Abstract
Insulin-like growth factor 1 (IGF-1) is a potent enhancer of tissue regeneration, and its overexpression in muscle injury leads to hastened resolution of the inflammatory phase. Here, we show that monocytes/macrophages constitute an important initial source of IGF-1 in muscle injury, as conditional deletion of the IGF-1 gene specifically in mouse myeloid cells (ϕIGF-1 CKO) blocked the normal surge of local IGF-1 in damaged muscle and significantly compromised regeneration. In injured muscle, Ly6C+ monocytes/macrophages and CD206+ macrophages expressed equivalent IGF-1 levels, which were transiently upregulated during transition from the inflammation to repair. In injured ϕIGF-1 CKO mouse muscle, accumulation of CD206+ macrophages was impaired, while an increase in Ly6C+ monocytes/macrophages was favored. Transcriptional profiling uncovered inflammatory skewing in ϕIGF-1 CKO macrophages, which failed to fully induce a reparative gene program in vitro or in vivo, revealing a novel autocrine role for IGF-1 in modulating murine macrophage phenotypes. These data establish local macrophage-derived IGF-1 as a key factor in inflammation resolution and macrophage polarization during muscle regeneration.
Introduction
Although the regenerative capacity of injured skeletal muscle is relatively robust due to an abundant muscle progenitor cell pool, more sustained damage in cases of major tissue trauma or progressive degenerative disease compromises muscle healing and leads to severe physical disability. Impaired resolution of inflammation in damaged muscle tissues poses a major impediment to regeneration, leading to fibrotic scar formation and loss of functionality. Thus, modulation of the inflammatory response in muscle damage or disease represents a promising therapeutic approach.
Macrophages are central regulators of inflammation in tissue injury. Heterogeneity and plasticity are macrophage hallmarks: maturation and differential activation (polarization) of monocytes infiltrating the damaged muscle generates functionally diverse macrophage subpopulations, which act in a coordinated and balanced manner to mediate the tissue repair process.1,2 For simplicity, these are often categorized as either M1 or M2 based on the Th1/Th2 classification system although in vivo macrophages demonstrate a spectrum of phenotypes which are dynamic and generated from exposure to combinations of stimuli in varied tissue and disease environments. Classically activated (M1) macrophages are induced by inflammatory cytokines such as interferon gamma (IFNγ) and in turn produce potent inflammatory cytokines and reactive oxygen intermediates. In the context of murine muscle injury, these macrophages amplify inflammatory signals, recruit additional inflammatory cells, phagocytose tissue debris, and apoptotic neutrophils, and produce cytokines that activate muscle progenitor (satellite) cells.1,3 Like their monocyte precursors, they express high levels of Ly6C. Later stages of regeneration are dominated by macrophages with anti-inflammatory and promyogenic functions, often collectively referred to as M2 or alternatively activated. Human M2 polarized macrophages promote myoblast fusion and the growth of myofibers in vitro.3 These immunomodulatory macrophages suppress inflammation by producing IL-10 and TGFβ, promoting remodeling of the extracellular matrix, and stimulating angiogenesis.4 Depletion of macrophages at late stages of repair also reduce myofiber growth,5 suggesting they produce cytokines and growth factors that support myogenesis.3,6 In the mouse, these muscle macrophages are similar to other M2 macrophages in their wound-healing and immunosuppressive functions. However, they are heterogeneous, with fractional expression of M2 surface markers CD206, CX3CR1, and CD163,1,5,7 markers that are more broadly represented on macrophages present in other tissues and diseases.8 These markers can also be highly expressed on resident populations. Further work is therefore needed to fully characterize the macrophage phenotypes in resting and regenerating muscle.
The regulation of macrophage polarization is a complex process involving multiple cellular effectors and intracellular pathways. In acute injury, activated macrophages are sequentially recruited with the inflammatory M1 population preceding the pro-regenerative M2 population. In damaged muscle, the M1 population must be tightly regulated as it releases inflammatory cytokines and reactive oxygen species, causing myofiber lysis,9 and exacerbating injury. Indeed, prolongation of the M1 phase due to abrogation of M2 polarization profoundly affects regeneration,10 while the administration of cytokines such as IL-10 to deactivate the M1 macrophage population reduces pathology in mouse muscular dystrophy models.11 Thus, timely resolution of the M1 phase and transition to M2 macrophages is critical to the outcome of tissue damage: dysregulation of this process may underlie more complex muscle diseases and contribute to pathology as well as prevention of repair.
Insulin-like growth factor 1 (IGF-1) is a peptide hormone that mediates proliferation, differentiation, and survival upon binding its receptor, which is expressed by all cells. The actions of IGF-1 in myogenesis are well defined. In vitro, it promotes both proliferation and differentiation of myoblasts, induces myofiber hypertrophy, and protects from atrophy. In vivo, IGF-1 also increases myoblast proliferation12 and myofiber protein synthesis.13 These functions are essential to the development and maintenance of skeletal muscle, demonstrated by disruption of IGF-1 signaling in postnatal myofibers resulting in severe hypoplasia,14 whereas supplemental IGF-1 induces muscle hypertrophy15 and protects against age-related atrophy.16
IGF-1 also plays an important role in muscle regeneration. It is strongly upregulated following exercise or injury and reduced IGF-1 levels correlate to poor repair.6,17 Distinct IGF-1 propeptides differ in their N terminal signal peptides (class 1 or class 2) and C-terminal E-peptides generated by alternative splicing (Ea and Eb; Figure 1). Eb and Ea peptide-containing IGF-1 propeptides are sequentially produced in response to stretch injury,18 and in vitro evidence has suggested distinct actions for the two propeptides (mitogenic versus differentiative).19,20 Transgenic or viral administration of IGF-1Ea propeptide enhances the repair process and hastens the restoration of contractile function,21 increasing myofiber hypertrophy and accelerating the course of inflammation.21,22
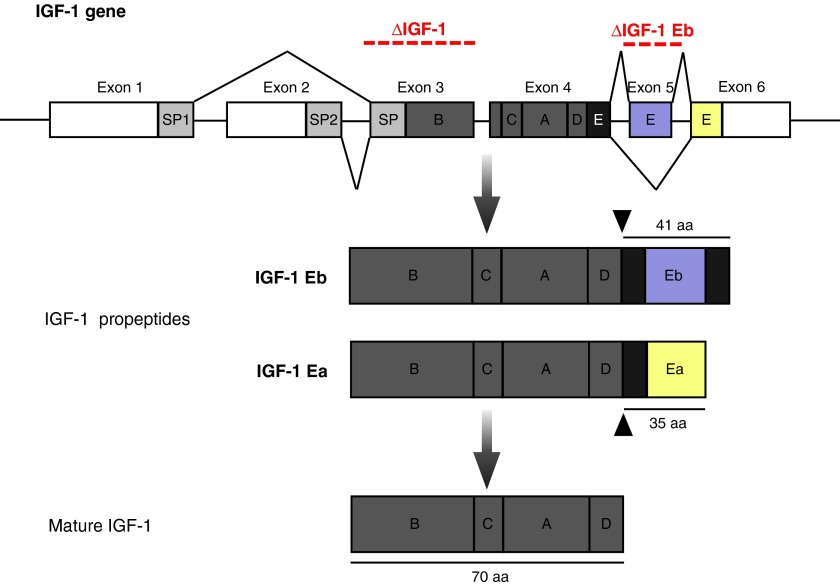
Figure 1.
Structure of the rodent IGF-1 gene. The IGF-1 gene is comprised of six exons and five introns. Exons 3 and 4 encode the mature protein. Differential splicing on the 3′-end of the gene gives rise to a short Ea-peptide (dark gray box) when exon 5 is spliced out (35aa) and a longer Eb-peptide (light gray box), when exon 5 is included. IGF-1 propeptides contain either the Ea- or the Eb-peptide, which can be cleaved to produce the mature IGF-1 protein. Regions spanning exon 3 and exon 5 deleted in ϕIGF-1 CKO mice (ΔIGF-1) and IGF-1Eb KO (ΔIGF-1Eb), respectively, are indicated.
Endogenous tissue IGF-1 is expressed by satellite cells, myofibers, fibroblasts, endothelial cells, and inflammatory cells. Macrophages are the predominant inflammatory cells involved in skeletal muscle repair and abrogation of their migration to the site of injury results in reduced muscle IGF-1 levels.6 Thus, infiltrating macrophages have been proposed as an important source of IGF-1 in regenerating skeletal muscle.6,23 Indeed in the kidney, an increase in macrophage presence caused by administration of macrophage colony-stimulating factor (MCSF) was associated with higher tissue IGF-1 levels and enhanced regeneration after ischemia/reperfusion injury24 and in the injured lung, IGF-1 localizes to Arginase 1–positive M2 macrophages.25 In vitro, macrophages also express significant quantities of IGF-1 propeptides in murine26 and human macrophages,27 upregulated by stimulation with M2-polarizing stimuli IL-4, IL-13, or MCSF.
