Abstract
Tissue injury transiently silences miRNA-dependent posttranscriptional gene silencing in its effort to unleash adult tissue repair. Once the wound is closed, miRNA biogenesis is induced averting neoplasia. In this work, we report that Dicer plays an important role in reestablishing the barrier function of the skin post-wounding via a miRNA-dependent mechanism. MicroRNA expression profiling of skin and wound-edge tissue revealed global upregulation of miRNAs following wound closure at day 14 post-wounding with significant induction of Dicer expression. Barrier function of the skin, as measured by trans-epidermal water loss, was compromised in keratinocyte-specific conditional (K14/Lox-Cre) Dicer-ablated mice because of malformed cornified epithelium lacking loricrin expression. Studies on human keratinocytes recognized that loricrin expression was inversely related to the expression of the cyclin-dependent kinase inhibitor p21Waf1/Cip1. Compared to healthy epidermis, wound-edge keratinocytes from Dicer-ablated skin epidermis revealed elevated p21Waf1/Cip1 expression. Adenoviral and pharmacological suppression of p21Waf1/Cip1 in keratinocyte-specific conditional Dicer-ablated mice improved wound healing indicating a role of Dicer in the suppression of p21Waf1/Cip1. This work upholds p21Waf1/Cip1 as a druggable target to restore barrier function of skin suffering from loss of Dicer function as would be expected in diabetes and other forms of oxidant insult.
Introduction
Wound healing is a dynamic process aimed at restoring injured cellular structures and tissue components. Following injury, a major goal of the cutaneous healing process is rapid re-epithelialization toward re-establishing barrier function of skin by the formation of a cornified cell envelope. However, this function is severely compromised in non-healing or chronic cutaneous wounds such as diabetic foot, venous, or pressure ulcers.1 The barrier function of the skin is mainly attributed to the upper cornified epithelium that is formed as a result of terminal differentiation of the basal layer of the keratinocytes. Although disruption of skin barrier function caused by dysregulated keratinocytes differentiation is noted in several pathological conditions such as atopic dermatitis, psoriasis, and ichthyosis vulgaris,2,3,4 the molecular mechanisms of restoring skin barrier function post-wounding remain elusive. MicroRNAs (miRNAs) dictate the functional fate of coding genes by post-transcriptional gene silencing.5,6 Control of gene expression by miRNA has been implicated in the regulation of development and in mature cell maintenance.7,8,9,10 Emerging evidence indicates that miRNAs generated by the cleavage of the RNAase-III enzyme Dicer play important roles in regulating a wide array of cellular functions. Although the involvement of miRNA in the pathogenesis of chronic wounds has been demonstrated,11,12,13 the significance of Dicer remains unknown. In mouse embryogenesis, expression of Dicer is crucial because its ablation results in early embryonic lethality.14 The Dicer-deficient embryo is aborted around embryonic day 7.5 (E7.5) prior to the onset of skin development.15
Downregulation of Dicer expression is known to occur in a number of pathophysiological conditions and has important bearing on health outcomes. In Caenorhabditis elegans, Dicer is downregulated with age.16,17 Dicer downregulation can be caused by oxidant insult resulting in premature senescence and other pathological conditions.18 Attenuated Dicer expression in retinal cells cause blindness.19,20 Dysregulation of Dicer is associated with numerous pathological conditions, including various geriatric disorder such as diabetes, cardiovascular disease, neurodegenerative disease, and cancer.14,21,22,23 Attenuated Dicer expression in the cord blood of infants predispose them to severe infection by the respiratory syncytial virus.24 Knockdown of Dicer in epithelial cells of the small intestine and podocytes leads to cytoskeletal disorganization, dedifferentiation, and compromised barrier function.25,26
p21Waf1/Cip1 is a critical regulator of cellular differentiation.27 The complexity of p21Waf1/Cip1 function is reflected in its role in self-renewal and differentiation of keratinocytes. Following keratinocytes division, p21Waf1/Cip1-dependent growth arrest initiates the differentiation process.28 Differentiation of the keratinocyte stem cell pool is also enabled by p21Waf1/Cip1.29 Once differentiation is initiated, it is critical, however, that the p21Waf1/Cip1 protein be downregulated to enable the later phases of differentiation to progress. In this context, the factors that govern such dynamic control of p21Waf1/Cip1 expression remain obscure. Dysregulated high p21Waf1/Cip1 is known to derail differentiation through downregulated expression of loricrin. The current state of evidence supports that p21Waf1/Cip1 drives a negative regulatory loop that needs to disengaged for keratinocyte differentiation to progress.30
In this work, we demonstrate a critical role of keratinocyte Dicer in re-establishing barrier function of the wounded skin. Keratinocyte Dicer deficiency, as may be expected following UV exposure, diabetes, aging, or any other oxidant insult, may compromise skin barrier function through dysregulation of p21Waf1/Cip1 (ref. 31).
Results
Wound healing is associated with increased miRNA biogenesis
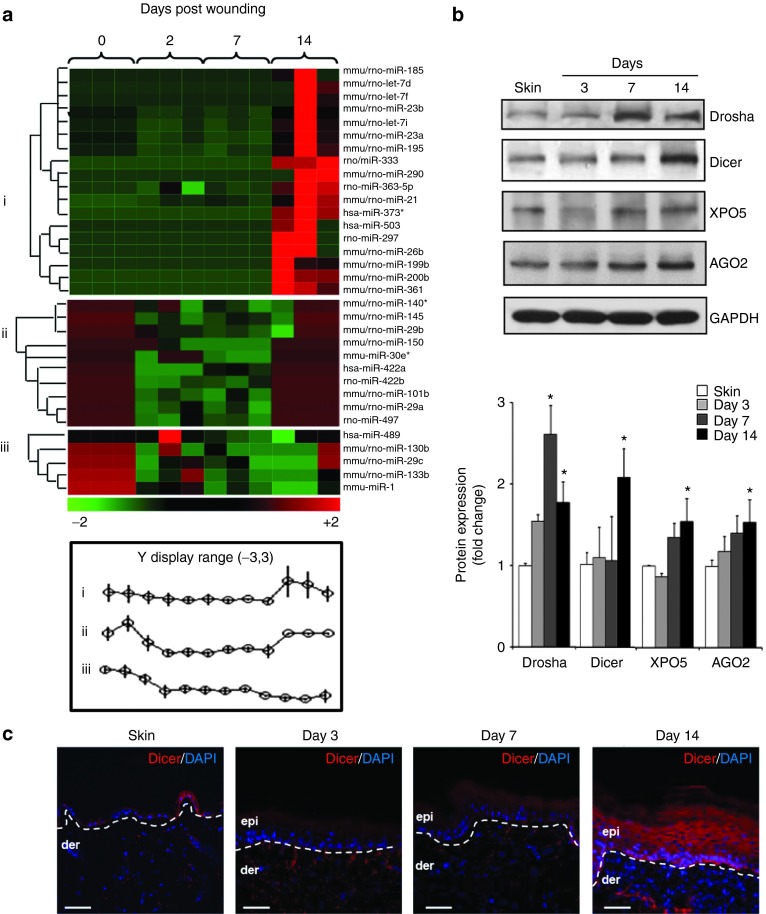
To determine the changes in miRNA expression in response to wounding, edge tissues from full-thickness excisional wounds containing both epidermis and dermis were subjected to miRNA expression profiling followed by hierarchical clustering (GEO accession number: GSE55661). Three distinct clusters of miRNA expression patterns were identified: (i) miRNA that did not change after wounding but were highly upregulated in the late phase (day 14) post-wounding; (ii) miRNAs whose expression were initially high, then the expression were downregulated and again upregulated at day 14, and (iii) miRNAs whose expression were initially high and then their expression dropped down and remained low even in the late phase (day 14) post-wounding (Figure 1a). Line graphs show average changes in miRNA expression profile (Figure 1a). Unlike responses in earlier time points where miRNA expression in the wound-edge tissue was acutely downregulated, a number of miRNA were upregulated at day 14 post-wounding. Validation of miRNA data was performed using real-time PCR (Supplementary Figure S1a). Western blot analyses of Drosha, Dicer, Exportin 5 (XPO5), and Argonaute 2 (AGO2) from the wound-edge tissue also showed increased expression at day 14 post-wounding (Figure 1b) indicative of bolstered miRNA biogenesis at the later phase of healing process (Supplementary Figure S1b). Immunohistochemical staining revealed copious epidermal localization of Dicer throughout all epidermal layers at day 14 post-wounding (Figure 1c and Supplementary Figure S1c).
