SUMMARY
Communication between cells in a multicellular organism occurs by the production of ligands (proteins, peptides, fatty acids, steroids, gases, and other low-molecular-weight compounds) that are either secreted by cells or presented on their surface, and act on receptors on, or in, other target cells. Such signals control cell growth, migration, survival, and differentiation. Signaling receptors can be single-span plasma membrane receptors associated with tyrosine or serine/threonine kinase activities, proteins with seven transmembrane domains, or intracellular receptors. Ligand-activated receptors convey signals into the cell by activating signaling pathways that ultimately affect cytosolic machineries or nuclear transcriptional programs or by directly translocating to the nucleus to regulate transcription.
In a multicellular organism, communication between cells is required to coordinate growth, migration, survival, and differentiation. Cells produce various ligands that bind to receptors on/in other cells, activating signaling pathways.
1. INTRODUCTION
Cells within multicellular organisms need to communicate with each other to coordinate their growth, migration, survival, and differentiation. They do so by direct cell–cell contact and secretion or release of molecules that bind to and activate receptors on the surface of or inside target cells. Such factors can stimulate the producer cell itself (autocrine stimulation), cells in the immediate vicinity (paracrine stimulation), or cells in distant organs (endocrine stimulation). The signaling induced within target cells is important during embryonic development, as well as in the adult, where it controls cell proliferation, differentiation, the response to infection, and numerous organismal homeostatic mechanisms.
Many cell-surface receptors contain an extracellular ligand-binding region, a single transmembrane segment, and an intracellular effector region, which may or may not have an associated enzyme activity. Some receptors contain multiple subunits that together form the ligand-binding site. Others, including those encoded by the largest gene family in the human genome, consist of a polypeptide that spans the cell membrane seven times. Finally, there are receptors that are located intracellularly and are activated by ligands that cross the cell membrane, such as steroid hormones. Below, we describe the major families of ligands and receptors and the signal transduction mechanisms they activate.
2. CELL-SURFACE RECEPTORS
2.1. Receptors with Associated Protein Kinase Activity
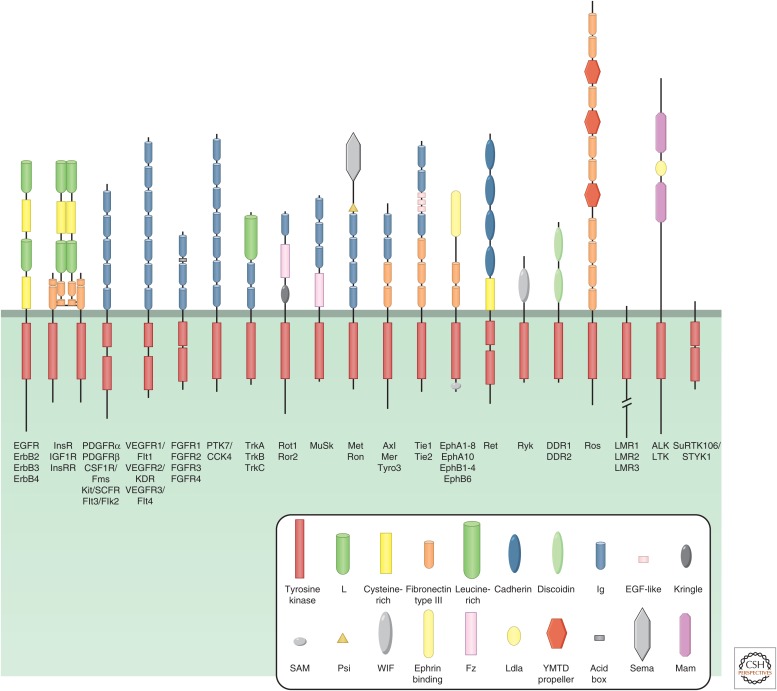
Several types of cell-surface receptors contain or are associated with kinase activities that respond to the binding of a ligand. Perhaps best understood are receptors with intrinsic protein tyrosine kinase domains. This receptor tyrosine kinase (RTK) family has more than 50 human members (Lemmon and Schlessinger 2010). RTKs have important roles in the regulation of embryonic development, as well as in the regulation of tissue homeostasis in the adult. Each has an extracellular, ligand-binding region, which consists of different combinations of various domains, such as Ig-like, fibronectin type III, and cysteine-rich domains. This is linked to a single transmembrane segment and an intracellular region that includes a tyrosine kinase domain. Based on their structural features, RTKs can be divided into 20 subfamilies (Fig. 1), a well-studied example being the epidermal growth factor (EGF) receptors (EGFRs).
Figure 1.
Receptor tyrosine kinase (RTK) families. The 20 subfamilies of human RTKs and their characteristic structural domains are shown. The individual members of each family are listed below. (From Lemmon and Schlessinger 2010; adapted, with permission.)
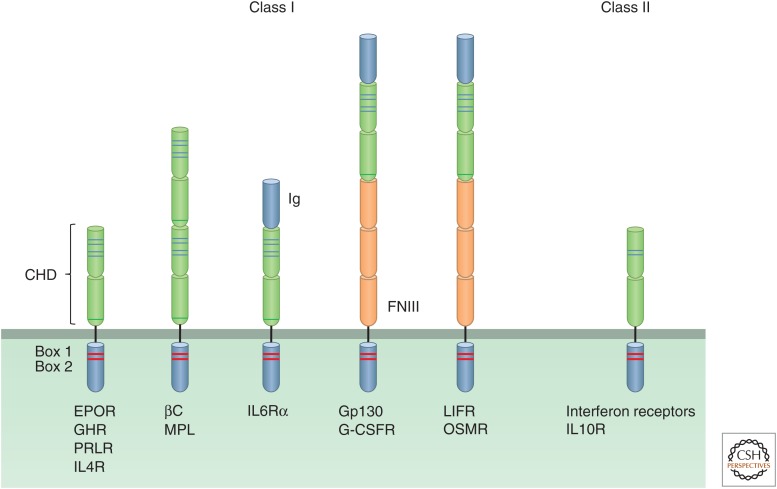
Members of the cytokine receptor family in contrast lack intrinsic kinase activity but associate with intracellular kinases. They have important roles in the regulation of the immune system and also promote cell differentiation. Cytokine receptors can be divided into two classes. The extracellular domains of class I cytokine receptors contain cytokine receptor homology domains (CHDs) consisting of two tandem fibronectin type III domains with a characteristic WSXWS motif in the second (Liongue and Ward 2007). Based on the number of CHDs and the presence of other types of domains, the class I cytokine receptors can be divided into five groups (Fig. 2), the growth hormone (GH) receptor being typical of the first group. Interferon receptors are typical of the 12-member class II cytokine receptor family, which also have extracellular regions based on tandem fibronectin domains but differ from those of class I receptors (Fig. 2) (Renauld 2003). Both classes of cytokine receptors have conserved box 1 and box 2 regions in their intracellular regions, which bind to JAK family tyrosine kinases that are activated upon ligand binding. The multisubunit antigen receptors on B cells and T cells (Zikherman and Weiss 2009) and the Fc receptors present on macrophages, mast cells, basophils, and other immune cells (Nimmerjahn and Ravetch 2008) are also associated with intracellular tyrosine kinases; activation of these receptors involves tyrosine phosphorylation by members of the Src family, followed by docking and activation of SH2-domain-containing Syk/Zap70 tyrosine kinases (see Samelson 2011; Cantrell 2014). Although receptors that have intrinsic tyrosine kinase activity and those associated with tyrosine kinases are structurally different and bind ligands of different kinds, the principles underlying their activation and the intracellular signals they initiate are similar (see below).
