SUMMARY
Histone posttranslational modifications represent a versatile set of epigenetic marks involved not only in dynamic cellular processes, such as transcription and DNA repair, but also in the stable maintenance of repressive chromatin. In this article, we review many of the key and newly identified histone modifications known to be deregulated in cancer and how this impacts function. The latter part of the article addresses the challenges and current status of the epigenetic drug development process as it applies to cancer therapeutics.
Gene expression is regulated, in part, by histone modifications (e.g., acetylation and methylation). The activation of oncogenes or the inactivation of tumor-suppressor genes can be caused by their dysregulation.
OVERVIEW
Cancer is a diverse collection of diseases characterized by the dysregulation of important pathways that control normal cellular homeostasis. This escape from normal control mechanisms leads to the six hallmarks of cancer, which include sustaining proliferative signaling, evading growth suppressors, resisting cell death, enabling replicative immortality, inducing angiogenesis, and activating invasion and metastasis (Hanahan and Weinberg 2011). The systematic investigation of the acquired and inherited molecular alterations in the genomes of somatic cells has revealed a great deal about the genetic basis for cancer initiation, progression, and maintenance. This progress has afforded a number of promising and effective opportunities for therapeutic intervention.
The epigenetic mechanisms that govern transcriptional regulation and the corresponding dysregulation in cancer, initially less well studied, have increasingly become the focus of cancer researchers, although the transgenerational effects remain largely unexplored. “Epigenetic” transcriptional control can occur through DNA methylation, covalent histone modification, the reading of these modifications by protein recognition modules, histone exchange, and alteration by ATP-dependent chromatin remodelers and via the effects of noncoding RNA. Because DNA methylation in cancer is addressed elsewhere, this article focuses on many of the covalent histone modifications that are altered in cancer, particularly the well-studied acetylation and methylation modifications.
Collectively, the combination of histone marks found in a localized region of chromatin function through multiple mechanisms as part of a “chromatin-based signaling” system (Jenuwein and Allis 2001; Schreiber and Bradley 2002). The landscape of known histone and nonhistone modifications that affect chromatin-based processes continues to expand. The diverse set of observed histone adornments includes phosphorylation, citrullination, sumoylation, adenosine diphosphate (ADP) ribosylation, deimination, and crotonylation. The various known epigenetic mechanisms work in a concerted and interdependent fashion to regulate gene expression. In cancers, their misregulation can result in the inappropriate activation of oncogenes or, conversely, the inappropriate inactivation of tumor suppressors. There is also a growing understanding and appreciation for the genetic basis contributing to epigenetic changes observed in cancer. This adds to the complexity of cancer etiology and, perhaps, offers important general insights into the epigenetic basis of human disease.
This article speaks of some of the challenges faced by the epigenetic drug discovery process, searching for molecules targeting epigenetic regulators and also supporting an emerging role of epigenetic alterations that occur during chemotherapy, thought to contribute to drug resistance. Finally, it highlights some of the more recent progress toward developing therapeutic agents in this promising target space.
1. INTRODUCTION
DNA within cells is packaged as chromatin, a dynamic structure composed of nucleosomes as the fundamental building blocks. Histones are the central component of the nucleosomal subunit, forming an octamer containing the four core histone proteins (H3, H4, H2A, H2B) around which is wrapped a 147-base-pair segment of DNA. Each of the largely globular histone proteins possesses a characteristic side chain, or tail, which is densely populated with basic lysine and arginine residues. The histone tails are subject to extensive covalent posttranslational modifications (PTMs) that cooperate to govern the chromatin state. Some PTMs can alter the charge density between histones and DNA, impacting chromatin organization and underlying transcriptional processes, but they can also serve as recognition modules for specific binding proteins that, when bound, may then signal for alterations in chromatin structure or function.
Alterations in the patterns of histone PTMs have been extensively linked to cancer, both at the global level across the genome (Seligson et al. 2005; Bannister and Kouzarides 2011) and at specific gene loci, using ChIP-chip (chromatin immunoprecipitation with DNA microarray analysis) and ChIP-sequencing (parallel sequencing technologies coupled to chromatin immunoprecipitation) technology. These findings have come on the back of the earlier, more established findings linking aberrant DNA methylation to cancer, discovered in the early 1980s (see Baylin and Jones 2014). In addition to recent PTM mapping projects, sequencing efforts have also now identified many of the enzymes responsible for placing (“writers”) and removing (“erasers”) such epigenetic marks (Fig. 1). Mutations in such enzymes turn out to be among the most frequently mutated targets in cancers (Shen and Laird 2013). Collectively, these findings show interplay between cancer genetics and epigenetics, adding to the complexity in our understanding of the oncogenic process. Advances in whole genome/exome sequencing of patient tumors have allowed for the identification of possible key epigenetic drivers of cancer. These epigenetic drivers may silence one or more tumor-suppressor genes and/or activate oncogenes, thus providing an alternative mechanism by which oncogenic reprogramming of the genome may occur (Shen and Laird 2013). Genomic studies have clearly implicated dysregulation of chromatin modifiers as drivers in many types of cancer (Garraway and Lander 2013) and recurrent mutations occur in the genes that encode the enzymes, which add, remove, and interpret the covalent histone modifications. Intriguingly, certain chromatin modifiers have been identified in cancer, for both increased and decreased levels of functionality, suggesting they operate both as tumor-suppressor genes and oncogenes. The loss-of-function mutations in affected tumors are often heterozygous, suggesting that haploinsufficiency for these chromatin-modifying enzymes drives the cancer, whereas total absence of function is cell lethal. As a result, this class of enzymes has broad appeal as potential therapeutic targets with both gain-of-function aberrations associated with oncogenic behavior and monoallelic loss-of-function changes rendering tumor cells particularly vulnerable to further inhibition.
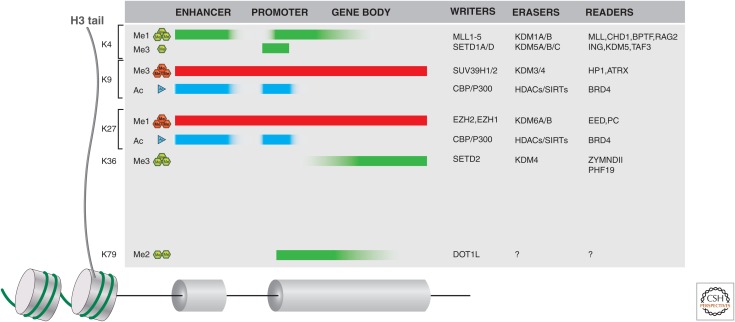
Figure 1.
Histone writers, erasers, and readers in cancer. Histone H3 tail lysine residues, frequently subject to posttranslational modifications (PTMs), are indicated along the left side. The typical distribution of these H3 PTMs is also indicated along the length of gene loci (including distal enhancers) as shaded blocks. Green (methylation) or cyan (acetylation) indicates histone marks associated with active genes, whereas red shading is indicative of silent genes. A few examples of writers, erasers, and readers that may propagate a mark or act as an effector protein are listed on the right side of the figure. For a more complete listing of these proteins, see Appendices A–D at the end of this article.
An additional level of epigenetic control, and with it additional complexity, is brought by the wide variety of proteins that act at the chromatin level through domains that “read” histone and even DNA PTMs. Direct aberrations in reader proteins have recently been shown to drive oncogenic transformation in some contexts (French et al. 2001, 2008). The discovery of potent and selective small-molecule inhibitors of prototypic members of readers from the bromodomain family, which recognize acetyl-lysine marks, suggests that these may represent attractive points of therapeutic intervention in cancers arising from other genetic and epigenetic drivers (elaborated in Qi 2014; Schaefer 2014; also see Filippakopoulos et al. 2010; Gallenkamp et al. 2014). In summary, the ensemble of chromatin regulators and adaptors possess a diverse set of specialized domains that serve to bind to and recognize histone modifications either individually or in specific combinations. These protein-binding modules play an important role in directing the appropriate transcriptional machinery to locations on chromatin.
This article focuses on highlighting what is known about the well-studied PTMs in cancer. Recent findings on some newer PTMs also illustrate the pace of progress in the field and serve to highlight the complexity of the epigenome, its regulation and dysregulation in cancer, and the nature of its regulators and interacting proteins. The second part of this article instructs the reader about the drug-discovery process as it applies to the search for effective therapeutics that interfere with epigenetic targets or reverse epigenetic adaptions contributing to drug resistance during cancer treatment.
2. HISTONE MODIFICATIONS
It is well noted that PTM of histones mediates a variety of critical biological processes, generally via chromatin modification that is conducive to the expression or repression of target genes. The bulk of the literature has focused on acetylation, methylation, and phosphorylation. However, in addition to these well-published modifications, histones may also be modified in other ways, some of which are illustrated in Figure 6 of Allis et al. (2014) or included in Zhao and Garcia (2014). These include citrullination, ubiquitination, ADP-ribosylation, deamination, formylation, O-GlcNAcylation, propionylation, butyrylation, crotonylation, and proline isomerization (Chen et al. 2007; Martin and Zhang 2007; Ruthenburg et al. 2007; Tan et al. 2011; Herranz et al. 2012; Tweedie-Cullen et al. 2012). It is generally accepted that the sum of all these PTMs largely determines the chromatin structure and, hence, biological outcome. The contextual reading and interpretation of the collection of histone modifications is critical, as the same combination of marks may result in distinct biologic outcomes at different genes within the same (or different) cells. This may, at least in part, be due to different reader recognition, DNA-binding proteins, and/or chromatin conformations. Experimentally, the modification and the reader cannot be readily separated for a specific output, and this is an important limitation of our current ability to readily translate combinatorial PTMs. Furthermore, the enzymes that modulate the addition or removal of these moieties are not fully understood (see Cheng 2014; Marmorstein and Zhou 2014; Seto and Yoshida 2014) or, in many cases, yet to be identified.
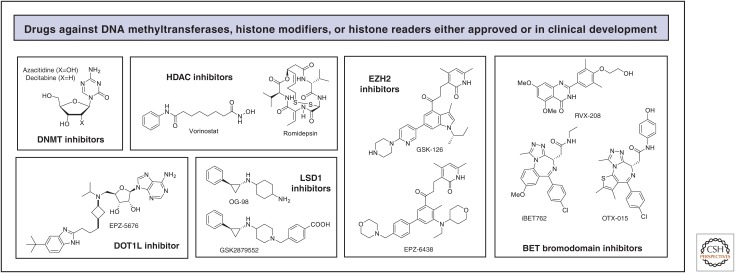
Figure 6.
Representative structures of drugs targeting DNA methylation, histone modifications, and histone readers in oncology.
2.1. Histone Acetylation
The addition of an acetyl group can occur at multiple lysine residues on histone tails. This has the potential to broadly influence the compaction state of chromatin by neutralizing the basic charge at unmodified lysine residues (Kouzarides 2007), weakening the electrostatic interaction between negatively charged DNA and histones. An increasing body of data, however, suggests that this may be an oversimplification of the consequences of this modification and likely belies the importance of specific acetylation events. Another recently discovered, generalized function of histone acetylation may be to regulate intracellular pH (pHi) (McBrian et al. 2013); this is corroborated by the fact that many tumors show low pHi and display concomitant reduced levels of histone acetylation, which correlates with a poor clinical outcome. From a functional perspective, we know that histone acetylation is largely associated with active transcription, particularly localized at enhancers, promoters, and the gene body (Di Cerbo and Schneider 2013). Altered global levels of histone acetylation, particularly acetylation of H4 at lysine (K)16, have been linked to a cancer phenotype in a variety of cancers (Fraga et al. 2005) and have even been found to be of potential prognostic value (Seligson et al. 2009). When hyperacetylation occurs, specifically involving proto-oncogenes, gene expression may be activated, whereas hypoacetylation of tumor suppressors often localizes to promoters, co-occurring with DNA methylation, causing the genes to be silenced (see Baylin and Jones 2014).
The enzymes that catalyze the addition of acetyl groups to histone lysine residues are the lysine (K) acetyltransferases (KATs), commonly referred to as histone acetyltransferases (HATs). These enzymes can also acetylate a broad range of nonhistone proteins, including p53, Rb, and MYC. The opposing histone deacetylases (HDACs) are responsible for their removal. Although HATs are generally associated with transcriptional activity, both HAT and HDAC activity together are required for properly regulated gene expression (Struhl 1998). This is partly to do with their activity often being an integral part of multisubunit chromatin-modifying complexes. As a result, genetic or epigenetic aberrations affecting HAT and HDAC expression, translation, protein stability, or domain function can have chromatin regulatory consequences beyond changes in the histone acetylation states. In addition to the structural effects acetylation has on chromatin, it can also function in chromatin as a signal that is recognized by a specific protein module (i.e., “reader”), such as the bromodomain. Epigenetic-based drugs, targeting acetyl readers, are thus likely to be of clinical relevance in the treatment of cancer. Given that acetylation modifications are reversible, the pharmacological intervention of HATs, HDACs, and acetyllysine readers represent known, in the case of HDACs, or viable strategies of therapeutic value in the treatment of cancer.
2.1.1. Histone Acetylation Writers
In humans, there are three major families of HATs: the Gcn5-related N-acetyltransferase family (GNAT), the MYST family (MOZ, Ybf2, Sas2, TIP60), and the orphan family (CBP/EP300 and nuclear receptors), whose structure and mechanism of action are elaborated in Marmorstein and Zhou (2014). A variety of studies have implicated HATs as both oncogenes and tumor suppressors, suggesting that the balance of acetylation is critical. Many of the HAT mutations frequently detected in a variety of cancers are captured in Appendix A at the end of this article (Di Cerbo and Schneider 2013). Interestingly, alterations in HAT levels, both upward (Chen et al. 2012b; Hou et al. 2012) and downward (Seligson et al. 2009), often occur without DNA mutation in cancers and are associated with poor outcome.
Somatic mutations in a single allele of the p300 and CBP genes have been identified in multiple cancers. The resulting loss of heterozygosity implicates them as tumor suppressors (Muraoka et al. 1996; Gayther et al. 2000; for more on the characterization of tumor-suppressor genes, see Baylin and Jones 2014). It is increasingly evident that CBP and/or EP300 are important in cancers (e.g., transgenic mice deficient in CBP or EP300 can develop hematologic malignancies [Iyer et al. 2004]). Another facet of HAT function revealed by the many mutations in CBP and EP300 is the protein’s ability to acetylate the nonhistone transcription factors, p53 and BCL6; a lack of P53 and BCL6 acetylation abrogates their transcriptional activator and repressor functions (Fig. 7A of Busslinger and Tarakhovsky 2014), making the resultant cells more tumorigenic via altered pathways that tolerate DNA damage yet evade apoptosis and cell-cycle arrest (Pasqualucci et al. 2011).
Enhanced HAT activity, conversely, has an overall oncogenic effect in cancers and usually occurs as the result of chromosomal translocations with diverse fusion partners, such as mixed lineage leukemia (MLL)-CBP, MLL-EP300, MOZ-EP300, or MOZ-CBP in hematological malignancies (Krivtsov and Armstrong 2007). The oncogenic effect arises when the translocations generate chimeric oncoproteins. This may allow HATs to abnormally acetylate the genomic targets of its fusion partner. Another mechanism by which HATs, such as EP300/CBP, are oncogenic is when they are recruited by more common fusion proteins, such as acute myloid leukemia (AML)1-ETO, to serve as transcriptional coactivators (Wang et al. 2011). Overall, a HAT’s oncogenic or tumor-suppressor effects in cancer are dependent on its dose; overexpression correlates with oncogenic potential, whereas loss of expression results in loss of acetylation capacity. This, therefore, suggests that HATs could be a good drug target, although, to date, progress toward producing a viable HAT inhibitor has lagged behind the development of inhibitors of its counterpart enzyme, HDACs, described in the following section.
2.1.2. Histone Acetylation Erasers
Alterations in HDACs, the enzymes that remove acetyl groups from histone lysine residues, have been observed in cancer. There are four major families of HDACs, termed class I, II, III, and IV. Class I, II, and IV are Zn2+-dependent, whereas class III/Sirtuins are nicotinamide adenine dinucleotide (NAD)-dependent (see Fig. 1 in Seto and Yoshida 2014). The contributions of specific HDAC subtypes to individual cancers are currently not fully understood (see Appendix B at the end of the article) (Barneda-Zahonero and Parra 2012). This may be, in part, because of the relatively low substrate specificity shown by the HDACs themselves; each individual enzyme is capable of deacetylating multiple divergent histone sites. Interestingly, mutations in HDACs are rare, but overexpression of HDACs is frequently observed in cancer patients (Dell’Aversana et al. 2012).
The class I HDACs that are misregulated, usually by overexpression or mistargeting, are found in a variety of human cancers. The frequency with which they are deregulated in cancer, perhaps, reflects their normal function in such a wide range of tissues, but in cancer, this is often associated with poor prognosis (Nakagawa et al. 2007). The role of HDACs in cancer, as with HATs, may not be confined to histones. The growing list of reported HDAC targets that become deacetylated include α-tubulin, HSP90 and cortactin (HDAC6), p53 (HDAC5), and ERRα (HDAC8); HDACs also directly act on proteins involved in tumor migration, metastasis, and growth (see Appendix B at the end of the article). The example of HDAC2 in lung cancer is a case in point; HDAC2 directly deacetylates p53 and the CDKN1B/1C/2A proteins, attenuating a cell’s ability to activate the apoptotic machinery or regulate cell cycle (Jung et al. 2012; Reichert et al. 2012). This, therefore, makes it hard to dissect what the exact epigenetic effects of HDAC deregulation are, and thus how HDAC inhibitors interfere with their activity.
Another mode by which HDACs are implicated in cancer is through the abnormal recruitment of HDAC-containing complexes to promoters as a consequence of chromosomal translocation, observed in leukemias (Mercurio et al. 2010). For example, the promyelocytic leukemia (PML)-retinoic acid receptor α (RARα) translocation, which is the driver for many acute promyelocytic leukemia cases, represses many RAR target genes by the aberrant recruitment of the N-CoR/HDAC repressor complexes (Minucci and Pelicci 2006).