Figure 1.
Excisional wound induces miRNA biogenesis at the later phase of healing. (a) Excisional wound-edge tissue from C57BL/6 mice was studied at day 0 (skin), 2, 7, and 14 day post-wounding. Profiling of miRNAs expression was performed using FlexmiR mouse miRNA panel (Luminex Technology). The heat map demonstrates miRNA that were down- (green) or upregulated (red) at a specific time post-wounding. Three clusters of expression pattern were identified compared to day 0 sample, i.e., nonwounded skin. (b) Representative western blot of Drosha, Dicer, XPO5, and AGO2 from skin and wound-edge tissue at day 3, 7, and 14 post-wounding. The quantification of the signal was normalized by GAPDH, and the results were expressed as fold induction compared with that of control (skin). Data were expressed as mean ± SD (n = 4; *P < 0.001. (c) Serial wound cross-sections were stained with anti-Dicer (red) antibody for visualization of Dicer in the wound-edge epidermis. The sections were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). The dermal–epidermal junction is indicated by a dashed white line in each panel. Bar = 50 μm.
Keratinocyte-specific depletion of Dicer in mice
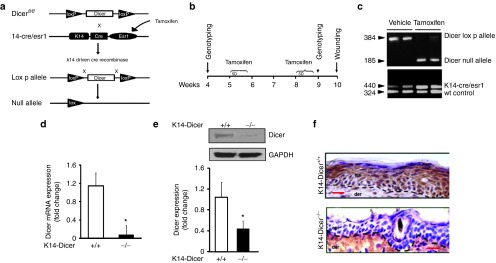
Mice carrying floxed Dicer1 (Dicerfl/fl) allele32 were crossed with mice expressing a tamoxifen-inducible Cre recombinase protein containing a mutated murine estrogen receptor ligand-binding domain under the control of a murine K14 promoter (Figure 2a). Keratinocyte-specific Dicer ablation was achieved by intraperitoneal administration of Tamoxifen for 5 consecutive days, 2 weeks apart (Figure 2b,c). Dicer deletion post tamoxifen treatment in the epidermis was confirmed by qRT-PCR (Figure 2d), western blot (Figure 2e), and immunohistochemistry (Figure 2f).
Figure 2.
Conditional deletion of Dicer in mouse epidermis. (a) Deletion of Dicer gene flanked by LoxP sites (Dicerfl/fl) was induced using a tamoxifen-inducible Cre recombinase protein fused to estrogen-receptor (ER) ligand-binding domain under keratin 14 promoter (K14-CreER). When crossed with strain containing Dicerfl/fl, the offspring produces tamoxifen-induced, Cre-targeted keratinocyte-specific deletion of Dicer. (b) Experimental setup and tamoxifen treatment protocol. K14-CreER Dicerfl/fl mice were injected (i.p) once daily for 5 consecutive days with either tamoxifen (80 mg/kg) or vehicle (corn oil) on weeks 5 and 8. On week 9, genotyping was performed followed by wounding on week 10. (c) PCR showing presence of Dicer-null fragments in tamoxifen-treated animals. (d) Epidermis and dermis were separated using dispase digestion. Dicer mRNA expression in epidermis of vehicle- (Dicer+/+) or tamoxifen-treated (Dicer-/-) mice. (e) Representative western blot of Dicer protein in vehicle- or tamoxifen-treated epidermal tissue. The quantification of the signal was normalized by GAPDH, and the result was expressed as mean ± SD (n = 4; *P < 0.001). (f) Immunohistochemical localization of Dicer in the epidermis. Counterstaining was performed using hematoxylin. The dermal–epidermal junction is indicated by a dashed black line in each panel. Bar = 20 μm.
Compromised wound closure in keratinocyte-specific Dicer-ablated mice post-wounding
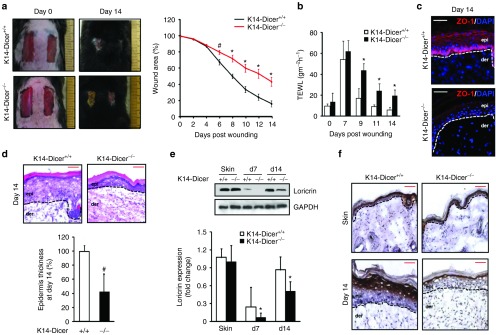
K14-Dicer–ablated mice showed compromised wound closure when compared with the control (Figure 3a). At day 14 post-wounding, the K14-Dicer-/- mice revealed defective wound epithelium or “leaky skin” as evident from high trans-epidermal water loss (TEWL; Figure 3b). This observation is indicative of compromised barrier function of the skin in a situation where re-epithelialization is structurally complete (Supplementary Figure S2a). Furthermore, immunohistochemistry of the tight junction proteins showed significant downregulation of ZO-1 (Figure 3c) but not ZO-2 (Supplementary Figure S2b) in K14-Dicer-/- mice indicating that Dicer level in the keratinocytes is critical for the restoration of barrier function following wounding. K14-Dicer–deficient mice also featured a thin epithelium as measured as an average thickness of five measurements from the re-epithelialized epidermis above the wound bed indicating stalled keratinocytes differentiation (Figure 3d).
Figure 3.
Wound closure is compromised in K14-Dicer-/- mice. (a) Representative photographs of excisional wound in Dicerfl/fl mice treated with vehicle (Dicer+/+) or tamoxifen (Dicer-/-) at day 0 and day 14 post-wounding. Wound closure presented as percentage of the initial wound area. (n = 4, #P < 0.05; *P < 0.001). (b) Trans-epidermal water loss (TEWL) from mice treated with vehicle (K14-Dicer+/+) or tamoxifen (K14-Dicer-/-). Data were presented as mean ± SD. (n = 6; #P < 0.05; *P < 0.001). The TEWL in C57BL/6 mice was found to be 8.5 ± 1.4 and 7.3 ± 1.9 gm-2h-1 in normal skin and at day 14 post-wounding. (c) Immunohistochemical localization of ZO-1 in the epidermis (red). The sections were counterstained with DAPI (blue). The dermal–epidermal junction is indicated by a dashed white line in each panel. Bar = 50 μm. (d) Wound cross-section harvested at day 14 post-wounding was stained with hematoxylin and eosin for visualization of the epidermis. Bar = 50 μm. The bar graph presents the epidermal thickness. Data were expressed as mean ± SD (n = 3; #P < 0.05). (e) Representative western blot of loricrin from skin and wound-edge tissue at day 0, 7, and 14 post-wounding. Data were normalized against GAPDH, and the results were expressed as fold induction compared with that of controls (K14 Dicer+/+). Data were expressed as mean ± SD (n = 3; *P < 0.001). (f) Immunohistochemical localization of loricrin in the epidermis. Counterstaining was performed using hematoxylin. The dermal–epidermal junction is indicated by dashed black line in each panel. Bar = 50 μm.