Figure 2.
Cytokine receptor families. The structural features of the five subfamilies of class I and class II cytokine receptors are depicted. The characteristic cytokine homology domains (CHDs) with their four cysteine residues (blue lines) and a WW motif (green line), as well as the box 1 and box 2 regions (red bands), Ig-like domains, and FNIII domains are shown.
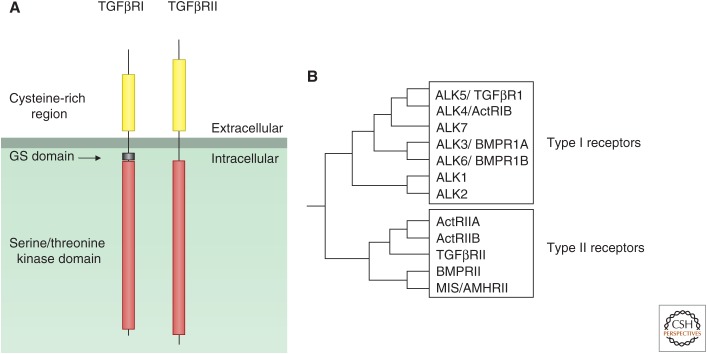
There is also a family of receptors that have intrinsic serine/threonine kinase domains, and these respond to members of the transforming growth factor β (TGFβ) family (see Moustakas and Heldin 2009; Wrana 2013). The human genome has only 12 genes encoding receptors of this type (Fig. 3). These receptors have rather small cysteine-rich extracellular domains; their intracellular domains are most often also small and consist mainly of the kinase domains. TGFβ receptors mediate signaling events during embryonic development. Because they often inhibit cell growth, they also exert a controlling function on the immune system and other tissues.
Figure 3.
Serine/threonine kinase receptors. (A) The structural features of type I (TGFβRI) and type II (TGFβRII) serine/threonine kinase receptors. (B) The different members of the type I and type II receptor subfamilies and their evolutionary relations. Act, Activin; ALK, activin-receptor-like kinase.
2.2. Ligands
Each of these different receptor types responds to a subfamily of structurally similar ligands. The ligands are normally small monomeric, dimeric, or trimeric proteins, often derived by proteolytic processing from larger precursors, some of which are transmembrane proteins. There is not a strict specificity in ligand–receptor interactions within the families; normally each ligand binds to more than one receptor, and each receptor binds more than one ligand. Although it is rare that ligands for completely different types of receptors bind to kinase-associated receptors, examples do exist.4
2.3. Activation by Dimerization
A common theme for activation of kinase-associated receptors is ligand-induced receptor dimerization or oligomerization (Heldin 1995). The juxtaposition of the intracellular kinase domains that occurs as a consequence allows autophosphorylation in trans within the complex. For RTKs, the autophosphorylation has two important consequences: it changes the conformation of the kinase domains, leading to an increase in their kinase activities; and it produces docking sites (phosphorylated sequences) for intracellular signaling molecules containing SH2 or PTB domains (see Lee and Yaffe 2014). The autophosphorylation that controls the kinase activity occurs on residues in various regions of the receptors. Most RTKs are phosphorylated in the activation loops of the kinases, that is, a flexible loop of the carboxy-terminal domain, which can fold back and block the active site of the kinase when unphosphorylated. Exceptions are Ret and the four members of the EGFR family that can be activated without phosphorylation in their activation loops (Lemmon and Schlessinger 2010). Several RTKs are activated by phosphorylation in the juxtamembrane region, including MuSK, Flt3, Kit, and Eph family members. Moreover, Tie2, platelet-derived growth factor (PDGF) β receptor (PDGFRβ) and Ron have been shown to be activated by phosphorylation of their carboxy-terminal tails. In each case, autophosphorylation causes a change in conformation that opens up the active site of the kinase. Interestingly, a phosphorylation-independent mechanism for activation of EGFR kinase activity has been elucidated: the carboxy-terminal lobe of one kinase domain makes contact with the amino-terminal lobe of the other kinase domain in the dimer and thereby activates it (Zhang et al. 2006). The juxtamembrane region of the receptor, which in other RTKs has an inhibitory role, is needed to stabilize this dimeric interaction (Jura et al. 2009; Red Brewer et al. 2009).
