-regulatory regions act as context-dependent regulators on which selection may act efficiently (for review, see Wray 2007). To test for lineage-specific selection on genes with divergent cis-acting variants between the subspecies, a gene-set approach was employed (Bullard et al. 2010; Fraser et al. 2010, 2011). Under a neutral model, an equal number of genes will be up- and down-regulated by cis variants. If a gene set associated with a biological function deviates from the null expectation by presenting a significant directional bias, we can infer lineage-specific selection. We tested this by grouping genes with only cis-acting variants by Gene Ontology (GO) terms (see Supplemental Methods). Three nonindependent biological process GO terms were identified with significant enrichment for biased directionality: (1) transcription, DNA-templated (GO:0006351, P = 0.0004); (2) positive regulation of transcription from RNA polymerase II promoter (GO:0045944, P = 0.02); and (3) regulation of transcription, DNA-templated (GO:0006355, P = 0.02). These inter-related gene sets collectively include 410 genes with putative evidence of selection and show biased directionality toward up-regulation in M. m. musculus (or down-regulation in M. m. domesticus).
Misexpression in hybrids
Crosses between M. m. domesticus (LEWES/EiJ) and M. m. musculus (PWK/PhJ) result in fertile hybrid males when the mother is M. m. domesticus and sterile hybrid males when the mother is M. m. musculus. To identify differences in expression between fertile and sterile hybrids and to identify misexpression, we summed reads mapping to both the M. m. domesticus and M. m. musculus allele for each sample and then for each genotype (see Supplemental Material). Total read counts for fertile and sterile hybrids are strongly correlated with the read counts of both subspecies (Supplemental Fig. S2).
First, we compared expression patterns on the X Chromosome between sterile and fertile mice. Previous work suggests a large role for the M. m. musculus X Chromosome in hybrid male sterility (Oka et al. 2004, 2014; Storchova et al. 2004; Good et al. 2008a, 2010; Bhattacharyya et al. 2013, 2014). Genes remaining in the analysis after filtering for low read counts were distributed across the X Chromosome. In fertile hybrids, the number of genes expressed above and below the level seen in M. m. domesticus was nearly equal, while in sterile hybrids the majority of genes were expressed above the level seen in M. m. musculus (Fisher's exact test, P < 0.0001) (Fig. 2; Supplemental Table S3). We next compared fold changes of X-linked genes with autosomal genes. Fold changes were calculated between both subspecies and between the sterile and fertile hybrids for 10,264 genes. The ratio of genes overexpressed on the X versus the autosomes in the sterile hybrid was significant (Fisher's exact test, P < 0.0001) (Supplemental Table S4), while there was no significant difference between these ratios in the fertile hybrid (Fisher's exact test, P = 1.0) (Supplemental Table S4). Together, these results suggest that the X Chromosome in the sterile hybrid is uniquely overexpressed compared to the fertile hybrid and to the autosomes. Overexpression of genes on the X Chromosome in sterile hybrids is consistent with previous work based on microarrays (Good et al. 2010). It is also consistent with expression studies of germ cells that were sorted by developmental stage (Campbell et al. 2013), indicating that overexpression of genes on the X is not an artifact of differences in the cellular composition of the testes of sterile and fertile mice (see Discussion).
Figure 2.
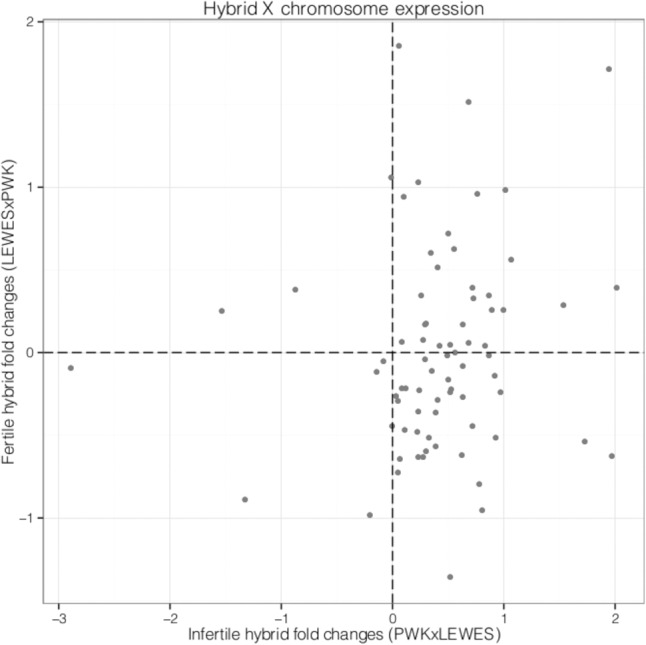
Expression on the X Chromosome in reciprocal hybrids. Each point represents one gene.
Next, we focused on patterns of expression of autosomal genes. Comparing the number of reads mapping to a gene in the hybrid and in the pure subspecies allowed us to identify misexpressed genes and to infer the mode of inheritance for expression for each gene (Supplemental Fig. S2). Genes that showed less than a 1.25-log2 fold change between the hybrid and both subspecies were considered “similar” regardless of significance (Gibson et al. 2004; McManus et al. 2010). Since this is a conservative cut-off, we found that most genes showed similar levels of expression in hybrids and in pure subspecies (86%, or 8834 genes, and 90%, or 9300 genes, of genes in the sterile and fertile hybrid, respectively) (Supplemental Table S5). While the number of genes categorized as similar in this analysis is higher than in previous studies, this is unsurprising given the short divergence time between M. m. musculus and M. m. domesticus. Genes that did not demonstrate conserved expression patterns were divided into dominant, additive, and misexpressed (see Supplemental Methods and Supplemental Table S5). Where 28 genes were misexpressed in the fertile hybrid, 63 genes were misexpressed in the sterile hybrid (Supplemental Table S5). In the fertile hybrid, an equal number of genes were misexpressed above and below the level of both subspecies, while in the sterile hybrid, significantly more genes were overexpressed (Fisher's exact test, P = 0.0006) (Supplemental Table S6). Eleven misexpressed genes were shared between the sterile and fertile hybrid, all of which were overexpressed.