In the cornified epithelium (CE), the cornified envelop proteins play a major role in establishing skin barrier function. Loricrin accounts for up to 80% of all CE proteins and contributes to mechanical properties of the skin.33,34,35 Severely compromised loricrin levels were noted in the repaired skin of K14-Dicer–ablated mice (Figure 3e and Supplementary Figure S2c). Immunohistochemistry of loricrin (Figure 3f), filaggrin, and involucrin (Supplementary Figure S2d) on day 14 wound-edge tissue demonstrated lower levels in the repaired epidermis of K14-Dicer–ablated mice.
Elevated p21Waf1/Cip1 expression in skin epidermis following Dicer ablation
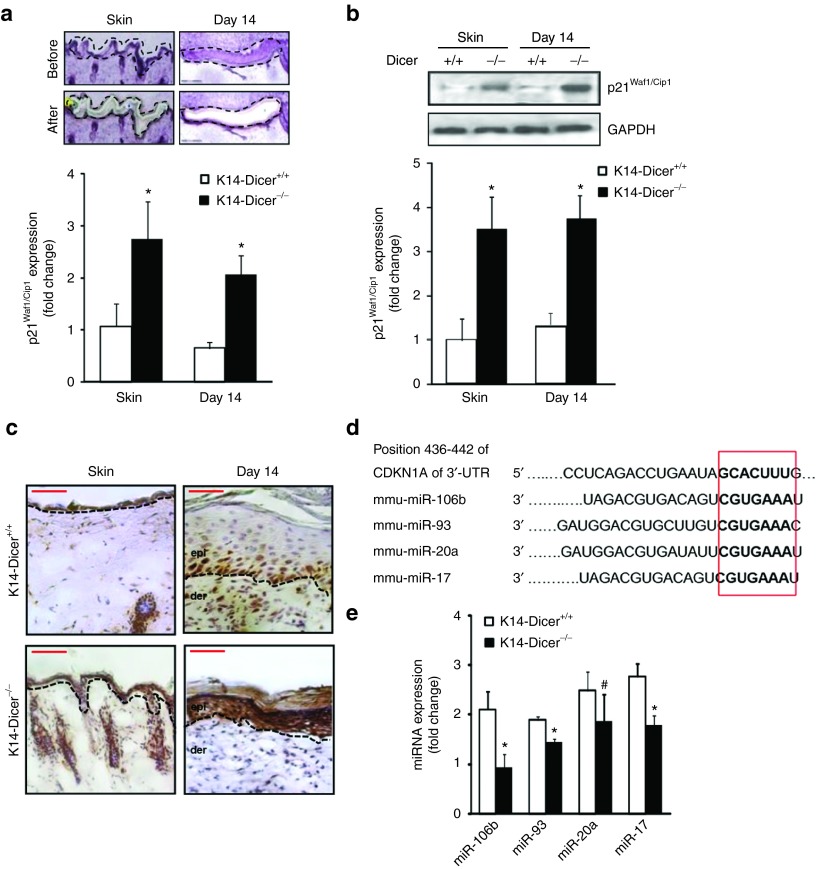
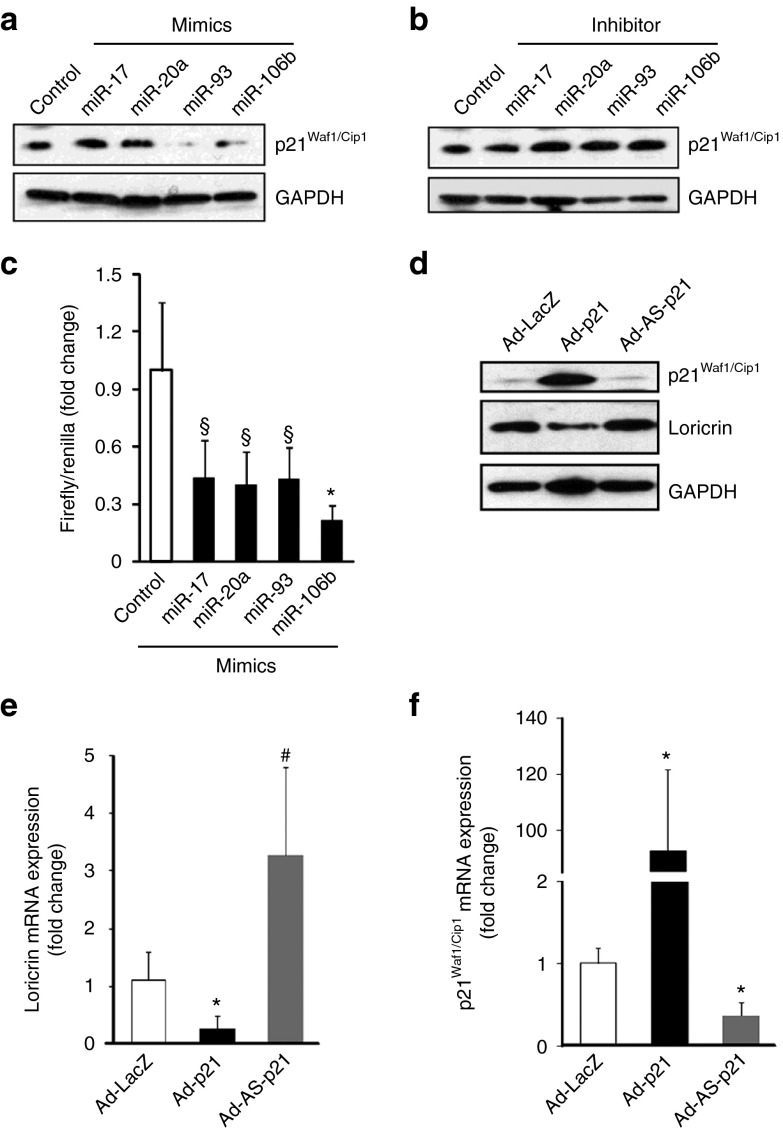
To test the plausible involvement of p21Waf1/Cip1 in loricrin expression, a key driver in keratinocytes differentiation,27 p21Waf1/Cip1 expression was quantified in laser microdissected wound-edge keratinocytes from K14-Dicer–ablated skin and wound-edge tissue at day 14 post-wounding. p21Waf1/Cip1 expression was significantly increased in skin and day 14 wound-edge tissue in K14-Dicer-/- mice (Figure 4a, Supplementary Figure S3) as compared to K14-Dicer+/+ mice. This effect was specific for the ablation of Dicer as it was not repressed in Dicerfl/fl mice treated with tamoxifen (data not shown). Elevated expression of p21Waf1/Cip1 in the posthealed (day 14) epidermis of K14-Dicer-/- mice was confirmed by western blot and immunohistochemistry (Figure 4b,c). In silico studies using TargetScan, PicTar, miRDB, miRanda, and Diana-MicroT algorithms revealed that the 3′-untranslated regions (3′-UTRs) of p21Waf1/Cip1 contain binding sites of 266 miRNA, out of which 28 miRNAs are currently validated and 11 of them are conserved.36 The four most conserved miRNAs known to alter p21Waf1/Cip1 expression by binding to its 3′-UTR are represented by miR-106b, miR-93, miR-20a, and miR-17 (Figure 4d). Expression of these miRNAs was significantly downregulated in the wound-edge tissue of K14-Dicer–ablated mice (Figure 4e). Taking the epidermal as well as dermal component of the wound-edge tissue into consideration, our data suggest that miRNAs are involved in desilencing of p21Waf1/Cip1 in the epidermis of K14-Dicer-/- mice (Figure 4e).