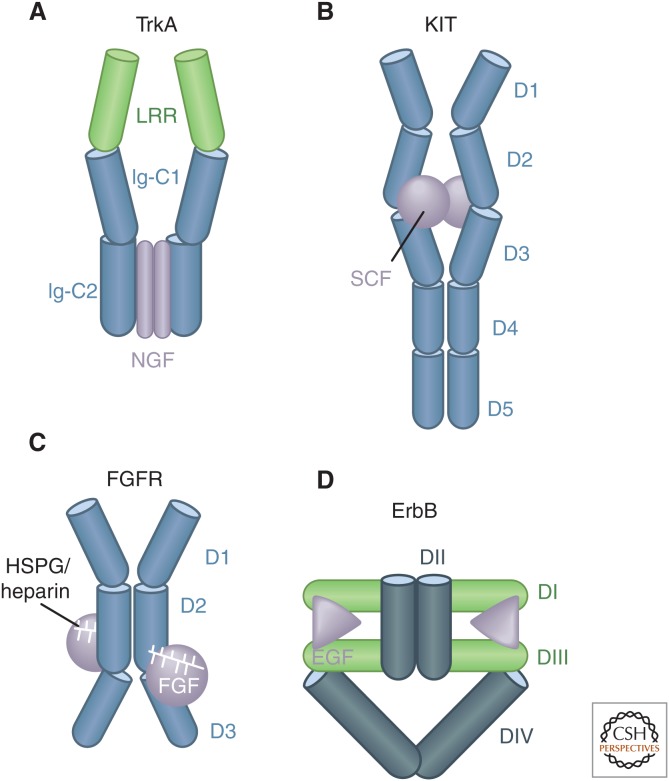
Different ligands use different mechanisms to cause receptor dimerization. Thus, GH is a monomeric ligand that dimerizes its receptors by inducing formation of an asymmetric complex containing one ligand and two receptors (De Vos et al. 1992). EGF is also a monomeric factor, but each molecule binds to a single receptor molecule, causing a conformational change that promotes direct binding of two ligand-bound receptors (Fig. 4) (Burgess et al. 2003). Similarly, dimeric ligands can induce symmetric 2:2 ligand–receptor complexes. In the case of nerve growth factor (NGF), the ligand itself is solely responsible for the dimerization (Wehrman et al. 2007); in the case of Kit and the PDGFRs, the ligand-induced receptor dimerization is stabilized by direct interactions between the receptors (Yuzawa et al. 2007). In other cases, accessory molecules are needed to stabilize dimerization; that is, heparin or heparan sulfate stabilizes fibroblast growth factor (FGF)–induced receptor dimers by interacting with both FGF and FGF-receptor subunits (Lemmon and Schlessinger 2010). Recent work on EGFR has shown that its activation requires an interaction between the amino-terminal parts of the transmembrane helices of the dimeric receptors, which promotes an antiparallel interaction between the cytoplasmic juxtamembrane segments and release of inhibition by the membrane (Arkhipov et al. 2013; Endres et al. 2013).
Figure 4.
Schematic illustration of different modes of dimerization of RTKs. (A) Some dimeric ligands, such as nerve growth factor (NGF), bind to receptors in a symmetric manner, but the receptors do not contact each other. (B) Other dimeric ligands, such as stem cell factor (SCF), also bind to RTKs in a symmetric manner, but the receptor dimer is in addition stabilized by direct receptor–receptor interactions. (C) In the case of fibroblast growth factor (FGF), a ternary complex involving the ligand, the receptor, and heparin/heparin sulfate stabilizes the receptor dimer. (D) In the case of members of the epidermal growth factor (EGF) receptor family such as ErbB, ligand binding induces a conformational change in the extracellular domain of the receptor that promotes direct receptor–receptor interactions. (From Lemmon and Schlessinger 2010; adapted, with permission.)
In addition to homodimerization of receptors, heterodimerization also often occurs. This is particularly common among cytokine receptors; a ligand-specific receptor subunit often interacts with one or more common subunits, such as gp130, βc, or γc, to form a heterodimer, heterotrimer, or even more complicated multimer (Wang et al. 2009). Similarly, members of different RTK subfamilies often form heterodimers. For instance, one member of the EGFR subfamily, ErbB2, cannot bind to ligand itself, but acts in heterodimers with other members of the family (Yarden and Sliwkowski 2001). In the PDGFR family, different dimeric isoforms of the ligand induce formation of different dimeric complexes of PDGFα and PDGFβ receptors (Heldin 1995). Because the downstream signaling pathways that are activated to a large extent depend on the specific docking of SH2- or PTB-domain-containing signaling molecules (see below and Lee and Yaffe 2014), differences in the autophosphorylation patterns of homodimeric versus heterodimeric receptor complexes will give rise to different combinatorial signals.
Serine/threonine kinase receptors present another variation on this theme. These receptors are activated by ligand-induced assembly of two type I and two type II receptors into heterotetrameric receptor complexes, in which constitutively active type II receptors phosphorylate type I receptors in serine- and glycine-rich sequences just upstream of the kinase domains. In the TGFβ type I receptor, this causes a change in conformation that prevents the inhibitory interaction of the receptor with the immunophilin FK506-binding protein (FKBP12) and activates its kinase (Kang et al. 2009).
Note that certain receptors are present in dimeric complexes even in the absence of ligand; examples include the erythropoietin cytokine receptor, and the insulin and insulin-like growth factor-1 (IGF1) receptors, RTKs that actually occur as disulfide-bonded dimers. Here, ligand binding induces conformational changes that lead to activation of the receptor-associated kinase. Importantly, there are indications that reorientation of the intracellular regions of the receptors relative to each other in the dimer is important for their activation (Jiang and Hunter 1999).
2.4. Signaling Downstream from Tyrosine-Kinase-Associated Receptors via SH2- or PTB-Domain-Containing Molecules
An important aspect of signaling by tyrosine-kinase-associated receptors is the formation of multiprotein signaling complexes (Pawson 2004). As mentioned above, SH2- or PTB-domain-containing proteins bind to specific phosphorylated tyrosine residues in the receptors themselves or in scaffolding proteins associated with the receptors (see Lee and Yaffe 2014). Examples of the latter include the four members of the insulin receptor substrate (IRS) family, which bind to insulin and IGF1 receptors; FRS2-family protein molecules, which bind to FGF and NGF receptors; Gab molecules, which bind to several RTKs; and SLP76 and LAT, which bind to antigen receptors.
Some of the proteins that bind to these sequences contain intrinsic enzymatic activities, for example, the tyrosine kinases of the Src family, the tyrosine phosphatases of the SHP family, GTPase-activating proteins (GAPs) that regulate Ras family GTPases, the ubiquitin ligase Cbl, and phospholipase C (PLC) γ. Others are proteins that form stable complexes with enzymes, for example, the regulatory p85 subunit of type 1 phosphoinositide 3-kinase (PI3K), which forms a complex with the catalytic p110 subunit, and Grb2, which forms a complex with SOS1, a guanine nucleotide exchange factor (GEF) that stimulates Ras. Cytokine receptors and some RTKs also activate STAT molecules, which translocate to the nucleus, where they act as transcription factors (see Harrison 2012). Finally, SH2-/SH3-domain-containing adaptor molecules (e.g., Shc, Nck, and Crk) bind to certain tyrosine phosphorylated sites in the receptors or associated molecules and form bridges to other signaling molecules, including the enzymes mentioned above.
Many of the signaling proteins that bind to the receptors also contain domains that mediate interactions with other molecules. These include SH3 domains, which bind proline-rich motifs in proteins; PDZ domains, which bind to the carboxy-terminal tails of proteins containing valine residues; and pleckstrin homology (PH) domains, which bind lipids present in membranes. Thus, large complexes of signaling proteins are transiently formed and mediate signaling to cytoplasmic machinery (e.g., enzymes controlling the cytoskeleton and cell migration), as well as to the nucleus, where transcription is affected.