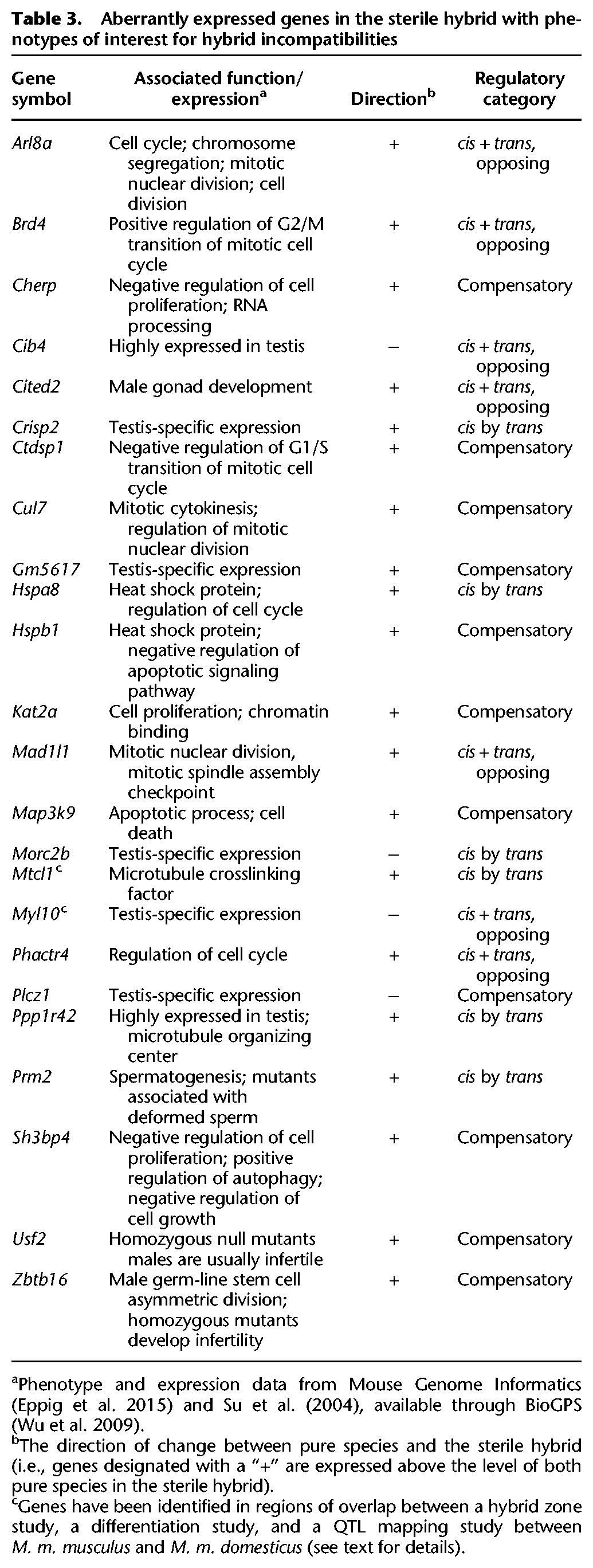
Genes that are over- or underexpressed in the sterile hybrid to the exclusion of the fertile hybrid are of interest as potential candidates for hybrid incompatibilities. First, we identified genes for which the number of reads mapping to the fertile and sterile hybrid was significantly different. Then, we eliminated genes with less than a 1-log2 fold difference between the sterile hybrid and both subspecies. A 1-log2 fold change corresponds to an expression difference that is twofold higher or lower, so differences between the sterile hybrid and each subspecies at this threshold may be biologically meaningful. We identified 202 genes at a 5% false discovery rate (FDR) with these criteria, hereafter referred to as genes with “aberrant expression” for simplicity. These 202 genes were enriched for 39 nonindependent GO terms at a 5% FDR, the most highly significant of which were (1) positive regulation of gene expression (FDR q-value = 0.0115), (2) positive regulation of RNA metabolic process (FDR q-value = 0.0139), and (3) regulation of cell migration (FDR q-value = 0.0236) (Eden et al. 2009). Of these aberrantly expressed genes, 17 were associated with only a cis-regulatory change and thus could be included in the test for positive selection. Remarkably, 12 of these 17 genes were identified as targets of positive selection in the analysis above, representing a highly significant overenrichment of positively selected genes among those associated with hybrid sterility (Fisher's exact test, P < 0.0001) (Supplemental Table S7).
A subset of the genes that are aberrantly expressed uniquely in the sterile hybrid are associated with male reproductive phenotypes or cell-cycle control in laboratory mice or are highly expressed in the testis relative to other tissues, making them potential candidates for reproductive incompatibilities between the subspecies (Table 3) (phenotype and expression data collected from Su et al. 2004; Wu et al. 2009; Eppig et al. 2015). Notably, five genes (Adgrg1, Itpka, Mtcl1, Myl10, and Micall2) have been identified in regions of overlap between the results of a genome-wide differentiation study between the subspecies (Phifer-Rixey et al. 2014), a QTL mapping study on measures of hybrid male sterility (White et al. 2011), and in regions of low introgression across the M. m. musculus and M. m. domesticus hybrid zone (Janoušek et al. 2012).
Table 3.