Figure 4.
Keratinocyte-specific Dicer ablation results in elevated p21Waf1/Cip1 expression in epidermis. (a) Hematoxylin-stained wound cross-section before and after laser capture microdissection of the epidermis. p21Waf1/Cip1 mRNA expression in epidermis of vehicle and tamoxifen treated mice at day 0 (skin) and day 14 post-wounding. (b) Representative western blot of p21Waf1/Cip1 in vehicle- or tamoxifen-treated skin and wound-edge tissue. Data were normalized against GAPDH. Result was expressed as mean ± SD (n = 4; *P < 0.001). (c) Immunohistochemical localization of p21Waf1/Cip1 (brown) in the epidermis. Counterstaining was performed using hematoxylin. The dermal–epidermal junction is indicated by dashed black line in each panel. Bar = 50 μm. (d) The conserved seed sequences of miR-106b, miR-93, miR-20a, and miR-17 that binds with the 3′-UTR of p21Waf1/Cip1. (e) miRNA expression of miR-106b, miR-93, miR-20a, and miR-17 in wound-edge tissue at day 14 post-wounding in vehicle- and tamoxifen-treated mice (n = 4; #P < 0.05; *P < 0.001).
p21Waf1/Cip1 represses loricrin expression
To determine whether miRNAs were involved in the suppression of p21Waf1/Cip1, in vitro experiments were performed with the four most conserved miRNA targeting 3′-UTR of p21Waf1/Cip1 such as miR-106b, miR-93, miR-20a, and miR-17 mimics and inhibitors (Supplementary Figure S4a–d). Delivery of miR mimics suppressed, while inhibitors significantly upregulated p21Waf1/Cip1 expression (Figure 5a,b) as well as its 3′-UTR reporter luciferase activity (Figure 5c).
Figure 5.
Overexpression of p21Waf1/Cip1 attenuates loricrin expression in human keratinocytes. Western blot analysis of p21Waf1/Cip1 expression after transfection of miR-106b, miR-93, miR-20a, and miR-17 (a) mimic and (b) inhibitor; (c) miRNA target reporter luciferase assay after miR-106b, miR-93, miR-20a, and miR-17 mimic delivery in HaCaT cells. Open and solid bars represent control mimic and miR-106b, miR-93, miR-20a, and miR-17 mimic-delivered cells, respectively. Results were normalized with Renilla luciferase. Data were expressed as mean ± SD (n = 6). §P < 0.01; *P < 0.001 compared to control. (d) Representative western blot of loricrin and p21Waf1/Cip1 protein in HaCaT cells after treating with Ad-LacZ, Ad-p21, and Ad-AS-p21. Quantitative PCR of (e) loricrin and (f) p21Waf1/Cip1 protein in HaCaT cells after treating with Ad-LacZ, Ad-p21, and Ad-AS-p21. Data were expressed as mean ± SD (n=5). #P < 0.05; *P < 0.001 compared to control.
To test whether upregulation of epidermal p21Waf1/Cip1 is causatively linked to downregulation of loricrin and ZO-1 resulting in impaired barrier function, human epidermal keratinocytes (HaCaT) were studied. Manipulation of gene expression using sense and anti-sense Ad-p21Waf1/Cip1, respectively, resulted in lowered and elevated expression of loricrin compared to the Ad-LacZ–infected controls (Figure 5d–f). Moreover, treatment with 10 µmol/l Butyrolactone I in 10% dimethyl sulfoxide, a pharmacological inhibitor of p21Waf1/Cip1, resulted in elevated expression of ZO-1 in HaCat cells (Supplementary Figure S4e), suggesting that ZO-1 expression is related to the terminal differentiation process.
Silencing p21Waf1/Cip1 accelerates wound closure in K14-Dicer–ablated mice
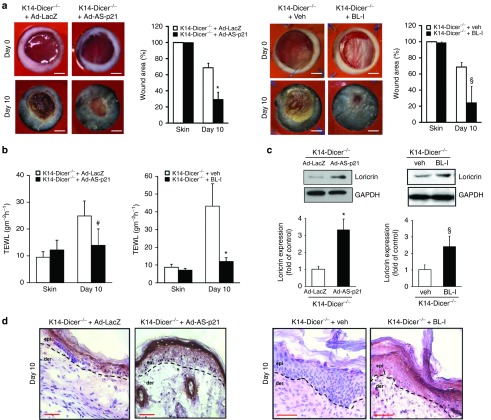
Evidence supporting that p21Waf1/Cip1 may repress loricrin, a key driver of keratinocytes differentiation in the healing wound, led to the hypothesis that silencing of induced p21Waf1/Cip1 in the healing epidermis of K14-Dicer-/- mice will spare loricrin and restore wound closure. Anti-sense Ad-p21Waf1/Cip1 and 10 µmol/l of Butyralactone was delivered to the wound-site of K14-Dicer-/- mice intradermally on day 3 post-wounding. The Butyralactone injection was done every alternate day from day 3 post-wounding onwards. TEWL was recorded every alternate day, and the animals were sacrificed when a significant difference in TEWL was observed (day 10 post-wounding). p21Waf1/Cip1 silencing by antisense adenovirus delivery and pharmacological inhibitor lowered p21Waf1/Cip1 expression at the wound-edge tissue (Supplementary Figure S5a,b) compare to its respective control and accelerated the wound healing (Figure 6a) with establishment of proper barrier function as evident from the TEWL data (Figure 6b). Restoration of the CE following silencing of p21Waf1/Cip1 in K14-Dicer-/- was evident from the western blot analysis of loricrin (Figure 6c) that was validated by immunohistochemistry of loricrin (Figure 6d) filaggrin and involucrin (Supplementary Figure S5c).
Figure 6.
Inhibition of p21Waf1/Cip1 expression in wounds accelerates closure in keratinocyte-specific Dicer null mice. (a) Representative digital images of 6 mm stented punch wound in K14-Dicer-/- mice treated with adenoviral (Ad-LacZ and Ad-AS-p21) and pharmacological (vehicle and Butyralactone I) p21Waf1/Cip1 inhibitor at day 0 and day 14 post-wounding. Wound closure curve presented as percentage of the initial wound area ± SD (n = 4; *P < 0.001). Bar =2 mm. (b) Trans-epidermal water loss from K14 Dicer-null mice treated with adenoviral (Ad-LacZ and Ad-AS-p21) and pharmacological (vehicle and Butyralactone I) p21Waf1/Cip1 inhibitor. Data were presented as mean ± SD (n = 4; #P < 0.05). (c) Representative western blot of loricrin from the wound-edge tissue at day 10 post-wounding. Data were normalized against GAPDH, and the results were expressed as fold induction compared with that of control. Data were expressed as mean ± SD (n = 3; *P < 0.001). (d) Immunohistochemical localization of loricrin (brown) in the epidermis. Counterstaining was performed using hematoxylin. The dermal–epidermal junction is indicated by dashed black line in each panel. Bar = 50 μm.