Note that the kinase domains of certain RTKs (e.g., ErbB3, CCK4, EphB6, and Ryk) lack critical catalytic residues normally found in kinases and thus have no, or very low, kinase activity. However, these receptors still have important roles in signal transduction, acting as phosphorylatable scaffolds in heterodimers with other receptors.
There are also indications that the EGFRs can translocate to the nucleus to regulate transcription (Lin et al. 2001). However, the functional significance of this needs to be further clarified.
2.5. Downstream Signaling by Serine/Threonine Kinase Receptors
Serine/threonine kinase receptors activate members of the Smad transcription factor family (Wrana 2013); similar to STATs, these molecules oligomerize after phosphorylation and activation and then translocate to the nucleus, where they induce transcription. In addition, serine/threonine kinase receptors can activate non-Smad pathways. The TGFβ type I receptor, for example, can autophosphorylate on tyrosine residues, which bind to the adaptor Shc, leading to activation of Ras and the ERK1/2 mitogen-activated protein kinase (MAPK) pathway (see Morrison 2012). Moreover, the p38 MAPK pathway is activated via binding of the ubiquitin ligase TRAF6 to the TGFβ type I receptor (Kang et al. 2009). In addition to activating the type I receptor, the TGFβ type II receptor also contributes to signaling by phosphorylating the polarity complex protein PAR6 (Ozdamar et al. 2005; McCaffrey and Macara 2012).
2.6. Interaction with Coreceptors
Several RTKs form complexes with nonkinase receptors, such as the hyaluronan receptor CD44 and integrins, which bind to extracellular matrix molecules. Such interactions can modulate their signaling. Similarly, the heparan sulfate proteoglycan agrin activates MuSK receptors by binding to low-density lipoprotein (LDL)-receptor-related protein 4 (LRP4); thus, LRP4 acts as a coreceptor for MuSK (Kim et al. 2008). Another example is glial-ceU-derived neurotrophic factor (GDNF) receptor α, a glycosylphosphatidylinositol (GPI)-anchored molecule needed for GDNF to bind to and activate Ret receptors (Schlee et al. 2006).
2.7. Feedback and Amplification Mechanisms
Signaling via kinase-associated receptors is controlled by multiple feedback mechanisms to ensure an appropriate level. Thus, many tyrosine kinase and cytokine receptors bind protein tyrosine phosphatases (PTPs), including SHP1 and SHP2, that contain SH2 domains. Through dephosphorylation of specific tyrosine residues, PTPs modulate signaling quantitatively as well as qualitatively (Lemmon and Schlessinger 2010).
Another example is activation of the small G protein Ras, which occurs by docking of the GEF SOS1 in complex with the adaptor Grb2 (see Lee and Yaffe 2014). PDGFRβ and possibly other receptors simultaneously bind the Ras-GAP molecules via another phosphorylated tyrosine residue, which counteracts Ras activation by promoting hydrolysis of bound GTP.
In addition, another downstream target of receptor signaling, protein kinase C (PKC), is involved in feedback control of certain RTKs, including EGFR, the insulin receptor, Met, and Kit. Activation of PLCγ leads to production of diacylglycerol (DAG) and increases in intracellular calcium levels, which activate the classical isoforms of PKC (see Newton et al. 2014). Phosphorylation of the receptors by PKC subsequently inhibits the kinase activity of the receptors.
After the induction of immediate-early genes encoding various effector molecules by signaling to the nucleus, RTKs induce the expression of delayed-early genes, many of which encode proteins that suppress signaling. Examples include NAB2, which binds to and inhibits EGFR; FOSL1, which binds to and inhibits AP1 transcription factors; Id2, which inhibits the TCF transcription complex; DUSPs, which dephosphorylate and inactivate MAPKs; and ZFP36, which recognizes AU-motifs in the 3′ ends of mRNA molecules and causes their degradation (Amit et al. 2007). Similarly, TGFβ receptors and cytokine receptors induce Smad7 (Moustakas and Heldin 2009) and SOCS proteins (Yoshimura et al. 2003), respectively, which exert negative-feedback control by promoting ubiquitin-dependent degradation of receptors by targeting receptor-containing vesicles to lysosomes (see below).
2.8. Endocytosis of Kinase-Associated Receptors
After ligand binding, kinase-associated receptors are internalized into the cell via clathrin-dependent or clathrin-independent pathways (Zwang and Yarden 2009). Whereas internalization of RTKs is induced by ligand binding, internalization of serine/threonine receptors is constitutive and independent of ligand binding. Internalization has both positive and negative effects on signaling. Thus, when present in endosomes, the receptors are still active; in some cases, internalization is even necessary for the receptors to interact with signaling molecules on intracellular endosomes (Miaczynska et al. 2004). Examples include TGFβ receptors, which need to be internalized to interact with Smad molecules presented to the receptors by SARA and endofin proteins. These proteins reside on endosomes, their FYVE domains binding to phosphatidylinositol 3-phosphate (PI3P), a phospholipid that is enriched in endosomal membranes (Tsukazaki et al. 1998). Signaling is interrupted when the pH of the endosomes becomes so low that the ligand dissociates; at this stage, receptors are dephosphorylated and recycle back to the cell surface. Alternatively, the receptors are recognized by components of the endosomal sorting complex required for transport (ESCRT) machinery, which facilitates translocation to multivesicular bodies and degradation in lysosomes (Raiborg and Stenmark 2009). The latter route is promoted by ubiquitylation of the receptors by Cbl or other ubiquitin ligases.
2.9. Kinase-Associated Receptors and Disease
Because tyrosine-kinase-associated receptors often stimulate cell proliferation and survival, overactivity of these receptors is linked to the development of cancer and other diseases characterized by excess cell proliferation, such as inflammatory and fibrotic conditions. There are several examples of gain-of-function mutations in RTKs that occur in malignancies (Lengyel et al. 2007; Sever and Brugge 2014). Point mutations in Kit and PDGFRα have been found in gastric intestinal stromal tumors, and the kinase domains of PDGFRs and FGFRs occur as constitutively active cytoplasmic fusion proteins in several rare leukemias. Moreover, the ERBB2 gene is amplified in ∼20% of breast cancers, and a mutated version of the EGFR gene is amplified in ∼30% of glioblastoma cases.
Because overactivity of these receptors is common in malignancies, several antagonists have been developed, including inhibitory antibodies, ligand traps, and low-molecular-weight kinase inhibitors. Several of these are now used routinely in the clinic or are undergoing clinical trials.