Here, we identify both Ly6C+ inflammatory monocytes/macrophages and CD206+ macrophages as early sources of local, transient IGF-1, required for efficient skeletal muscle repair in vivo. IGF-1 expression by macrophages also modulates their own transition to a pro-repair phenotype through an autocrine loop which limits inflammation and promotes muscle reconstruction. We further delineate the IGF-1Ea propeptide as the active isoform in muscle regeneration. Modulation of macrophage phenotype by IGF-1 sheds new light on basic mechanisms of inflammation control in muscle injury and presents a therapeutically relevant intervention point for promoting effective tissue regeneration.
Results
A transient surge in macrophage IGF-1 expression at the transition from inflammation to repair
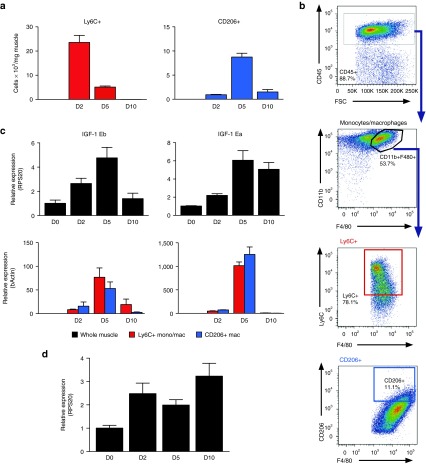
In the setting of cardiotoxin (CTX)-injured skeletal muscle, IGF-1 expression is induced in multiple resident cell types including myoblasts, endothelial cells, and fibroblasts, reaching highest levels at day 10 postinjury (Supplementary Figure S1). We compared the dynamics of IGF-1 expression in CTX-injured mouse muscle and infiltrating macrophages and correlated IGF-1 levels to the accumulation of macrophage populations (Figure 2). Monocytes/macrophages isolated from injured muscle were distinguished by Ly6C and CD206 expression, as illustrated in Figure 2b and Supplementary Figure S2a. The CD45+CD11b+F4/80+Ly6C+ cells also contain monocytes, and we refer to them as inflammatory Ly6C+ monocytes/macrophages, while we refer to the CD45+CD11b+F4/80+CD206+ cells as CD206+ macrophages. Consistent with previous studies,5,28 Ly6C+ inflammatory monocytes/macrophages were observed in injured muscle during the first days after injury, reaching peak numbers at day 2 (Figure 2a), whereas CD206+ macrophages increased significantly at day 5 as the Ly6C+ population declined. By day 10, almost all macrophages were CD206+; however, the overall number of macrophages in the muscle decreased concomitant with the resolution of inflammation.
Figure 2.
The surge in monocyte/macrophage IGF-1 production correlates with transition from inflammatory to repair macrophage phenotypes following muscle injury. (a) Kinetics of Ly6C+ monocyte/macrophage and CD206+ macrophage accumulation in the quadricep muscle is presented at various time points after CTX injury in WT muscle. (b) For analysis of monocyte/macrophage subpopulations, mononuclear cells were isolated from quadricep muscles and analyzed by flow cytometry. Monocytes/macrophages were defined as CD45+CD11b+F4/80+ as shown in the first two images, and then, the inflammatory monocyte/macrophages were discriminated by Ly6C expression while a mature reparative macrophage population was defined by CD206 expression. Isotype controls were used as negative gating controls to define positive signals and are shown in Supplementary Figure S2. (c) IGF-1Ea and IGF-1Eb propeptide expression was determined by qPCR of whole muscle (black bars) as well as Ly6C+ (red bars) and CD206+ (blue bars) macrophages isolated from CTX-injured muscle at 0, 2, 5, and 10 days after CTX injury. (d) MCSF expression measured by qPCR in whole muscle at 0, 2, 5, and 10 days after CTX injury. n = 7–8. Data represent mean ± SEM. CTX, cardiotoxin; WT, wild type.
High IGF-1 production has been associated with M2 macrophage phenotypes, modeled in vitro by stimulation with IL-4 and IL-13 causing a strong upregulation of IGF-1 propeptides in murine26 and human macrophages.27 Using probes that distinguish between IGF-1Ea and IGF-1Eb propeptide transcripts, we revealed temporally distinct expression patterns in whole CTX-injured muscle which we compared to expression in macrophages fluorescence activated cell sorted (FACS)-isolated from injured muscle (Figure 2c). IGF-1Ea expression was strongly upregulated at day 5 in both Ly6C+ and CD206+ cells (21-fold and 18-fold, respectively) but effectively suppressed in both populations by day 10, when it remained elevated in whole muscle samples, presumably due to expression in fibroblasts and other resident cells at this time point (Supplementary Figure S1). Isolated Ly6C+ and CD206+ populations expressed lower levels of IGF-1Eb transcripts at 2 days after injury, increasing at day 5 (ninefold and threefold for Ly6C+ and CD206+, respectively) and then declining at day 10. Thus, transient expression of IGF-1Ea in macrophages correlates with CD206+ macrophage accumulation in regenerating muscle.
The IGF-1Eb propeptide has been implicated in muscle growth and differentiation.19 The upregulation of IGF-1Eb propeptide observed during muscle regeneration prompted us to examine the effects of IGF-1Eb ablation on muscle repair by generating IGF-1Eb knockout (KO) mice, driving deletion of a floxed exon 5 allele (Figure 1) crossed into a CMV Cre mouse line. The resulting offspring were born in Mendelian ratios, developed normally, and lacked an overt phenotype. IGF-1Eb KO mice showed no obvious abnormalities in muscle repair after CTX injury (Supplementary Figure S3), indicating that the IGF-1Eb propeptide is dispensable for muscle regeneration.29
MCSF is a strong positive regulator of IGF-1 in macrophages30 and administration of MCSF during ischemic kidney injury increased tissue IGF-1 levels,24 suggesting that it regulates macrophage IGF-1 expression during repair. We therefore compared IGF-1 expression kinetics to those of MCSF in the same samples (Figure 2d). MCSF mRNA was induced by 2 days after injury as was IGF-1; however, expression did not further increase at 5 days when we observed the peak of IGF-1.
In summary, monocytes/macrophages contribute significantly to IGF-1 levels in skeletal muscle during the initial stages of regeneration (days 2–5) but not during the later phase (day 10). IGF-1 expression patterns are similar in Ly6C+ and CD206+ monocyes/macrophages, suggesting that the regulation of monocye/macrophage IGF-1 propeptide expression is related to the stage of muscle repair rather than to the polarization state. Indeed, the upregulation in monocye/macrophage expression of IGF-1Ea 5 days after injury correlates with the transition from inflammation to regeneration, suggesting a role for monocye/macrophage-derived IGF-1 at this time.
Impaired muscle regeneration in the absence of monocye/macrophage-derived IGF-1
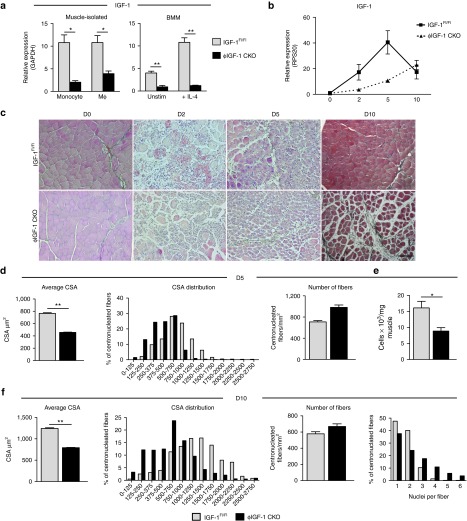
To determine the importance of monocye/macrophage-derived IGF-1Ea expression in muscle repair, we produced a monocye/macrophage-restricted knockout of the IGF-1 gene by crossing a mouse carrying a floxed exon 3 in the IGF-1 gene locus (encoding the main body of the protein, Figure 1)31 with the lysozyme M (LysM) Cre line that expresses Cre in the myeloid lineage.32,33 Substantial recombination and deletion of IGF-1 exon 3 in monocytes and macrophages was confirmed by qPCR on cells freshly isolated from injured muscle as well as bone marrow–derived macrophage (BMM) cultures (Figure 3a). To confirm the specificity of deletion, we measured LysM and Cre expression in myoblasts, endothelial cells, inflammatory cells, and all remaining cells isolated from injured muscle (Supplementary Figure S4a,b). As expected LysM and Cre expression was restricted to the inflammatory cell fraction, although very minor expression was detected in the fraction negative for all markers (CD45-α7β1-CD31-) likely to be fibroblasts. We measured IGF-1 expression in this negative fraction (Supplementary Figure S4d) but did not observe a significant reduction in IGF-1 transcripts in LysMcre/+;IGF-1 Exon 3Flox/Flox (ϕIGF-1 CKO) compared to IGFFl/Fl controls. Homozygous conditional ϕIGF-1 CKO mice were not born in Mendelian ratio (of 195 pups born, only 4.6% were ϕIGF-1 CKOs when 25% was expected), underscoring an important role for IGF-1 in embryonic myeloid lineages. Surviving homozygotes had slightly reduced serum IGF-1 levels (78.2% of wild-type levels; Supplementary Figure S5a); however, ϕIGF-1 CKO homozygous neonates grew and developed normally, and no differences were observed in organ weights compared to littermate controls (Supplementary Figure S4b,c). No abnormalities were detected in the ratios of white blood cells present in peripheral blood, nor in Ly6C+ and Ly6C- monocyte populations under homeostatic conditions (Supplementary Figure S5d–f).