Discussion
Dicer, an endoribonuclease belonging to the RNase III family, supports RNA interference by cleaving dsRNA into 20–25 nucleotide fragments, which then binds to the 3′-UTR of target mRNAs resulting in mRNA degradation.18,37,38 Dicer also catalyzes the synthesis of small interfering RNAs which are directly implicated in gene silencing and epigenetic regulation of both somatic as well as germline cells.39 It plays a pivotal role in fine-tuning the transcriptome, thereby determining the cell fate by altering the miRNA pool.40 Two independent works have reported that neonatal mice with specific epidermal Dicer ablation lose weight and die without developing a hair coat in spite of having identical phenotype when compared to control littermates at birth.7,8 Hair follicles, underdeveloped and misaligned, that failed to invaginate into the dermis were characteristics of newborn Dicer-deficient mice. The follicular evagination result in physical perturbation which led to compromised epidermal barrier function,7 thus underscoring the significance of miRNA biogenesis during epidermal differentiation. The skin epidermis undergoes a tightly controlled differentiation program that involves the conversion of epidermal keratinocytes into rigid cell remnants without any intracellular organelles called corneocytes. Dysregulation of the balance between keratinocyte proliferation, differentiation, and death leads to skin diseases such as psoriasis, atopic dermatitis, and some ichthyosis as characterized by epidermal defects which are accompanied by dysregulated miRNA biogenesis.
p21Waf1/Cip1 drives cell differentiation in multiple tissue types including squamous epithelia.41,42 In such cases, induction of p21Waf1/Cip1 is transient peaking at 4–12 hours after differentiation is triggered and returning to baseline within 24 hours. This lowering of p21Waf1/Cip1 expression is required for later stages of keratinocyte differentiation to occur. Although downregulation of p21Waf1/Cip1 hinder smooth muscle cell migration,43 p21Waf1/Cip1 knockout is known to enhance migration and proliferative potential of keratinocytes.28,44 Persistent p21Waf1/Cip1 expression suppresses differentiation.28,30 Such temporal regulation of p21Waf1/Cip1 is critical in re-establishing the barrier function of the skin. It is estimated that p21Waf1/Cip1 can be directly targeted by 28 miRNAs.36 Several of these miRNAs are depleted in response to Dicer knockdown thus desilencing p21Waf1/Cip1. Indeed, p21Waf1/Cip1 is reportedly silenced by several conserved miRNAs such as miR-93, miR-20a/b, miR-17, and miR-106a/b, which share the same seed sequence.41,42 Expression of all these miRNAs were found to be significantly downregulated at the wound-edge tissue of K14 directed Dicer-ablated mouse. Of note, the miR-103/107 family attenuates miRNA biosynthesis by targeting Dicer.45 miR-103/107 also targets caveolin-1, a critical regulator of the insulin receptor that maintains glucose homeostasis. The expression of miR-103/107 is upregulated in obese mice that serves as one of the established model for type 2 diabetes.46 These observations lead to the notion that nonhealing diabetic wounds will have attenuated Dicer expression caused by elevated miR-103/107 expression.
In summary, this work demonstrates that miRNA biogenesis at the wound-edge tissue is bolstered at the later phase of wound healing with concomitant induction of Dicer. Such induction of Dicer is primarily localized in keratinocytes. Keratinocyte-specific ablation of Dicer severely compromised epidermal differentiation following wounding. Desilencing of p21Waf1/Cip1 expression caused by depletion of miRs targeting the p21Waf1/Cip1 gene is recognized as the underlying mechanism. Elevated p21Waf1/Cip1 expression in the epidermis disrupted the barrier function of the skin. Small-molecule inhibitors of p21Waf1/Cip1, such as those used in therapeutics for chemotherapy-resistant kidney cancer,47 may help restore barrier function of the repaired skin. Thus, p21Waf1/Cip1 is recognized as a druggable target to restore barrier function of skin suffering from loss of Dicer function as may be expected following UV exposure, diabetes, aging, or any other oxidant insult.31
Materials and Methods
Cell and cell culture. Immortalized human keratinocytes (HaCaT) were grown in Dulbecco's low-glucose modified Eagle's medium (Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 0.1 mg/ml streptomycin (Invitrogen Life Technologies) as described previously.48 The cells were maintained in a standard culture incubator with humidified air containing 5% CO2 at 37 °C. In some experiment, HaCaT cells were treated with Butyrolactone I (Santa Cruz Biotechnology, Dallas, TX; sc-201533), a commercially available p21 inhibitor at a dose of 10 µmol/l as described.49
Transfection of miRNA mimic and inhibitors. Cells were seeded in a 12-well plate at 0.05 × 106 cells/well (50% confluence) 1 day prior to transfection. 50 mmol/l miRNA mimic and 100 mmol/l miRNA inhibitor were employed to transfect cells using DharmaFECTT 1 transfection reagent (Dharmacon RNA Technologies, Lafayette, CO). Nontargeting miRNA mimics or inhibitors (Dharmacon RNA Technologies) were transfected in the cells to serve as negative controls, respectively.
miRNA target reporter luciferase assay. HaCaT cells, transfected with control and miRs mimic for 48 hours, were transfected with 100 ng of pLuc-p21 3′-UTR plasmid (Origene, Rockville, MD) or control construct using Lipofectamine LTX/Plus reagent (Invitrogen) according to the manufacturer's protocol. 10 ng of Renilla plasmid was cotransfected for normalization. Luciferase activity was determined 24 hours after transfection using the dual-luciferase reporter assay system (Promega, Madison, WI). Data are presented as ratio of Firefly to Renilla luciferase assay.
Infection of adenovirus encoding sense and antisense p21Waf1/Cip1. HaCaT cells were infected with adenovirus encoding sense and antisense p21Waf1/Cip1 (a kind gift from Dr Bert Vogelstein) as described previously.50,51 Briefly, HaCaT cells were grown in standard 12-well plates to 75% confluence. Next, cells were transfected with 250 multiplicity of infection of ad-p21, Ad-AS-p21, or Ad-LacZ as control in 750 μl of media. Subsequently, 750 μl of additional media was added after 4 hours. Cells were harvested for protein and RNA extraction after 72 hours postinfection.