The serine/threonine kinase receptors often relay growth inhibitory and apoptotic signals and therefore have tumor-suppressive effects. Thus, loss-of-function mutations of TGFβ type I and type II receptors have been observed in some cancers (e.g., colorectal carcinomas).
2.10. Receptors Activated by Proteolytic Cleavage
The highly conserved Notch family of receptors consists of four members, which have important roles during embryonic development and tissue renewal (Kopan and Ilagan 2009). The Notch receptors are single-pass transmembrane proteins with large extracellular domains containing multiple EGF-like repeats. They are cleaved extracellularly by furin-like proteases during transit to the plasma membrane to create a heterodimer held together by noncovalent interactions (Kopan 2012).
There are five canonical Notch ligands (Delta-ligand-like 1, 3, and 4, and Jagged 1 and 2). These are also single-pass transmembrane proteins with extracellular EGF-like regions and other domains present on neighboring cells. The Notch receptor is normally triggered upon cell–cell contact. Notch activation involves a series of proteolytic events. Ligand binding induces cleavage of Notch by ADAM metalloproteases at a site about 12 amino acid residues outside the transmembrane domain. Removal of the extracellular domain allows an intramembrane cleavage by γ-secretase, which causes release of the intracellular domain (ICD) of Notch. Because the ICD has a nuclear localization sequence, it translocates to the nucleus, where it, together with the DNA-binding protein RBPjκ/CBF-1 and certain coactivators and corepressors (together referred to as CSL), regulates transcription of target genes via its transactivation domain. Because the Notch ligands are membrane-associated, signaling may also be induced in the ligand-bearing cells after binding to Notch.
Some RTK receptors (Ni et al. 2001) and the type I TGFβ receptor (Mu et al. 2011) are also subjected to sequential cleavage by metalloproteases and γ-secretase. The intracellular region of the receptors can then translocate to the nucleus to regulate transcription. Meanwhile, the extracellular portion of the receptor is liberated and can negatively regulate signaling by acting as a decoy for ligand (Ancot et al. 2009). In addition, some RTKs are classified as “dependence receptors.” When they are not occupied by ligand, they can be cleaved by caspases (a group of proteases that control apoptosis) to generate fragments with apoptotic activity. For example, fragments of EGFR, ErbB2, Ret, Met, TrkC, ALK, and EphA4 have apoptotic effects, which contrast with the normal antiapoptotic effects of the full-length receptors stimulated by their ligands (Ancot et al. 2009).
2.11. G-Protein-Coupled Receptors
Approximately 2% of all genes in the human genome encode G-protein-coupled receptors (GPCRs), which represent the largest family of cell-surface molecules involved in signal transmission. They are so called because their signals are transduced by heterotrimeric G proteins; members of the GPCR family regulate a wide range of key physiological functions, including neurotransmission, blood pressure, cardiac activity, vascular integrity, hemostasis after tissue injury, glucose and lipid metabolism, sensory perception, regulation of endocrine and exocrine gland function, immune responses, multiple developmental processes, and stem cell function and maintenance (Pierce et al. 2002; Dorsam and Gutkind 2007). Reflecting this remarkable multiplicity of activities, GPCR dysregulation contributes to some of the most prevalent human diseases. Indeed, GPCRs are the direct or indirect target of >50% of all available medicines (Flower 1999; Pierce et al. 2002).
2.12. A Shared Heptahelical Structure Transduces Signals Initiated by Highly Diverse Molecular Agonists
GPCRs are characterized by the presence of an extracellular amino terminus, an intracellular carboxy-terminal tail, and a shared structural core composed of seven transmembrane α helices that weave in and out of the membrane, thus forming three intracellular and three extracellular loops (Fig. 5) (Pierce et al. 2002). They are regulated by a diverse array of agonists, as small as photons, ions, nucleotides, amino acids, biogenic amines, bioactive lipids, and glucose metabolites, and as large as chemokines, glycoproteins, and proteases. They can be grouped into three classes and subfamilies of these according to the structural features involved in ligand–receptor recognition and subsequent stimulation (Fig. 5).
Figure 5.
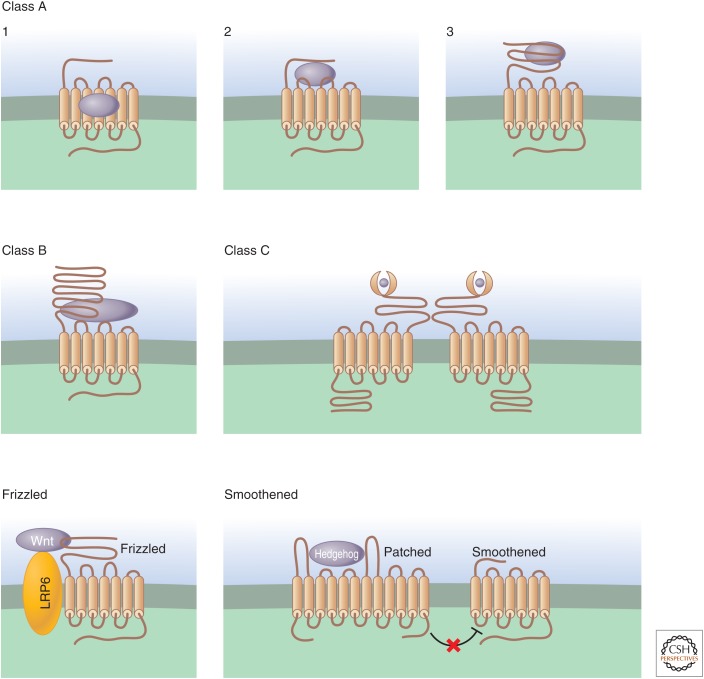
GPCRs use distinct structural features for ligand recognition. GPCRs have been classified based on their sequence similarities and ligand-binding properties. In class A GPCRs, the largest group, the ligand-binding site is deep within the transmembrane domains in subfamily 1. It involves interactions with the amino terminus, the extracellular loops, and the transmembrane domains in subfamily 2. The ligand-binding site is in the long extracellular domain in subfamily 3. Class B GPCRs are activated by high-molecular-weight hormones, which bind to the ligand-binding site within the long amino-terminal region, as well as some of the extracellular loops. Class C GPCRs are characterized by a very long amino terminus that shares some sequence similarity with periplasmic bacterial proteins; activation involves obligatory dimerization. The Frizzled family of receptors contains the Frizzled and “Smoothened” subfamilies, which are structurally distinct and have complex mechanisms of agonist activation. Wnt binds to and activates Frizzled through an interaction with a cysteine-rich amino-terminal region, whereas low-density lipoprotein-receptor-related protein 5 (LRP5) or LRP6 (single-transmembrane-span proteins) acts as a coreceptor (Lim and Nusse 2013). When Hedgehog binds to Patched, a negative regulatory effect of Patched on Smoothened activity is relieved, and Smoothened regulates both G-protein-dependent and -independent signals (Ingham 2012).