The class III HDACs, the Sirtuins (SIRT1-7), are also capable of deacetylating a combination of histones (SIRT1-3, 6, 7) and nonhistone proteins (SIRT1-3, 5, 7), as well as ADP-ribosylating (SIRT4), and desuccinylating various proteins (SIRT5) (see Appendix B at the end of the article). They are distinguished from the other classes of HDACs by a distinct catalytic mechanism of action (Seto and Yoshida 2014). The bulk of efforts has focused on studying SIRT1, but has, to date, failed to clarify whether SIRT1 is a tumor suppressor or oncogene, suggesting that these activities may be contextual (Stunkel and Campbell 2011). SIRT1 is overexpressed in many tumors (leukemia, lymphoma, prostate, liver, breast, ovarian, gastric, colorectal, and melanoma) but significantly reduced in others (bladder, colon, glioma). It may be that SIRT1 plays an oncogenic role by inactivating other tumor suppressors (e.g., HIC1) and/or activating tumor promoting genes (e.g., through N-Myc stabilization, or p53) or other proteins (cortactin).
SIRT1 levels are elevated in drug-resistant SK-N-SH neuroblastoma cells, a result that prompted the evaluation of class I/II (Vorinostat) and class III (Cambinol) HDAC inhibitors in animal models of neuroblastoma (Lautz et al. 2012). In neuroblastoma, N-Myc induces SIRT1 transcription, which, in turn, enhances N-Myc stability in a feed-forward loop. It is believed that SIRT1 represses MKP3 phosphatase, leading to elevated ERK phosphorylation/activation and then to N-Myc phosphorylation, a more stable form of N-Myc. The antitumor efficacy of cambinol, a SIRT1/2 inhibitor, in both wild-type and doxorubicin-resistant neuroblastoma is consistent with a tumor-promoting role for SIRT1 and/or SIRT2. Gene silencing and use of pharmacological inhibitors in MCF7 breast tumor cells indicate that blockade of both SIRT1 and SIRT2 may be required to induce apoptosis. Other dual SIRT1/2 inhibitors have shown efficacy in xenograft models (melanoma, Burkitt’s lymphoma). In genetically engineered mouse models, SIRT1/PTEN-null transgenic mice spontaneously develop aggressive prostate and thyroid carcinomas. In contrast, SIRT+/–/p53+/– mice develop tumors in multiple organs and SIRT-null mice form prostatic intraepithelial neoplasia.
SIRT3-5 are localized primarily to the mitochondria (although SIRT3 has been reported to deacetylate histones) and can modify a number of substrates involved in energy metabolism via deacetylation, ADP-ribosylation, or desuccinnylation (see Appendix B at the end of the article). It is believed that these Sirtuins act as metabolic sensors to adjust mitochondrial energy production during stress or energy deprivation (Haigis and Sinclair 2010). SIRT3, and perhaps also SIRT6, have been identified as tumor suppressors that regulate glycolysis in tumor cell (Haigis et al. 2012; Sebastian et al. 2012), with reduced expression in human breast cancer tissue versus normal. Its loss correlates with hypoxia-induced factor (HIF)α stabilization, up-regulation of HIF1α target genes, increased glycolysis, tumor cell proliferation, and fibroblast transformation (Bell et al. 2011), whereas overexpression has the opposite effect (Finley et al. 2011; Sebastian et al. 2012). Similarly, depletion of SIRT6 leads to tumorigenesis, with transformed SIRT6-deficient MEFs displaying increased glycolysis and tumor growth. Conditional SIRT6 knockout in vivo increases the number, size, and aggressiveness of tumors. These data suggest that SIRT6 plays a role in both the establishment and maintenance of cancer (Sebastian et al. 2012).
SIRT7 deacetylates H3K18 and is highly expressed in many cancers (see Appendix A at the end of the article) (Van Damme et al. 2012; Paredes et al. 2014). Hypoacetylation of H3K18, which is indicative of high SIRT7 activity, causes tumor-suppressor gene repression, and is associated with cancer progression and poor prognosis (Seligson et al. 2005; Barber et al. 2012; Paredes et al. 2014).
Broad-spectrum (class I/II) HDAC (e.g., vorinostat, romidepsin) and DNA methylation inhibitors (Dacogen, Vidaza) were the first types of epigenetically targeted therapies approved by the FDA for the treatment of hematological malignancies (discussed in more depth in Baylin and Jones 2014). Data are evolving that suggest that these therapies may be even more effective in combination with standards of care or even with each other (e.g., HDACi + DNA methylation inhibitor), and perhaps be used to ameliorate chemoresistance (discussed further in Sec. 3.1). However, it is abundantly clear that the functions of each specific HDAC isoform are much more complicated and different than anticipated. This is, in part, due to their contextual function in various protein complexes and ability to modify both histone and nonhistone proteins. In the near future, it is expected that more HDAC isoform-specific small molecules (especially inhibitors) will emerge to provide more clarity as to the roles of each HDAC in tumorigenesis and the therapeutic value of more advanced HDAC modulators.
2.1.3. Histone Acetylation Readers
Proteins that read histone acetylated lysines can do so via the bromodomain motif. There are more than 40 bromodomain-containing proteins, which share a high level of sequence homology and structural similarity within the domain. The bromodomain was the first histone-binding module and, consequently, is the most prominent and thoroughly studied histone recognition domain. It is intrinsic to a number of histone-modifying writers (e.g., p300 and MLL) and remodelers (e.g., SMARCA2), as well as other proteins associated with chromatin function and transcriptional control. Its occurrence in such a vast array of chromatin-associating proteins has made it an obvious druggable motif in the search for new epigenetic drugs. The primary structural feature of the bromodomain is a hydrophobic acetyllysine-binding pocket surrounding, most typically, an asparagine residue, which engages in a hydrogen-bonding interaction with the modified histone substrate (for details, see Fig. 6 of Marmorstein and Zhou 2014).
The BET (bromodomain and extraterminal domain) subset of bromodomain proteins consists of four family members, BRD2, BRD3, BRD4, and BRDT, with a common architecture and structural design. BRD4 is a protein that, when bound to acetylated histones, can activate transcription (see Sec. 6 of Busslinger and Tarakhovsky 2014). The reversal of cancer cell phenotype (i.e., the promotion of differentiation and growth impairment using bromodomain-specific inhibitors, such as JQ1 and I-BET) provided the first proof of concept that this histone mark reader could act as a potential therapeutic target for the treatment of cancers, such as leukemias (described in Qi 2014; Schaefer 2014). The inhibitors were effectively shown to phenocopy BRD4 small hairpin RNA knockdown experiments. These approaches were applied to murine MLL-AF9/NRasG12D leukemia models, MLL-fusion cancer cell lines, and patient-derived cells, leading to differentiation and growth impairment (Fig. 2A) (Dawson et al. 2011; Zuber et al. 2011). Further testing of the effects of selective BET inhibitors in other cancer models continues to reinforce the notion that BET proteins are potential therapeutic targets in other cancers, including nuclear protein in testis (NUT)-midline carcinoma, multiple myeloma, lymphoma, lung cancer, and neuroblastoma (Delmore et al. 2011; Mertz et al. 2011; Lockwood et al. 2012; Wyce et al. 2013). The NUT-midline carcinoma, for example, has a translocation, which fuses the BRD3 or 4 protein to the NUT transcriptional regulator. This fusion produces an oncoprotein that, through binding to acetylated histones, is thought to promote transcription of proliferation genes (e.g., MYC). Although work on BET inhibitors continues to be the main focus of research, it is interesting that a dual-drug strategy involving HDAC inhibitors coupled to the JQ1 BET inhibitor has shown positive results as a therapeutic approach for the treatment of AML (Fiskus et al. 2014).
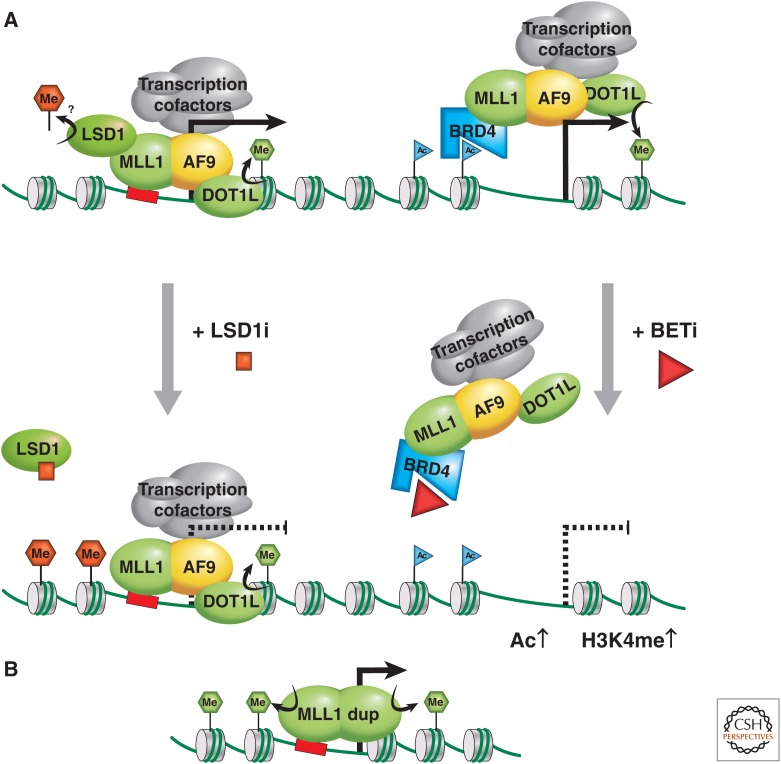
Figure 2.
MLL as an oncogene. (A) The MLL1-AF9 (or ENL) fusion oncoprotein activates transcription via two possible mechanisms; the left mechanism attributes recruitment of MLL1-AF9 to chromatin via the MLL1 portion, and transcription is activated via association with cofactors, including the DOT1 methyltransferase, methylating H3K79 (green hexagon) and the pTEFb complex, which modifies RNA Polymerase II into the active elongating form. (B) The partial duplication of an MLL gene can result in duplication of an internal region that includes chromatin binding features and protein–protein interaction domains, providing oncogenic methyltransferase H3K4me3 activity and increased transcriptional activation. LSD1 may be involved via the MLL supercomplex, or a transcription elongation complex contributing to oncogenic activity through H4K4me2 or H3K9me2 demethylase activity. LSD1 inhibition somehow reduces the oncogenic program, promoting differentiation. In the mechanism on the right, recruitment of MLL1-AF9 to chromatin is attributed to its association with BRD-2, -3, or -4 via acetylated chromatin. This mechanism of gene activation can therapeutically be targeted via treatment with BET inhibitors.
BRD7 and, more recently, BRD9 bromodomain proteins have been identified as components of chromatin-remodeling switch/sucrose nonfermenting (SWI/SNF) complexes, increasingly found to be mutated in a variety of human cancers (Kaeser et al. 2008; Kadoch et al. 2013). BRD7 has specifically been linked to the control of p53 and BRCA1 transcriptional pathways (Drost et al. 2010; Harte et al. 2010). In the case of p53, BRD7 is thought to help recruit the P300 HAT to p53 target genes, activating transcription of senescence genes (Drost et al. 2010). In the case of estrogen receptor (ER)-α response genes, BRD7 is believed to function in the recruitment of the BRCA1 transcriptional activator (Harte et al. 2010).
CBP and EP300, previously described in the context of their HAT activity, also possess bromodomains and transcription factor binding domains. These multifunctional proteins are critical as mediators of histone marks at transcriptional enhancers (Fig. 1) (elaborated in Pirrotta 2014) and required for transcriptional activation. This is illustrated by the loss of expression at MYB target genes, controlling the proliferation and differentiation of hematopoietic stem and progenitor cells, because MYB is mutated at its P300 interacting domain and thus is no longer able to activate gene expression (Sandberg et al. 2005). It should be noted that transcriptional activation occurs in a three-dimensional context, in which looping together of enhancer and promoter regions is thought to ensure proper expression. This relies on correct local histone signatures written by HATs and lysine methyltransferases (KMTs) to facilitate an open chromatin structure and the recruitment of the full transcription machinery (Fig. 1), often through histone PTM reader proteins, such as BRD4 (reviewed in Ong and Corces 2011; exemplified in Fig. 1 of Qi 2014).
P300 was also found to mark binding sites of leukemic fusion proteins, such as AML1-ETO and PML-RARα, and to directly acetylate AML1-ETO fusion proteins, indicating it may have particular importance in contributing to oncogenic transcriptional programs in leukemia (Wang et al. 2011; Saeed et al. 2012). The specific function of the bromodomains in this context has not been definitively shown, but the recent identification of potent and selective chemical probe molecules should afford a similar opportunity to interrogate the function of these domains as has occurred for the BET proteins (Jennings et al. 2014). One would anticipate that, by virtue of BRD proteins being involved in multiple complexes, their mutation must have far-reaching oncogenic effects.
2.2. Histone Methylation
Methylation of lysine and arginine residues on histone tails represents a complex and more subtle chromatin modification than does acetylation. Multiple methylation states exist for both lysine and arginine residues, but when modified, they retain their fundamental basic character (Bannister and Kouzarides 2011). The suggestion that individual methylation states for each histone lysine or arginine can be meaningfully recognized generates enormous functional complexity. This modification is tightly regulated by a number of methyltransferase writers and demethylase eraser enzymes that act in concert to place and remove specific methyl marks critical for gene expression, cell fate, and genomic stability. Individual methyltransferases are highly specific with regard to both the lysine residue on which they operate and the degree of methylation. Appendix C at the end of this article captures some of the important information about lysine and arginine methyltransferases in cancer. Appendix D at the end of this article summarizes some of the important demethylases implicated in cancer. It is important to note that, whereas many of the methyltransferases and demethylases are amplified, overexpressed, deleted, misregulated, rearranged, or mutated, their direct causality on cancer as a result of changes to lysine methylation has not been broadly shown (Black et al. 2012). Additionally, cancer associations have been observed both for elevations and reductions in histone methylation (illustrated in Fig. 7 of Busslinger and Tarakhovsky 2014). The literature has much more on histone KMTs, as elaborated in this section, partly because of their earlier discovery. Discussion of protein arginine methyltransferases (PRMTs) in cancer is discussed in Section 1.4 on PTM cross talk and further in Cheng (2014).
2.2.1. Histone H3K4 Writers
The MLL family of KMTs is implicated in many forms of cancer, either by loss of function or, in the case of MLL1, through dysregulation following translocation or rearrangement (see Appendix C at the end of this article). Members of the MLL (KMT2) methyltransferases specifically methylate histone H3 at lysine 4. MLL1 is frequently translocated in myeloid and lymphoid leukemias, in which such rearrangements account for ∼80% of infant leukemias and 5%–10% of adult leukemias (Smith et al. 2011; Zhang et al. 2012a; Li et al. 2013a). The MLL1 fusion proteins generally do not retain the catalytic SET methyltransferase domain, but do retain their DNA-binding motifs that target Hox genes. The chimeric proteins inappropriately recruit epigenetic factors to MLL targets, altering the transcriptional control of critical genes, such as HOXA9. Constitutive HOXA9 expression prevents differentiation in myeloid lineages, thus, contributing to the maintenance of a multipotent phenotype. The fusion partners also frequently play a role in transcriptional elongation at the target genes. The partial duplication of MLL1’s set KMT domain further corroborates a role for MLL1 in cancer, showing in this case, increased HoxA gene expression associated with increased H3K4me3 marks at the promoter, albeit by an unknown mechanism (Fig. 2B) (Dorrance et al. 2006). In the case of MLL2/3 loss-of-function mutations, the molecular consequences are, at present, unclear, although increasing evidence suggests that cancers may alter H3K4 methylation states as a common method of achieving a growth advantage.
2.2.2. Histone Lysine Erasers
LSD1 (KDM1A) was the first reported lysine demethylase (KDM) enzyme specific to H3K4 and H3K9 residues (Shi and Tsukada 2013). It is a classic oncogene, based on the numerous reports of its overexpression in many types of cancer (see Appendix D at the end of this article). This overexpression, coupled with the fact that LSD1 is part of the MLL supercomplex and found at MLL target genes in MLL-fusion instances of AML, implicates it in the oncogenic gene expression program of these leukemia stem cells. Promising results have been obtained using a small molecule inhibitor of LSD1 in both human and murine AML cell lines with the MLL-AF9 translocation, in which induction of differentiation of otherwise proliferative undifferentiated tumor cells was observed (Fig. 2A) (Harris et al. 2012). The similar use of an LSD1 inhibitor in PML-RARα translocated AML, but this time in combination with all-trans-retinoic acid (ATRA), produced a superior antileukemic effect to the single-agent approach (Schenk et al. 2012). ATRA alone can enhance differentiation of tumor cells through the dissociation of RARα-recruited repressor complexes at RARα target genes. The combination therapy of LSD1 inhibitor plus ATRA caused ATRA-insensitive cancer cells (i.e., a drug-resistant population) to become differentiated. Although these two results show great therapeutic promise, the mechanism by which LSD1 modulates expression is not clear at present.
Apart from KDM1 (i.e., LSD1 and -2), all the other known KDMs are members of the larger JmjC domain family of demethylases. The characterization of many of these proteins is in progress through both functional studies and the development of small molecule inhibitors targeted against them and stand to be invaluable for our understanding and treatment of cancer. One interesting finding made by Sharma et al. (2010) was the discovery of an underlying epigenetic basis for transient and reversible drug resistance that develops in certain cancer cell populations during treatment with cancer drugs. The H3K4-specific JmjC domain demethylase, KDM5A, in particular, was implicated in the development of drug resistance. Further research should be able to confirm whether epigenetic mechanisms, such as histone demethylation truly represent a broad and adaptive mechanism by which cancer cells can avoid eradication during chemotherapy (Sharma et al. 2010). The related KDM5B has also been associated with divergent roles in cancer: in metastatic melanoma, it has a putative tumor-suppressive function (Roesch et al. 2006; Roesch et al. 2008); in breast cancer, a proproliferative role (Mitra et al. 2011); and in prostate cancer, overexpression has been observed.