Aberrantly expressed genes in the sterile hybrid with phenotypes of interest for hybrid incompatibilities
Compensatory evolution is associated with misexpression in sterile hybrids
If cis and trans changes interact epistatically to result in hybrid incompatibilities, we expect divergence between subspecies that involves both cis and trans changes to be associated with novel expression patterns in the sterile hybrid. Genes with both cis and trans changes in opposing directions should be particularly enriched if the breakdown of co-adapted regulatory machinery contributes to misexpression in sterile hybrids. To test this hypothesis, we examined the regulatory categories associated with genes that were misexpressed in sterile hybrids (genes with a >1.25-log2 fold change between the sterile hybrid and both subspecies) (Supplemental Table S8). A number of the misexpressed genes could not be analyzed for regulatory divergence due to low read counts. Of the genes that remained in the analysis, there was a nonrandom association between cis and trans variants acting in opposing directions and misexpression in the sterile hybrid compared to genes where cis or trans variants acted alone or in the same direction (Fisher's exact test, P < 0.0001) (Table 4). Genes categorized as strictly compensatory, where there was no significant difference in expression between subspecies despite significant differences between alleles in the hybrid, were the most enriched in the misexpressed gene set (Fisher's exact test, P = 0.0004) (Supplemental Table S9). Far fewer misexpressed genes were retained for analysis from the fertile hybrid (17 genes total). No regulatory category was enriched in the misexpressed gene set of the fertile hybrid, although this may be due to lack of power given the low number of genes tested (Fisher's exact test, P = 1.0) (Supplemental Table S10).
Table 4.
Numbers of misexpressed genes in different regulatory categories
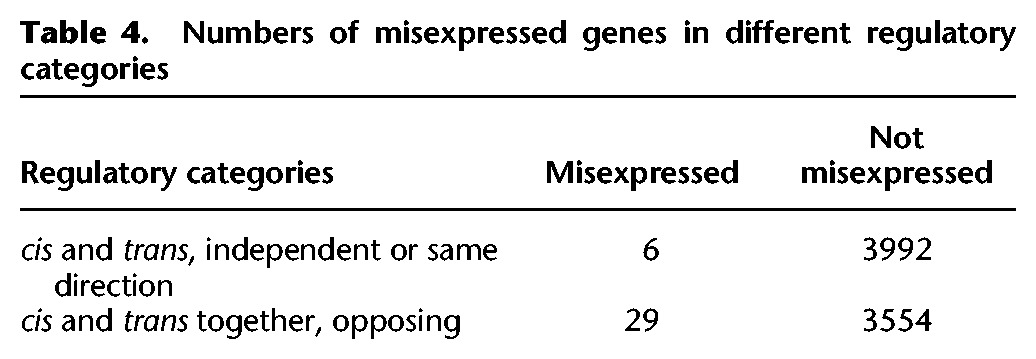
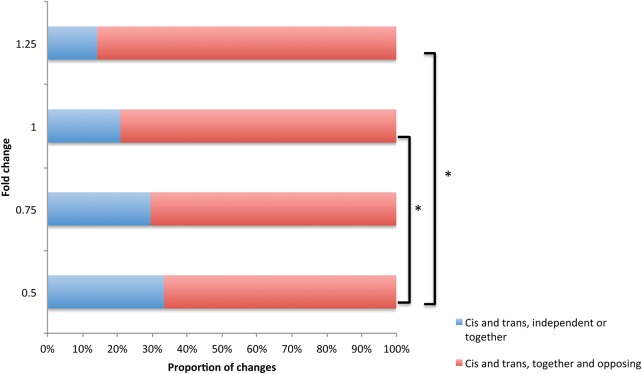
Next, we repeated this analysis using the previously described “aberrantly expressed” genes (Supplemental Table S11) (i.e., a more relaxed cut-off in which expression was at least 1-log2 fold different between the sterile hybrid and both subspecies). As above, genes for which cis and trans variants acted in opposition were enriched compared to genes for which cis and trans variants acted independently or in the same direction (Fisher's exact test, P < 0.0001) (Supplemental Table S12). Likewise, strictly compensatory changes again were especially enriched in this differentiated gene set (Fisher's exact test, P < 0.0001) (Supplemental Table S13). Finally, to further investigate the relationship between compensatory evolution and misexpression in the sterile hybrid, genes were binned based on log2 fold changes between the sterile hybrid and both subspecies. As fold change increased, the proportion of genes where cis and trans variants act in opposition increased (Fig. 3).
Figure 3.
The relationship between the magnitude of expression differences and the number of genes in different regulatory categories. Larger fold changes between both subspecies and the sterile hybrid are associated with a greater proportion of genes where cis and trans variants act in opposition to one another. (*) P < 0.05, Fisher's exact test.
Expression comparisons between multiple subspecies lines
The findings described above were based on a small number of wild-derived inbred lines. This limits the extent to which our conclusions speak to regulatory divergence between M. m. musculus and M. m. domesticus in general as opposed to regulatory divergence between these particular lines. To expand this analysis and look more generally at expression divergence between the subspecies, we took advantage of data from a recent study that analyzed the testis transcriptomes from seven lines of M. m. domesticus and eight lines of M. m. musculus (Phifer-Rixey et al. 2014). While Phifer-Rixey et al. (2014) included more lines, coverage per line was lower than in our analysis. Still, overlap between the two data sets is high: 77% of the genes in Phifer-Rixey et al. (2014) were represented in our data. We reanalyzed the data of Phifer-Rixey et al. (2014) for this subset of 9779 genes that were shared between the two studies.
Importantly, genes that were differentially expressed in the data of Phifer-Rixey et al. (2014) overlap significantly with genes that have significant parental ratios in our analysis (hypergeometric test, P = 1.749 × 10−16). Genes categorized as cis and cis + trans where variants act in the same direction were particularly enriched in this overlap, making up 57% of the genes found to be differentially expressed between M. m. musculus and M. m. domesticus in both analyses (P < 0.0001). Conversely, genes where cis and trans variants act in opposing directions (cis × trans and a subset of cis + trans categories) showed the lowest proportion of overlap.
We also reanalyzed the data from Phifer-Rixey et al. (2014) to see if our conclusions about cis changes subject to positive selection were general. Genes with significantly different expression between M. m. musculus and M. m. domesticus in Phifer-Rixey et al. (2014) that overlapped with genes identified in our analysis as divergent in cis alone were categorized based on directionally. Genes in the three sets we identified as targets of selection (biological process GO terms GO:006351, GO:0045944, and GO:0006355; see results above) were then subjected to a hypergeometric test as in the previous analysis. Despite the reduction in genes represented in each gene set, all three sets maintained biased directionality at a 10% FDR in this new analysis based on a larger number of inbred lines.