In class A GPCRs, the ligand-binding site is deep within the transmembrane domains in subfamily 1, which includes most receptors for small molecules, such as neurotransmitters, lipid mediators, and odorants. Subfamily 2 members are activated by protein ligands, such as chemokines and the tethered ligand resulting from thrombin-mediated cleavage of the protease-activated receptor 1 (PAR1) receptor. Subfamily 3 members have a very long extracellular domain, which binds to leuteinizing hormone (LH; also known as lutropin), thyroid-stimulating hormone (TSH), and follicle-stimulating hormone (FSH) (Kornbluth and Fissore 2014). This subfamily also includes LGR5, LGR6, and LGR7, which are GPCR-like receptors involved in adult stem cell specification and function (Hsu et al. 2000; Leushacke and Barker 2011). The class B GPCRs are activated by high-molecular-weight hormones (e.g., glucagon, secretin, and vasoactive intestinal peptide [VIP]), whereas class C GPCRs include metabotropic glutamate receptors, calcium-sensing receptors, γ-amino butyric acid (GABA) B receptors, and receptors for taste compounds. Although many class A and class B GPCRs can form homo- and heterodimers (Terrillon and Bouvier 2004), dimer formation is obligatory for class C GPCRs (Kniazeff et al. 2011). The Frizzled family of receptors comprises the “Frizzled” and “Smoothened” subfamilies, which are structurally distinct and have complex mechanisms of agonist activation (Ingham 2012; Lim and Nusse 2013).
2.13. GPCR Activation, Trafficking, and G-Protein-Coupling Specificity
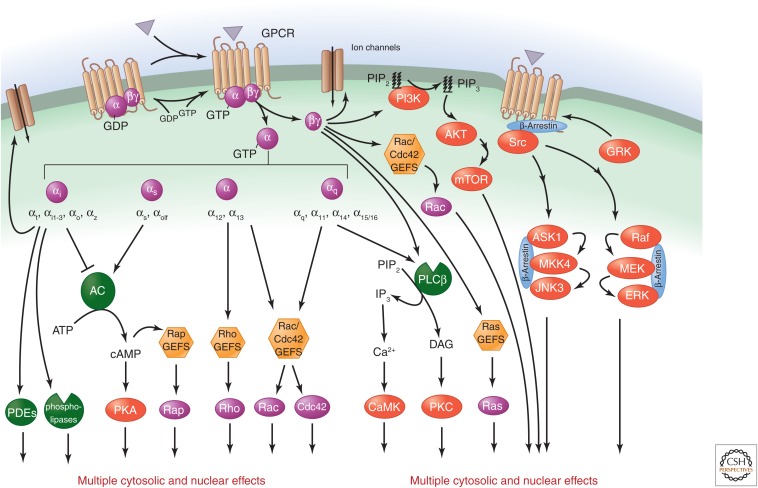
The heterotrimeric G proteins that relay signals from GPCRs are associated with the underside of the plasma membrane and are composed of an α subunit and a βγ dimer. Agonist-activated GPCRs act as GEFs that catalyze the exchange of GDP bound to the α subunit for GTP, causing release of Gβγ (Lee and Yaffe 2014). A single ligand-bound GPCR can activate several G proteins, providing the first layer of signal amplification. The GTP-bound Gα subunits and Gβγ subunits can then promote the activation of a variety of downstream effectors, stimulating a network of signaling events that is highly dependent on the G-protein-coupling specificity of each receptor (Fig. 6). The human G-protein α subunits are encoded by 16 distinct genes and can be divided into four subfamilies: Gαs (Gαs and Gαolf), Gαi (Gαt, Gαgust, Gαi1-3, Gαo, and Gαz), Gαq (Gαq, Gα11, Gα14, and Gα15/16), and Gα12 (Gα12 and Gα13) (Fig. 6) (Cabrera-Vera et al. 2003). A single GPCR can couple to either one or more than one family of Gα subunits. Five different β subunits and 12 γ subunits that form functional βγ dimers have been described.
Figure 6.
Regulation of classical second messenger systems and Ras and Rho GTPases by GPCRs. Agonist-activated GPCRs promote the dissociation of GDP bound to the α subunit pf heterotrimeric G proteins and its replacement by GTP. Gα and Gβγ subunits can then activate numerous downstream effectors. The 16 human G protein α subunits can be divided into the four subfamilies shown, and a single GPCR can couple to one or more families of Gα subunits. Downstream effectors regulated by their targets include a variety of second messenger systems, as well as members of the Ras and Rho families of small GTP-binding proteins, which, in turn, control the activity of multiple MAPKs, including ERK, JNK, p38, and ERK5. G-protein-dependent activation of these by GPCRs and β-arrestin-mediated G-protein-independent activation of ERK and JNK can have multiple effects in the cytosol. MAPKs also translocate to the nucleus, where they regulate gene expression. Activation of the PI3K–Akt and mTOR pathways plays a central role in the regulation of cell metabolism, migration, growth, and survival by GPCRs.
Studies of the structural perturbations caused by ligand binding to class A GPCRs reveal that agonist binding causes a contraction of a ligand-binding pocket located within the transmembrane α-helical regions. In particular, upon ligand binding, the transmembrane (TM) α helix 6 moves outward from the center of the transmembrane bundle, loses contact with TM3, and moves closer to TM5. The consequent conformational changes in the intracytoplasmic loops lead to the formation of a new pocket between TM3, TM5, and TM6, which binds to the carboxyl terminus of the Gα subunits, leading to G-protein activation by promoting the release of GDP and its exchange for GTP (Kobilka 2011). Ligands that bind and stabilize the receptors in conformations other than this fully activated state act as partial agonists, and they provoke a more limited G-protein activation and hence a restricted response. Ligands that stabilize the inactive conformation of the GPCR act as classical antagonists or inverse agonists. There is now a great interest in the development of novel drugs that act as allosteric modulators by binding at a site distinct from that to which the natural GPCR ligands bind. This new generation of pharmacological agents changes the receptor conformation, thereby modifying the affinity and/or efficacy of the endogenous ligands (May et al. 2007). Note that the nature of the agonist binding can impact receptor conformation, thus biasing the choice of G protein and hence the signaling output.