H3K9 is another histone residue in which methylation is aberrantly regulated in multiple cancers. This could be explained by alterations in H3K9-specific KMTs or opposing KDM enzymes (see Appendices C and D). With respect to the G9a KMT, both deletion or lowered expression and increased expression have been observed in cancer (see Appendix C at the end of this article). For H3K9-specific demethylases, KDM3A and KDM4 are often amplified or highly expressed, whereas attenuated expression of KDM4A has been observed in cancer (see Appendix D at the end of this article). One mechanism by which KDM3 and KDM4 could become overexpressed is by the targeted gene-activating effect of the HIF, itself induced by tumorigenic hypoxic conditions.
2.2.3. Histone H3K27 Writers and Erasers
EZH2 is the catalytic component of the Polycomb repressive complex 2 (PRC2) responsible for the di- and trimethylation of H3K27 (H3K27me2 and -me3) via its SET domain. EZH2 was among the earliest methyltransferase to have putative oncogenic capabilities because it was shown to be overexpressed or amplified in multiple cancers, including breast, prostate, and bladder (Bracken et al. 2003). More recently, altered EZH2 functionality was reported as oncogenic. Intriguingly, it is also believed to act as a tumor suppressor in some cancers, suggesting the balance of resulting H3K27 methylation is key.
Change-of-function mutations within the SET domain of EZH2 are oncogenic, giving rise to H3K27me3 accumulation. Such mutations modify EZH2’s H3K27 substrate preference, increasing catalytic activity at H3K27me2 substrates (not increased for H3K27me1 substrates vs. wild type) (see Appendix C at the end of the article). These heterozygous oncogenic gain-of-function mutations strongly implicate the catalytic activity in the malignant transformation process, and support consequent gene silencing as being the driver for genesis or maintenance of tumor cells. Moreover, diffuse large B-cell lymphoma tumors harboring such EZH2 mutations are extremely sensitive to EZH2 inhibition with small molecule inhibitors, suggesting promise for their clinical translation and offering a path toward molecular stratification of patients on the basis of the mutational status. More compelling evidence that accumulation of H3K27me3 is central to EZH2’s oncogenic effect in medulloblastoma is that EZH2 overexpression or amplification was found to be mutually exclusive to inactivating mutations in the reversing H3K27 demethylase UTX (KDM6A) (Robinson et al. 2012).
A potential tumor-suppressor role for EZH2, or PRC2, has been shown by loss-of-function mutations in myeloid malignancies (Khan et al. 2013). This was further supported by the observation that a K27M point mutation in the histone H3 gene (H3F3A) in pediatric glioblastoma resulted in lowered or absent H3K27me3 (Venneti et al. 2013). This indicated that a loss of a lysine residue at position 27 of H3 abrogated the capacity for chromatin to appropriately repress PRC2-mediated gene expression. EZH2’s repressive effects are not only modulated by the opposing UTX demethylase, but also by the activity of the SWI/SNF chromatin-remodeling complex. SWI/SNF, in fact, critically depends on EZH2 activity, as shown in cell lines and mouse models in which inactivation of EZH2 blocked tumor formation driven by SNF5 loss (Wilson et al. 2010). Collectively, these results indicate that correct genomic and developmental gene repression programs are largely ensured by H3K27 methylation. The writers and erasers of this modification, as well as other epigenetic modulators, are central to maintaining the balance of H3K27 methylation and represent viable drug-discovery targets in cancer. It is interesting to note that continued endeavors to epigenomically map cancers are suggesting that the correct balance of H3K27 methylation may be affected by other chromatin PTMs, namely DNA methylation in cancer, but also the interplay with other histone PTMs, such as acetylation (see Pirrotta 2014).
2.2.4. Other Histone Lysine Writers and Erasers
H3K36-specific methyltransferase writers are implicated in a variety of cancers (see Appendix C at the end of this article). These enzymes are members of the NSD (nuclear receptor-binding SET domain) family (i.e., KMT3B/3F/3G). In AML, NSD1 (KMT3B) is fused to nucleoporin 98 (NUP98), resulting in enhanced H3K36me3 at critical HOXA gene loci, accompanied by increased transcription (Wang et al. 2007). The loss of gene repression may partly be achieved by NSD1/H3K36me3 preventing PRC2 complex access. Similarly, NSD2 (WHSC1, KMT3G) is translocated in 20% of multiple myelomas and has been reported to act as a potent coactivator of NF-κβ, which plays an important role in cancer progression. The investigators suggest that NSD2 is recruited to NF-κβ target gene promoters on pathway induction, resulting in an elevation of histone H3K36me2 and H3K36me3 marks at their promoters, directly implicating NSD2 methyltransferase activity in gene deregulation (Yang et al. 2012).
Another common site of histone lysine methylation is at H4K20, which when monomethylated (H4K20me1) is associated with transcriptional repression and when dimethylated (H4K20me2) is linked to DNA repair pathways. The presence of H4K20me3, accompanied by the loss of H4K16 acetylation, constitutes a common H4 cancer “epigenetic signature” predominantly at DNA repetitive sequences (Fraga et al. 2005). This suggests that one or more of the known histone H4K20 methyltransferases—that is, SUV420H1 and SUV420H2 (H4K20me2 and H4K20me3) or SETD8 (HK20me1) (KMT5A)—could be targets for disruption in cancer cells.
2.3. “Atypical” Histone Modifications
The growing base of knowledge regarding some of these less prevalent or newer PTMs, namely, histone deamination/citrullination, ubiquitination, ADP-ribosylation, deamination, N6-formylation, and O-GlcNAcylation, are discussed below.
2.3.1. Histone Arginine Citrullination
Histone arginine residues may become methylated like lysines, and this modification is catalyzed by the PRMT family of enzymes. Histone arginine residues, unmodified or monomethylated, may also be modified by hydrolysis to Citrulline, a process termed citrullination or deimination. This removes the positive charge, as does histone acetylation, and is enzymatically catalyzed by the partitioning and anchoring domain (PADs or PADIs). Of the PAD family, only PAD4 has a nuclear localization signal and been clearly shown to deiminate/citrullinate histones H3 (Arg residues 2, 8, 17, 26), H2A, H4 (Arg3), and H1R54 (Wang et al. 2004; Tanikawa et al. 2012; Christophorou et al. 2014), although a recent report showed PAD2 as deiminating H3R26 (Zhang et al. 2012b). The citrullination of histone tails is associated with the decondensation of chromatin, but this is tightly regulated to occur only in naïve pluripotent (i.e., embryonic stem or inner cell mass cells) cells, conducive to the transcriptional activation of key stem-cell genes, or in myeloid lineages as part of the immune response to inflammatory stimuli. The H1-specific deimination at stem-cell loci seems to be one mechanism by which PAD4 effects chromatin decondensation through the weakening and eventual displacement of H1 on chromatin in ground-state pluripotent cells (Christophorou et al. 2014). Citrullination of H3R8, in some instances, may also contribute to chromatin decondensation, possibly through the interference of HP1a binding to H3K9me3 (Sharma et al. 2012). Given that PAD4 is highly expressed in many tumor tissues and cell lines (e.g., non-small-cell lung carcinoma [NSCLC], ovarian, breast, and hepatocellular carcinomas) compared with normal tissues or more-benign hyperplastic tissues (Chang et al. 2009), it will be interesting to see whether PAD4 contributes to tumorigenesis. It is conceivable that this may occur by promoting chromatin decondensation, resulting in the activation of pluripotent stem cell genes, or interference of HP1b heterochromatin, or via its gene-repressive effect via involvement in the p53 pathway described below.
Citrullination may have a gene-repressive effect, reported first in conjunction with HDAC1-mediated histone deacetylation of estrogen-regulated genes (Cuthbert et al. 2004; Wang et al. 2004; Denis et al. 2009). Depletion of PAD4 in colorectal tumor cells has an antitumorigenic effect, increasing the expression of p53 target genes (p21, GADD45, PUMA) and inducing cell-cycle arrest and apoptosis (Li et al. 2008). As p53 and PAD4 directly interact, it is not certain whether PAD4 acts in these cells via a scaffolding effect, enzymatic deiminase activity, or a combination of both.
Based on the accumulated evidence, there is interest in developing inhibitors of PADs (especially PAD2 and PAD4) for the treatment of cancer, and possibly also inflammatory diseases. Currently, PAD inhibitors are in early preclinical development, so more time will be required to understand the therapeutic potential of these targeted epigenetic therapies.
2.3.2. Histone Lysine ADP-Ribosylation
During the cell cycle, DNA is being damaged and repaired by an elaborate set of mechanisms that have evolved to conserve genomic integrity. Although cancers may result from improper/incomplete DNA repair, targeted inhibition of DNA repair mechanisms may also be exploited to kill cancer cells. One clinical paradigm is to interfere with the tumor’s ability to repair its DNA following treatment with a cytotoxic chemotherapy (i.e., chemosensitization). c
ADP-ribosylation of lysine residues is a relatively rare histone modification, occurring in <1% of all histone proteins, but is observed particularly in instances of single-DNA-strand breaks (Boulikas 1989). Some of the NAD+-dependent Poly(ADP-ribose) polymerases (PARPs), more recently referred to as ADP-ribosyltransferases (ART), ADP-ribosylate histone and nonhistone proteins, as do some of the Sirtuins (Stunkel and Campbell 2011). PARP1 (ARTD1), the best-characterized histone ADP-ribosylating enzyme, is activated by environmental stresses, such as DNA damage, and has been reported to ADP-ribosylate the amino-terminal tails of all core histones, specifically at H2AK13, H2BK30, H3K27, H3K37, and H4K16 (Messner et al. 2010). This PTM is thought to cause chromatin decondensation and recruit DNA repair machinery. Interestingly, acetylation of H4K16 inhibits ADP-ribosylation by PARP1 (Messner et al. 2010). This is the first direct piece of evidence pointing to PTM cross talk between lysine ADP-ribosyl and acetylation marks. Interestingly, a group of macrodomain-containing proteins have been shown as mono-ADP-ribosylhydrolases and define a class of enzymes that renders mono-ADP-ribosylation a reversible modification (Rosenthal et al. 2013).
Responding to DNA damage repair is crucial for maintaining genomic stability and, typically, tumors are deficient in a DNA repair pathway. It is still early for deciphering a functional understanding of the biological role of histone ADP-ribosylation and individual PARP enzymes in chromatin function. Nonetheless, observed histone ADP-ribosylation and PARP1 detected during DNA repair has led to the use of PARP1 inhibitors in the treatment of breast and ovarian cancers. PARP1 inhibitors (olaparib) induce synthetic lethality (i.e., when two or more mutations together cause cell lethality) in cancer cells already mutated for BRCA1 or -2 genes. The BRCA proteins are essential components of the double-strand break (DSB) repair pathway through homologous recombination (discussed in Sec. 1.4.3), and in their absence, cancer cells become reliant on PARP1-dependent DNA repair pathways. Thus, the use of PARP1 inhibitors in BRCA mutant cancers results in tumor cell death, whereas normal cells survive owing to their still-intact BRCA1-dependent DNA damage repair pathway.
The positive progression of several PARP inhibitors through clinical trials (breast, colorectal, and ovarian cancers), although yet unapproved, lends credence to a parallel clinical approach with histone ADP-ribosylation inhibitors.
2.3.3. Other Histone PTMs with Implications in Cancer
A relatively new entry to histone PTMs is histone lysine deamination. This PTM already appears to play a known role in cancer progression and metastasis via genetic studies of its writing enzyme, lysyl oxidase–like 2 enzyme (LOXL2). Histone H3K4me3 can be deaminated by LOXL2 and this enzymatic activity is required to silence the tumor-suppressor gene, CDH1, and induce the epithelial to mesenchymal transition, commonly associated with metastasis, in breast tumor cells (Herranz et al. 2012).
N6-formylation of histone lysine residues is one of the rarest histone PTMs, found at very low abundancy (0.04%–0.1% of all lysines in acid-soluble chromatin proteins) (Jiang et al. 2007). The linker histone H1 is N-formylated most frequently (13 residues), but core histones also contain multiple sites of Lys formylation (19 sites in total for H2A, H2B, H3, and H4) (Jiang et al. 2007; Wisniewski et al. 2008). Lysine formylation increases during oxidative stress (Jiang et al. 2007), which has been associated with various disease states, including cancer. The biological relevance of N6-lysine histone formylation may lie in its potential to disrupt gene expression mediated by other histone lysine marks. For example, N-formylation is observed at H3K79, which could possibly interfere with methylation at that site catalyzed by DOT1L, a target that has been linked to MLL-rearranged leukemias. However, specific cross talk between lysine N-formylation and acetylation or methylation remains unverified and there is no clear disease linkage established with histone formylation.
Histone PTM is not restricted to Arg and Lys residues. Histone Ser or Thr phosphorylation has long been known to be an important PTM involved in the cellular response to DNA damage (Rossetto et al. 2012). For example, H2A.X is phosphorylated at Ser 139 shortly after DNA damage, demarcating the region of chromatin around the DNA lesion.
Recently, O-GlcNAcylation (addition of β-N-acetylglucosamine) of Ser and Thr residues on core histones H2A at Thr101, H2B at Ser36, and H4 at Ser47 was reported, although more residues are thought to be subject to this modification (Sakabe et al. 2010). This PTM increases during heat shock and the mitotic phase of the cell cycle. The known sites of O-GlcNAcylation are also modified by phosphorylation, the latter of which are potentially important for H2A-H2B dimerization. This suggests that O-GlcNAcylation may interfere with residues that are potentially to be phosphorylated, hence, explaining the observed increase in chromatin condensation when the O-GlcNAc transferase (OGT) enzyme is overexpressed or O-GlcNAcylation is triggered following heat shock (Hanover 2010). Thus, the increase in histone O-GlcNAcylation during heat shock, concurrent with DNA condensation, suggests a possible role in DNA damage.
Recently, OGT was found to operate in a ten eleven translocation (TET)2-dependent manner, particularly at transcriptional start sites. This suggests there is a combinatorial effect of DNA 5-hydroxymethylation and O-GlyNAcylation in regulating gene expression (Chen et al. 2013). Collectively, these findings suggest a possible role in transcriptional regulation, DNA damage, and thus cancer, particularly, given that the OGT partner, TET2, is a known tumor suppressor involved in myeloproliferative diseases (see Baylin and Jones 2014).
2.4. Cross Talk among Histone Modifications
As noted above, histones can be decorated with a wide variety of PTMs, presenting numerous combinatorial patterns (Jenuwein and Allis 2001). These can occur on any of the four core histones in the histone octamer (two copies each of histones H2A, H2B, H3, and H4) and may even differ between tails of the same histone within a nucleosome or between nucleosomes (Fig. 3) (Ruthenburg et al. 2007). There is no doubt that PTMs are implicated in cancer. Although single marks have been generalized as activating (e.g., H3K9ac, H3K4me3) or repressive (H3K9me3, H3K27me3) (Ruthenburg et al. 2007; Zhu et al. 2013), it is clear that certain PTMs can affect the ability of other marks to either be put down or read (more extensively reviewed by Ruthenburg et al. 2007; Suganuma and Workman 2008). Extracting which are the important modifications that drive tumorigenesis versus which are passengers is a key endeavor for the field. It is complicated by the fact that multiple interactions occur between PTMs, often referred to as cross talk. A few examples that have a bearing on our understanding of the epigenetic role in cancer are described in this section.
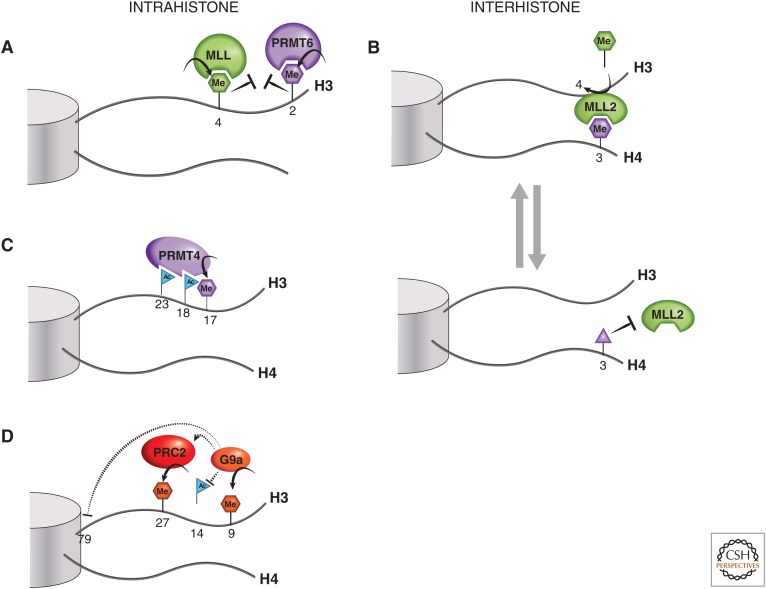
Figure 3.
Histone tail cross talk. (A) H3R2me2a and H3K4 methylation are examples of mutually exclusive intrahistone H3 PTMs. (B) An example of an interhistone cross talk is when H4R3me0 or H4R3me2a marks (purple hexagon) are converted into H4R3me2s (purple triangle), which is thought to block the binding of MLL2 via its PHD4-6 domain, thus preventing the methyltransferase activity of MLL2 at H3K4. (C) PRMT4, a histone arginine methyltransferase, is thought to partially rely on H3 acetylation at K18 and K23 for recruitment to H3 and subsequent dimethylation of the nearby R17 residue. (D) An illustration of the increasingly complex picture of histone H3 tail cross talk, involving H3K9, H3K27, and H3K79 methylation and H3K14 acetylation. See text for a more detailed explanation.