The general concordance between these data sets suggests that many of the conclusions described above do not simply represent line effects but instead characterize regulatory divergence between these two subspecies more generally.
Discussion
We characterized regulatory divergence in testis between Mus musculus domesticus and Mus musculus musculus as well as aberrant expression associated with sterility in hybrids. We identified evidence of widespread compensatory evolution consistent with stabilizing selection as well as evidence for lineage-specific positive selection on a subset of genes related to transcriptional regulation. Lastly, we identified genes with aberrant expression unique to sterile hybrids. These sterility-associated genes were nonrandomly associated with cis and trans changes that act in opposition to one another, consistent with the idea that regulatory changes might underlie Dobzhansky-Muller incompatibilities and be important in speciation.
Regulatory divergence between M. m. domesticus and M. m. musculus
A large number of genes in this study showed evidence of gene expression divergence between M. m. domesticus and M. m. musculus. To mitigate the potential effects of inbreeding, we crossed two different inbred lines within each subspecies to create heterozygous individuals against which inter-subspecific hybrids could be compared. This approach, which is rarely used in studies of expression evolution, eliminates differences in gene expression that arise between subspecies as a result of differences in inbreeding depression, and it eliminates expression differences between the subspecies and hybrids as a result of heterosis. We also compared our results to an independent expression study that included more inbred lines (Phifer-Rixey et al. 2014). Without population level sampling, it is impossible to distinguish between line-specific effects and subspecific differences. However, by characterizing the intersection between these two data sets, we identified patterns that are more likely to be representative of subspecific differences. The high correspondence between the two studies despite their differences in depth and breadth suggests that we have captured a large proportion of subspecific divergence.
The majority of the regulatory divergence between M. m. musculus and M. m. domesticus was the consequence of cis variants, either alone or together with one or more trans variants. Conversely, regulatory divergence due to trans variants alone was relatively rare, accounting for only a small proportion of genes tested. Comparisons between the median expression differences associated with variants acting in cis or trans alone revealed that cis variants were of greater magnitude. Consistent with the results presented here, divergence in cis has been demonstrated to be more common than divergence in trans in insects and nematodes (Gordon and Ruvinsky 2012) and was previously shown to contribute to a larger proportion of differentially expressed genes in the liver between the house mouse subspecies M. m. castaneus (CAST/EiJ) and M. m. domesticus (C57BL/6J) (Goncalves et al. 2012). Similarly, Crowley et al. (2015) found allelic imbalance consistent with cis regulatory effects in 85% of testable genes in comparisons between mouse subspecies. These results stand in contrast to those of McManus et al. (2010) and Coolon et al. (2014), both of whom found a large proportion of expression divergence to be the result of trans differences in Drosophila crosses. Elevated trans divergence in these two studies may be due to demographic or biological differences between species or to differences in the experimental methods (e.g., the use of whole files versus specialized tissue types, number of replicates, etc.).
Studies in yeast and flies suggest that cis-regulatory divergence typically contributes more to differences between species than to differences within species (Tirosh et al. 2009; Emerson et al. 2010) and increases consistently and proportionately with divergence time (Coolon et al. 2014). While cis-regulatory variation is substantial in natural populations (Osada et al. 2006; Campbell et al. 2008; Genissel et al. 2008; Gruber and Long 2009; Lemmon et al. 2014), trans-acting variation contributes more to polymorphic expression variation within species (Lemos et al. 2008; Wittkopp et al. 2008; Coolon et al. 2014). M. m. domesticus and M. m. musculus diverged roughly 350,000 years ago and still share some ancestral variation. Thus, some of the regulatory differences observed between inbred strains could still be polymorphic in one or both subspecies. Finally, overlap between our data and those of Phifer-Rixey et al. (2014) is greatest for genes associated with cis changes and cis + trans changes (where variants act in the same direction), suggesting that these two regulatory categories may contribute disproportionately to regulatory divergence between subspecies compared to within-subspecies variation.
Stabilizing selection has been identified as a dominant force underlying gene expression evolution (Gilad et al. 2006). A widespread reduction in gene expression variation compared to neutral expectations based on intra- and inter-specific comparisons (Rifkin et al. 2003; Lemos et al. 2005) and mutation accumulation lines (Denver et al. 2005) suggests that changes in expression are frequently deleterious. The apparent reduction in expression divergence in these studies compared to neutral expectations could be the outcome of two separate processes: the elimination of cis- and trans-acting variants through purifying selection or compensatory evolution between regulatory elements that conserves expression levels. Our results favor the latter explanation. We identified a significantly greater proportion of instances where cis and trans variants acted in opposition than expected under neutrality, consistent with widespread lineage-specific compensatory evolution.
What drives this compensatory evolution? One possibility is that selection initially favors a mutation acting in trans, perhaps because selection favors a change in expression of some downstream gene. If the initial trans change is highly pleiotropic, it may alter the expression of other downstream genes in a suboptimal way. Selection would then favor the restoration of optimal expression levels at these genes through compensatory cis changes (Goncalves et al. 2012; Coolon et al. 2014).
Against this background of widespread compensatory evolution involving changes in both cis and trans, we also found evidence for lineage-specific positive selection on a subset of cis-only changes. Selection is predicted to act efficiently on cis-regulatory variants (Wray 2007), and simulations suggest that natural selection is more likely to drive cis-regulatory divergence than trans-regulatory divergence (Emerson et al. 2010). In our study, hundreds of genes related to transcriptional regulation with cis changes showed biased directionality. It is clear from this result that positive, directional selection is contributing to a nonnegligible proportion of regulatory divergence.