Figure 3.
Monocyte/macrophage-derived IGF-1 regulates muscle regeneration. (a) Myeloid deletion of IGF-1 was confirmed in ϕIGF-1 CKO mice by qPCR of monocytes and macrophages isolated from CTX-injured muscle as well as BMM cultures. Monocytes (CD45+CD11b+F480lo/-Ly6G-) and macrophages (CD45+CD11b+F480hi) isolated from injured ϕIGF-1 CKO muscle were compared to IGF-1Fl/Fl controls, n = 4. BMM prepared from ϕIGF-1 CKO and IGF-1Fl/Fl controls were stimulated with IL-4 to induce IGF-1 upregulation and mRNA levels measured after 12 hours (n =4). (b) Quadricep muscles of control IGF-1Fl/Fl and ϕIGF-1 CKO mice were injured with CTX and analyzed for IGF-1 expression by qPCR 0, 2, 5, and 10 days after injury. Data represent mean ± SEM of four muscles. (c) Representative images of Trichrome-stained TA muscle sections from IGF-1Fl/Fl and ϕIGF-1 CKO mice 0, 2, 5, and 10 days after CTX injury. Bar = 100 μm. (d–f) Quantification of regeneration in the TA at 5 and 10 days postinjury. (d) Regeneration parameters at 5 days after injury; the mean CSA of regenerating (centrally-nucleated) myofibers, the distribution plot of myofiber CSAs and number of fibers per unit area. (e) Myoblasts were isolated from injured muscle 5 days after injury using flow cytometry, identified as shown in Supplementary Figure S1a. The data are presented as the number of myoblasts per milligram of muscle in IGF-1Fl/Fl and ϕIGF-1 CKO mice. (f) The mean CSA, CSA distribution plot, and number of fibers per square millimeter of section at 10 days after injury. At the very right, the number of nuclei per myofiber is shown for the 10-day time point. *P ≤ 0.05; **P ≤ 0.01 compared to IGF-1Fl/Fl control by Mann–Whitney test. BMM, bone marrow–derived macrophages; CSA, cross-sectional area; CTX, cardiotoxin; Mϕ, macrophage; TA, tibialis anterior.
Quantitative RT-PCR for IGF-1 transcription was performed on quadricep muscles at 0, 2, 5, and 10 days after CTX-induced injury on IGFFl/Fl and ϕIGF-1 CKO mice (Figure 3b). In comparison to IGFFl/Fl muscle, upregulation of IGF-1 transcript levels in ϕIGF-1 CKO muscle was abrogated at 2 and 5 days after injury but returned to normal levels by 10 days. Since LysM Cre is expressed by neutrophils as well as monocytes and macrophages,32,33 we cannot exclude the possibility that this cell type contributes to IGF-1 production during the first 24 hours postinjury when it is present in high numbers.6 However, monocytes and macrophages predominate during the peak of IGF-1 expression at 2 and 5 days after injury.
Histological analysis of CTX-injured tibialis anterior (TA) muscles showed comparable fiber necrosis, tissue dysmorphology and inflammatory infiltrates in ϕIGF-1 CKO and IGFFl/Fl controls 2 days after injury (Figure 3c). By day 5, ϕIGF-1 CKO muscles displayed impaired regeneration, as nascent myofibers were substantially smaller than those observed in control muscles (Figure 3c). The difference was even more evident 10 days after injury, when ϕIGF-1 CKO muscles retained broader interstitial spaces, fat deposits, and had smaller fibers compared to IGFFl/Fl muscle (Figure 3c). Quantitation of regenerating (centrally nucleated) fiber cross-sectional area (CSA) revealed a 40% reduction in average ϕIGF-1 KO CSA at the 5-day time point, while the number of fibers per square millimeter was increased, indicating a problem in fiber growth rather than formation of myotubes. Analysis of ϕIGF-1 CKO CSA distribution showed a general shift toward smaller fibers (Figure 3d). Analysis of activation and expansion of myoblasts at day 5 by flow cytometric measurement of α7β1 integrin+ cells revealed that ϕIGF-1 CKO mice had only half the number of myoblasts per milligram of muscle when compared with IGFFl/Fl controls (Figure 3e). By 10 days, the average CSA of ϕIGF-1 CKO muscle increased (792 μm2 compared to 457 μm2), although still 36% lower than the IGFFl/Fl control (Figure 3f) with increased interstitial space taking the place of new fibers. Although the regenerating ϕIGF-1 CKO fibers appeared unable to expand in area, they had a greater number of nuclei associated with each fiber than controls (Figure 3f), suggesting the process of myoblast fusion was not disrupted.
These data demonstrate that monocytes/macrophages are an important source of IGF-1 at the initial stage of damage and during the transition to the reparative phase. Although it was not possible to specifically delete the IGF-1Ea propeptide without affecting IGF-1Eb propeptide expression due to the exon structure of the IGF-1 gene, absence of the IGF-1Eb peptide did not affect muscle repair (Supplementary Figure S3), and we therefore deduce that the regenerative deficit in the ϕIGF-1 CKO mice derives principally from absence of the IGF-1Ea propeptide. Ablation of the surge in IGF-1Ea expression disrupts regeneration, affecting myoblast expansion and myofiber growth, the consequences of which are still apparent by 10 days after injury when macrophages are no longer present in the injury site.
Reduced accumulation of CD206+ macrophages in injured ϕIGF-1 CKO muscle
IGF-1 has a well-documented role in myogenesis, and the reduced levels of IGF-1 in the ϕIGF-1 CKO muscle likely impair regeneration in part because of compromised paracrine signaling in the myoblasts and newly formed fibers.34 This growth factor also acts by autocrine signaling, which prompted us to examine the absence of IGF-1 on the myeloid population itself.
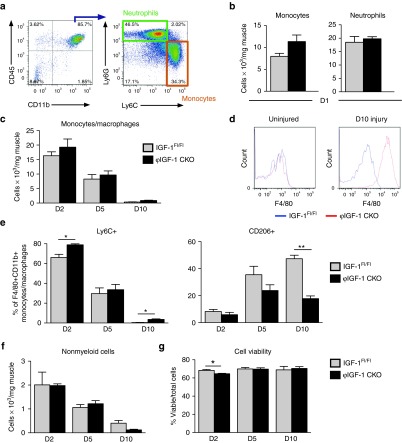
Analysis of myeloid cell infiltration in CTX-injured IGFFl/Fl control and ϕIGF-1 CKO muscles revealed increased monocyte (CD45+CD11b+Ly6G-Ly6Chi) recruitment to the muscle (42% when compared to IGFFl/Fl controls) but did not affect accumulation of neutrophils (CD45+CD11b+Ly6G+Ly6C+; Figure 4a,b) at their maximum accumulation 24 hours after injury.6 Modestly increased monocyte/macrophage numbers at 2 and 5 days after injury in ϕIGF-1 CKO muscle became significant at the 10-day time point, 2.7-fold higher in ϕIGF-1 CKO muscle than those in IGFFl/Fl control muscle at the completion of regeneration (Figure 4c). At this time point, ϕIGF-1 CKO macrophage numbers maintained high expression of F4/80, which can indicate an activated state consistent with incomplete regeneration (Figure 4c,d). Accumulation of nonmyeloid hematopoetic cells (CD45+CD11b-) was also unaffected in ϕIGF-1 CKO muscle until day 10, when their numbers were reduced relative to controls (Figure 4f). Monocyte/macrophage viability was similar in ϕIGF-1 CKO and IGFFl/Fl control muscle despite a small yet significant difference at day 2 (Figure 4g), indicating that the myeloid ablation of IGF-1 did not have a large autocrine impact on cell survival.