2.4.1. Histone Lysine and Arginine Methylation Cross Talk
There are instances in which histone PTMs can act as exclusion marks, preventing the occurrence of other marks and/or binding of chromatin readers (Migliori et al. 2010). For instance, the presence of H3K4me3 appears to inhibit the deposition of the H3R2me2a mark by PRMT6 (Guccione et al. 2007; Hyllus et al. 2007). Conversely, the H3R2me2a mark prevents methylation of H3K4 by an MLL complex, making the coexistence of these marks mutually exclusive (Fig. 3A). Another example of exclusion marks is seen when PRMT7 promotes symmetric dimethylation of H4R3 (H4R3me2s) and inhibits expression of MLL2-dependent target genes (Dhar et al. 2012). PRMT7 knockdown causes an increase in MLL2-catalyzed H3K4 methylation, suggesting an inverse relationship between these methylation sites. It has been hypothesized that the H4R3me2s mark may block binding by tandem PHD4-6 reader domain(s) within MLL2, which normally recognize H4R3me0 or H4R3me2a, and are required for its methyltransferase activity (Fig. 3B) (Dhar et al. 2012). In this way, the tails of two histones, H3 and H4, could interact contextually to dictate target gene expression.
2.4.2. Cis Histone Cross Talk
Histone lysine acetylation and arginine methylation can also act cooperatively to localize and activate other methyltransferases. A sequential process has been proposed in which estrogen stimulation, CBP/EP300, first acetylates H3K18, then H3K23, and, finally, PRMT4 is attracted to the histone tail in which it dimethylates H3R17 (Daujat et al. 2002). In vitro histone H3K18/K23 acetylation was shown to tether recombinant PRMT4 to the H3 tail to efficiently catalyze arginine dimethylation (Fig. 3C) and activate estrogen-dependent genes.
Histone methylation and phosphorylation can interact in a regulatory fashion. One key example is when chromodomain recognition of a methylated lysine can be disrupted by an adjacent phosphorylation event. H3K9me3 is important for recruiting heterochromatin protein 1 (HP1) to distinct chromosomal regions, required for heterochromatin formation, and thereby regulating gene expression (Stewart et al. 2005). H3 serine 10 phosphorylation adjacent to the trimethylated H3 (H3K9me3S10ph) by Aurora B causes dissociation of HP1 from heterochromatin during M phase and allows for mitotic progression (Fischle et al. 2005; Hirota et al. 2005). This apparently occurs by a disruption of the chromodomain binding of HP1 to H3K9me3 (see Fig. 12 of Allis et al. 2014).
With the advent of modern mass spectrometry and corresponding analytical tools, the entire panoply of histone marks may be interrogated simultaneously from cell extracts. An example is provided by the mass spectrometric analysis of HEK293 cells when G9a/GLP-1 (EHMT1/EHMT2) is knocked down, revealing not only a reduction in the expected methyltransferase products, H3K9me1 and H3K9me2 (H3K9me2 being predominant), but also an increase in H3K79me2, a mark associated with gene expression, added by DOT1L (Plazas-Mayorca et al. 2010). In addition, the greatest reduction in H3K9me2 was seen on peptides containing H3K14 acetylation. The biological significance of these mark changes is unknown, but it infers cross talk between G9a/GLP-1, H3K9me2, H3K79me2, and H3K14ac (Fig. 3D). It is conceivable that H3K9me2 and H3K14ac are mutually exclusive marks representing a form of conditional epigenetic switch. The situation is further complicated by PRC2 recruitment being shown as dependent on G9a and GLP-1 association, affecting, therefore, the degree of H3K27me3 at target genes, which mediates gene silencing (Mozzetta et al. 2014). This shows that there is additional interplay between H3K27me3 and H3K9 methylation, which are both typically repressive marks.
Ultimately, the challenge of relating changes in primary chromatin definitively to cancer, and then determining how cross talk contributes to cancer are key questions in the field. A clear understanding of these marks is limited both by the technologies available with which to interrogate this, as well as our ability to interpret the role that the readers themselves play on the biologic output of chromatin modifications.
2.4.3. DNA Damage Repair: Ubiquitination and Other Cross-Talking PTMs
The repair of DNA damage is an essential cellular function, requiring a rapid dynamic response. It is important to note that repair has to occur in a chromatin context and involves extensive chromatin remodeling. The loss of total repair functionality is cell lethal; however, in cancer, there is often partial loss of function that is accompanied by an increase in genomic instability and mutation rates. PTMs of histone proteins around the DNA damage site and recruited repair proteins is a central means by which the process is regulated. In fact, our current knowledge of mammalian DNA repair in a chromatin context, illustrates the central involvement of histone PTMs as signals in the recruitment or binding inhibition of repair proteins, involving extensive cross talk. Ubiquitination, in particular, represents one of the first key modifications activating the repair pathway. Ubiquitination occurs at histone lysine residues, as do the already discussed acetylation and methylation modifications.
One of the earliest stages of the DNA damage response is when H2AX undergoes monoubiquitination at Lys119/Lys120, mediated by the RNF2-BMI1 complex (components of the PRC1 complex). This is necessary for the recruitment of early sensors of DNA damage. They include the ATM protein, which phosphorylates histone variant H2AX to form γ-H2AX, MRN complex at broken DNA ends, and MDC1 (Fig. 4B) (Ginjala et al. 2011; Pan et al. 2011; reviewed in Panier and Durocher 2013). As RNF2-BMI1 complex-depleted cells have an impaired DNA repair capability and increased sensitivity to ionizing radiation (Pan et al. 2011), it is tempting to speculate that pharmacological inhibitors of H2AX monoubiquitination may act as radio-sensitizing agents in cancer. Monoubiquitination of H2A at Lys119, incidentally, is also involved in transcriptional repression via PRC proteins, such as the RING1A and 1B ubiquitin ligases (de Napoles et al. 2004; Fang et al. 2004; Endoh et al. 2008; Trojer et al. 2011). BMI-1 functions as an oncogene in medulloblastoma and other malignancies, although its deregulation seems to mostly impact its function as a stem-cell factor (see Grossniklaus and Paro 2014). Given BMI-1’s dual role, it is hard to separate its role when considering using it as a drug target, as both functions must be taken into consideration. RING1B has been proposed to control the balance between early myeloid progenitors and mature myeloid populations (Cales et al. 2008); potential dysfunction could lead to myeloid dysfunction and/or malignancies. Indeed, RING1B deficiency was found to result in an accelerated onset of hematopoietic neoplasia in the absence of the tumor suppressor, Ink4a (Cales et al. 2008).
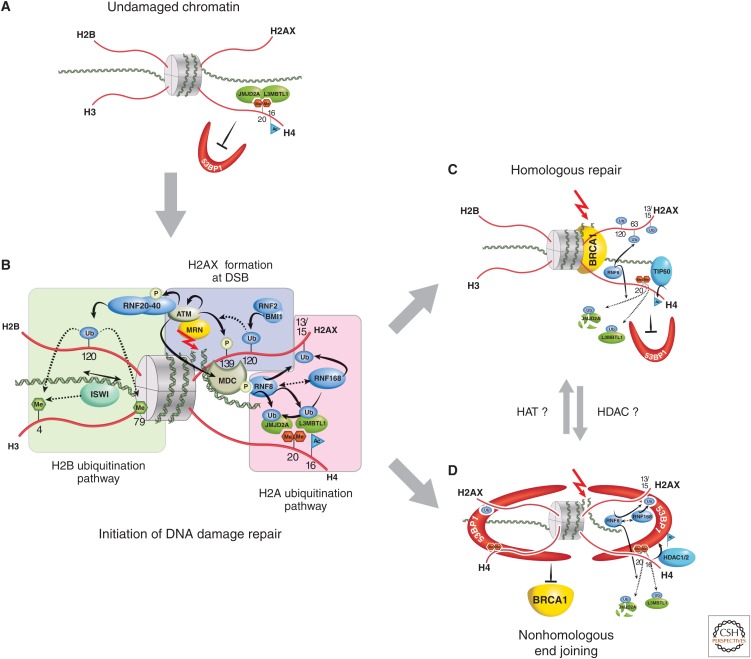
Figure 4.
Role of histone PTMs in DNA damage repair. (A) Before DNA damage, L3MBTL1 and JMJD2A (possibly also JMJD2B) bind, via their Tudor domains, to H4K20me2 (red hexagons), hindering access by DNA repair proteins, such as 53BP1. (B) When double-strand DNA breaks occur (red arrow), the DNA damage–sensing proteins, such as MRN, act to initiate cascades of protein and histone tail PTMs (blue-shaded area). In particular, the phosphorylation of H2A at S139 represents the conversion of H2A into H2A.X. Subsequent chromatin-remodeling events occur as a consequence of H2B ubiquitination (green-shaded area), whereas further ubiquitination events on H2A.X alter local chromatin structure (pink-shaded area). This latter RNF8/RNF168-catalyzed H2A ubiquitination pathway also polyubiquitinates JMJD2A/B for degradation, whereas L3MBTL1 is removed by RNF8-mediated ubiquitination. This then allows DSBs in DNA to be repaired by homologous recombinant repair (HRR), where joining occurs between two similar or identical strands of DNA (C); or by nonhomologous end joining (NHEJ), in which the two DNA ends are joined directly, usually with no sequence homology, although, in some cases, regions of microhomology are used (D). (C) During HRR, TIP60 acetylates H4K16, RNF8 ubiquitinates H2AK63, and selectively allows for BRCA1 binding but not 53BP1. (D) If NHEJ is required, H4K16 is deacetylated (presumably by HDAC1,2) and H2AK15 is ubiquitinated. 53BP1 binds to both H4K20me2 and H2AK15ub, whereas BRCA1 is excluded. To illustrate 53BP1 oligmer formation, only histone H4 and H2AX tails are illustrated in this panel.
Monoubiquitination of H2B at Lys120 is also induced at sites of DNA damage and required for DNA DSB repair (reviewed in Prinder et al. 2013). The ubiquitin ligase, RNF20 (really interesting new gene [RING]-finger), when phosphorylated by ATM, likely mediates H2B ubiquitination. Ubiquitination of histone H2B, in turn, promotes methylation of H3K4 and H3K79, which is critical for remodeling chromatin to allow access to the DNA repair machinery. The remodeling is partly facilitated by the SNF2h subunit of the ISWI complex, likely through interaction with methylated H3K4, as SNF2h depletion reduces repair, at least by the homologous repair (HR) pathway (Fig. 4B). This H2B ubiquitination branch of the DNA repair pathway is a necessary part of either NHEJ or HR mechanisms at DSB (Pinder et al. 2013).
The HR pathway involves polyubiquitination of H2A or H2AX at Lys63 by the RNF8 and RNF168 ubiquitin ligases at sites of DNA DSBs, acting to recruit DNA repair factors, such as BRCA1 (Doil et al. 2009; Campbell et al. 2012). Repair by NHEJ involves histone lysine PTM cross talk to dictate 53BP1 protein recruitment, which interacts with the nucleosome in an oligovalent trans-histone fashion (Fig. 4D). Focal accumulation of 53BP1 at DSBs depends on the specific interaction of its tandem Tudor domain with dimethylated H4K20 (H4K20me2) and ubiquitinylated H2AK15. Rapid induction of H4K16 deacetylation after DSBs facilitates 53BP1-induced DNA damage signaling and DSB repair (Hsiao and Mizzen 2013). In the absence of DNA damage, TIP60 acetyltransferase-mediated H4K16 acetylation inhibits the interaction between 53BP1 and H4K20me2 (Tang et al. 2013). Following the initiation of DSB repair, the acetylation status of histone H4K16, written by TIP60 or removed by HDAC1/2, in fact, may act as a switch between BRCA1 and 53BP1 localization to DSB chromatin by affecting the binding affinity of the 53BP1 Tudor domain for H4K20me2 (Tang et al. 2013). These histone modifications (H4K16ac, H4K20me2) could function to balance 53BP1 DSB chromatin occupancy for NHEJ, versus BRCA1 DSB localization and HRR (Fig. 4).
In summary, DSB repair is a complex process involving many proteins and PTMs. When mutated, many of these factors contribute to cancer: among them are TIP60, BRCA1, and 53BP1, documented to have tumor-suppressor function, thus making these and other epigenetic modifying proteins involved in the DNA repair pathway potential cancer drug targets. Conversely, tumors may be made more susceptible to standard cytotoxic therapies if their DNA damage repair pathways are perturbed by targeted pharmacological agents, such as epigenetic therapies.
3. DRUG-DISCOVERY CHALLENGES FOR HISTONE-MODIFYING TARGETS
Two possible modes by which epigenetic drug therapy may be able to halt or even prevent the oncogenic process are (1) repressing oncogenes and/or activating tumor-suppressor genes that are deregulated by epigenetic processes (Baylin and Ohm 2006; Jones and Baylin 2007) and (2) overcoming resistance to chemotherapy. The “epigenetic” drugs that are currently being developed primarily target histone-modifying enzymes, histone readers, or other chromatin-associated proteins.
3.1. Epigenetics and Drug Resistance
There is evidence to suggest there is an epigenetic basis for resistance to cancer chemotherapy. Because resistance develops to virtually all forms of cancer therapy, overcoming drug resistance remains the biggest obstacle to improving a positive and durable outcome cancer treatment. Examples of epigenetic alterations being attributed to resistance include the aforementioned KDM5A in Section 2.2.2 and overexpression of EZH2, observed during cisplatin chemotherapy; EZH2 overexpression results in increased gene repression and enhanced cell proliferation (Hu et al. 2010; Crea et al. 2011). Clearly, more research is required to understand how epigenetic alterations mechanistically contribute to drug resistance and how to apply epigenetic drug therapy appropriately to patients, depending on an individual’s drug and/or (epi)genomic profile.
3.2. Approved Epigenetic Drugs
The earliest clinical entries to epigenetic drugs were actually not developed with a focus on histone-modifying targets, but were discovered to interact with chromatin modifiers and express their biological effects through these targets after the fact. Dimethylsulfoxide and the later agent HMBA, for example, although explored therapeutically since the 1950s, were only recently shown as inhibitory to a subset of bromodomains (including the BET family).
Among the agents specifically designed against histone-modifying targets that have been clinically approved thus far are the HDAC inhibitors (see Figs. 5 and 6). These drugs (Zolinza, Istodax) broadly inhibit HDACs and show single-agent activity only in a limited set of patients with cutaneous T-cell lymphoma. A large number of clinical trials are now ongoing with these and newer, more subtype-selective HDAC inhibitors (some examples are found in Fig. 8 of Seto and Yoshida 2014) in combination with standard care drugs, in the hopes of expanding the utility of these treatments. Given the early successes with epigenetic therapy, particularly when used in combination (i.e., HDAC and DNA methylation inhibitors, Vidaza and Dacogen) for the treatment of myelodysplastic syndromes and AML, there has been great interest in producing selective agents that target other histone-modifying proteins.
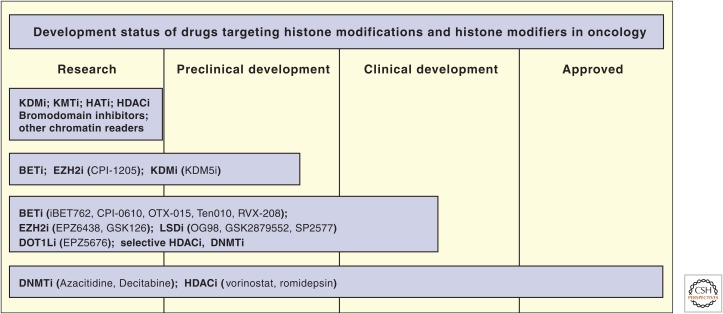
Figure 5.
The development status of epigenetic drugs. KDMi, lysine demethylase inhibitor; KMTi, lysine methyltransferase inhibitor; HATi, histone acetyltransferase inhibitor; HDACi, histone deacetylase inhibitor; BETi, bromodomain and extracarboxy terminal domain inhibitor; DNMTi, DNA methyltransferase inhibitor.
3.3. The Epigenetic Drug-Discovery Process
The enthusiasm for developing novel epigenetic therapeutics has been tempered by the challenges presented in the epigenetic-targeted drug-discovery process (Table 1). Contemporary drug discovery, with some variation, is most commonly seen to consist of three phases: target identification/validation, lead generation, and lead optimization. Optimized lead compounds are then entered into a development pipeline, again consisting of multiple stages. These include the preclinical (or early development) stage, clinical stage (which is typically further subdivided into Phases 1, 2, and 3), and the registration/approval phase. From beginning to end, this discovery/development process can take more than a decade despite myriad efforts on the part of academics, industry, and the regulatory agencies to streamline the process.
Table 1.
Epigenetic drug-discovery challenges
| Issues | |
| Target selection |
|
| Chemistry |
|
| Assay development |
|
| In vivo biology |
|
| Toxicology |
|
| Clinical |
|
Modified, from Campbell and Tummino 2014, with permission of the American Society for Clinical Investigation.
Epigenetic drug discovery presents some unique challenges at each stage of discovery and development. For example, in the target identification/validation stage, we still have yet to fully “map” the epigenetic landscape of all cell types, normal or tumorigenic, on a structural, biochemical, and functional level. This fundamental lack of knowledge introduces a number of pragmatic and scientific issues, which must be considered in the drug-discovery process. For example, challenges include identifying the most disease-relevant targets, understanding the associated biology, developing biologically relevant assays with limited tools (e.g., proteins, antibodies, etc.), and finding suitable small molecule activators or inhibitors with few chemical starting points. Cancer relevance for a given target (e.g., a particular KMT like MLL1) is often determined from genomic analyses of patient tumors; this may reveal mutations, translocations, rearrangements, fusions, amplifications, or copy number changes. Additional experimental data is also used to corroborate their role in cancer, such as testing for synthetic lethality or gene silencing. Experimental data using RNA silencing, although beneficial in these analyses, must be interpreted with care, especially for epigenetic enzymes, as knockdown can disrupt protein complexes and not necessarily be directly associated with the catalytic activity. Use of catalytically inactive “dominant-negative” mutants in concert with selective small molecule activators/inhibitors can be more informative. Clinical relevance of a target in oncology, especially in the absence of tool compounds that modify it, may be inferred by correlating a target’s expression with mortality, disease-free survival, and/or development of resistance to chemotherapy.