Misexpression in sterile hybrids
In crosses between M. m. musculus (PWK/PhJ) females and M. m. domesticus (LEWES/EiJ) males, hybrid males have significantly smaller testes and lower sperm counts compared to hybrid males in the reciprocal cross (Good et al. 2008a) (see Supplemental Table S16 for phenotypes of the mice in this study). We took advantage of the asymmetrical nature of hybrid male sterility in this cross to identify genes that were uniquely misexpressed in sterile hybrids. This approach allowed us to separate misexpression that was associated with hybridization from misexpression that was associated with sterility. For example, the 28 genes that were misexpressed in fertile hybrids (Supplemental Table S5) can be excluded as contributing to reproductive isolation.
Despite the power of this approach, it is important to recognize that it does not allow us to directly identify genes causing sterility. The set of genes that are misexpressed only in sterile hybrids is expected to include causative genes, but it may also include genes that are misexpressed as downstream effects of genes causing sterility. The latter category is likely inflated by differences in the cellular composition of testes in fertile and sterile animals. Testes contain a heterogeneous mixture of cell types; sterile and fertile hybrids contain different proportions of somatic, mitotic, early meiotic, and postmeiotic cells. For example, in the well-studied cross between M. m. domesticusC57BL/6J and M. m. musculusPWD in which Prdm9 is implicated in hybrid male sterility, essentially complete meiotic arrest occurs in pachytene with spermatocytes undergoing apoptosis (Mihola et al. 2009). Nonetheless, several lines of evidence suggest that differences in cellular composition are not the main cause of the expression differences we have identified here. First, in contrast to the cross between M. m. domesticusC57BL/6J and M. m. musculusPWD, meiotic arrest is incomplete in the cross performed here. Cells from all stages of spermatogenesis can be found in the testes of sterile males, although the proportions differ in sterile and fertile animals. Second, we would expect to see a greater effect of cellular composition on X-linked gene expression than autosomal expression in whole testis, since transcription on the X Chromosome is largely silenced from pachytene through the later stages of spermatogenesis (Turner 2007). Despite this expectation, there is close agreement between our finding of X-linked overexpression and the results of Campbell et al. (2013), who studied X-linked expression in flow-sorted germ cells for the same genotypes. Campbell et al. (2013) showed that the overexpression of the X Chromosome in sterile hybrid males from this cross is not an artifact of changes in cellular composition but reflects major shifts in gene expression in sterile animals that occur in individual cell types. Third, all patterns of allele-specific expression documented here are robust to cellular composition since they were determined only in fertile F1 males, which have the same cellular composition as the parents. Finally, the strong association between cis-trans compensatory evolution and misexpression (Table 4) would not be expected if cellular composition is the primary driver of differences in gene expression between fertile and sterile hybrids (unless different cell types showed differences in the amount of compensatory regulatory evolution, a pattern that is not seen) (Supplemental Methods and Supplemental Tables S17, S18).
Numerous studies have established a central role for the X Chromosome in hybrid male sterility in house mice. Quantitative trait locus mapping of sterility phenotypes (White et al. 2011; Bhattacharyya et al. 2014), phenotyping of introgression lines (Oka et al. 2004, 2014; Good et al. 2008b; Campbell et al. 2013), and studies of introgression across the hybrid zone (e.g., Tucker et al. 1992; Dod et al. 1993; Payseur et al. 2004) have all suggested that loci on the M. m. musculus X Chromosome contribute to postzygotic isolation between the subspecies. Misexpression of M. m. musculus X-linked genes in sterile hybrids is associated with disruption of meiotic sex chromosome inactivation (MSCI), the process of transcriptional silencing of X and Y Chromosomes during spermatogenesis (Good et al. 2010; Bhattacharyya et al. 2013, 2014; Campbell et al. 2013). The up-regulation of X-linked genes in sterile hybrids seen here is consistent with this earlier work.
Previous studies have also implicated numerous autosomal loci in reproductive isolation in mice (e.g., Forejt and Iványi 1974; Mihola et al. 2009; White et al. 2011; Forejt et al. 2012; Janoušek et al. 2012; Phifer-Rixey et al. 2014). In particular, Mihola et al. (2009) characterized a gene on Chromosome 17, Prdm9, which interacts with the M. m. musculus X Chromosome to drive hybrid sterility; however, sterility in the cross studied here is not associated with known variants of this locus (Good et al. 2008b, 2010). Nonetheless, we identified 202 autosomal genes with aberrant expression only in the sterile hybrid, consistent with the idea that autosomal genes contribute to hybrid male sterility. Interestingly, these were enriched for GO categories associated with gene regulation. Several of the aberrantly expressed genes where cis and trans variants act in opposition are known from previous work to play a role in spermatogenesis, cell cycle control, or to be expressed mainly in the testis (Table 3). Candidates of particular interest which deserve follow-up in future studies are Myl10 and Mtcl1, both of which were identified in regions of overlap between our study, a study identifying peaks of differentiation between the subspecies, a QTL mapping study on markers of hybrid sterility, and regions of low introgression across the hybrid zone (White et al. 2011; Janoušek et al. 2012; Phifer-Rixey et al. 2014). More detailed characterization of the phenotype of hybrid sterility in this cross will be useful for elucidating the role of particular genes.
We also found a highly significant overrepresentation of genes showing positive selection among those that were aberrantly expressed only in sterile hybrids. Because the test we used was restricted to those genes showing cis changes alone, the nature and identity of the interacting loci, if any, are unknown. Nonetheless, an emerging pattern from studies of the genetics of postzygotic isolation is that most of the identified genes show signatures of positive selection (Presgraves 2010). Our results are certainly consistent with this emerging picture and further suggest that selection on regulatory changes contributes to the evolution of reproductive isolation.
Previous studies have identified an association between cis and trans changes favoring the expression of the opposite allele and misexpression in hybrids (Landry et al. 2005; Tirosh et al. 2009; McManus et al. 2010; Schaefke et al. 2013). Landry et al. (2005) first identified an association between compensatory coevolution between cis and trans elements and misexpression in hybrids. While this initial study made powerful predictions as to how regulatory divergence could result in reproductive incompatibilities between species, a phenotypic association with this pattern that is separable from expression differences associated with hybridization has been lacking until now.