Figure 4.
Impaired accumulation of CD206+ macrophages in injured ϕIGF-1 CKO mice. (a) Representation of the gating strategy used to identify monocytes (CD45+CD11b+Ly6G-Ly6Chi) and neutrophils (CD45+CD11b+Ly6G+). (b) Monocyte and neutrophil recruitment to quadricep muscles of ϕIGF-1 CKO mice, and IGF-1Fl/Fl controls was measured 1 day after CTX injury. (c) Accumulation of macrophages (CD45+CD11b+F4/80+) was quantified at 2, 5, and 10 days. (d) Histogram showing F4/80 expression on ϕIGF-1 CKO (red line) and IGF-1Fl/Fl control (blue line) macrophages (CD45+CD11b+F4/80+) isolated 10 days after injury or from uninjured muscle. (e) To follow changes in macrophage phenotypes, the proportion of Ly6C+ monocytes/macrophages and CD206+ macrophages were measured at 2, 5, and 10 days after CTX injury in ϕIGF-1 CKO and control muscle. (f) Accumulation of nonmyeloid hematopoietic cells (CD45+CD11b-) in injured muscle. (g) Viability of cells isolated from injured muscle showed was determined using a ViCell counter. Data represent mean ± SEM of 4–5 mice. *P ≤ 0.05; ** P ≤ 0.01 compared to IGF-1Fl/Fl by Mann–Whitney test.
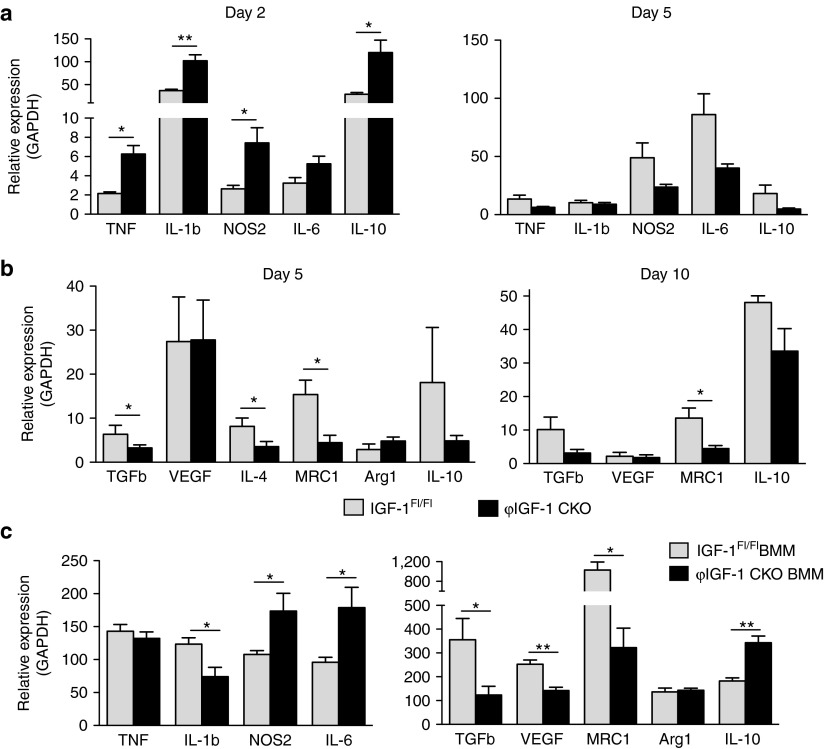
However, polarization was affected by IGF-1 ablation: at day 2 after injury significantly more Ly6C+ monocytes/macrophages were present in ϕIGF-1 CKO muscle (Figure 4e), which normalized to control levels by day 5. Impairment in the generation of CD206+ macrophages in ϕIGF-1 CKO was evident at day 5 and was more severe at day 10. Reduced CD206+ macrophages at days 5 and 10 was accompanied by increased numbers of persistently activated Ly6C+ inflammatory monocytes/macrophages in ϕIGF-1 CKO muscle, indicating inflammatory skewing in the myeloid response to injury.
Compromised gene expression in ϕIGF-1 CKO macrophages
Infiltrating monocytes/macrophages respond to damage signals by producing inflammatory cytokines to amplify the inflammatory signal and activate repair response. Expression of TNF, NOS2, and particularly high levels of IL-1β detected in control monocytes/macrophages at day 2 (Figure 5a) were significantly increased in ϕIGF-1 CKO monocytes/macrophages. Expression of IL-10, a potent immunosuppressive cytokine that is upregulated by activated inflammatory macrophages as part of a self-limiting feedback loop to prevent excessive inflammation,35 was increased fourfold in ϕIGF-1 CKO muscle monocytes/macrophages compared to IGFFl/Fl controls at day 2 (Figure 5a), presumably as a regulatory response to the increased production of inflammatory mediators in ϕIGF-1 CKO monocytes/macrophages. This is likely the cause of the suppression of Th1 cytokine overproduction in ϕIGF-1 CKO monocytes/macrophages observed at day 5. Consistent with an impaired transition to a reparative phenotype, ϕIGF-1 CKO macrophages expressed significantly less TGFβ1, IL-4, and MRC1 at day 5, which was exacerbated at day 10 (Figure 5b). Expression of Arginase 1 characteristic of IL-4 induced M2a type polarization was not altered, nor was VEGFα, indicating that only selected M2 transcripts are inhibited in the absence of IGF-1.
Figure 5.
Dysregulated inflammatory gene expression in ϕIGF-1 CKO monocytes/macrophages. (a, b) Monocytes/macrophages were FACS isolated from injured ϕIGF-1 CKO muscle at days 2, 5, and 10 days after induction of CTX injury for gene expression analyses. (a) Inflammatory gene expression was measured in the cells at 2 and 5 days after injury corresponding to peak expression. (b) Expression of M2 genes was measured in isolated ϕIGF-1 CKO monocytes/macrophages 5 and 10 days after injury and compared to IGF-1Fl/Fl controls. (c) BMM were prepared from ϕIGF-1 CKO and IGF-1Fl/Fl and analyzed for gene expression changes by qPCR. For the measurement of inflammatory M1 genes, BMM were polarized with IFNγ/LPS. For the measurement of M2 genes, BMM were stimulated with IL-4 for 12 hours. Data represent mean ± SEM of six tests. For cell isolation experiments, n = 4; *P ≤ 0.05, **P ≤ 0.01 compared to IGF-1Fl/Fl by Mann–Whitney test. BMM, bone marrow–derived macrophages.
Autocrine loop in bone marrow-derived ϕIGF-1 CKO macrophages
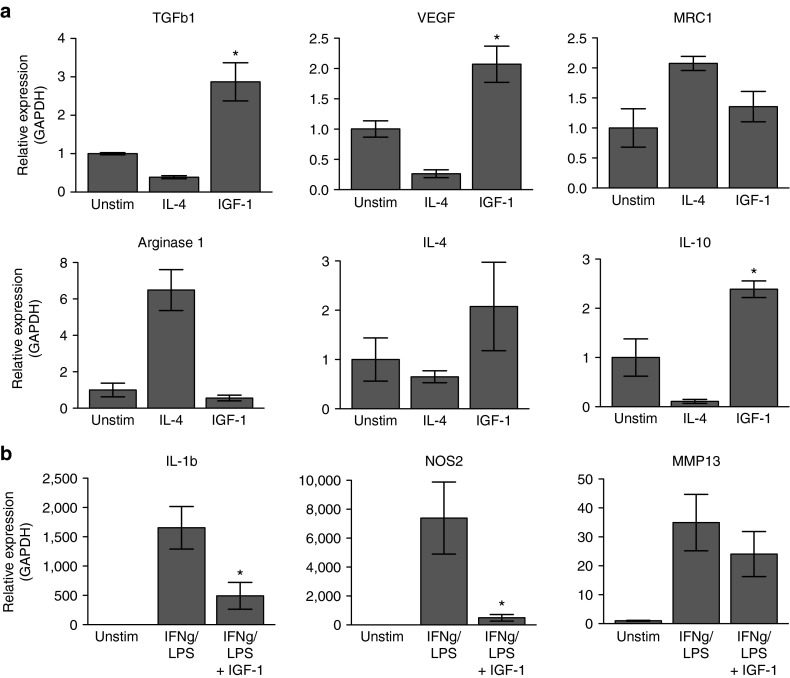
The gene expression changes in ϕIGF-1 CKO macrophages may result from abrogated autocrine IGF-1 signaling which directly interferes with macrophage gene expression programs or from disrupted cross-talk with IGF-1 responsive cells in the muscle bed. To distinguish between these possibilities, we examined the response of ϕIGF-1 KO and IGFFl/Fl BMM cultures to M1 (IFNγ/LPS) and M2 (IL-4) polarizing stimuli in vitro. Expression of inflammatory cytokines NOS2 and IL-6 were significantly increased in ϕIGF-1 CKO BMM (Figure 5c), suggesting that autocrine IGF-1 restricts their expression. Interestingly, IL-1β was downregulated and TNF was unchanged in ϕIGF-1 CKO BMM. This defines a subset of cytokines repressed by autocrine IGF-1 in macrophages. Examining gene expression in IL-4 polarized macrophages, we observed a clear impairment in the upregulation of M2 products TGFβ1, VEGFα, and MRC1. As in muscle macrophages, Arginase 1 was not affected and IL-10 expression was increased in ϕIGF-1 CKO BMM relative to controls, indicating that autocrine IGF-1 signaling is required for specific components of the M2 gene program.