With modern targeted chemotherapies, the ultimate goal is to tailor drugs to patients most likely to respond. Success in this endeavor requires a thorough understanding of the patient, their tumor(s), and the underlying biology driving tumor growth and metastasis. Epigenetic targets, as well as being a good focus for drug development, are increasingly proving to be good biomarkers of cancer etiology and chemotherapy response. This may be determined by tracking various histone mark changes, target gene, and/or downstream gene expression patterns (e.g., looking for overexpression, wild-type expression, mutation, translocation, or partnering as a fusion protein, as illustrated in scenarios of Fig. 2). The presence or absence of a mutated or fusion protein, thus, can provide a binary patient-tailoring marker. For instance, EZH2-activating mutations are observed in subsets of lymphoma, providing a biomarker for this type of lymphoma (Yap et al. 2011; Majer et al. 2012; McCabe et al. 2012), whereas MLL-fusion proteins in rearranged leukemias have altered DOT1L-dependent H3K79 methylation profiles, which is again a biomarker for MLL-rearranged cancers (Bernt and Armstrong 2011). However, aside from EZH2, very few epigenetic activating mutations or translocations have been found in human tumors; many epigenetic gene alterations are predicted to result in loss of function (i.e., loss of tumor-suppressor function, which is extensively discussed in Baylin and Jones 2014), or alteration of function, the latter of which does not constitute a straightforward overexpression consequence. This makes epigenetic targeting and patient tailoring quite challenging—that is, how does one “reactivate” the target when deleted, truncated, or conformationally unfavorable?
Once a compelling target has been identified and selected, a series of assays are constructed to screen compounds and ultimately optimize drug-like properties. An initial high-throughput screening assay is typically biochemical and -analytical in nature, investigating disruption or enhancement of protein–protein interactions and catalytic activity. It is critical that these biochemical assays reflect (or at least, generally, predict) the cellular context of the active process one wishes to modulate. For example, chromatin-modifying proteins can exist in heterogeneous multimeric complexes (e.g., PRC1, PRC2, COMPASS) such that partners and enzymatic substrates may be recruited or exchanged to elicit specific biological responses. It is therefore conceivable that different results may be obtained when screening with various protein forms, such as the apoprotein lacking its nonpolypeptide moiety versus the complexed form, or using various substrate forms (i.e., peptide vs. protein/histone, core histones, or nucleosomes, as discussed in Sec. 16.2 of Patel 2014). The choice of enzymatic substrates is further complicated by the fact that many epigenetic enzymes can posttranslationally modify nonhistone targets. Among the many known examples are HDAC6, SIRT1, SIRT2, SET7/9, SETD8, SMYD2, PRMT5, and EHMT2/G9a) (Hubbert et al. 2002; Huang et al. 2006; Shi et al. 2007; Pradhan et al. 2009; Karkhanis et al. 2011; Stunkel and Campbell 2011).
The presence of “tool compounds” or chemical probes to understand the translation from biochemical to cellular assays brings us a step closer to the elucidation of in vivo biological function. This collection of specific epigenetic probes is currently quite limited; however, efforts to expand the collection of such tools continue to progress. A tool compound, such as the S-adenosyl methionine (SAM) analog, Sinefungin, for example, can be useful for SAM-dependent enzyme assays on lysine and arginine histone methyltransferase enzymes, but is poorly cell permeable. Other tools that can be used are the recently described selective, cell-active inhibitors of EZH2, DOT1L, SMYD2, G9a/GLP1, PRMT3, JMJD3, LSD1, BET (BRD2-4), L3MBTL1, L3MBTL3, CBP/EP300, PCAF, and BAZ bromodomains (Arrowsmith et al. 2012; Muller and Brown 2012; James et al. 2013; Gallenkamp et al. 2014). It is anticipated that these and future chemical tools will greatly facilitate assay development and our understanding of target biology. However, a cautionary note is warranted: as powerful as probes may be in revealing the mechanistic and phenotypic consequences of inhibiting specific domains of chromatin modifiers and their readers, such data is clearly dependent on the quality of the probes, the thoughtful design of experiments, and the cellular assays designed to monitor their readout, such that the outcome can be most directly linked to the specific target. As more data is generated and more targets are credentialed, probes must continue to be reviewed for specificity and cellular target engagement (i.e., ability to specifically modulate the downstream target-dependent biology, such as a histone mark change) to ensure that the biology observed is properly interpreted.
The challenge of identifying chemical probe molecules of sufficient potency, selectivity, and cell-permeability suggests that traditional chemical libraries may be deficient in the most relevant chemotypes and screening methodologies were not optimal. As such, new chemical diversity is being explored, particularly, using small molecular weight fragment approaches, and generating substrate or cofactor analogs, such as SAM- and Lys/Arg-peptide mimetics for use with histone methyltransferase targets. A growing collection of crystal structures has been solved to facilitate drug design, some of which are covered in the structural articles of this collection (see Cheng 2014; Marmorstein and Zhou 2014; Patel 2014; Seto and Yoshida 2014). However, still too few structures represent protein complexes or have a small molecule modulator bound, a problem discussed in Section 16.2 of Patel (2014).
Epigenetic-targeted drug discovery is in its infancy, but steadily evolving to overcome the unique challenges of prosecuting its targets. To be successful, ideally, a comprehensive “epigenetics toolbox” must be assembled to test and better understand the dominant biology, relevant histone marks (or nonhistone PTM), structural biology, appropriate in vitro assays, pharmacodynamics of lead drug and target interactions, and patient tailoring biomarkers (perhaps, a major challenge with today’s pharma-favoring genetic features to reflect a real epigenetic basis for sensitivity). The possibility of “antitarget” action must also be determined (i.e., action with another target), which might contribute to clinically unmanageable toxicities.
To accelerate the production of epigenetic drug development tools, a new paradigm has emerged whereby private-public partnerships, such as the Structural Genomics Consortium, have joined forces to generate necessary epigenetic chemical probes, assays, antibodies, and X-ray crystal structures. Other large group efforts are performing sequencing (exome and next-generation) of patient tumors and performing synthetic lethal screens in genomically characterized tumor cell lines. This is helping to identify epigenetic target opportunities and determine patient populations sensitive to therapeutic intervention. It is anticipated that substantial progress will be made in the coming years as a result of knowledge sharing between these pooled collaborative efforts. Such progress is occurring at an increasing rate and, as reflected in Figure 5, early stage drugs targeting histone modifications and modifiers are now progressing through to clinical phases of development. Beyond the initial DNA methyltransferase and HDAC drugs already approved, new chemical entities targeting the BET bromodomains have been entered into a number of clinical studies in a variety of cancers by multiple sponsors (Figs. 5 and 6). Similarly, small molecule inhibitors of the histone methyltransferases, DOT1L and EZH2, have advanced to the clinic, as well as inhibitors of the histone demethylase LSD1. Data from these and subsequent studies will be critical for refining our understanding of the role of histone modifications and histone-modifying enzymes (and their readers) in cancer initiation, cancer progression, and, ultimately, translating to new and improved cancer therapies.
APPENDICES
Appendix A.
HAT mutations in cancer
| Gene | Common name | Tumor types | Cell type | Tissue type | Mutation types | Fusion proteins |
|---|---|---|---|---|---|---|
| MYST family | ||||||
| KAT5 | TIP60 | Colorectal, head and neck, stomach | Somatic | Epithelial | Missense, frameshift nonsense | |
| KAT7 | HBOI | Lung, colorectal, breast, prostate, ovarian, sarcoma | Somatic | Epithelial | Amplification, missense, splice | |
| KAT6A/MYST3 | MOZ | Colorectal, lung, breast, acute myelogenous leukemia | Somatic | Epithelial, leukemia/lymphoma | Nonsense, missense amplification, deletion, translocation | MOZ-CBP, MOZ-EP300, MOZ-TIF2, MOZ-NCOA3, MOZ-ASXL2 |
| KAT6B/MYST4 | MORF | Colorectal, glioblastoma, lung, ovarian, acute myelogenous leukemia | Somatic | Epithelial, leukemia/lymphoma | Nonsense, missense amplification, deletion translocation | MORF-CBP |
| MYST1 | MOF | Lung, colorectal, medulloblastoma | Somatic | Epithelial | Missense, nonsense, deletion | |
| GNAT family | ||||||
| KAT2A | GCN5 | Breast, colorectal, prostate, lung, kidney, sarcoma | Somatic | Epithelial | Deletion, amplification | |
| KAT2B | PCAF | Lung, kidney, sarcoma, colorectal | Somatic | Epithelial | Missense, frameshift, deletion, amplification | |
| Orphan family | ||||||
| EP300 | p300 | Colorectal, breast, pancreatic, AML, ALL, DLBCL (10%), NHL (7%), FL (8.7%) | Somatic | Epithelial, leukemia lymphoma | Translocation, nonsense, frameshift, missense, other | p300-MOZ, MLL-p300 |
| CREBBP | CBP | ALL (18.3%), AML, DLBCL (29%), NHL (21%) | Somatic | Leukemia lymphoma | Translocation, non-sense, frameshift, missense, other | CBP-MOZ, CBP-MORF, MLL-CBP |
| CREBBP | CBP | Hematological (Rubstein- Taybe syndrome) | Germline | Leukemia/lymphoma | Deletion | |
| NCOAI/KATI3A | SRCI | Lung, colorectal | Somatic | Epithelial | Missense, deletion | PAX3-NCOAI |
| NCOA3/KATI3B | CRC-3/ACTR | Colorectal, ovarian, lung | Somatic | Epithelial | Nonsense, missense, amplification, in frame insertion | NCOA3-MOZ |
| KATI3D | CLOCK | Colorectal, glioblastoma, lung | Somatic | Epithelial | Missense, nonsense, amplification, other | |
| KAT4 | TAFI | Lung, colorectal, breast, glioblastoma, ovarian, kidney | Somatic | Epithelial | Missense, nonsense, splice | |
Adapted from Di Cerbo and Schneider 2013, by permission of Oxford University Press.
NHL, B-cell non-Hodgkin lymphoma; AML, acute myelogenous leukemia; ALL, acute lymphocytic leukemia; DLBCL, diffuse large B-cell lymphoma.
Appendix B.
Histone deacetylases in cancer
| Name | HDAC class | Substrate(s) | Links to cancer |
|---|---|---|---|
| HDAC1 | I | H4K16, H3K56 | Overexpressed in ALL, CLL, Hodgkin’s lymphoma, renal cell cancer, breast, gastric, pancreatic, and colorectal (associated with poor prognosis), prostate (high grade, hormone-resistant), lung, and hepatocellular carcinoma (advanced stage) |
| HDAC2 | I | H4K16, H3K56 | Overexpressed in ALL, Hodgkin’s lymphoma, renal cell cancer, lung, colorectal (higher in polyps), cervical, gastric, and prostate (associated with advanced stage and/or poor prognosis); HDAC2-inactivating mutation in MSI+ colon, gastric, and endometrial tumors |
| HDAC3 | I | H3K9,K14 H4K5 H4K12 | Overexpression in CLL, Hodgkin’s lymphoma, renal cell cancer, gastric, colorectal, prostate correlated with poor prognosis (coincident with HDAC1, -2 overexpression); higher expression correlated with poor 5-year event-free survival |
| HDAC8 | I | ERRα, | Overexpressed in ALL, pediatric neuroblastoma (associated with advanced stage disease; poor survival) |
| HDAC4 | IIA | Not defined | High expression in breast cancer (relative to renal, bladder, and colorectal tumors); associated with prednisone-poor response in ALL |
| HDAC5 | IIA | p53 (Sen et al. 2013) | High expression in colorectal cancer (relative to bladder, renal, breast cancer) and medulloblastoma (associated with poor overall survival); underexpressed in AML blasts |
| HDAC7 | IIA | Not defined | High expression in CLL, colorectal (relative to bladder, renal, breast cancer), pancreatic cancer, and childhood ALL (associated with poor survival) |
| HDAC9 | IIA | Not defined | Lower expression in higher grade astrocytoma and glioblastoma (vs. lower grade astrocytoma and normal brain), lung tumors (vs. nontumor epithelial cells, although no correlation with disease-free survival) (Okudela et al. 2013), higher expression observed in medulloblastoma, CLL, Philadephia-negative chronic myeloproliferative neoplasms, and childhood ALL associated with poorer overall survival |
| HDAC6 | IIB | α-tubulin, HSP90, cortactin | Overexpressed in CLL, oral squamous cell cancer (higher in advanced stage), breast cancer (but correlated with better survival; sensitivity to endocrine treatment), DLBCL and AML; expression progressively increased with progression of Philadephia-negative chronic myeloproliferative neoplasms |
| HDAC10 | IIB | Not defined | Reduced expression linked to poor prognosis in lung cancer; overexpressed in CLL; low expression correlated to better survival in advanced stage primary neuroblastoma (Oehme et al. 2013) |
| HDAC11 | IV | Not defined | Overexpressed in mixed lobular and ductal breast carcinoma (vs. normal breast tissue) (Deubzer et al. 2013); elevated in Mantle cell lymphoma (Shah et al. 2012) and Philadephia-negative chronic myeloproliferative neoplasms |
| SIRT1 | III | H4K16, H3K9, p53, p73, PTEN, FOX01, FOX03a, FOX04, NICD, MEF2, HIF-1α, HIF-2α, TAF(I)68, SREBP-1c, β-catenin, RelA/p65, PGC1α, BMAL1, Per2, Ku70, XPA, SMAD7, cortactin, IRS-2, APE1, PCAF, TIP60, p300, SUV39H1, AceCS1, PPARγ, ER-α, ERRα, AR, LXR | Overexpressed in AML, CLL, colon carcinoma, prostate, ovarian, gastric, melanoma, and a subset of HCC (Chen et al. 2012a); underexpressed in bladder, colon, glioma, prostate (?), ovarian tumors (?) |
| SIRT2 | III | H4K16, H3K56, α-tubulin, ATP-citrate lyase (Lin et al. 2013), SETD8 (Serrano et al. 2013), CDK9 (Zhang et al. 2013) | Higher nuclear expression in GBM (vs. astrocytoma or normal by IHC) (Imaoka et al. 2012) and grade 3 ER+ breast cancer (associated with reduced disease-free survival and increased frequency of relapse, although the reverse trend was observed in Grade 2 tumors) (McGlynn et al. 2014); however, decreased expression of total SIRT2 in NSCLC (Li et al. 2013b), breast, GBM, HCC as compared with normal tissue (Park et al. 2012) |
| SIRT3 | III | H4K16, H3K56, H3K9, GDH, IDH2, HMGCS2, SOD2, AceCS2, Ku70, LCAD, SdhA, PDHA1, and PDP (Fan et al. 2014) | Overexpressed in CLL (Van Damme et al. 2012); higher levels associated with node-positive breast cancer |
| SIRT4 | III | GDH (ADP-ribosylation) | Underexpressed in AML blasts |
| SIRT5 | III | Cytochrome c, CPS1 HMGCS2, and SOD1 (desuccinylation) (Rardin et al. 2013) | SIRT5 suggested to desuccinylate and activate SOD1, inhibiting lung tumor growth in vitro (Lin et al. 2013) |
| SIRT6 | III | H3K9, H3K56 | Overexpressed in CLL (Van Damme et al. 2012) |
| SIRT7 | III | H3K18 , p53 | Low levels of H3K18ac predict higher risk of prostate cancer, poor prognosis in pancreatic adenocarcinoma; overexpressed in CLL (Van Damme et al. 2012) and hepatoceullar cancer; higher levels associated with node-positive breast cancer |
Adapted from Witt et al. 2009; Bosch-Presegue and Vaquero 2011; Stunkel and Campbell 2011; Barneda-Zahonero and Parra 2012; Houtkooper et al. 2012; Bernt 2013; Gong and Miller 2013; Paredes et al. 2014.
HDAC, histone deacetylase; ALL, acute lymphoblastic leukemia; CLL, chronic lymphocytic leukemia; AML, acute myelogenous leukemia; HCC, hepatocellular carcinoma; ER-α, estrogen receptor α; GBM, glioblastoma multiforme; GDH, glutamate dehydrogenase; IHC, immunohistochemistry; NSCLC, non-small-cell lung carcinoma.
Appendix C.