Wound models. Male C57BL/6 mice were obtained from Harlan Laboratory (Indianapolis, IN). Mutant mice carrying floxed Dicer1 (Dicerfl/fl) allele was provided as a gift by Dr Elaine Fuchs.32 Keratinocyte-specific Dicer–ablated mouse (K14-Dicer-/-) was generated by crossing Dicerfl/fl mouse with mouse having Cre recombinase protein joined to estrogen-receptor ligand-binding domain under keratin 14 promoter obtained from Jackson Laboratories (Bar Harbor, ME) (STOCK Tg(KRT14-cre/ERT)20Efu/J; stock no:005107). For wounding, two 8 × 16 mm full-thickness excisional wounds were created on the dorsal skin, equidistant from the midline and adjacent to the four limbs.52 The primary advantages of the use of such large wounds are that contraction alone may not wholly close the wound. Furthermore, the longer drawn wound healing process clearly separates the different phases of wound healing thereby allowing appropriate opportunity to temporally resolve underlying mechanisms. To maximize the uniform adenovirus delivery in the wound-edge tissue, two 6-mm-diameter full-thickness excisional wounds were developed on the dorsal skin of mice with a 6-mm disposable biopsy punch and splinted with a silicon sheet to prevent contraction (that were not practical in 8 × 16 mm full-thickness excisional wounds considering murine dimensions) thereby allowing wounds to heal through granulation and re-epithelialization a process similar to that in humans.53,54,55 All animal studies were performed in accordance with protocols approved by the Laboratory Animal Care and Use Committee of The Ohio State University. During the wounding procedure, mice were anesthetized by low-dose isoflurane inhalation as per standard recommendation. Each wound was digitally photographed at the time point indicated. Wound area was calculated using ImageJ software (NIH, Bethesda, MD).
The animals were euthanized at the indicated post-wounding time point, and wound-edge tissues (1 mm away from the wound, snap frozen) or the wound tissues in optimal cutting temperature compound (OCT) were harvested.
In vivo adenoviral delivery of plasmids and Butyrolactone I. In vivo dermal delivery of adenovirus was achieved by intradermal injection of antisense p21Waf1/Cip1 adenovirus. Briefly, antisense p21Waf1/Cip1 along with the LacZ control construct at titer 1 × 107 colony-forming unit/ml (50 μl per wound) was intradermally injected into the skin 1 mm away from the wound-edge 2 days before inducing 6-mm-diameter full-thickness cutaneous wound on C57BL/6 mice under anesthesia as described above. The injection procedure was repeated on days 0 and 3 post-wounding. Each wound was digitally photographed at the time point indicated. In another set of experiment, 50 µl of 10 µmol/l Butyrolactone I in 10% dimethyl sulfoxide was injected intradermally at the wound site. 10% dimethyl sulfoxide was used as vehicle control.
RNA extraction miRNA profiling and quantitative real-time PCR. RNA from mouse wound-edge tissue or cells were isolated using miRVana miRNA Isolation Kit according to the manufactures protocol (Ambion Life Technologies). The RNA quality was assessed using Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Comprehensive profiling of all the mouse/rat miRs annotated in the miRBase 8.0 was performed using the FlexmiR miRNA mouse/rat panel array (Luminex, Austin, TX). All steps were followed according to the manufacturer's instructions. The final reading was obtained from a Luminex 200 analyzer, and the Luminex IS Software Version 2.3. Data Normalization was performed by bead to bead background subtraction and quantile normalization. Data analysis was performed using Genespring GX (Agilent Technologies). All differentially expressed miRNA were identified using a two-tailed t-test where significance level was set at P < 0.05 with Benjamin–Hochberg correction for false discovery rate.56,57 All wound-responsive miRNA that were significantly changed (P < 0.05 with Benjamin–Hochberg correction) were subjected to hierarchical clustering. For determination of miR expression, specific TaqMan assays for miRs and the TaqMan miRNA reverse transcription kit were used, followed by real-time PCR using the Universal PCR Master Mix (Applied Biosystems, Foster City, CA).58 mRNA was quantified by real-time or quantitative (Q) PCR assay using the double-stranded DNA binding dye SYBR Green-I as described previously.58 The data were normalized against U6 (miRNA) and GAPDH (mRNA).The corresponding sequences of primers have been presented in Supplementary Table S1.
Western blots. Western blot was performed using primary antibodies against Drosha (Abcam; ab12286; 1:300), Exportin5 (Abcam; ab131281; 1:500), Dicer (Abcam; ab13502; 1:300), Ago-2 (Abcam; ab32381; 1:500), Loricrin (Covance Biolegend, San Diego, CA; PRB-155P; 1:1,000), p21Waf1/Cip1 (Santa Cruz; sc-397; 1:200), and ZO-1 (Invitrogen; 61–7300; 1:1,000). Signal was visualized using corresponding horseradish peroxidase–conjugated secondary antibody (Amersham; 1:3,000) and ECL Plus Western Blotting Detection Reagents (Amersham GE Healthcare, Pittsburgh, PA). GAPDH (Sigma-Aldrich, St. Louis, MO; G9295; 1: 15,000) served as loading control.
Immunohistochemistry. Immunostaining of Dicer, K14, loricrin, filaggrin, involucrin, p21Waf1/Cip1 ZO-1, and ZO-2 was performed on cryosections of wound sample using specific antibodies as described previously.59 Briefly, OCT embedded tissue were cryosectioned at 10 μm thick, fixed with cold acetone, blocked with 10% normal goat serum, and incubated with specific antibodies against Dicer (Abcam; ab13502; 1:200), loricrin (Covance; PRB-145P; 1:400), filaggrin (Covance; PRB-417P; 1:500), involucrin (Abcam; ab68; 1:200), p21Waf1/Cip1 (Santa Cruz; sc-397; 1:200), ZO-1 (Invitrogen; 61–7,300; 1:200), ZO-2 (Invitrogen; 38–9,100; 1:200), and Keratin14 (Covance; PRB-155P; 1:400), overnight at 4 °C. Signal was visualized by subsequent incubation with either biotinylated-tagged or fluorescence-tagged appropriate secondary antibodies (Alexa 568-tagged α-mouse; Alexa 488-tagged α-rabbit, 1:200; Alexa 568-tagged α-rabbit, 1:200) for 3,3'-diaminobenzidine and immunofluroscence staining.
Trans-epidermal water loss (TEWL). TEWL serves as a reliable index to evaluate the skin barrier function in vivo.60 TEWL was measured from the wounds using DermaLab TEWL Probe (cyberDERM, Broomall, PA).61 The data were expressed in gm-2h-1.
Laser capture microdissection of epidermis. Laser capture microdissection was performed using the laser microdissection system from PALM Technologies (Bernreid, Germany) as described previously by our group.50,62 For epidermal laser capture microdissection captures, sections were stained with hematoxylin for 30 seconds, subsequently washed with DEPC-H2O and dehydrated in ethanol as described.63 Epidermal fraction was identified based on the histology. Epidermal fraction were typically cut and captured under a 20× ocular lens. The samples were catapulted into 25 μl of cell direct lysis extraction buffer (Invitrogen). Approximately 10,00,000 μm2 of tissue area was captured into each cap, and the lysate was then stored at −80 °C for further processing.50,62
Statistical analyses. In vitro data are reported as mean ± SD of three to six experiments as indicated in respective figure legends. For animal studies, data are reported as mean ± SD of at least four to six animals as indicated. Comparison between two groups was tested using Student's t-test (two-tailed), whereas the comparisons among multiple groups were tested using analysis of variance. P < 0.05 was considered statistically significant.