To determine whether exogenous IGF-1 directly alters gene expression, we stimulated BMM with recombinant IGF-1 and monitored the induction of M1 and M2 genes by qPCR (Figure 6). IGF-1 stimulated expression of the pro-reparative factors IL-4, VEGFα, and TGFβ1 characteristic of M2 polarization, whereas Arginase 1 was unaffected by IGF-1 stimulation (Figure 6a). In contrast, M1-specific genes activated by IFNγ/LPS stimulation (IL-1β, NOS2, MMP13; Figure 6b) were significantly repressed in the presence of exogenous IGF-1. These results confirm the direct effect of IGF-1 on the truncation of inflammation and the induction of M2 effector molecules in BMM.
Figure 6.
Macrophage gene regulation by IGF-1 in vitro. (a) Bone marrow–derived macrophages were stimulated with mature, recombinant IGF-1 for 12 hours and expression of M2 genes assessed by qPCR. Treatment with IL-4 was performed for comparison. *P ≤ 0.05, for IGF-1-treated cells compared to unstimulated cells. (b) Macrophages M1 polarized with LPS and IFNγ were subsequently exposed to recombinant IGF-1 for 8 hours and inflammatory gene expression evaluated by qPCR, normalized to GAPDH. *P ≤ 0.05, for IGF-1/IFNγ-treated cells compared to IFNγ-treated cells. Data represent mean ± SEM of six tests.
Discussion
The importance of macrophage polarization in the resolution of tissue injury is well established, but the mechanistic underpinnings of this process have not been delineated, impeding therapeutic intervention. Here, we document a local transient induction of IGF-1 expression by monocytes/macrophages in response to muscle injury that regulates myoblast proliferation and myofiber growth, in that inactivation of the IGF-1 gene in monocytes/macrophages was associated with a decrease in myoblast numbers and a striking reduction in myofiber CSA. The same phenotype has been documented in damaged muscle with abrogation of IGF-1 signaling by administration of a neutralizing antibody17 or deletion of the IGF-1 receptor in satellite cells.36 In the absence of monocyte/macrophage IGF-1 expression, whole muscle IGF-1 levels were restored by the later stages of regeneration but without correcting aberrant muscle architecture and fat deposition. Taken together, these observations underscore a role for monocyte/macrophage-derived IGF-1 in orchestrating the early events of muscle regeneration.
These data also point to promotion of skeletal muscle regeneration by monocytes/macrophages through fiber hypertrophy at the expense of hyperplasia, as reduced fiber CSA in ϕIGF-1 CKO muscle by day 10 was accompanied by a net increase in the average number of nuclei associated with each regenerated fiber. In contrast, muscle-specific IGF-1R deletion reduced both the number of nuclei per fiber and fiber CSA after injury,34 suggesting that residual IGF-1 expressed by other cell types in the ϕIGF-1 CKO muscle was sufficient to maintain satellite cell proliferation and myoblast fusion. It also likely that monocytes/macrophages secrete additional growth-promoting factors in response to autocrine IGF-1 that are necessary for full recovery of muscle mass after injury. Taken together, the two studies are in accordance with the idea that monocytes/macrophages are both an important source of IGF-1 in muscle regeneration, stimulating muscle repair directly as well as indirectly through autocrine feedback loops.
Myeloid deletion of IGF-1 also causes changes in monocyte/macrophage dynamics, observed as a bias toward Ly6C+ monocytes/macrophages. These results highlight a productive autocrine loop that reinforces the progression from inflammation to the resolution of muscle damage. IGF-1 targets key genes in the polarization program as macrophages lacking IGF-1 (ϕIGF-1 CKO) exhibited altered gene expression programs, exaggerated inflammatory gene expression (TNF, IL-1β, and NOS2). In isolated macrophage cultures a similar pattern of increased inflammation and hindered expression of pro-reparative mediators was observed, confirming an autocrine role for IGF-1. Although the immunosuppressive nature of IGF-1 has been documented (reviewed in ref. 37), its action on macrophage polarization has not been previously reported. The normal surge in IGF-1 expression in injured muscle coincides with the transition from inflammatory monocytes/macrophages to reparative macrophages and likely reinforces the progression to tissue healing by suppressing inflammation and inducing M2 genes previously implicated in the resolution phase of muscle regeneration.3,38 It is possible that IGF-1 also alters the balance between macrophage phenotypes by regulating the trafficking of specific subsets; however, this has not been addressed in our study.
As in other tissues, the temporal sequence of different macrophage populations in skeletal muscle is presumably tied to different requirements at the various phases of tissue repair. Although it remains to be elucidated how IGF-1 expression is regulated in macrophages during skeletal muscle repair, its induction is independent of the polarization state: intramuscular Ly6C+ and CD206+ monocytes/macrophages produced similar amounts of IGF-1Ea and IGF-1Eb propeptide, each with its own expression pattern. These results concur with the findings of Lu et al.6 who reported high levels of IGF-1 expression in both Ly6C- and Ly6C+ macrophages, although more prominently by the Ly-6C-subset, measured at a single time point after barium chloride injury. Our observations run counter to the accepted paradigm in which high production of IGF-1 is considered a feature of M2 polarization. This conclusion is based on in vitro data,39,40 which do not adequately model the complex kinetics of macrophage activation states in vivo, resulting from multiple interacting cues. Indeed, colocalization of IGF-1 with Arginase 1 but not IL-10 expression in lung macrophages was not accompanied by direct comparison to inflammatory macrophages.25
Macrophages present in diseased and regenerating muscle are heterogeneous with distinct gene programs regulating their diverse functions.41,42 IGF-1 administered to BMM in vitro induced upregulation of IL-10 and TGFβ1 and hindered expression of inflammatory mediators such as NOS2 and IL-1β, similar to the polarization achieved in vitro by IL-10, TGFβ1 stimulation, or interaction with Treg cells.11,43 Referred to as wound healing or M2c macrophages, these are associated with the resolution phase of regeneration42 and are involved in the termination of inflammation, support of angiogenesis and reinstatement of homeostasis. A similar phenotype is observed in mature resident macrophages44; however, the relationship between the two has not been specifically studied in skeletal muscle. In contrast, an alternative M2 phenotype, M2a, is described to produce IL-4 and Arginase 1 that block the production of NOS2 and promote the production of ornithine and polyamines, a phenotype not directly induced by IGF-1, although its suppression of inflammatory signals may also allow for further polarization of macrophages by other M2-inducing stimuli in the regenerating muscle bed.
Research on the IGF-1Eb propeptide has heretofore been restricted to ex vivo or overexpression experimentation.45 Enhanced skeletal muscle satellite cell activation and proliferation has been ascribed to a derivative synthetic 24aa Eb peptide moiety referred to as MGF,46 which has not been detected in vivo. Moreover, the mitogenic effects of synthetic MGF in vitro are contested,19,29,47,48 and it remains unclear whether an equivalent peptide is derived from cleavage of endogenous IGF-1Eb propeptide. Although genetic deletion of IGF-1Eb had negligible effects on muscle regeneration in vivo, it might promote tissue growth in other physiological contexts.