The Dobzhansky-Muller model of postzygotic isolation is one of the cornerstones in our understanding of the genetics of speciation (Coyne and Orr 2004). Despite the fact that gene regulation necessarily involves interactions between loci, there have been few systematic attempts to link disruptions in gene regulation across the genome to phenotypes underlying reproductive isolation (Turner et al. 2014). Here, we showed that genes that are misexpressed uniquely in sterile hybrid males are associated with opposing changes in cis and trans. Strictly compensatory changes (i.e., where expression levels in both subspecies are the same) were particularly enriched in genes with aberrant or misexpression. These results provide strong evidence that compensatory regulatory evolution may underlie Dobzhansky-Muller incompatibilities and contribute to reproductive isolation between M. m. musculus and M. m. domesticus.
Methods
Samples
M. m. musculus was represented by whole testis from the wild-derived inbred strains PWK/PhJ and CZECHII/EiJ (hereafter, M. m. musculusPWK and M. m. musculusCZII), and M. m. domesticus was represented by whole testis from the LEWES/EiJ and WSB/EiJ strains (hereafter, M. m. domesticusLEWES and M. m. domesticusWSB).
Hybrids were generated from reciprocal crosses between M. m. musculusPWK and M. m. domesticusLEWES. Male hybrids in this cross are sterile when the mother is M. m. musculusPWK and fertile when the mother is M. m. domesticusLEWES. To circumvent the problem of inbreeding depression in pure species, we crossed M. m. musculusPWK females to M. m. musculusCZII males and M. m. domesticusLEWES females to M. m. domesticusWSB males.
Sequencing and mapping
For each sample, 100-bp paired-end reads were sequenced from mRNA on the Illumina HiSeq 2000 platform. A mean of 7.5 Gb of sequence was obtained for each sample.
Subspecies were mapped with the program TopHat (Kim et al. 2013) to the appropriate pair of reference genomes (either M. m. musculusPWK and M. m. musculusCZII or M. m. domesticusLEWES and M. m. domesticusWSB) as well as to the opposite maternal reference (M. m. domesticusLEWES or M. m. musculusPWK). Hybrids were mapped to M. m. musculusPWK and M. m. domesticusLEWES. Only reads that mapped preferentially to one subspecies were retained for further analysis. See Supplemental Methods for information on the reference genomes used for mapping.
On average, a greater proportion of reads mapped to M. m. musculusPWK per sample than to M. m. domesticusLEWES (see Supplemental Table S1). This difference may be due to real differences in allelic expression or due to a mapping bias; to account for the difference in the number of allele-specific reads across samples, reads were later randomly down-sampled across samples (see below).
Regulatory divergence
An equal number of reads from each parental sample were combined to create a mixed parental pool comparable to allele-specific counts in fertile hybrids. Down-sampling was chosen to equalize power across comparisons as described in Coolon et al. (2014). Reads were then pooled for the following categories: (1) M. m. musculus subspecies reads; (2) M. m. domesticus subspecies reads; (3) fertile hybrid M. m. musculus allelic reads; and (4) fertile hybrid M. m. domesticus allelic reads. Genes with fewer than 20 reads for any sample or allele were excluded. Genes were sorted into regulatory categories based on a binomial test between reads mapping to each parent, a binomial test between reads mapping to each allele in the fertile hybrid, and a Fisher's exact test comparing these values (see Supplemental Methods for details on regulatory divisions) (Wittkopp et al. 2004; McManus et al. 2010). As described by Goncalves et al. (2012), cis + trans can further be subdivided into genes where cis and trans are acting in the same direction (hybrid ratio < pure species ratio) or opposite directions (hybrid ratio > pure species ratio).
Inheritance patterns
After reads were mapped and counted, reads mapping to M. m. domesticusLEWES and M. m. musculusPWK were combined for each sample for total hybrid counts. Mapped reads from pure species and hybrids were down-sampled to an equivalent number per sample and then pooled by genotype (metaseqR) (Moulos and Hatzis 2014).
Testing for enrichment of opposing or reinforcing cis and trans changes
The expected numbers of cis and trans changes acting in the same or opposing directions were calculated based on the proportion of negative and positive cis and trans changes (Supplemental Table S15). Expected numbers were calculated by multiplying the proportion of directional independent cis and trans changes together and then in opposition by the total number of genes with divergence in both cis and trans.
Data access
The sequencing data generated for this study have been submitted to the NCBI BioProject (http://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA286765.
Supplementary Material
Acknowledgments
We thank M. Bomhoff for early computational analysis and support. We thank M. Phifer-Rixey, K. Bi, and members of the Nachman lab for thoughtful comments on this manuscript. We thank our reviewers for their constructive comments on this manuscript. This work was supported by National Institutes of Health (grant R01 GM074245 to M.W.N.) and the Museum of Vertebrate Zoology's Louise Kellogg Fund (to K.L.M.). This work used the Extreme Science and Engineering Discovery Environment (XSEDE), supported by National Science Foundation grant number ACI-1053575.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.195743.115.