2.14. Regulation of Classical Second Messenger Systems by G Proteins and Their Linked GPCRs
Many of the immediate actions of GPCRs involve the rapid generation of second messengers (Fig. 6). Gαs stimulates adenylyl cyclases, which increases the cytosolic levels of cAMP, whereas Gαi inhibits adenylyl cyclases and hence decreases cAMP levels as a result of tonic phosphodiesterase activity. The different adenylyl cyclase isoforms appear to be distinctly regulated by Gαs and Gαi, as well as by Gβγ subunits, intracellular calcium, and PKCs (Taussig and Gilman 1995). Thus, the effects of different GPCR agonists on the intracellular levels of cAMP are highly dependent on the adenylyl cyclases expressed in a given cell type. Gαt and Gαgust (also known as transducin and gustducin, respectively) activate phosphodiesterases in the visual system and gustative papillae, respectively, thus decreasing the cytoplasmic levels of cGMP by converting it to GMP (Cabrera-Vera et al. 2003). Members of the Gαq family bind to and activate phospholipase Cβ, which cleaves phosphatidylinositol 4,5-bisphosphate (PIP2) into DAG and inositol 1,4,5-trisphosphate (IP3). The latter causes an increase in cytosolic calcium levels by promoting the release of calcium from intracellular stores and also subsequent calcium influx into the cells, whereas DAG activates PKC (Hubbard and Hepler 2006; Julius and Nathans 2012; Sassone-Corsi 2012; Newton et al. 2014).
The released Gβγ dimers can independently activate many signaling molecules, including PLCβ, adenylyl cyclases, and ion channels (particularly the GIRK family of potassium channels). Although, in principle, the activation of any G protein by GPCRs should result in the release of Gβγ dimers and hence activation of their downstream targets, Gi and Go are often the most highly expressed G proteins and represent the most frequent source of free Gβγ complexes.
The targets of diffusible second messengers activated by G proteins include a large number of ion channels, calcium-sensitive enzymes, and kinases such as PKA, PKC, cGMP-dependent kinase (PKG), and calcium-/calmodulin-dependent kinases (CaMKs), which are stimulated by cAMP, calcium/DAG, cGMP, and calcium, respectively. Heterotrimeric G proteins can also regulate other effectors, including signaling molecules that activate kinase cascades and small GTPases.
2.15. GPCRs Regulate a Network of Ras- and Rho-Related GTPases, MAPK Cascades, and PI3K-Regulated Signaling Circuits
GPCRs stimulate pathways that control cell migration, survival, and growth in part by activating MAPKs, a group of highly related proline-targeted serine/threonine kinases that link multiple cell-surface receptors to transcription factors. MAPKs include ERK1/2, JNK1-3, p38α-δ, and ERK5 MAPKs (Gutkind 1998; Morrison 2012).
The small GTPase Ras, tyrosine kinases, PI3Ks, PKCs, and arrestins can act downstream from GPCRs to promote the activation of ERK1/2 in a cell-specific fashion (for review, see Gutkind 2000). The JNK cascade is activated downstream from the small G proteins Rac, Rho, and Cdc42 (Coso et al. 1995). Indeed, Rac and Cdc42 can mediate signaling of Gβγ dimers and Gα12, Gα13, Gαq, and Gαi to JNK (Gutkind 2000; Yamauchi et al. 2000). Many GPCRs coupled to Gi activate Rac and JNK through the direct interaction of Gβγ subunits with the P-REX1/2 family of Rac-GEFs (Welch et al. 2002; Rosenfeldt et al. 2004). Similarly, Gαq activates Rho GTPases, and hence JNK, through direct interaction with p63-RhoGEF and Trio (Lutz et al. 2007). Gα12 and Gα13 bind to and act on three GEFs—p115-RhoGEF, PDZ-RhoGEF, and LARG—to promote the activation of Rho downstream from GPCRs (Hart et al. 1998; Fukuhara et al. 2001). Additional GEFs can also contribute to this network. How GPCRs activate p38 and ERK5 is much less clear, but, in general, these MAPKs are activated primarily by Gαq, Gα12/13, and Gβγ dimers (Gutkind 2000).
Although human cancer-associated viruses express constitutively active viral GPCRs, emerging data from deep sequencing studies have revealed that a large fraction of human malignancies harbour mutations in GPCRs and G-protein α subunits (O’Hayre et al. 2013). This has increased the interest in the molecular mechanisms by which G proteins and GPCRs control normal and cancer cell growth. Recent findings suggest that although GPCRs can stimulate multiple diffusible-second-messenger-generating systems, their ability to promote normal and aberrant cell proliferation often relies on the persistent activation of Rho GTPases and MAPK cascades based on the direct interaction of Gα subunits with RhoGEFs. The MAPKs regulate the activity of nuclear transcription factors and coactivators, such as Jun, Fos, and YAP (Yu et al. 2012; Vaqué et al. 2013).
Activation of the PI3K–Akt and mTOR pathways plays a central role in cell metabolism, migration, growth, and survival (Zoncu et al. 2011). PI3K generates 3′-phosphorylated inositol phosphates that participate in activation of the kinase Akt and mTOR, which relay downstream signals (Hemmings and Restuccia 2012; Laplante and Sabatini 2012). PI3Kγ shows restricted tissue distribution and is activated specifically by GPCRs by the direct interaction of its catalytic (p110γ) and regulatory (p101) subunits with Gβγ subunits (Lopez-Ilasaca et al. 1997). PI3Kγ is involved in the chemokine-induced migration of leukocytes and plays significant roles in innate immunity (Costa et al. 2011). In cells lacking PI3Kγ expression, GPCRs can use PI3Kβ to stimulate phosphatidylinositol-(3,4,5)-tris-phosphate (PIP3) synthesis (Ciraolo et al. 2008).
2.16. GPCR-Interacting Proteins in Receptor Compartmentalization, Trafficking, and G-Protein-Independent Signaling
GPCRs interact with a diverse array of proteins, which regulate compartmentalization to plasma membrane microdomains, endocytosis, trafficking between intracellular compartments and the plasma membrane, and G-protein-independent signaling (see below). These include receptor-activity-modifying proteins (RAMPs), GPCR-associated sorting proteins (GASPs), Homer, β-arrestins, arrestin-domain-containing proteins (ARRDCs), and DEP-domain proteins (Ballon et al. 2006; Magalhaes et al. 2012).