Our results do not rule out the role of other myeloid cell types at earlier time points in muscle regeneration. As Cre-mediated deletion of the IGF-1 gene locus was driven by a LysM promoter, which is expressed in neutrophils present within 24 hours after injury,32,33 we cannot rule out their early contribution to the IGF-1 pool in regenerating muscle. However, as these cells are present in relatively low numbers at the time points used in the study (2, 5, and 10 days) reduction of IGF-1 expression in injured ϕIGF-1 CKO muscle is largely attributable to monocytes/macrophages. Indeed, several studies have blocked the recruitment of macrophages to injured muscle and regardless of the method employed, myofiber growth was retarded, regeneration was impaired, and was typically associated with fat deposits and reduced local IGF-1 levels.6,49
What is the likely source of IGF-1 late in regeneration, present even in ϕIGF-1 CKO muscle? IGF-1 is highly expressed by interstitial fibroblasts, which expand in numbers at this time and are known to be high producers of IGF-1 in other tissues.50 Although macrophage-derived IGF-1 induces myofibroblast activation and proliferation in vitro,51 fibroblast numbers were not reduced in the ϕIGF-1 CKO setting and may be the main source of IGF-1 at the 10 day time point. Additionally, myoblasts upregulate IGF-1 as they differentiate so the maturing myofibers and may be a significant source of IGF-1 as the muscle returns to homeostasis.
Our findings have important clinical implications for regenerative medicine. Current treatments for diseases with a high inflammatory burden, such as muscular dystrophy, could lead to indiscriminate macrophage depletion, compromising resolution of tissue damage. Macrophages have been cited as protagonists of tissue inflammation and repair in cardiovascular disease, working in organ networks that can lead to derailed infarct healing exacerbated by age-related comorbidity factors that is unlikely to benefit from broad immunosuppressive regimes. Rather, a controlled modulation of immune function will be required to achieve the physiological stability required for durable prevention of disease progression. Thus, monocyte/macrophage IGF-1 represents a potentially powerful point of clinical intervention for modulation of aberrant immune responses in a variety of degenerative diseases.
Materials and Methods
Animals. Mice were housed in a clean, temperature controlled (22 °C) mouse facility and maintained on a 12 hour light/dark cycle with free access to standard chow and drinking water. Mice were sacrificed by asphyxiation with CO2 and cervical dislocation. If blood samples were taken, the mice were anesthetized, the chest was opened then blood was taken directly from the heart and kept at 4 °C over night. Blood serum was separated by centrifugation of blood samples (4 °C, 14,000 rpm for 20 minutes) and stored at −80 °C until needed. All animal experiments were approved by the European Molecular Biology Laboratory Monterotondo Ethical Committee and were in accordance with national and European regulations.
Generation of IGF-1 floxed alleles. Homologous recombination was used to insert LoxP sites flanking exon 3 (gene ablation)31 or exon 5 (IGF-1Eb propeptide) of the IGF-1 gene using RecE-RecT recombination (Figure 1a). A LoxP-flanked PGK-neomycin-resistence cassette was inserted 5′ of exon of interest, used for selection and later removed by electroporating positive clones with a Cre-recombinase-expressing plasmid, leaving a single 5′ LoxP site. The FRT-flanked PGK-neomycin-resistence cassette and an additional LoxP site were inserted 3′ of the targeted exon. Each targeting construct was confirmed by sequencing analysis.
Generation of IGF-1 knockout mice. To generate the conditional macrophage IGF-1 knockout (ϕIGF-1 CKO), mice carrying a floxed exon 3 allele (46) were crossed with the myeloid-specific LysM-Cre line32 (on a C57/Bl6 background) and further crossed to obtain ubiquitous homozygous deletion of the targeted exon. To generate the IGF-1Eb KO, at least two correctly targeted ES cell clones per construct were microinjected into C57/Bl6 J host blastocysts to obtain chimeric mice. Chimeric males were mated to C57/Bl6 J females and germline transmission of the mutated allele to their offspring was confirmed by PCR and Southern blotting analysis. Mice carrying floxed exon 5 allele were crossed with the ubiquitous CMV-Cre line52 (on a C57/Bl6 background) and further crossed to obtain ubiquitous homozygous deletion of the targeted exon. For both IGF-1 floxed alleles, no phenotypic differences were observed in two independently generated lines. For most experiments, LysM-Cre mice were included as controls, with no discernable phenotype.
IGF-1 serum concentration measurements. To determine circulating IGF-1 levels, the OCTEIA Rat/Mouse IGF-1 IEMA for the quantitative determination of IGF-1 in rat and mouse serum was used (iDS, Boldon, UK) according to the manufacturer's instructions. Results were calculated by logarithmic plotting of the mean absorbance of each calibrator against concentration of rat IGF-1 calibrators.
Muscle regeneration. Mice were anesthetized with isofluorane and the TA and quadriceps from 2–3-month-old mice were injected with 4 and 8 injections of 5 µl CTX, respectively (Latoxan, Valence, France, diluted to 10 µmol/l in phosphate buffered saline (PBS). Muscle samples were taken various times postinjury. For histology, at least three mice were analyzed per time point, while 4–8 mice were analyzed per time point for flow cytometry and cell sorting experiments.
BMM culture. Femurs and tibiae were collected from mice then crushed in the presence of PBS + 1% fetal bovine serum (FBS), filtered through a 70 μm filter (BD Bioscience, Oxford, UK) and pelleted. The bone marrow cells were plated on plastic tissue culture dishes at a density of 1.5 × 106 cells/10 cm dish in RPMI medium supplemented with 20 ng/ml macrophage colony-stimulating factor (Sigma, St Louis, MO) and 50 µmol/l β-mercaptoethanol for 6 days to derive macrophages. M-CSF was reduced to 10 ng/ml for 12 hours then depleted from the medium for another 12 hours and serum reduced to 2%. Stimulation of macrophages was performed with 50 ng/ml rIGF-1 (Peprotech, Rocky Hill, NJ) or 5 U/ml IL-4 (Peprotech) for 8 hours before RNA isolation. Some cells were treated overnight with 100 U/ml IFNγ (Peprotech) then for 4 hours with 100 U/ml IFNγ + 1 µg/ml LPS (Sigma) before stimulation with rIGF-1 for 8 hours. For RNA isolation, cells were washed in PBS and lysed in TRIzol Reagent (Invitrogen, Carlsbad, CA).
RNA isolation. Total RNA was extracted from snap-frozen tissues and cultured cells with TRIzol Reagent (Invitrogen) according to the manufacturer's instructions. For FACS-sorted cells total RNA was prepared using the RNeasy micro and mini kits (Qiagen, Manchester, UK), depending on the number of isolated cells.
Quantitative PCR. cDNA was prepared from 1 µg of RNA with a Quantitect reverse transcription kit (Qiagen). Quantitative PCR was performed with Taqman probes run on the 7500 real-time RT-PCR system (Life Technologies, Monza, Italy) and quantified by the delta ΔcT method normalized to house keepings genes as indicated. Taqman primer and probe sets were as follows: Arg1, Mm00475988_m1; mouse ACTB (actin beta) endogenous control; Csf1, Mm00432686_m1; Emr1, Mm01233106_m1; mouse GAPDH endogenous control; IGF-1Ea, Mm00710307_m1; IGF-1Eb, Mm01228180_m1; total IGF-1; IL-1b, Mm01336189_m1; Mm-00439560_m1; IL-4, Mm00445259_m1; IL-10, Mm00439614_m1; IL-12p40, Mm00434174_m1; MRC1, Mm03024053_m1; NOS2, Mm01288989_m1; TGFβ1, Mm00485148_m1; TNF, Mm00443258_m1; Rps20, Mm02342828_g1; VEGFα, Mm00437306_m1; YM1, Mm04213363_u1; Cre, custom primers and probe, For - GCG GTC TGG CAG TAA AAA CTA TC Rev - GTG AAA CAG CAT TGC TGT CAC TT Probe - AAA CAT GCT TCA TCG TCG GTC CGG.
Histology. For paraffin sections, tissues were fixed in 4% paraformaldehyde for 30 minutes, dehydrated in series of ethanol dilutions, passed though xylene, xylene/paraffin, and embedded in paraffin. Transverse cross sections were cut at 8 µm and stained with Trichrome (Sigma, Dorset, UK). Morphometric analysis was performed on digital images taken by a camera (Nikon DXM 1200F) connected to a Leica PM RBE microscope. Nonoverlapping images were taken of the entire cross-sectional area of the muscle using ImagePro Plus 4.0 (Microsoft) software and tiled to reconstitute the whole image of the muscle. CSA was measured by tracing individual myofibers using FIJI.