SUPPLEMENTARY MATERIAL Figure S1. Validation of miRNA array data, wound re-epithelialization at different time points and dicer antibody validation data. Figure S2. Compromised barrier function in K14-Dicer-/- mice skin post-wounding. Figure S3. Mosaic image showing localization of p21Waf1/Cip1 in epidermis of control and K14-Dicer-/- mice. Figure S4. Quantitative PCR of miR-17, miR-20a, miR-93, and miR-106b after delivery of mimic and inhibitor. Figure S5. Restoration of skin barrier function post-wounding in K14-Dicer-/- mice after treatment with anti-sense p21Waf1/Cip1 adenovirus. Table S1. Primer sequences.
Acknowledgments
We thank Ohio State University Laboratory Animal Resources for care of mice in accordance with National Institutes of Health (NIH) guidelines. This study was supported in part by the NIH RO1 grants GM069589, GM077185, and NR013898 to C.K.S. and in part by NIH DK076566 to S.R. S.G.
S.R. and C.K.S. conceived and designed the study; S.G., Y.C.C., S.K., and J.B. collected and analyzed data for this study and participated in the preparation of the manuscript; S.G., S.R., and C.K.S. wrote the manuscript.
The authors state no conflict of interest.
Supplementary Material
References
- Herschthal, J and Kirsner, RS (2008). Current management of venous ulcers: an evidence-based review. Surg Technol Int 17: 77–83. [PubMed] [Google Scholar]
- Hoffjan, S and Stemmler, S (2007). On the role of the epidermal differentiation complex in ichthyosis vulgaris, atopic dermatitis and psoriasis. Br J Dermatol 157: 441–449. [DOI] [PubMed] [Google Scholar]
- Tschachler, E (2007). Psoriasis: the epidermal component. Clin Dermatol 25: 589–595. [DOI] [PubMed] [Google Scholar]
- Gruber, R, Elias, PM, Crumrine, D, Lin, TK, Brandner, JM, Hachem, JP et al. (2011). Filaggrin genotype in ichthyosis vulgaris predicts abnormalities in epidermal structure and function. Am J Pathol 178: 2252–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pressman, S, Bei, Y and Carthew, R (2007). Posttranscriptional gene silencing. Cell 130: 570. [DOI] [PubMed] [Google Scholar]
- Sundaram, GM, Common, JE, Gopal, FE, Srikanta, S, Lakshman, K, Lunny, DP et al. (2013). ‘See-saw' expression of microRNA-198 and FSTL1 from a single transcript in wound healing. Nature 495: 103–106. [DOI] [PubMed] [Google Scholar]
- Andl, T, Murchison, EP, Liu, F, Zhang, Y, Yunta-Gonzalez, M, Tobias, JW et al. (2006). The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol 16: 1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, R, Pasolli, HA, Landthaler, M, Hafner, M, Ojo, T, Sheridan, R et al. (2009). DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci USA 106: 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, M and Schratt, GM (2009). microRNA involvement in developmental and functional aspects of the nervous system and in neurological diseases. Neurosci Lett 466: 55–62. [DOI] [PubMed] [Google Scholar]
- Fineberg, SK, Kosik, KS and Davidson, BL (2009). MicroRNAs potentiate neural development. Neuron 64: 303–309. [DOI] [PubMed] [Google Scholar]
- Pastar, I, Khan, AA, Stojadinovic, O, Lebrun, EA, Medina, MC, Brem, H et al. (2012). Induction of specific microRNAs inhibits cutaneous wound healing. J Biol Chem 287: 29324–29335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastar, I, Ramirez, H, Stojadinovic, O, Brem, H, Kirsner, RS and Tomic-Canic, M (2011). Micro-RNAs: new regulators of wound healing. Surg Technol Int XXI: 51–60. [PubMed] [Google Scholar]
- Shilo, S, Roy, S, Khanna, S and Sen, CK (2007). MicroRNA in cutaneous wound healing: a new paradigm. DNA Cell Biol 26: 227–237. [DOI] [PubMed] [Google Scholar]
- Bernstein, E, Kim, SY, Carmell, MA, Murchison, EP, Alcorn, H, Li, MZ et al. (2003). Dicer is essential for mouse development. Nat Genet 35: 215–217. [DOI] [PubMed] [Google Scholar]
- Yi, R and Fuchs, E (2010). MicroRNA-mediated control in the skin. Cell Death Differ 17: 229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibáñez-Ventoso, C, Yang, M, Guo, S, Robins, H, Padgett, RW and Driscoll, M (2006). Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell 5: 235–246. [DOI] [PubMed] [Google Scholar]
- Kato, M, Chen, X, Inukai, S, Zhao, H and Slack, FJ (2011). Age-associated changes in expression of small, noncoding RNAs, including microRNAs, in C. elegans. RNA 17: 1804–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, MA, Raghavan, P, Thomou, T, Boucher, J, Robida-Stubbs, S, Macotela, Y et al. (2012). Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metab 16: 336–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko, H, Dridi, S, Tarallo, V, Gelfand, BD, Fowler, BJ, Cho, WG et al. (2011). DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 471: 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarallo, V, Hirano, Y, Gelfand, BD, Dridi, S, Kerur, N, Kim, Y et al. (2012). DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell 149: 847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm, M and Slack, F (2005). A developmental timing microRNA and its target regulate life span in C. elegans. Science 310: 1954–1957. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou, C, Muljo, SA, Kung, AL, Ganesan, S, Drapkin, R, Jenuwein, T et al. (2005). Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19: 489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanceta, J, Prough, RA, Liang, R and Wang, E (2010). MicroRNA group disorganization in aging. Exp Gerontol 45: 269–278. [DOI] [PubMed] [Google Scholar]
- Inchley, CS, Sonerud, T, Fjærli, HO and Nakstad, B (2011). Reduced Dicer expression in the cord blood of infants admitted with severe respiratory syncytial virus disease. BMC Infect Dis 11: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, LB, Schug, J, Vourekas, A, McKenna, JB, Bramswig, NC, Friedman, JR et al. (2010). MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology 139: 1654–64, 1664.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey, SJ, Jarad, G, Cunningham, J, Goldberg, S, Schermer, B, Harfe, BD et al. (2008). Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J Am Soc Nephrol 19: 2150–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devgan, V, Nguyen, BC, Oh, H and Dotto, GP (2006). p21Waf1/Cip1 suppresses keratinocyte differentiation independently of the cell cycle through transcriptional up-regulation of the IGF-I gene. J Biol Chem 281: 30463–30470. [DOI] [PubMed] [Google Scholar]
- Missero, C, Di Cunto, F, Kiyokawa, H, Koff, A and Dotto, GP (1996). The absence of p21Cip1/WAF1 alters keratinocyte growth and differentiation and promotes ras-tumor progression. Genes Dev 10: 3065–3075. [DOI] [PubMed] [Google Scholar]
- Topley, GI, Okuyama, R, Gonzales, JG, Conti, C and Dotto, GP (1999). p21(Waf1/Cip1) functions as a suppressor of malignant skin tumor formation and a determinant of keratinocyte stem-cell potential. Proc Natl Acad Sci USA 96: 9089–9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cunto, F, Topley, G, Calautti, E, Hsiao, J, Ong, L, Seth, PK et al. (1998). Inhibitory function of p21Cip1/WAF1 in differentiation of primary mouse keratinocytes independent of cell cycle control. Science 280: 1069–1072. [DOI] [PubMed] [Google Scholar]