Selected histone methyltransferases in cancer
| Name | Synonyms | Histone target(s) | Links to cancer |
|---|---|---|---|
| KMT1A | SUV39H1 | H3K9 | Overexpressed in colorectal cancer (Kang et al. 2007), associated with transcriptional repression |
| KMT1C | G9a, EHMT2 | H3K9 | Overexpressed in lung cancers (Watanabe et al. 2008; Chen et al. 2010, for advanced lung cancer), regulation of centrosome duplication via chromatin structure (Kondo et al. 2008) |
| KMT1F | SETDB2 | H3K9 | Involved in chromosome segregation (Falandry et al. 2010) |
| KMT2A | MLL | H3K4 | Rearranged and translocated in leukemias (Zhang et al. 2012a) |
| KMT2B | MLL2 | H3K4 | Frequently mutated in non-Hodgkin lymphoma (Morin et al. 2011), critical role in lymphomagenesis (Chung et al. 2012), role in multiple myeloma and Kabuki syndrome |
| KMT2C | MLL3 | H3K4 | Germline mutation in colorectal cancer and AML (Li et al. 2013a); mutations in glioblastoma, melanoma, pancreatic carcinoma, and colorectal cancer |
| KMT2D | MLL4 | H3K4 | Regulates cell-cycle progression and viability in colon cancer (Ansari et al. 2012) |
| KMT2E | MLL5 | H3K4 | Implicated as a tumor suppressor, MLL5 expression positive prognostic in AML (Damm et al. 2011) |
| KMT3A | SETD2 | H3K36 | Mutations in high-grade gliomas (Fontebasso et al. 2013), tumor suppressor in breast cancer and renal cell carcinoma (Hakimi et al. 2012) |
| KMT3B | NSD1 | H3K36 | Mutated in AML, myeloma, and lung cancers, NUP98-NSD1 translocation linked to tumorogenesis in AML (Wang et al. 2007) |
| KMT3C | SMYD2 | H3K36 | Overexpression in esophageal squamous cell carcinoma correlated with poor survival (Komatsu et al. 2009) |
| KMT3E | SMYD3 | H3K4, H4K5 | Overexpressed in liver, breast, and rectal carcinomas (Van Aller et al. 2012) |
| KMT3F | WHSC1L1/NSD3 | H3K36 | Overexpressed in CML, bladder, lung, and liver cancers (Kang et al. 2013); amplified in human breast cancer cell lines (Angrand et al. 2001) |
| KMT3G | WHSC1/NSD2 | H3K36 | Implicated in constitutive NF-κB signaling for cancer cell proliferation, survival, and tumor growth (Yang et al. 2012), overexpressed in myeloma because of t(4;14) chromosomal translocation, modulates cMYC in myeloma (Min et al. 2012) |
| KMT4 | DOT1L | H3K79 | Implicated in MLL-rearranged leukemias (Krivtsov et al. 2008; Bernt and Armstrong 2011) |
| KMT5A | SETD8/PR-SET7 | H4K20 | Overexpressed in bladder cancer, NSCLC, small cell lung cancer, pancreatic cancer, hepatocellular carcinoma, and chronic myelogenous leukemia (Takawa et al. 2012) |
| KMT6 | EZH2 | H3K27 | Catalytic component of PRC2 complex (Kuzmichev et al. 2002), overexpressed, is a marker for advanced and metastatic breast and prostate cancer (Chase and Cross 2011), essential for glioblastoma cancer stem-cell maintenance (Suva et al. 2009), somatic mutations in follicular and diffuse large B-cell lymphomas (Morin et al. 2010) |
| KMT7 | SET7/SET9/SETD7 | H3K4 | Regulation of the estrogen receptor (Subramanian et al. 2008) |
| PRDM14 | Unknown | Amplified and overexpressed in breast cancer (Nishikawa et al. 2007; Moelans et al. 2010), overexpressed in lymphoid neoplasms, and implicated in initiation of lymphoblastic leukemia (Dettman et al. 2011) | |
| PRMT4 | CARM1 | H3R17, H3R26 | Deregulated in melanoma (Limm et al. 2013), methylation CBP/P300 required for estrogen-induced targeting to chromatin (Ceschin et al. 2011) |
| PRMT5 | H4R3, H3R8 | Essential component of HIF-1 signaling (Lim et al. 2012); silences the tumor suppressor ST7 and is overexpressed in GBM (Yan et al. 2014), non-Hodgkin’s lymphoma (Chung et al. 2013), and melanoma (Nicholas et al. 2013) | |
| PRMT6 | H3R2, H3R42 | Overexpressed in prostate carcinoma (Vieira et al. 2014); PRMT6 silencing reduces PELP1-mediated ER activation, proliferation, and colony formation in breast tumor cells (Mann et al. 2014) |
AML, acute myelogenous leukemia; CML, chronic myelogenous leukemia; MLL, mixed lineage leukemia;
NSCLC, non-small-cell lung carcinoma; PRC2, Polycomb repressive complex 2; HIF-1, hypoxia-inducible factor 1; GBM, glioblastoma multiforme; ER, estrogen receptor.
Appendix D.
Histone demethylases in cancer
| Name | Synonyms | Targets | Links to cancer |
|---|---|---|---|
| KDM1A | LSD1, AOF2 | H3K4me2/me1, H3K9me2/me1 | Overexpressed in prostate carcinoma, bladder cancer (Kauffman et al. 2011), ER-negative breast cancer (Lim et al. 2010), neuroblastoma (Schulte et al. 2009); inhibition in animal models of engrafted AML (Schenk et al. 2012) |
| KDM1B | LSD2, AOF1 | H3K4me2/me1 | |
| KDM2A | FBXL11A, JHDM1A | H3K36me2/me1 | Overexpressed in NSCLC; KDM2A knockdown inhibits NSCLC tumor growth in mouse xenografts (Wagner et al. 2013) |
| KDM2B | FBXL10B, JHDM1B | H3K36me2/me1, H3K4me3 | Required for initiation and maintenance of AML (He et al. 2011) |
| KDM3A | JMJD1A, JHDM2A | H3K9me2/me1 | High expression correlated with bad prognosis in colorectal cancer (Uemura et al. 2010), overexpressed in renal cell carcinoma (Guo et al. 2011) |
| KDM3B | JMJD1B, JHDM2B | H3K9me2/me1 | |
| KDM4A | JMJD2A, JHDM3A | H3K9me3/me2, H3K36me3/me2 | Required for proliferation of breast cancer cells (Lohse et al. 2011), attenuated expression in bladder cancer (Kauffman et al. 2011), required for latency and replication of viruses that cause cancer (Chang et al. 2011) |
| KDM4B | JMJD2B | H3K9me3/me2, H3K36me3/me2 | Overexpressed in gastric cancer (Li et al. 2011), required for proliferation and formation of metastasis in breast cancer cells (Kawazu et al. 2011) |
| KDM4C | JMJD2C, GASC1 | H3K9me3/me2, H3K36me3/me2 | Overexpressed in breast cancer (Liu et al. 2009), esophageal cancer (Yang et al. 2000), MALT lymphoma (Vinatzer et al. 2008), AML (Hélias et al. 2008), and lung sarcomatoid carcinoma (Italiano et al. 2006) |
| KDM4D | JMJD2D | H3K9me3/me2/me1, H3K36me3/me2 | Required for cell proliferation and survival in colon carcinoma cells (Kim et al. 2012a; Kim et al. 2012b) |
| KDM4E | JMJD2E | H3K9me3/me2 | |
| KDM5A | Jarid1A, RBP2 | H3K4me3/me2 | Involved in drug resistance (Sharma et al. 2010) |
| KDM5B | Jarid1B, PLU1 | H3K4me3/me2 | Tumor-suppressive function in metastatic melanoma cells (Roesch et al. 2006, 2008), proproliferative in breast cancer (Mitra et al. 2011), and overexpressed in prostate cancer |
| KDM5C | Jarid1C, SMCX | H3K4me3/me2 | Inactivating mutations found in clear cell renal carcinoma (Dalgliesh et al. 2010) |
| KDM5D | Jarid1D, SMCY | H3K4me3/me2 | |
| KDM6A | UTX, MGC141941 | H3K27me3/me2 | Tumor-suppressive function (Tsai et al. 2010) |
| KDM6B | JMJD3, KIAA0346, PHF8, KIAA1111, ZNF422 | H3K27me3/me2, H3K9me2/me1, H4K20me1 | Overexpressed in Hodgkin’s lymphoma (Anderton et al. 2011) |
| KDM7 | KIAA1718, JHDM1D (KDM7A) | H3K9me2/me1, H3K27me2/me1 | Increased expression of KDM7A suppresses tumor growth in HeLa and B16 xenograft models (Osawa et al. 2011) |
| KDM8 | JMJD5, FLJ13798 | H3K36me2 | Overexpressed in breast tumors; may play a critical role in regulation of cell cycle by activating the cyclin A1 locus via demethylation of H3K36me2 (Hsia et al. 2010) |
Adapted, with permission, from Hoffmann et al. 2012, © Elsevier.
From ER, estrogen receptor; AML, acute myloid leukemia; NSCLC, non-small-cell lung carcinoma; KDM2A, lysine demethylase 2A; MALT, mucosa-associated lymphoid tissue.
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, Danny Reinberg, and Monika Lachner
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this subject collection.
- *.Allis CD, Jenuwein T, Reinberg D. 2014. Overview and concepts. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018739. [DOI] [Google Scholar]
- Anderton JA, Bose S, Vockerodt M, Vrzalikova K, Wei W, Kuo M, Helin K, Christensen J, Rowe M, Murray PG, et al. 2011. The H3K27me3 demethylase, KDM6B, is induced by Epstein-Barr virus and over-expressed in Hodgkin’s Lymphoma. Oncogene 30: 2037–2043. [DOI] [PubMed] [Google Scholar]
- Angrand PO, Apiou F, Stewart AF, Dutrillaux B, Losson R, Chambon P. 2001. NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics 74: 79–88. [DOI] [PubMed] [Google Scholar]
- Ansari KI, Kasiri S, Mishra BP, Mandal SS. 2012. Mixed lineage leukaemia-4 regulates cell-cycle progression and cell viability and its depletion suppresses growth of xenografted tumour in vivo. Br J Cancer 107: 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. 2012. Epigenetic protein families: A new frontier for drug discovery. Nat Rev Drug Discov 11: 384–400. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res 21: 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K, et al. 2012. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 487: 114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barneda-Zahonero B, Parra M. 2012. Histone deacetylases and cancer. Mol Oncol 6: 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Baylin SB, Jones PA. 2014. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Ohm JE. 2006. Epigenetic gene silencing in cancer—A mechanism for early oncogenic pathway addiction? Nat Rev Cancer 6: 107–116. [DOI] [PubMed] [Google Scholar]
- Bell EL, Emerling BM, Ricoult SJ, Guarente L. 2011. SirT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene 30: 2986–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt M. 2013. HDAC expression patterns in pediatric ALL. Leuk Res 37: 1191–1192. [DOI] [PubMed] [Google Scholar]
- Bernt KM, Armstrong SA. 2011. A role for DOT1L in MLL-rearranged leukemias. Epigenomics 3: 667–670. [DOI] [PubMed] [Google Scholar]
- Black JC, Van Rechem C, Whetstine JR. 2012. Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol Cell 48: 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch-Presegue L, Vaquero A. 2011. The dual role of sirtuins in cancer. Genes Cancer 2: 648–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulikas T. 1989. DNA strand breaks alter histone ADP-ribosylation. Proc Natl Acad Sci 86: 3499–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. 2003. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 22: 5323–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Busslinger M, Tarakhovsky A. 2014. Epigenetic control of immunity. Cold Spring Harb Perspect Biol 6: a019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cales C, Roman-Trufero M, Pavon L, Serrano I, Melgar T, Endoh M, Perez C, Koseki H, Vidal M. 2008. Inactivation of the polycomb group protein Ring1B unveils an antiproliferative role in hematopoietic cell expansion and cooperation with tumorigenesis associated with Ink4a deletion. Mol Cell Biol 28: 1018–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RM, Tummino PJ. 2014. Cancer epigenetics drug discovery and development: The challenge of hitting the mark. J Clin Invest 124: 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell SJ, Edwards RA, Leung CC, Neculai D, Hodge CD, Dhe-Paganon S, Glover JN. 2012. Molecular insights into the function of RING finger (RNF)-containing proteins hRNF8 and hRNF168 in Ubc13/Mms2-dependent ubiquitylation. J Biol Chem 287: 23900–23910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceschin DG, Walia M, Wenk SS, Duboe C, Gaudon C, Xiao Y, Fauquier L, Sankar M, Vandel L, Gronemeyer H. 2011. Methylation specifies distinct estrogen-induced binding site repertoires of CBP to chromatin. Genes Dev 25: 1132–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang X, Han J, Pang L, Zhao Y, Yang Y, Shen Z. 2009. Increased PADI4 expression in blood and tissues of patients with malignant tumors. BMC Cancer 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PC, Fitzgerald LD, Hsia DA, Izumiya Y, Wu CY, Hsieh WP, Lin SF, Campbell M, Lam KS, Luciw PA, et al. 2011. Histone demethylase JMJD2A regulates Kaposi’s sarcoma-associated herpesvirus replication and is targeted by a viral transcriptional factor. J Virol 85: 3283–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase A, Cross NC. 2011. Aberrations of EZH2 in cancer. Clin Cancer Res 17: 2613–2618. [DOI] [PubMed] [Google Scholar]
- Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng J, Gu W, Zhao Y. 2007. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics 6: 812–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MW, Hua KT, Kao HJ, Chi CC, Wei LH, Johansson G, Shiah SG, Chen PS, Jeng YM, Cheng TY, et al. 2010. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res 70: 7830–7840. [DOI] [PubMed] [Google Scholar]
- Chen HC, Jeng YM, Yuan RH, Hsu HC, Chen YL. 2012a. SIRT1 promotes tumorigenesis and resistance to chemotherapy in hepatocellular carcinoma and its expression predicts poor prognosis. Ann Surg Oncol 19: 2011–2019. [DOI] [PubMed] [Google Scholar]
- Chen YF, Luo RZ, Li Y, Cui BK, Song M, Yang AK, Chen WK. 2012b. High expression levels of COX-2 and P300 are associated with unfavorable survival in laryngeal squamous cell carcinoma. Eur Arch Otorhinolaryngol 270: 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Chen Y, Bian C, Fujiki R, Yu X. 2013. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 493: 561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Cheng X. 2014. Structural and functional coordination of DNA and histone methylation. Cold Spring Harb Perspect Biol 6: a018747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou MA, Castelo-Branco G, Halley-Stott RP, Oliveira CS, Loos R, Radzisheuskaya A, Mowen KA, Bertone P, Silva JC, Zernicka-Goetz M, et al. 2014. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature 507: 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YR, Schatoff E, Abdel-Wahab O. 2012. Epigenetic alterations in hematopoietic malignancies. Int J Hematol 96: 413–427. [DOI] [PubMed] [Google Scholar]
- Chung J, Karkhanis V, Tae S, Yan F, Smith P, Ayers LW, Agostinelli C, Pileri S, Denis GV, Baiocchi RA, et al. 2013. Protein arginine methyltransferase 5 (PRMT5) inhibition induces lymphoma cell death through reactivation of the retinoblastoma tumor suppressor pathway and polycomb repressor complex 2 (PRC2) silencing. J Biol Chem 288: 35534–35547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crea F, Nobili S, Paolicchi E, Perrone G, Napoli C, Landini I, Danesi R, Mini E. 2011. Epigenetics and chemoresistance in colorectal cancer: An opportunity for treatment tailoring and novel therapeutic strategies. Drug Resist Updat 14: 280–296. [DOI] [PubMed] [Google Scholar]
- Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD, Tempst P, Bannister AJ, et al. 2004. Histone deimination antagonizes arginine methylation. Cell 118: 545–553. [DOI] [PubMed] [Google Scholar]
- Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, et al. 2010. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463: 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm F, Oberacker T, Thol F, Surdziel E, Wagner K, Chaturvedi A, Morgan M, Bomm K, Gohring G, Lubbert M, et al. 2011. Prognostic importance of histone methyltransferase MLL5 expression in acute myeloid leukemia. J Clin Oncol 29: 682–689. [DOI] [PubMed] [Google Scholar]
- Daujat S, Bauer UM, Shah V, Turner B, Berger S, Kouzarides T. 2002. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr Biol 12: 2090–2097. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. 2011. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478: 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Aversana C, Lepore I, Altucci L. 2012. HDAC modulation and cell death in the clinic. Exp Cell Res 318: 1229–1244. [DOI] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146: 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. 2004. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7: 663–676. [DOI] [PubMed] [Google Scholar]
- Denis H, Deplus R, Putmans P, Yamada M, Metivier R, Fuks F. 2009. Functional connection between deimination and deacetylation of histones. Mol Cell Biol 29: 4982–4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettman EJ, Simko SJ, Ayanga B, Carofino BL, Margolin JF, Morse HC 3rd, Justice MJ. 2011. Prdm14 initiates lymphoblastic leukemia after expanding a population of cells resembling common lymphoid progenitors. Oncogene 30: 2859–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deubzer HE, Schier MC, Oehme I, Lodrini M, Haendler B, Sommer A, Witt O. 2013. HDAC11 is a novel drug target in carcinomas. Int J Cancer 132: 2200–2208. [DOI] [PubMed] [Google Scholar]
- Dhar SS, Lee SH, Kan PY, Voigt P, Ma L, Shi X, Reinberg D, Lee MG. 2012. Trans-tail regulation of MLL4-catalyzed H3K4 methylation by H4R3 symmetric dimethylation is mediated by a tandem PHD of MLL4. Genes Dev 26: 2749–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cerbo V, Schneider R. 2013. Cancers with wrong HATs: The impact of acetylation. Brief Funct Genomics 12: 231–243. [DOI] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136: 435–446. [DOI] [PubMed] [Google Scholar]
- Dorrance AM, Liu S, Yuan W, Becknell B, Arnoczky KJ, Guimond M, Strout MP, Feng L, Nakamura T, Yu L, et al. 2006. Mll partial tandem duplication induces aberrant Hox expression in vivo via specific epigenetic alterations. J Clin Invest 116: 2707–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, Mantovani F, Tocco F, Elkon R, Comel A, Holstege H, Kerkhoven R, Jonkers J, Voorhoeve PM, Agami R, et al. 2010. BRD7 is a candidate tumour suppressor gene required for p53 function. Nat Cell Biol 12: 380–389. [DOI] [PubMed] [Google Scholar]
- Endoh M, Endo TA, Endoh T, Fujimura Y, Ohara O, Toyoda T, Otte AP, Okano M, Brockdorff N, Vidal M, et al. 2008. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development 135: 1513–1524. [DOI] [PubMed] [Google Scholar]
- Falandry C, Fourel G, Galy V, Ristriani T, Horard B, Bensimon E, Salles G, Gilson E, Magdinier F. 2010. CLLD8/KMT1F is a lysine methyltransferase that is important for chromosome segregation. J Biol Chem 285: 20234–20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Shan C, Kang HB, Elf S, Xie J, Tucker M, Gu TL, Aguiar M, Lonning S, Chen H, et al. 2014. Tyr phosphorylation of PDP1 toggles recruitment between ACAT1 and SIRT3 to regulate the pyruvate dehydrogenase complex. Mol Cell 53: 534–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Chen T, Chadwick B, Li E, Zhang Y. 2004. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J Biol Chem 279: 52812–52815. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. 2010. Selective inhibition of BET bromodomains. Nature 468: 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, et al. 2011. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell 19: 416–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438: 1116–1122. [DOI] [PubMed] [Google Scholar]