References
- Bell GD, Kane NC, Rieseberg LH, Adams KL. 2013. RNA-seq analysis of allele-specific expression, hybrid effects, and regulatory divergence in hybrids compared with their parents from natural populations. Genome Biol Evol 5: 1309–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya T, Gregorova S, Mihola O, Anger M, Sebestova J, Denny P, Simecek P, Forejt J. 2013. Mechanistic basis of infertility of mouse intersubspecific hybrids. Proc Natl Acad Sci 110: E468–E477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya T, Reifova R, Gregorova S, Simecek P, Gergelits V, Mistrik M, Martincova I, Pialek J, Forejt J. 2014. X chromosome control of meiotic chromosome synapsis in mouse inter-subspecific hybrids. PLoS Genet 10: e1004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton-Davidian J, Fel-Clair F, Lopez J, Alibert P, Boursot P. 2005. Postzygotic isolation between the two European subspecies of the house mouse: estimates from fertility patterns in wild and laboratory-bred hybrids. Biol J Linnean Soc 84: 379–393. [Google Scholar]
- Bullard JH, Mostovoy Y, Dudoit S, Brem RB. 2010. Polygenic and directional regulatory evolution across pathways in Saccharomyces. Proc Natl Acad Sci 107: 5058–5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CD, Kirby A, Nemesh J, Daly MJ, Hirschhorn JN. 2008. A survey of allelic imbalance in F1 mice. Genome Res 18: 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P, Good JM, Nachman MW. 2013. Meiotic sex chromosome inactivation is disrupted in sterile hybrid male house mice. Genetics 193: 819–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36. [DOI] [PubMed] [Google Scholar]
- Combes MC, Hueber Y, Dereeper A, Rialle S, Herrera JC, Lashermes P. 2015. Regulatory divergence between parental alleles determines gene expression patterns in hybrids. Genome Biol Evol 7: 1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolon JD, McManus CJ, Stevenson KR, Graveley BR, Wittkopp PJ. 2014. Tempo and mode of regulatory evolution in Drosophila. Genome Res 24: 797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR, Hirschhorn JN, Altshuler D, Lander ES. 2002. Detection of regulatory variation in mouse genes. Nature Genet 32: 432–437. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. 2004. Speciation. Sinauer & Associates, Sunderland, MA. [Google Scholar]
- Crowley J, Zhabotynsky V, Sun W, Huang S, Pakatci IK, Kim Y, Wang JR, Morgan AP, Calaway JD, Aylor DL, et al. 2015. Analyses of allele-specific gene expression in highly divergent mouse crosses identifies pervasive allelic imbalance. Nature Genet 47: 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Morris K, Streelman JT, Kim SK, Lynch M, Thomas WK. 2005. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nature Genet 37: 544–548. [DOI] [PubMed] [Google Scholar]
- Dod B, Jermiin LS, Boursot P, Chapman VH, Nielsen JT, Bonhomme F. 1993. Counterselection on sex chromosomes in the Mus musculus European hybrid zone. J Evol Biol 6: 529–546. [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Hsieh LC, Sung HM, Wang TY, Huang CJ, Lu HH, Lu MY, Wu SH, Li WH. 2010. Natural selection on cis and trans regulation in yeasts. Genome Res 2: 826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE, Mouse Genome Database Group. 2015. The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res 28: D726–D736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forejt J. 1985. Chromosomal and genic sterility of hybrid type in mice and men. Exp Clin Immunogenet 2: 106–119. [PubMed] [Google Scholar]
- Forejt J. 1996. Hybrid sterility in the mouse. Trends Genet 12: 412–417. [DOI] [PubMed] [Google Scholar]
- Forejt J, Iványi P. 1974. Genetic studies on male sterility of hybrids between laboratory and wild mice (Mus musculus L.). Genet Res 24: 189–206. [DOI] [PubMed] [Google Scholar]
- Forejt J, Pialek J, Trachtulec Z. 2012. Hybrid male sterility genes in the mouse subspecific crosses In Evolution of the house mouse, pp. 482–503. Cambridge University Press, New York. [Google Scholar]
- Fraser HB, Moses A, Schadt EE. 2010. Evidence for widespread adaptive evolution of gene expression in budding yeast. Proc Natl Acad Sci 107: 2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Babak T, Tsang J, Zhou Y, Zhang B, Mehrabian M, Schadt EE. 2011. Systematic detection of polygenic cis-regulatory evolution. PLoS Genet 7: e1002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genissel A, McIntyre LM, Wayne ML, Nuzhdin SV. 2008. Cis and trans regulatory effects contribute to natural variation in transcriptome of Drosophila melanogaster. Mol Biol Evol 25: 101–110. [DOI] [PubMed] [Google Scholar]
- Gibson G, Riley-Berger R, Harshman L, Kopp A, Vacha S, Nuzhdin S, Wayne M. 2004. Extensive sex-specific nonadditivity of gene expression in Drosophila melanogaster. Genetics 167: 1791–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Oshlack A, Rifkin SA. 2006. Natural selection on gene expression. Trends Genet 22: 456–461. [DOI] [PubMed] [Google Scholar]
- Goncalves A, Leigh-Brown S, Thybert D, Stefflova K, Turro E, Flicek P, Brazma A, Odom DT, Marioni JC. 2012. Extensive compensatory cis–trans regulation in the evolution of mouse gene expression. Genome Res 22: 2376–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good JM, Handel MA, Nachman MW. 2008a. Asymmetry and polymorphism of hybrid male sterility during the early stages of speciation in house mice. Evolution 62: 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good JM, Dean MD, Nachman MW. 2008b. A complex genetic basis to X-linked hybrid male sterility between two species of house mice. Genetics 179: 2213–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good JM, Giger T, Dean MD, Nachman MW. 2010. Widespread over-expression of the X chromosome in sterile F1 hybrid mice. PLoS Genet 6: e1001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon KL, Ruvinsky I. 2012. Tempo and mode in evolution of transcriptional regulation. PLoS Genet 8: e1002432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graze RM, McIntyre LM, Main BJ, Wayne ML, Nuzhdin SV. 2009. Regulatory divergence in Drosophila melanogaster and D. simulans, a genomewide analysis of allele-specific expression. Genetics 183: 547–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber JD, Long AD. 2009. Cis-regulatory variation is typically polyallelic in Drosophila. Genetics 181: 661–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. 2007. The locus of evolution: evo devo and the genetics of adaptation. Evolution 61: 995–1016. [DOI] [PubMed] [Google Scholar]