Cell Isolation and flow cytometry. For cell isolation from tissue, injured muscles were placed in warmed DMEM (Life Technologies, Paisley, UK), and matrix, fibrotic tissue, and nerves were removed carefully. The muscles were chopped into small pieces and enzymatic disaggregation was performed, first using freshly prepared 4 mg/ml collagenase type II (Worthington, Lakewood, NJ) for 30 minutes (37 °C), then using 1 mg/ml collagenase/dispase (Roche, West Sussex, UK) for 20 minutes (37 °C). Disaggregation was stopped with 5ml of ice-cold horse serum (heat-inactivated; Life Technologies, Paisley, UK) and cells filtered through a 40 µm cell strainer (BD Bioscience). For blood cell isolation, the blood was collected in heparin-coated tubes and red blood cells lysed with Geys solution. An aliquot of the cell suspension was used for cell quantitation on a ViCell counter (Beckman Coulter). Isolated cells were mixed with cold PBS plus 1% FCS (Gibco) and blocked with purified anti CD16/32 (eBioscience, Hatfield, UK) for 10 minutes on ice. For monocyte/macrophage analysis and sorting, cells were stained with the following antibodies for 10 minutes on ice: CD45-APC/Cy7 (eBioscience), CD11b-PE (eBioscience), F4/80-biotin (AbD serotec, Kidlington, UK), Ly6C-APC (eBioscience), and CD206-Alexa 488 (Biolegend, London, UK). The macrophage population was very heterogeneous with only a proportion expressing Ly6C or CD206. For analysis of monocyte and neutrophil populations cells were stained with CD45-APC/Cy7, CD11b-PE, F4/80-biotin, Ly6C-APC and Ly6G-FITC (eBioscience). To distinguish other cell populations in the muscle, a combination of CD45-APC/Cy7, a7b1-integrin-PE (RnD Systems, Abingdon, UK) and CD31-PerCPCy5.5 (Biolegend) were used. For biotinylated antibodies Streptavidin-PECy7 (eBioscience) was used as a secondary reagent. Sytox blue (Invitrogen) was used to exclude dead cells and debris and isotype controls used to set gates. Samples were acquired with a FACSCalibur and analyzed with FlowJo software (Tree Star, Ashland, OR). To quantitate the absolute number of a defined cell population, the percentage of the defined population out of the total number of live cells (determined by Sytox blue exclusion) was divided by 100 and multiplied by the viable cell count for that sample taken on the ViCell counter. This was divided by the muscle weight for that sample for normalization. For gene expression analysis, cells were sorted directly into RLT Buffer (Qiagen).
Statistical analysis. Statistical analyses were performed with GraphPad Prism 5 software (GraphPad Software, San Diego, CA). Data are expressed as ± SEM. For comparison of two groups, the Mann–Whitney U-test was performed and differences considered significant if P < 0.05. N numbers are specified in the figure legends. The growth curve in Supplementary Figure S4b was analyzed with a two-way paired analysis of variance test.
SUPPLEMENTARY MATERIAL Figure S1. Cellular sources of IGF-1 in regenerating muscle. Figure S2. Gating strategies and FMO controls used to define monocytes/macrophages isolated from muscle. Figure S3. IGF-1Eb propeptide is dispensable for muscle regeneration. Figure S4. Specificity of Lysozyme M Cre-mediated deletion of IGF-1. Figure S5. Phenotype of mice with myeloid-specific deletion of IGF-1.
Acknowledgments
We are grateful to Stephen Rothery and the Facility for Imaging by Light Microscopy at Imperial College London for excellent technical assistance and to members of the Rosenthal Laboratory for critical discussion. This work was supported by funds from European Molecular Biology Laboratory, EU grants to N.R.: Eumodic (EU Integrated Project No. LSHG-CT- 2006–037188); ENDOSTEM (EU Integrated Project HEALTH-2009-1.4-3); and British Heart Foundation grants to N.R. and MDS (PG/08/111 and PG/09/10, CH/08/002, RE/08/002, RG/08/007, SI/11/2/28875) and grants to A.M. from the Progetti di Ricerca di Interesse Nazionale (PRIN), Fondazione Telethon, the Association Francaise contre les Myopathies (AFM), and the Agenzia Spaziale Italiana. N.R. is an NH&MRC Australia Fellow.
Supplementary Material
References
- Tidball, JG and Villalta, SA (2010). Regulatory interactions between muscle and the immune system during muscle regeneration. Am J Physiol Regul Integr Comp Physiol 298: R1173–R1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, HB and Mosser, DM (2013). Extrinsic and intrinsic control of macrophage inflammatory responses. J Leukoc Biol 94: 913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saclier, M, Yacoub-Youssef, H, Mackey, AL, Arnold, L, Ardjoune, H, Magnan, M et al. (2013). Differentially activated macrophages orchestrate myogenic precursor cell fate during human skeletal muscle regeneration. Stem Cells 31: 384–396. [DOI] [PubMed] [Google Scholar]
- Ochoa, O, Sun, D, Reyes-Reyna, SM, Waite, LL, Michalek, JE, McManus, LM et al. (2007). Delayed angiogenesis and VEGF production in CCR2-/- mice during impaired skeletal muscle regeneration. Am J Physiol Regul Integr Comp Physiol 293: R651–R661. [DOI] [PubMed] [Google Scholar]
- Arnold, L, Henry, A, Poron, F, Baba-Amer, Y, van Rooijen, N, Plonquet, A et al. (2007). Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med 204: 1057–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H, Huang, D, Saederup, N, Charo, IF, Ransohoff, RM and Zhou, L (2011). Macrophages recruited via CCR2 produce insulin-like growth factor-1 to repair acute skeletal muscle injury. FASEB J 25: 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H, Melton, DW, Porter, L, Sarwar, ZU, McManus, LM and Shireman, PK (2014). Altered macrophage phenotype transition impairs skeletal muscle regeneration. Am J Pathol 184: 1167–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, LC, Jenkins, SJ, Allen, JE and Taylor, PR (2013). Tissue-resident macrophages. Nat Immunol 14: 986–995.24048120 [Google Scholar]
- Nguyen, HX and Tidball, JG (2003). Interactions between neutrophils and macrophages promote macrophage killing of rat muscle cells in vitro. J Physiol 547(Pt 1): 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffell, D, Mourkioti, F, Gambardella, A, Kirstetter, P, Lopez, RG, Rosenthal, N et al. (2009). A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc Natl Acad Sci USA 106: 17475–17480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalta, SA, Rinaldi, C, Deng, B, Liu, G, Fedor, B and Tidball, JG (2011). Interleukin-10 reduces the pathology of mdx muscular dystrophy by deactivating M1 macrophages and modulating macrophage phenotype. Hum Mol Genet 20: 790–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton-Davis, ER, Shoturma, DI and Sweeney, HL (1999). Contribution of satellite cells to IGF-I induced hypertrophy of skeletal muscle. Acta Physiol Scand 167: 301–305. [DOI] [PubMed] [Google Scholar]
- Bark, TH, McNurlan, MA, Lang, CH and Garlick, PJ (1998). Increased protein synthesis after acute IGF-I or insulin infusion is localized to muscle in mice. Am J Physiol 275(1 Pt 1): E118–E123. [DOI] [PubMed] [Google Scholar]
- Fernández, AM, Dupont, J, Farrar, RP, Lee, S, Stannard, B and Le Roith, D (2002). Muscle-specific inactivation of the IGF-I receptor induces compensatory hyperplasia in skeletal muscle. J Clin Invest 109: 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musarò, A, McCullagh, KJ, Naya, FJ, Olson, EN and Rosenthal, N (1999). IGF-1 induces skeletal myocyte hypertrophy through calcineurin in association with GATA-2 and NF-ATc1. Nature 400: 581–585. [DOI] [PubMed] [Google Scholar]
- Barton-Davis, ER, Shoturma, DI, Musaro, A, Rosenthal, N and Sweeney, HL (1998). Viral mediated expression of insulin-like growth factor I blocks the aging-related loss of skeletal muscle function. Proc Natl Acad Sci USA 95: 15603–15607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefaucheur, JP and Sébille, A (1995). Muscle regeneration following injury can be modified in vivo by immune neutralization of basic fibroblast growth factor, transforming growth factor beta 1 or insulin-like growth factor I. J Neuroimmunol 57: 85–91. [DOI] [PubMed] [Google Scholar]