- Fiskus W, Sharma S, Qi J, Valenta JA, Schaub LJ, Shah B, Peth K, Portier BP, Rodriguez M, Devaraj SG, et al. 2014. Highly active combination of BRD4 antagonist and histone deacetylase inhibitor against human acute myelogenous leukemia cells. Mol Cancer Ther 13: 1142–1154. [DOI] [PubMed] [Google Scholar]
- Fontebasso AM, Schwartzentruber J, Khuong-Quang DA, Liu XY, Sturm D, Korshunov A, Jones DT, Witt H, Kool M, Albrecht S, et al. 2013. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol 125: 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, et al. 2005. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 37: 391–400. [DOI] [PubMed] [Google Scholar]
- French CA, Miyoshi I, Aster JC, Kubonishi I, Kroll TG, Dal Cin P, Vargas SO, Perez-Atayde AR, Fletcher JA. 2001. BRD4 Bromodomain gene rearrangement in aggressive carcinoma with translocation t(15;19). Am J Pathol 159: 1987–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CA, Ramirez CL, Kolmakova J, Hickman TT, Cameron MJ, Thyne ME, Kutok JL, Toretsky JA, Tadavarthy AK, Kees UR, et al. 2008. BRD-NUT oncoproteins: A family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene 27: 2237–2242. [DOI] [PubMed] [Google Scholar]
- Gallenkamp D, Gelato KA, Haendler B, Weinmann H. 2014. Bromodomains and their pharmacological inhibitors. ChemMedChem 9: 438–464. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Lander ES. 2013. Lessons from the cancer genome. Cell 153: 17–37. [DOI] [PubMed] [Google Scholar]
- Gayther SA, Batley SJ, Linger L, Bannister A, Thorpe K, Chin SF, Daigo Y, Russell P, Wilson A, Sowter HM, et al. 2000. Mutations truncating the EP300 acetylase in human cancers. Nat Genet 24: 300–303. [DOI] [PubMed] [Google Scholar]
- Ginjala V, Nacerddine K, Kulkarni A, Oza J, Hill SJ, Yao M, Citterio E, van Lohuizen M, Ganesan S. 2011. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol Cell Biol 31: 1972–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F, Miller KM. 2013. Mammalian DNA repair: HATs and HDACs make their mark through histone acetylation. Mutat Res 750: 23–30. [DOI] [PubMed] [Google Scholar]
- *.Grossniklaus U, Paro R. 2014. Transcriptional silencing by Polycomb-group proteins. Cold Spring Harb Perspect Biol 6: a019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Luscher B, Amati B. 2007. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449: 933–937. [DOI] [PubMed] [Google Scholar]
- Guo X, Shi M, Sun L, Wang Y, Gui Y, Cai Z, Duan X. 2011. The expression of histone demethylase JMJD1A in renal cell carcinoma. Neoplasma 58: 153–157. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. 2010. Mammalian sirtuins: Biological insights and disease relevance. Annu Rev Pathol 5: 253–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Deng CX, Finley LW, Kim HS, Gius D. 2012. SIRT3 is a mitochondrial tumor suppressor: A scientific tale that connects aberrant cellular ROS, the Warburg effect, and carcinogenesis. Cancer Res 72: 2468–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi AA, Chen YB, Wren J, Gonen M, Abdel-Wahab O, Heguy A, Liu H, Takeda S, Tickoo SK, Reuter VE, et al. 2012. Clinical and pathologic impact of select chromatin-modulating tumor suppressors in clear cell renal cell carcinoma. Eur Urol 63: 848–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: The next generation. Cell 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Hanover JA. 2010. Epigenetics gets sweeter: O-GlcNAc joins the ‘histone code’. Chem Biol 17: 1272–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, Ciceri F, Blaser JG, Greystoke BF, Jordan AM, et al. 2012. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 21: 473–487. [DOI] [PubMed] [Google Scholar]
- Harte MT, O’Brien GJ, Ryan NM, Gorski JJ, Savage KI, Crawford NT, Mullan PB, Harkin DP. 2010. BRD7, a subunit of SWI/SNF complexes, binds directly to BRCA1 and regulates BRCA1-dependent transcription. Cancer Res 70: 2538–2547. [DOI] [PubMed] [Google Scholar]
- He J, Nguyen AT, Zhang Y. 2011. KDM2b/JHDM1b, an H3K36me2-specific demethylase, is required for initiation and maintenance of acute myeloid leukemia. Blood 117: 3869–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hélias C, Struski S, Gervais C, Leymarie V, Mauvieux L, Herbrecht R, Lessard M. 2008. Polycythemia vera transforming to acute myeloid leukemia and complex abnormalities including 9p homogeneously staining region with amplification of MLLT3, JMJD2C, JAK2, and SMARCA2. Cancer Genet Cytogenet 180: 51–55. [DOI] [PubMed] [Google Scholar]
- Herranz N, Dave N, Millanes-Romero A, Morey L, Diaz VM, Lorenz-Fonfria V, Gutierrez-Gallego R, Jeronimo C, Di Croce L, Garcia de Herreros A, et al. 2012. Lysyl oxidase-like 2 deaminates lysine 4 in histone H3. Mol Cell 46: 369–376. [DOI] [PubMed] [Google Scholar]
- Hirota T, Lipp JJ, Toh BH, Peters JM. 2005. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438: 1176–1180. [DOI] [PubMed] [Google Scholar]
- Hoffmann I, Roatsch M, Schmitt ML, Carlino L, Pippel M, Sippl W, Jung M. 2012. The role of histone demethylases in cancer therapy. Mol Oncol 6: 683–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Li Y, Luo RZ, Fu JH, He JH, Zhang LJ, Yang HX. 2012. High expression of the transcriptional co-activator p300 predicts poor survival in resectable non-small cell lung cancers. Eur J Surg Oncol 38: 523–530. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Pirinen E, Auwerx J. 2012. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol 13: 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia DA, Tepper CG, Pochampalli MR, Hsia EY, Izumiya C, Huerta SB, Wright ME, Chen HW, Kung HJ, Izumiya Y. 2010. KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. Proc Natl Acad Sci 107: 9671–9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao KY, Mizzen CA. 2013. Histone H4 deacetylation facilitates 53BP1 DNA damage signaling and double-strand break repair. J Mol Cell Biol 5: 157–165. [DOI] [PubMed] [Google Scholar]
- Hu S, Yu L, Li Z, Shen Y, Wang J, Cai J, Xiao L, Wang Z. 2010. Overexpression of EZH2 contributes to acquired cisplatin resistance in ovarian cancer cells in vitro and in vivo. Cancer Biol Ther 10: 788–795. [DOI] [PubMed] [Google Scholar]
- Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL. 2006. Repression of p53 activity by Smyd2-mediated methylation. Nature 444: 629–632. [DOI] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. 2002. HDAC6 is a microtubule-associated deacetylase. Nature 417: 455–458. [DOI] [PubMed] [Google Scholar]
- Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, Hsieh J, Bauer UM. 2007. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev 21: 3369–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaoka N, Hiratsuka M, Osaki M, Kamitani H, Kambe A, Fukuoka J, Kurimoto M, Nagai S, Okada F, Watanabe T, et al. 2012. Prognostic significance of sirtuin 2 protein nuclear localization in glioma: An immunohistochemical study. Oncol Rep 28: 923–930. [DOI] [PubMed] [Google Scholar]
- Italiano A, Attias R, Aurias A, Pérot G, Burel-Vandenbos F, Otto J, Venissac N, Pedeutour F. 2006. Molecular cytogenetic characterization of a metastatic lung sarcomatoid carcinoma: 9p23 neocentromere and 9p23∼p24 amplification including JAK2 and JMJD2C. Cancer Genet Cytogenet 167: 122–130. [DOI] [PubMed] [Google Scholar]
- Iyer NG, Ozdag H, Caldas C. 2004. p300/CBP and cancer. Oncogene 23: 4225–4231. [DOI] [PubMed] [Google Scholar]
- James LI, Korboukh VK, Krichevsky L, Baughman BM, Herold JM, Norris JL, Jin J, Kireev DB, Janzen WP, Arrowsmith CH, et al. 2013. Small-molecule ligands of methyl-lysine binding proteins: Optimization of selectivity for L3MBTL3. J Med Chem 56: 7358–7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings LE, Measures AR, Wilson BG, Conway SJ. 2014. Phenotypic screening and fragment based approaches to the discovery of small molecule bromodomain ligands. Future Med Chem 6: 179–204. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293: 1074–1080. [DOI] [PubMed] [Google Scholar]
- Jiang T, Zhou X, Taghizadeh K, Dong M, Dedon PC. 2007. N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proc Natl Acad Sci 104: 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. 2007. The epigenomics of cancer. Cell 128: 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Noh JH, Kim JK, Eun JW, Bae HJ, Xie HJ, Chang YG, Kim MG, Park H, Lee JY, et al. 2012. HDAC2 overexpression confers oncogenic potential to human lung cancer cells by deregulating expression of apoptosis and cell cycle proteins. J Cell Biochem 113: 2167–2177. [DOI] [PubMed] [Google Scholar]
- Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. 2013. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 45: 592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser MD, Aslanian A, Dong MQ, Yates JR 3rd, Emerson BM. 2008. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J Biol Chem 283: 32254–32263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MY, Lee BB, Kim YH, Chang DK, Kyu Park S, Chun HK, Song SY, Park J, Kim DH. 2007. Association of the SUV39H1 histone methyltransferase with the DNA methyltransferase 1 at mRNA expression level in primary colorectal cancer. Int J Cancer 121: 2192–2197. [DOI] [PubMed] [Google Scholar]
- Kang D, Cho HS, Toyokawa G, Kogure M, Yamane Y, Iwai Y, Hayami S, Tsunoda T, Field HI, Matsuda K, et al. 2013. The histone methyltransferase Wolf-Hirschhorn syndrome candidate 1-like 1 (WHSC1L1) is involved in human carcinogenesis. Genes Chromosomes Cancer 52: 126–139. [DOI] [PubMed] [Google Scholar]
- Karkhanis V, Hu YJ, Baiocchi RA, Imbalzano AN, Sif S. 2011. Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem Sci 36: 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman EC, Robinson BD, Downes MJ, Powell LG, Lee MM, Scherr DS, Gudas LJ, Mongan NP. 2011. Role of androgen receptor and associated lysine-demethylase coregulators, LSD1 and JMJD2A, in localized and advanced human bladder cancer. Mol Carcinog 50: 931–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazu M, Saso K, Tong KI, McQuire T, Goto K, Son DO, Wakeham A, Miyagishi M, Mak TW, Okada H. 2011. Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS One 6: e17830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SN, Jankowska AM, Mahfouz R, Dunbar AJ, Sugimoto Y, Hosono N, Hu Z, Cheriyath V, Vatolin S, Przychodzen B, et al. 2013. Multiple mechanisms deregulate EZH2 and histone H3 lysine 27 epigenetic changes in myeloid malignancies. Leukemia 27: 1301–1309. [DOI] [PubMed] [Google Scholar]
- Kim TD, Oh S, Shin S, Janknecht R. 2012a. Regulation of tumor suppressor p53 and HCT116 cell physiology by histone demethylase JMJD2D/KDM4D. PLoS One 7: e34618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TD, Shin S, Berry WL, Oh S, Janknecht R. 2012b. The JMJD2A demethylase regulates apoptosis and proliferation in colon cancer cells. J Cell Biochem 113: 1368–1376. [DOI] [PubMed] [Google Scholar]
- Komatsu S, Imoto I, Tsuda H, Kozaki KI, Muramatsu T, Shimada Y, Aiko S, Yoshizumi Y, Ichikawa D, Otsuji E, et al. 2009. Overexpression of SMYD2 relates to tumor cell proliferation and malignant outcome of esophageal squamous cell carcinoma. Carcinogenesis 30: 1139–1146. [DOI] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Ahmed S, Boumber Y, Sekido Y, Haddad BR, Issa JP. 2008. Downregulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells. PLoS One 3: e2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell 128: 693–705. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. 2007. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer 7: 823–833. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. 2008. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell 14: 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16: 2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautz TB, Naiditch JA, Clark S, Chu F, Madonna MB. 2012. Efficacy of class I and II vs class III histone deacetylase inhibitors in neuroblastoma. J Pediatr Surg 47: 1267–1271. [DOI] [PubMed] [Google Scholar]
- Li P, Yao H, Zhang Z, Li M, Luo Y, Thompson PR, Gilmour DS, Wang Y. 2008. Regulation of p53 target gene expression by peptidylarginine deiminase 4. Mol Cell Biol 28: 4745–4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhao L, Zang W, Liu Z, Chen L, Liu T, Xu D, Jia J. 2011. Histone demethylase JMJD2B is required for tumor cell proliferation and survival and is overexpressed in gastric cancer. Biochem Biophys Res Commun 416: 372–378. [DOI] [PubMed] [Google Scholar]
- Li WD, Li QR, Xu SN, Wei FJ, Ye ZJ, Cheng JK, Chen JP. 2013a. Exome sequencing identifies an MLL3 gene germ line mutation in a pedigree of colorectal cancer and acute myeloid leukemia. Blood 121: 1478–1479. [DOI] [PubMed] [Google Scholar]
- Li Z, Xie QR, Chen Z, Lu S, Xia W. 2013b. Regulation of SIRT2 levels for human non-small cell lung cancer therapy. Lung Cancer 82: 9–15. [DOI] [PubMed] [Google Scholar]
- Lim S, Janzer A, Becker A, Zimmer A, Schüle R, Buettner R, Kirfel J. 2010. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis 31: 512–520. [DOI] [PubMed] [Google Scholar]
- Lim JH, Choi YJ, Cho CH, Park JW. 2012. Protein arginine methyltransferase 5 is an essential component of the hypoxia-inducible factor 1 signaling pathway. Biochem Biophys Res Commun 418: 254–259. [DOI] [PubMed] [Google Scholar]
- Limm K, Ott C, Wallner S, Mueller DW, Oefner P, Hellerbrand C, Bosserhoff AK. 2013. Deregulation of protein methylation in melanoma. Eur J Cancer 49: 1305–1313. [DOI] [PubMed] [Google Scholar]
- Lin R, Tao R, Gao X, Li T, Zhou X, Guan KL, Xiong Y, Lei QY. 2013. Acetylation stabilizes ATP-citrate lyase to promote lipid biosynthesis and tumor growth. Mol Cell 51: 506–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Bollig-Fischer A, Kreike B, van de Vijver MJ, Abrams J, Ethier SP, Yang ZQ. 2009. Genomic amplification and oncogenic properties of the GASC1 histone demethylase gene in breast cancer. Oncogene 28: 4491–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. 2012. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci 109: 19408–19413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse B, Nielsen AL, Kristensen JB, Helgstrand C, Cloos PA, Olsen L, Gajhede M, Clausen RP, Kristensen JL. 2011. Targeting histone lysine demethylases by truncating the histone 3 tail to obtain selective substrate-based inhibitors. Angew Chem Int Ed Engl 50: 9100–9103. [DOI] [PubMed] [Google Scholar]
- Majer CR, Jin L, Scott MP, Knutson SK, Kuntz KW, Keilhack H, Smith JJ, Moyer MP, Richon VM, Copeland RA, et al. 2012. A687V EZH2 is a gain-of-function mutation found in lymphoma patients. FEBS Lett 586: 3448–3451. [DOI] [PubMed] [Google Scholar]
- Mann M, Zou Y, Chen Y, Brann D, Vadlamudi R. 2014. PELP1 oncogenic functions involve alternative splicing via PRMT6. Mol Oncol 8: 389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Marmorstein R, Zhou M-M. 2014. Writers and readers of histone acetylation: Structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol 6: a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y. 2007. Mechanisms of epigenetic inheritance. Curr Opin Cell Biol 19: 266–272. [DOI] [PubMed] [Google Scholar]
- McBrian MA, Behbahan IS, Ferrari R, Su T, Huang TW, Li K, Hong CS, Christofk HR, Vogelauer M, Seligson DB, et al. 2013. Histone acetylation regulates intracellular pH. Mol Cell 49: 310–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y, Smitheman KN, Ott HM, Pappalardi MB, Allen KE, et al. 2012. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc Natl Acad Sci 109: 2989–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn LM, Zino S, MacDonald AI, Curle J, Reilly JE, Mohammed ZM, McMillan DC, Mallon E, Payne AP, Edwards J, et al. 2014. SIRT2: Tumour suppressor or tumour promoter in operable breast cancer? Eur J Cancer 50: 290–301. [DOI] [PubMed] [Google Scholar]
- Mercurio C, Minucci S, Pelicci PG. 2010. Histone deacetylases and epigenetic therapies of hematological malignancies. Pharmacol Res 62: 18–34. [DOI] [PubMed] [Google Scholar]
- Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ 3rd. 2011. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci 108: 16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner S, Altmeyer M, Zhao H, Pozivil A, Roschitzki B, Gehrig P, Rutishauser D, Huang D, Caflisch A, Hottiger MO. 2010. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res 38: 6350–6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliori V, Phalke S, Bezzi M, Guccione E. 2010. Arginine/lysine-methyl/methyl switches: Biochemical role of histone arginine methylation in transcriptional regulation. Epigenomics 2: 119–137. [DOI] [PubMed] [Google Scholar]
- Min DJ, Ezponda T, Kim MK, Will CM, Martinez-Garcia E, Popovic R, Basrur V, Elenitoba-Johnson KS, Licht JD. 2012. MMSET stimulates myeloma cell growth through microRNA-mediated modulation of c-MYC. Leukemia 27: 686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minucci S, Pelicci PG. 2006. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer 6: 38–51. [DOI] [PubMed] [Google Scholar]
- Mitra D, Das PM, Huynh FC, Jones FE. 2011. Jumonji/ARID1 B (JARID1B) protein promotes breast tumor cell cycle progression through epigenetic repression of microRNA let-7e. J Biol Chem 286: 40531–40535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moelans CB, de Weger RA, Monsuur HN, Vijzelaar R, van Diest PJ. 2010. Molecular profiling of invasive breast cancer by multiplex ligation-dependent probe amplification-based copy number analysis of tumor suppressor and oncogenes. Mod Pathol 23: 1029–1039. [DOI] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. 2010. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 42: 181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, et al. 2011. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476: 298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozzetta C, Pontis J, Fritsch L, Robin P, Portoso M, Proux C, Margueron R, Ait-Si-Ali S. 2014. The histone H3 lysine 9 methyltransferases G9a and GLP regulate polycomb repressive complex 2-mediated gene silencing. Mol Cell 53: 277–289. [DOI] [PubMed] [Google Scholar]