- Fore-Pfliger, J (2004). The epidermal skin barrier: implications for the wound care practitioner, part I. Adv Skin Wound Care 17: 417–425. [DOI] [PubMed] [Google Scholar]
- Yi, R, O'Carroll, D, Pasolli, HA, Zhang, Z, Dietrich, FS, Tarakhovsky, A et al. (2006). Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet 38: 356–362. [DOI] [PubMed] [Google Scholar]
- Bickenbach, JR, Greer, JM, Bundman, DS, Rothnagel, JA and Roop, DR (1995). Loricrin expression is coordinated with other epidermal proteins and the appearance of lipid lamellar granules in development. J Invest Dermatol 104: 405–410. [DOI] [PubMed] [Google Scholar]
- Steinert, PM and Marekov, LN (1995). The proteins elafin, filaggrin, keratin intermediate filaments, loricrin, and small proline-rich proteins 1 and 2 are isodipeptide cross-linked components of the human epidermal cornified cell envelope. J Biol Chem 270: 17702–17711. [DOI] [PubMed] [Google Scholar]
- Steinert, PM and Marekov, LN (1999). Initiation of assembly of the cell envelope barrier structure of stratified squamous epithelia. Mol Biol Cell 10: 4247–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S, Huang, S, Ding, J, Zhao, Y, Liang, L, Liu, T et al. (2010). Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3' untranslated region. Oncogene 29: 2302–2308. [DOI] [PubMed] [Google Scholar]
- Guo, H, Ingolia, NT, Weissman, JS and Bartel, DP (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M and Doudna, JA (2009). A three-dimensional view of the molecular machinery of RNA interference. Nature 457: 405–412. [DOI] [PubMed] [Google Scholar]
- Duchaine, TF, Wohlschlegel, JA, Kennedy, S, Bei, Y, Conte, D Jr, Pang, K et al. (2006). Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354. [DOI] [PubMed] [Google Scholar]
- Ebert, MS and Sharp, PA (2012). Roles for microRNAs in conferring robustness to biological processes. Cell 149: 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska, I, Ball, AS, Diaz, RL, Magnus, JF, Kibukawa, M, Schelter, JM et al. (2008). MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol 28: 2167–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrocca, F, Visone, R, Onelli, MR, Shah, MH, Nicoloso, MS, de Martino, I et al. (2008). E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 13: 272–286. [DOI] [PubMed] [Google Scholar]
- Fukui, R, Shibata, N, Kohbayashi, E, Amakawa, M, Furutama, D, Hoshiga, M et al. (1997). Inhibition of smooth muscle cell migration by the p21 cyclin-dependent kinase inhibitor (Cip1). Atherosclerosis 132: 53–59. [DOI] [PubMed] [Google Scholar]
- Li, XL, Hara, T, Choi, Y, Subramanian, M, Francis, P, Bilke, S et al. (2014). A p21-ZEB1 complex inhibits epithelial-mesenchymal transition through the microRNA 183-96-182 cluster. Mol Cell Biol 34: 533–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello, G, Rosato, A, Ferrari, F, Manfrin, A, Cordenonsi, M, Dupont, S et al. (2010). A MicroRNA targeting dicer for metastasis control. Cell 141: 1195–1207. [DOI] [PubMed] [Google Scholar]
- Trajkovski, M, Hausser, J, Soutschek, J, Bhat, B, Akin, A, Zavolan, M et al. (2011). MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 474: 649–653. [DOI] [PubMed] [Google Scholar]
- Liu, R, Wettersten, HI, Park, SH and Weiss, RH (2013). Small-molecule inhibitors of p21 as novel therapeutics for chemotherapy-resistant kidney cancer. Future Med Chem 5: 991–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen, CK, Khanna, S, Babior, BM, Hunt, TK, Ellison, EC and Roy, S (2002). Oxidant-induced vascular endothelial growth factor expression in human keratinocytes and cutaneous wound healing. J Biol Chem 277: 33284–33290. [DOI] [PubMed] [Google Scholar]
- Fischer, A, Sananbenesi, F, Schrick, C, Spiess, J and Radulovic, J (2002). Cyclin-dependent kinase 5 is required for associative learning. J Neurosci 22: 3700–3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, S, Roy, S, Banerjee, J, Hussain, SR, Khanna, S, Meenakshisundaram, G et al. (2010). Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc Natl Acad Sci USA 107: 6976–6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna, S, Roy, S, Maurer, M, Ratan, RR and Sen, CK (2006). Oxygen-sensitive reset of hypoxia-inducible factor transactivation response: prolyl hydroxylases tune the biological normoxic set point. Free Radic Biol Med 40: 2147–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S, Khanna, S, Nallu, K, Hunt, TK and Sen, CK (2006). Dermal wound healing is subject to redox control. Mol Ther 13: 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y, Chen, L, Scott, PG and Tredget, EE (2007). Mesenchymal stem cells enhance wound healing through differentiation and angiogenesis. Stem Cells 25: 2648–2659. [DOI] [PubMed] [Google Scholar]
- Chen, L, Tredget, EE, Liu, C and Wu, Y (2009). Analysis of allogenicity of mesenchymal stem cells in engraftment and wound healing in mice. PLoS One 4: e7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L, Tredget, EE, Wu, PY and Wu, Y (2008). Paracrine factors of mesenchymal stem cells recruit macrophages and endothelial lineage cells and enhance wound healing. PLoS One 3: e1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S, Shah, H, Rink, C, Khanna, S, Bagchi, D, Bagchi, M et al. (2007). Transcriptome of primary adipocytes from obese women in response to a novel hydroxycitric acid-based dietary supplement. DNA Cell Biol 26: 627–639. [DOI] [PubMed] [Google Scholar]
- Roy, S, Khanna, S, Rink, C, Biswas, S and Sen, CK (2008). Characterization of the acute temporal changes in excisional murine cutaneous wound inflammation by screening of the wound-edge transcriptome. Physiol Genomics 34: 162–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, A, Ganesh, K, Khanna, S, Sen, CK and Roy, S (2014). Engulfment of apoptotic cells by macrophages: a role of microRNA-21 in the resolution of wound inflammation. J Immunol 192: 1120–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna, S, Roy, S, Park, HA and Sen, CK (2007). Regulation of c-Src activity in glutamate-induced neurodegeneration. J Biol Chem 282: 23482–23490. [DOI] [PubMed] [Google Scholar]
- Pinnagoda, J, Tupker, RA, Agner, T and Serup, J (1990). Guidelines for transepidermal water loss (TEWL) measurement. A report from the Standardization Group of the European Society of Contact Dermatitis. Contact Dermatitis 22: 164–178. [DOI] [PubMed] [Google Scholar]
- Roy, S, Elgharably, H, Sinha, M, Ganesh, K, Chaney, S, Mann, E et al. (2014). Mixed-species biofilm compromises wound healing by disrupting epidermal barrier function. J Pathol 233: 331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S, Patel, D, Khanna, S, Gordillo, GM, Biswas, S, Friedman, A et al. (2007). Transcriptome-wide analysis of blood vessels laser captured from human skin and chronic wound-edge tissue. Proc Natl Acad Sci USA 104: 14472–14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, DE, Roy, S, Radtke, J, Khanna, S and Sen, CK (2007). Laser microdissection and capture of pure cardiomyocytes and fibroblasts from infarcted heart regions: perceived hyperoxia induces p21 in peri-infarct myocytes. Am J Physiol Heart Circ Physiol 292: H1245–H1253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.