- McKoy, G, Ashley, W, Mander, J, Yang, SY, Williams, N, Russell, B et al. (1999). Expression of insulin growth factor-1 splice variants and structural genes in rabbit skeletal muscle induced by stretch and stimulation. J Physiol 516 (Pt 2): 583–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, SY and Goldspink, G (2002). Different roles of the IGF-I Ec peptide (MGF) and mature IGF-I in myoblast proliferation and differentiation. FEBS Lett 522: 156–160. [DOI] [PubMed] [Google Scholar]
- Barton, ER, DeMeo, J and Lei, H (2010). The insulin-like growth factor (IGF)-I E-peptides are required for isoform-specific gene expression and muscle hypertrophy after local IGF-I production. J Appl Physiol (1985) 108: 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertzer, JD and Lynch, GS (2006). Comparative evaluation of IGF-I gene transfer and IGF-I protein administration for enhancing skeletal muscle regeneration after injury. Gene Ther 13: 1657–1664. [DOI] [PubMed] [Google Scholar]
- Pelosi, L, Giacinti, C, Nardis, C, Borsellino, G, Rizzuto, E, Nicoletti, C et al. (2007). Local expression of IGF-1 accelerates muscle regeneration by rapidly modulating inflammatory cytokines and chemokines. FASEB J 21: 1393–1402. [DOI] [PubMed] [Google Scholar]
- Gow, DJ, Sester, DP and Hume, DA (2010). CSF-1, IGF-1, and the control of postnatal growth and development. J Leukoc Biol 88: 475–481. [DOI] [PubMed] [Google Scholar]
- Alikhan, MA, Jones, CV, Williams, TM, Beckhouse, AG, Fletcher, AL, Kett, MM et al. (2011). Colony-stimulating factor-1 promotes kidney growth and repair via alteration of macrophage responses. Am J Pathol 179: 1243–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F, Liu, Z, Wu, W, Rozo, C, Bowdridge, S, Millman, A et al. (2012). An essential role for TH2-type responses in limiting acute tissue damage during experimental helminth infection. Nat Med 18: 260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkins, S, Rebeiz, N, Biragyn, A, Reese, DL and Kelley, KW (1993). Murine macrophages express abundant insulin-like growth factor-I class I Ea and Eb transcripts. Endocrinology 133: 2334–2343. [DOI] [PubMed] [Google Scholar]
- Martinez, FO, Gordon, S, Locati, M and Mantovani, A (2006). Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol 177: 7303–7311. [DOI] [PubMed] [Google Scholar]
- Rigamonti, E, Touvier, T, Clementi, E, Manfredi, AA, Brunelli, S and Rovere-Querini, P (2013). Requirement of inducible nitric oxide synthase for skeletal muscle regeneration after acute damage. J Immunol 190: 1767–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornaro, M, Hinken, AC, Needle, S, Hu, E, Trendelenburg, AU, Mayer, A et al. (2014). Mechano-growth factor peptide, the COOH terminus of unprocessed insulin-like growth factor 1, has no apparent effect on myoblasts or primary muscle stem cells. Am J Physiol Endocrinol Metab 306: E150–E156. [DOI] [PubMed] [Google Scholar]
- Arkins, S, Rebeiz, N, Brunke-Reese, DL, Minshall, C and Kelley, KW (1995). The colony-stimulating factors induce expression of insulin-like growth factor-I messenger ribonucleic acid during hematopoiesis. Endocrinology 136: 1153–1160. [DOI] [PubMed] [Google Scholar]
- Temmerman, L, Slonimsky, E and Rosenthal, N (2010). Class 2 IGF-1 isoforms are dispensable for viability, growth and maintenance of IGF-1 serum levels. Growth Horm IGF Res 20: 255–263. [DOI] [PubMed] [Google Scholar]
- Clausen, BE, Burkhardt, C, Reith, W, Renkawitz, R and Förster, I (1999). Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res 8: 265–277. [DOI] [PubMed] [Google Scholar]
- von Brühl, ML, Stark, K, Steinhart, A, Chandraratne, S, Konrad, I, Lorenz, M et al. (2012). Monocytes, neutrophils, and platelets cooperate to initiate and propagate venous thrombosis in mice in vivo. J Exp Med 209: 819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavalli, MD, DiGirolamo, DJ, Fan, Y, Riddle, RC, Campbell, KS, van Groen, T et al. (2010). Distinct growth hormone receptor signaling modes regulate skeletal muscle development and insulin sensitivity in mice. J Clin Invest 120: 4007–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells, CA, Ravasi, T and Hume, DA (2005). Inflammation suppressor genes: please switch out all the lights. J Leukoc Biol 78: 9–13. [DOI] [PubMed] [Google Scholar]
- Zhang, L, Wang, XH, Wang, H, Du, J and Mitch, WE (2010). Satellite cell dysfunction and impaired IGF-1 signaling cause CKD-induced muscle atrophy. J Am Soc Nephrol 21: 419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, TJ (2010). Insulin-like growth factor-I regulation of immune function: a potential therapeutic target in autoimmune diseases? Pharmacol Rev 62: 199–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saclier, M, Cuvellier, S, Magnan, M, Mounier, R and Chazaud, B (2013). Monocyte/macrophage interactions with myogenic precursor cells during skeletal muscle regeneration. FEBS J 280: 4118–4130. [DOI] [PubMed] [Google Scholar]
- Wynes, MW and Riches, DW (2003). Induction of macrophage insulin-like growth factor-I expression by the Th2 cytokines IL-4 and IL-13. J Immunol 171: 3550–3559. [DOI] [PubMed] [Google Scholar]
- Arkins, S, Rebeiz, N, Brunke-Reese, DL, Biragyn, A and Kelley, KW (1995). Interferon-gamma inhibits macrophage insulin-like growth factor-I synthesis at the transcriptional level. Mol Endocrinol 9: 350–360. [DOI] [PubMed] [Google Scholar]
- Chazaud, B, Brigitte, M, Yacoub-Youssef, H, Arnold, L, Gherardi, R, Sonnet, C et al. (2009). Dual and beneficial roles of macrophages during skeletal muscle regeneration. Exerc Sport Sci Rev 37: 18–22. [DOI] [PubMed] [Google Scholar]
- Villalta, SA, Nguyen, HX, Deng, B, Gotoh, T and Tidball, JG (2009). Shifts in macrophage phenotypes and macrophage competition for arginine metabolism affect the severity of muscle pathology in muscular dystrophy. Hum Mol Genet 18: 482–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani, A, Biswas, SK, Galdiero, MR, Sica, A and Locati, M (2013). Macrophage plasticity and polarization in tissue repair and remodelling. J Pathol 229: 176–185. [DOI] [PubMed] [Google Scholar]
- Pinto, AR, Paolicelli, R, Salimova, E, Gospocic, J, Slonimsky, E, Bilbao-Cortes, D et al. (2012). An abundant tissue macrophage population in the adult murine heart with a distinct alternatively-activated macrophage profile. PLoS One 7: e36814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheny, RW Jr, Nindl, BC and Adamo, ML (2010). Minireview: Mechano-growth factor: a putative product of IGF-I gene expression involved in tissue repair and regeneration. Endocrinology 151: 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippou, A, Stavropoulou, A, Sourla, A, Pissimissis, N, Halapas, A, Maridaki, M et al. (2008). Characterization of a rabbit antihuman mechano growth factor (MGF) polyclonal antibody against the last 24 amino acids of the E domain. In Vivo 22: 27–35. [PubMed] [Google Scholar]
- Ates, K, Yang, SY, Orrell, RW, Sinanan, AC, Simons, P, Solomon, A et al. (2007). The IGF-I splice variant MGF increases progenitor cells in ALS, dystrophic, and normal muscle. FEBS Lett 581: 2727–2732. [DOI] [PubMed] [Google Scholar]
- Stavropoulou, A, Halapas, A, Sourla, A, Philippou, A, Papageorgiou, E, Papalois, A et al. (2009). IGF-1 expression in infarcted myocardium and MGF E peptide actions in rat cardiomyocytes in vitro. Mol Med 15: 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Shannon, V, Ochoa, O, Reyes-Reyna, SM, Sun, D, Michalek, JE, Kuziel, WA et al. (2007). Fat accumulation with altered inflammation and regeneration in skeletal muscle of CCR2-/- mice following ischemic injury. Am J Physiol Cell Physiol 292: C953–C967. [DOI] [PubMed] [Google Scholar]
- Horio, T, Maki, T, Kishimoto, I, Tokudome, T, Okumura, H, Yoshihara, F et al. (2005). Production and autocrine/paracrine effects of endogenous insulin-like growth factor-1 in rat cardiac fibroblasts. Regul Pept 124: 65–72. [DOI] [PubMed] [Google Scholar]
- Wynes, MW, Frankel, SK and Riches, DW (2004). IL-4-induced macrophage-derived IGF-I protects myofibroblasts from apoptosis following growth factor withdrawal. J Leukoc Biol 76: 1019–1027. [DOI] [PubMed] [Google Scholar]
- Schwenk, F, Baron, U and Rajewsky, K (1995). A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res 23: 5080–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.