- Muller S, Brown PJ. 2012. Epigenetic chemical probes. Clin Pharmacol Ther 92: 689–693. [DOI] [PubMed] [Google Scholar]
- Muraoka M, Konishi M, Kikuchi-Yanoshita R, Tanaka K, Shitara N, Chong JM, Iwama T, Miyaki M. 1996. p300 gene alterations in colorectal and gastric carcinomas. Oncogene 12: 1565–1569. [PubMed] [Google Scholar]
- Nakagawa M, Oda Y, Eguchi T, Aishima S, Yao T, Hosoi F, Basaki Y, Ono M, Kuwano M, Tanaka M, et al. 2007. Expression profile of class I HDACs in human cancer tissues. Oncology Rep 18: 769–774. [PubMed] [Google Scholar]
- Nicholas C, Yang J, Peters SB, Bill MA, Baiocchi RA, Yan F, Sif S, Tae S, Gaudio E, Wu X, et al. 2013. PRMT5 is upregulated in malignant and metastatic melanoma and regulates expression of MITF and p27(Kip1.). PLoS One 8: e74710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa N, Toyota M, Suzuki H, Honma T, Fujikane T, Ohmura T, Nishidate T, Ohe-Toyota M, Maruyama R, Sonoda T, et al. 2007. Gene amplification and overexpression of PRDM14 in breast cancers. Cancer Res 67: 9649–9657. [DOI] [PubMed] [Google Scholar]
- Oehme I, Linke JP, Bock BC, Milde T, Lodrini M, Hartenstein B, Wiegand I, Eckert C, Roth W, Kool M, et al. 2013. Histone deacetylase 10 promotes autophagy-mediated cell survival. Proc Natl Acad Sci 110: E2592–E2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okudela K, Mitsui H, Suzuki T, Woo T, Tateishi Y, Umeda S, Saito Y, Tajiri M, Masuda M, Ohashi K. 2013. Expression of HDAC9 in lung cancer—Potential role in lung carcinogenesis. Int J Clin Exp Pathol 7: 213–220. [PMC free article] [PubMed] [Google Scholar]
- Ong CT, Corces VG. 2011. Enhancer function: New insights into the regulation of tissue-specific gene expression. Nat Rev Genet 12: 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa T, Muramatsu M, Wang F, Tsuchida R, Kodama T, Minami T, Shibuya M. 2011. Increased expression of histone demethylase JHDM1D under nutrient starvation suppresses tumor growth via down-regulating angiogenesis. Proc Natl Acad Sci 108: 20725–20729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan MR, Peng G, Hung WC, Lin SY. 2011. Monoubiquitination of H2AX protein regulates DNA damage response signaling. J Biol Chem 286: 28599–28607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S, Durocher D. 2013. Push back to respond better: Regulatory inhibition of the DNA double-strand break response. Nat Rev Mol Cell Biol 14: 661–672. [DOI] [PubMed] [Google Scholar]
- Paredes S, Villanova L, Chua KF. 2014. Molecular pathways: Emerging roles of mammalian Sirtuin SIRT7 in cancer. Clin Cancer Res 20: 1741–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Zhu Y, Ozden O, Kim HS, Jiang H, Deng CX, Gius D, Vassilopoulos A. 2012. SIRT2 is a tumor suppressor that connects aging, acetylome, cell cycle signaling, and carcinogenesis. Transl Cancer Res 1: 15–21. [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, Kasper LH, Lerach S, Tang H, Ma J, et al. 2011. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 471: 189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Patel DJ. 2014. A structural perspective on readout of epigenetic histone and DNA methylation marks. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinder JB, Attwood KM, Dellaire G. 2013. Reading, writing, and repair: The role of ubiquitin and the ubiquitin-like proteins in DNA damage signaling and repair. Front Genet 4: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Pirrotta V. 2014. The necessity of chromatin: A view in perspective. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plazas-Mayorca MD, Bloom JS, Zeissler U, Leroy G, Young NL, DiMaggio PA, Krugylak L, Schneider R, Garcia BA. 2010. Quantitative proteomics reveals direct and indirect alterations in the histone code following methyltransferase knockdown. Mol Biosyst 6: 1719–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan S, Chin HG, Esteve PO, Jacobsen SE. 2009. SET7/9 mediated methylation of non-histone proteins in mammalian cells. Epigenetics 4: 383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Qi J. 2014. Bromodomain and extraterminal domain inhibitors (BETi) for cancer therapy: Chemical modulation of chromatin structure. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR, Guo A, Gut P, Sahu AK, Li B, et al. 2013. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab 18: 920–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert N, Choukrallah MA, Matthias P. 2012. Multiple roles of class I HDACs in proliferation, differentiation, and development. Cell Mol Life Sci 69: 2173–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, et al. 2012. Novel mutations target distinct subgroups of medulloblastoma. Nature 488: 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Becker B, Schneider-Brachert W, Hagen I, Landthaler M, Vogt T. 2006. Re-expression of the retinoblastoma-binding protein 2-homolog 1 reveals tumor-suppressive functions in highly metastatic melanoma cells. J Invest Dermatol 126: 1850–1859. [DOI] [PubMed] [Google Scholar]
- Roesch A, Mueller AM, Stempfl T, Moehle C, Landthaler M, Vogt T. 2008. RBP2-H1/JARID1B is a transcriptional regulator with a tumor suppressive potential in melanoma cells. Int J Cancer 122: 1047–1057. [DOI] [PubMed] [Google Scholar]
- Rosenthal F, Feijs KL, Frugier E, Bonalli M, Forst AH, Imhof R, Winkler HC, Fischer D, Caflisch A, Hassa PO, et al. 2013. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat Struct Mol Biol 20: 502–507. [DOI] [PubMed] [Google Scholar]
- Rossetto D, Avvakumov N, Côté J. 2012. Histone phosphorylation: A chromatin modification involved in diverse nuclear events. Epigenetics 7: 1098–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. 2007. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol 8: 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed S, Logie C, Francoijs KJ, Frige G, Romanenghi M, Nielsen FG, Raats L, Shahhoseini M, Huynen M, Altucci L, et al. 2012. Chromatin accessibility, p300, and histone acetylation define PML-RARα and AML1-ETO binding sites in acute myeloid leukemia. Blood 120: 3058–3068. [DOI] [PubMed] [Google Scholar]
- Sakabe K, Wang Z, Hart GW. 2010. β-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci 107: 19915–19920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg ML, Sutton SE, Pletcher MT, Wiltshire T, Tarantino LM, Hogenesch JB, Cooke MP. 2005. c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Dev Cell 8: 153–166. [DOI] [PubMed] [Google Scholar]
- *.Schaefer U. 2014. Pharmacological inhibition of bromodomain-containing proteins in inflammation. Cold Spring Harb Perspect Biol 6: a018671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk T, Chen WC, Göllner S, Howell L, Jin L, Hebestreit K, Klein HU, Popescu AC, Burnett A, Mills K, et al. 2012. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med 18: 605–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SLB, Bradley E. 2002. Signaling network model of chromatin. Cell Cycle 111: 771–778. [DOI] [PubMed] [Google Scholar]
- Schulte JH, Lim S, Schramm A, Friedrichs N, Koster J, Versteeg R, Ora I, Pajtler K, Klein-Hitpass L, Kuhfittig-Kulle S, et al. 2009. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: Implications for therapy. Cancer Res 69: 2065–2071. [DOI] [PubMed] [Google Scholar]
- Sebastian C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, et al. 2012. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 151: 1185–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. 2005. Global histone modification patterns predict risk of prostate cancer recurrence. Nature 435: 1262–1266. [DOI] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, McBrian MA, Mah V, Yu H, Tze S, Wang Q, Chia D, Goodglick L, Kurdistani SK. 2009. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol 174: 1619–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen N, Kumari R, Singh MI, Das S. 2013. HDAC5, a key component in temporal regulation of p53-mediated transactivation in response to genotoxic stress. Mol Cell 52: 406–420. [DOI] [PubMed] [Google Scholar]
- Serrano L, Martinez-Redondo P, Marazuela-Duque A, Vazquez BN, Dooley SJ, Voigt P, Beck DB, Kane-Goldsmith N, Tong Q, Rabanal RM, et al. 2013. The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev 27: 639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Seto E, Yoshida M. 2014. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb Perspect Biol 6: a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah BD, Martin P, Sotomayor EM. 2012. Mantle cell lymphoma: A clinically heterogeneous disease in need of tailored approaches. Cancer Control 19: 227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. 2010. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Azebi S, England P, Christensen T, Møller-Larsen A, Petersen T, Batsché E, Muchardt C. 2012. Citrullination of histone H3 interferes with HP1-mediated transcriptional repression. PLoS Genet 8: e1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Laird PW. 2013. Interplay between the cancer genome and epigenome. Cell 153: 38–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Shi YG, Tsukada Y. 2013. The discovery of histone demethylases. Cold Spring Harb Perspect Biol 5: a017947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O. 2007. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell 27: 636–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Lin C, Shilatifard A. 2011. The super elongation complex (SEC) and MLL in development and disease. Genes Dev 25: 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MD, Li J, Wong J. 2005. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol Cell Biol 25: 2525–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev 12: 599–606. [DOI] [PubMed] [Google Scholar]
- Stunkel W, Campbell RM. 2011. Sirtuin 1 (SIRT1): The misunderstood HDAC. J Biomol Screen 16: 1153–1169. [DOI] [PubMed] [Google Scholar]
- Subramanian K, Jia D, Kapoor-Vazirani P, Powell DR, Collins RE, Sharma D, Peng J, Cheng X, Vertino PM. 2008. Regulation of estrogen receptor α by the SET7 lysine methyltransferase. Mol Cell 30: 336–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Workman JL. 2008. Crosstalk among histone modifications. Cell 135: 604–607. [DOI] [PubMed] [Google Scholar]
- Suva ML, Riggi N, Janiszewska M, Radovanovic I, Provero P, Stehle JC, Baumer K, Le Bitoux MA, Marino D, Cironi L, et al. 2009. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res 69: 9211–9218. [DOI] [PubMed] [Google Scholar]
- Takawa M, Cho HS, Hayami S, Toyokawa G, Kogure M, Yamane Y, Iwai Y, Maejima K, Ueda K, Masuda A, et al. 2012. Histone lysine methyltransferase SETD8 promotes carcinogenesis by deregulating PCNA expression. Cancer Res 72: 3217–3227. [DOI] [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. 2011. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146: 1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. 2013. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol 20: 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanikawa C, Espinosa M, Suzuki A, Masuda K, Yamamoto K, Tsuchiya E, Ueda K, Daigo Y, Nakamura Y, Matsuda K. 2012. Regulation of histone modification and chromatin structure by the p53-PADI4 pathway. Nat Commun 3: 676. [DOI] [PubMed] [Google Scholar]
- Trojer P, Cao AR, Gao Z, Li Y, Zhang J, Xu X, Li G, Losson R, Erdjument-Bromage H, Tempst P, et al. 2011. L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Mol Cell 42: 438–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Wang JK, Chang HY. 2010. Tumor suppression by the histone demethylase UTX. Cell Cycle 9: 2043–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie-Cullen RY, Brunner AM, Grossmann J, Mohanna S, Sichau D, Nanni P, Panse C, Mansuy IM. 2012. Identification of combinatorial patterns of post-translational modifications on individual histones in the mouse brain. PLoS One 7: e36980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura M, Yamamoto H, Takemasa I, Mimori K, Hemmi H, Mizushima T, Ikeda M, Sekimoto M, Matsuura N, Doki Y, et al. 2010. Jumonji domain containing 1A is a novel prognostic marker for colorectal cancer: In vivo identification from hypoxic tumor cells. Clin Cancer Res 16: 4636–4646. [DOI] [PubMed] [Google Scholar]
- Van Aller GS, Reynoird N, Barbash O, Huddleston M, Liu S, Zmoos AF, McDevitt P, Sinnamon R, Le B, Mas G, et al. 2012. Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5 methylation. Epigenetics 7: 340–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme M, Crompot E, Meuleman N, Mineur P, Bron D, Lagneaux L, Stamatopoulos B. 2012. HDAC isoenzyme expression is deregulated in chronic lymphocytic leukemia B-cells and has a complex prognostic significance. Epigenetics 7: 1403–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT, Heguy A, Santi M, Thompson CB, Judkins AR. 2013. Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol 23: 558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira FQ, Costa-Pinheiro P, Ramalho-Carvalho J, Pereira A, Menezes FD, Antunes L, Carneiro I, Oliveira J, Henrique R, Jeronimo C. 2014. Deregulated expression of selected histone methylases and demethylases in prostate carcinoma. Endocr Relat Cancer 21: 51–61. [DOI] [PubMed] [Google Scholar]
- Vinatzer U, Gollinger M, Müllauer L, Raderer M, Chott A, Streubel B. 2008. Mucosa-associated lymphoid tissue lymphoma: Novel translocations including rearrangements of ODZ2, JMJD2C, and CNN3. Clin Cancer Res 14: 6426–6431. [DOI] [PubMed] [Google Scholar]
- Wagner KW, Alam H, Dhar SS, Giri U, Li N, Wei Y, Giri D, Cascone T, Kim JH, Ye Y, et al. 2013. KDM2A promotes lung tumorigenesis by epigenetically enhancing ERK1/2 signaling. J Clin Invest 123: 5231–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, et al. 2004. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 306: 279–283. [DOI] [PubMed] [Google Scholar]
- Wang GG, Cai L, Pasillas MP, Kamps MP. 2007. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol 9: 804–812. [DOI] [PubMed] [Google Scholar]
- Wang L, Gural A, Sun XJ, Zhao X, Perna F, Huang G, Hatlen MA, Vu L, Liu F, Xu H, et al. 2011. The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science 333: 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Soejima K, Yasuda H, Kawada I, Nakachi I, Yoda S, Naoki K, Ishizaka A. 2008. Deregulation of histone lysine methyltransferases contributes to oncogenic transformation of human bronchoepithelial cells. Cancer Cell Int 8: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, Koellhoffer EC, Pomeroy SL, Orkin SH, Roberts CW. 2010. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 18: 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Mann M. 2008. Nepsilon-formylation of lysine is a widespread post-translational modification of nuclear proteins occurring at residues involved in regulation of chromatin function. Nucleic Acids Res 36: 570–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt O, Deubzer HE, Lodrini M, Milde T, Oehme I. 2009. Targeting histone deacetylases in neuroblastoma. Curr Pharm Des 15: 436–447. [DOI] [PubMed] [Google Scholar]
- Wyce A, Ganji G, Smitheman KN, Chung CW, Korenchuk S, Bai Y, Barbash O, Le B, Craggs PD, McCabe MT, et al. 2013. BET inhibition silences expression of MYCN and BCL2 and induces cytotoxicity in neuroblastoma tumor models. PloS One 8: e72967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, Alinari L, Lustberg ME, Martin LK, Cordero-Nieves HM, Banasavadi-Siddegowda Y, Virk S, Barnholtz-Sloan J, Bell EH, Wojton J, et al. 2014. Genetic validation of the protein arginine methyltransferase PRMT5 as a candidate therapeutic target in glioblastoma. Cancer Res 74: 1752–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZQ, Imoto I, Fukuda Y, Pimkhaokham A, Shimada Y, Imamura M, Sugano S, Nakamura Y, Inazawa J. 2000. Identification of a novel gene, GASC1, within an amplicon at 9p23-24 frequently detected in esophageal cancer cell lines. Cancer Res 60: 4735–4739. [PubMed] [Google Scholar]
- Yang P, Guo L, Duan ZJ, Tepper CG, Xue L, Chen X, Kung HJ, Gao AC, Zou JX, Chen HW. 2012. Histone methyltransferase NSD2/MMSET mediates constitutive NF-κB signaling for cancer cell proliferation, survival, and tumor growth via a feed-forward loop. Mol Cell Biol 32: 3121–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap DB, Chu J, Berg T, Schapira M, Cheng SW, Moradian A, Morin RD, Mungall AJ, Meissner B, Boyle M, et al. 2011. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 117: 2451–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Park SH, Pantazides BG, Karpiuk O, Warren MD, Hardy CW, Duong DM, Park SJ, Kim HS, Vassilopoulos A, et al. 2013. SIRT2 directs the replication stress response through CDK9 deacetylation. Proc Natl Acad Sci 110: 13546–13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen A, Yan XM, Huang G. 2012a. Disordered epigenetic regulation in MLL-related leukemia. Int J Hematol 96: 428–437. [DOI] [PubMed] [Google Scholar]
- Zhang X, Bolt M, Guertin MJ, Chen W, Zhang S, Cherrington BD, Slade DJ, Dreyton CJ, Subramanian V, Bicker KL, et al. 2012b. Peptidylarginine deiminase 2-catalyzed histone H3 arginine 26 citrullination facilitates estrogen receptor α target gene activation. Proc Natl Acad Sci 109: 13331–13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Zhao Y, Garcia BA. 2014. Comprehensive catalog of currently documented histone modifications. Cold Spring Harb Persp Biol 10.1101/cshperspect.a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager PL, et al. 2013. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell 152: 642–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478: 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]