- Janoušek V, Wang L, Luzynski K, Dufková P, Vyskočilová MM, Nachman MW, Munclinger P, Macholán M, Piálek J, Tucker PK. 2012. Genome-wide architecture of reproductive isolation in a naturally occurring hybrid zone between Mus musculus musculus and M. m. domesticus. Mol Ecol 21: 3032–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S, et al. 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484: 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. 1975. Evolution at two levels in humans and chimpanzees. Science 188: 107–116. [DOI] [PubMed] [Google Scholar]
- Landry CR, Wittkopp PJ, Taubes CH, Ranz JM, Clark AG, Hartl DL. 2005. Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics 171: 1813–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon ZH, Bukowski R, Sun Q, Doebley JF. 2014. The role of cis regulatory evolution in maize domestication. PLoS Genet 10: e1004745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Meiklejohn CD, Cáceres M, Hartl DL. 2005. Rates of divergence in gene expression profiles of primates, mice, and flies: stabilizing selection and variability among functional categories. Evolution 59: 126–137. [PubMed] [Google Scholar]
- Lemos B, Araripe LO, Fontanillas P, Hartl DL. 2008. Dominance and the evolutionary accumulation of cis- and trans-effects on gene expression. Proc Natl Acad Sci 105: 14471–14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, Coolon JD, Duff MO, Eipper-Mains J, Graveley BR, Wittkopp PJ. 2010. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res 20: 816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. 2009. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323: 373–375. [DOI] [PubMed] [Google Scholar]
- Moulos P, Hatzis P. 2014. Systematic integration of RNA-Seq statistical algorithms for accurate detection of differential gene expression patterns. Nucleic Acids Res 43: e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A, Mita A, Sakurai-Yamatani N, Yamamoto H, Takagi N, Takano-Shimizu T, Toshimori K, Moriwaki K, Shiroishi T. 2004. Hybrid breakdown caused by substitution of the X chromosome between two mouse subspecies. Genetics 166: 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A, Aoto T, Totsuka Y, Takahashi R, Ueda M, Mita A. 2007. Disruption of genetic interaction between two autosomal regions and the X chromosome causes reproductive isolation between mouse strains derived from different subspecies. Genetics 175: 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A, Mita A, Takada Y, Koseki H, Shiroishi T. 2010. Reproductive isolation in hybrid mice due to spermatogenesis defects at three meiotic stages. Genetics 186: 339–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A, Takada T, Fujisawa H, Shiroishi T. 2014. Evolutionarily diverged regulation of X-chromosomal genes as a primal event in mouse reproductive isolation. PLoS Genet 10: e1004301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada M, Kohn MH, Wu C-I. 2006. Genomic inference of cis-regulatory nucleotide polymorphism underlying gene expression differences between Drosophila melanogaster mating races. Mol Biol Evol 23: 1585–1591. [DOI] [PubMed] [Google Scholar]
- Payseur BA, Krenz JG, Nachman MW. 2004. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution 58: 2064–2078. [DOI] [PubMed] [Google Scholar]
- Phifer-Rixey M, Bomhoff M, Nachman MW. 2014. Genome-wide patterns of differentiation among house mouse subspecies. Genetics 198: 283–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. 2010. The molecular evolutionary basis of species formation. Nat Rev Genet 11: 175–180. [DOI] [PubMed] [Google Scholar]
- Rifkin SA, Kim J, White KP. 2003. Evolution of gene expression in the Drosophila melanogaster subgroup. Nature Genet 33: 138–144. [DOI] [PubMed] [Google Scholar]
- Schaefke B, Emerson JJ, Wang TY, Lu MY, Hsieh LC, Li WH. 2013. Inheritance of gene expression level and selective constraints on trans- and cis-regulatory changes in yeast. Mol Biol Evol 30: 2121–2133. [DOI] [PubMed] [Google Scholar]
- Shen SQ, Turro E, Corbo JC. 2014. Hybrid mice reveal parent-of-origin and cis- and trans-regulatory effects in the retina. PLoS One 9: e109382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Ng DW, Zhang C, Comai L, Ye W, Chen ZJ. 2012. Cis- and trans-regulatory divergence between progenitor species determines gene-expression novelty in Arabidopsis allopolyploids. Nat Commun 3: 950. [DOI] [PubMed] [Google Scholar]
- Storchova R, Gregorová S, Buckiova D, Kyselova V, Divina P, Forejt J. 2004. Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm Genome 15: 515–524. [DOI] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci 101: 6062–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Levy AA, Barkai N. 2009. A yeast hybrid provides insight into the evolution of gene expression regulation. Science 324: 659–662. [DOI] [PubMed] [Google Scholar]
- Tucker PK, Sage RD, Warner J, Wilson AC, Eicher EM. 1992. Abrupt cline for sex chromosomes in a hybrid zone between two species of mice. Evolution 46: 1146–1163. [DOI] [PubMed] [Google Scholar]
- Tulchinsky AY, Johnson NA, Watt WB, Porter AH. 2014. Hybrid incompatibility arises in a sequence-based bioenergetic model of transcription factor binding. Genetics 198: 1155–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JM. 2007. Meiotic sex chromosome inactivation. Development 134: 1823–1831. [DOI] [PubMed] [Google Scholar]
- Turner LM, White MA, Tautz D, Payseur BA. 2014. Genomic networks of hybrid sterility. PLoS Genet 10: e1004162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyskočilová M, Pražanová G, Piálek J. 2009. Polymorphism in hybrid male sterility in wild-derived Mus musculus musculus strains on proximal chromosome 17. Mamm Genome 20: 83–91. [DOI] [PubMed] [Google Scholar]
- White MA, Steffy B, Wiltshire T, Payseur BA. 2011. Genetic dissection of a key reproductive barrier between nascent subspecies of house mice, Mus musculus domesticus and Mus musculus musculus. Genetics 169: 289–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. 2004. Evolutionary changes in cis and trans gene regulation. Nature 43: 85–88. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark A. 2008. Genetic basis of regulatory variation within and between Drosophila species. Nature Genet 40: 346–350. [DOI] [PubMed] [Google Scholar]
- Wray GA. 2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet 8: 206–216. [DOI] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, et al. 2009. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 10: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Adams KL. 2007. Extensive allelic variation in gene expression in Populus F1 hybrids. Genetics 177: 1987–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.