RAMPs are single-transmembrane-span proteins that associate with some class C GPCRs, such as the calcitonin receptor and calcitonin-like receptor. RAMPs facilitate the glycosylation of calcitonin family receptors in the endoplasmic reticulum, thereby facilitating their expression at the cell surface, and remain associated at the plasma membrane, where RAMPs contribute to ligand binding and receptor signaling (Bouschet et al. 2005). GASPs interact with the carboxy-terminal tail of many GPCRs and are primarily involved in their postendocytic sorting (Whistler et al. 2002). Homer 1a-c, Homer 2, and Homer 3 represent a class of proteins that harbor Enabled/VASP homology (EVH)–like domains and bind to metabotropic glutamate receptors (mGluRs) through a carboxy-terminal polyproline sequence (PPXXFP) (Bockaert and Pin 1999).
GPCRs can also associate with molecules containing protein–protein interaction domains, such as DEP, PDZ, WW, SH2, and SH3 domains (Lee and Yaffe 2014), as well as polyproline-containing regions (Bockaert and Pin 1999; Brzostowski and Kimmel 2001). These interactions facilitate the localization of GPCR-initiated signaling to specific cellular structures or membrane microdomains, including the neuronal synapse, and also determine the signaling output by favoring the activation of a subset of GPCR targets by increasing their local accumulation in the vicinity of the GPCR.
β-Arrestins were initially described as adaptor proteins promoting the endocytosis of activated GPCRs (see below) but are now believed to scaffold a wide variety of signaling complexes (Luttrell and Gesty-Palmer 2010; Rajagopal et al. 2010). In particular (Andreeva et al. 2007), they can interact with Src family kinases as well as multiple serine/threonine kinases, small GTPases and their GEFs, E3 ubiquitin ligases, phosphodiesterases, and transcription factors (Luttrell and Gesty-Palmer 2010; Rajagopal et al. 2010). β-Arrestins can act downstream from GPCRs within endocytic vesicles in a pathway leading to the activation of ERK1/2 and JNK, particularly in response to β-arrestin-biased agonists for some GPCRs, thus initiating intracellular signaling independently of the activation of heterotrimeric G proteins (Rajagopal et al. 2010). Interestingly, β-arrestins can form multimeric signaling complexes with ERK1/2 and JNK that are retained in the cytosol, thus restricting the nuclear translocation of these MAPKs, which instead act on cytosolic substrates (Fig. 6) (Rajagopal et al. 2010). Besides the best-studied β-arrestins, a family of α-arrestins that are conserved from budding yeast to humans has recently received increased attention because of their potential role in GPCR trafficking and degradation (Nabhan et al. 2010).
2.17. GPCR-Independent Activation of G Proteins
Heterotrimeric G proteins can be also activated in a GPCR-independent fashion by a family of proteins known as activators of G-protein-mediated signaling (AGS proteins) (Blumer et al. 2007). These proteins substitute for GPCRs by promoting nucleotide exchange on Gα subunits (e.g., AGS1), or can regulate the physical interaction and localization of G-protein subunits without affecting nucleotide exchange (e.g., AGS3, also known as LNG or PINS proteins) (Blumer et al. 2007). The latter play an important conserved role in cell polarity and polarized cell division and share the presence of a GoLoco motif by which they bind to Gα subunits to prevent nucleotide exchange (Willard et al. 2004).
2.18. GPCR Signal Termination
Considerable attention has focused on mechanisms of termination of GPCR signaling, because persistent activation occurs in many diseases (Pierce et al. 2002). This desensitization is highly regulated and occurs through several well-understood mechanisms, including GPCR-targeted kinases known as GPCR kinases (GRKs), and more general second-messenger-regulated kinases, such as PKC and PKA. PKC and PKA phosphorylation uncouples receptors from their respective G proteins, presumably by phosphorylating G-protein-interaction sites or by recruiting arrestins (Benovic et al. 1985), thereby forming a negative-feedback loop. Activation of PKA and PKC can also result in the heterologous desensitization of multiple GPCRs within a cell (Chuang et al. 1996). In contrast, GRKs phosphorylate only the activated or agonist-occupied form of the receptor, primarily in the carboxy-terminal tail, which then binds arrestin dimers that can prevent G-protein interaction and promote the removal of the receptor from the cell surface by endocytosis (Shenoy and Lefkowitz 2011). Internalized receptors can be recycled back to the plasma membrane or degraded in lysosomes, a process influenced by the ability of ligand-bound receptors to interact with ubiquitin ligases and a complex repertoire of sorting molecules (Hanyaloglu and von Zastrow 2008). Concomitantly, a family of regulators of GPCR signaling (RGS) molecules act as GAPs on GTP-bound G-protein α subunits, accelerating GTP hydrolysis and hence signal termination (Berman and Gilman 1998). Signaling and inactivation are intertwined, because molecules such as arrestins can also regulate GPCR signaling specificity and/or localization (Shenoy and Lefkowitz 2011), and many G-protein targets include RGS domains or act as GAPs, thus acting as direct effectors that concomitantly limit the duration of G-protein signaling.
2.19. GPCR Signal Integration
GPCRs are best known for their ability to control the activity of adenylyl cyclases, phosphodiesterases, phospholipases, ion channels, and ion transporters. The rapid regulation of these classical diffusible-second-messenger-generating systems and their direct molecular targets is now believed to represent a subset of the extensive repertoire of molecular mechanisms deployed by GPCRs in physiological and pathological contexts. Our recently gained understanding of GPCR signaling circuitries, including GEFs, Ras and Rho GTPases, MAPKs, PI3Ks, and their numerous downstream cytosolic and nuclear targets, provide a more global view of the general systems by which these receptors exert their numerous physiological and pathological roles. Indeed, the final biological outcome of GPCR activation most likely results from the integration of the network of GPCR-initiated biochemical responses in each cellular context. A new, systems-level understanding may provide a molecular framework for the development of novel approaches for therapeutic intervention in some of the most prevalent human diseases.
3. THE TNF RECEPTOR FAMILY
3.1. TNF Isoforms and TNF Receptors
Tumor necrosis factor (TNF) belongs to a 19-member family of structurally related factors that bind to the 29 members of the TNF receptor (TNFR) family. Most TNF and TNFR members are expressed in the immune system, where they regulate defense against infection (see Newton and Dixit 2012); however, some members are expressed elsewhere, regulating hematopoiesis and morphogenesis. Perturbation of signaling by TNFRs is implicated in several diseases, including tumorigenesis, bone resorption, rheumatoid arthritis, and diabetes (Aggarwal 2003; Croft 2009).
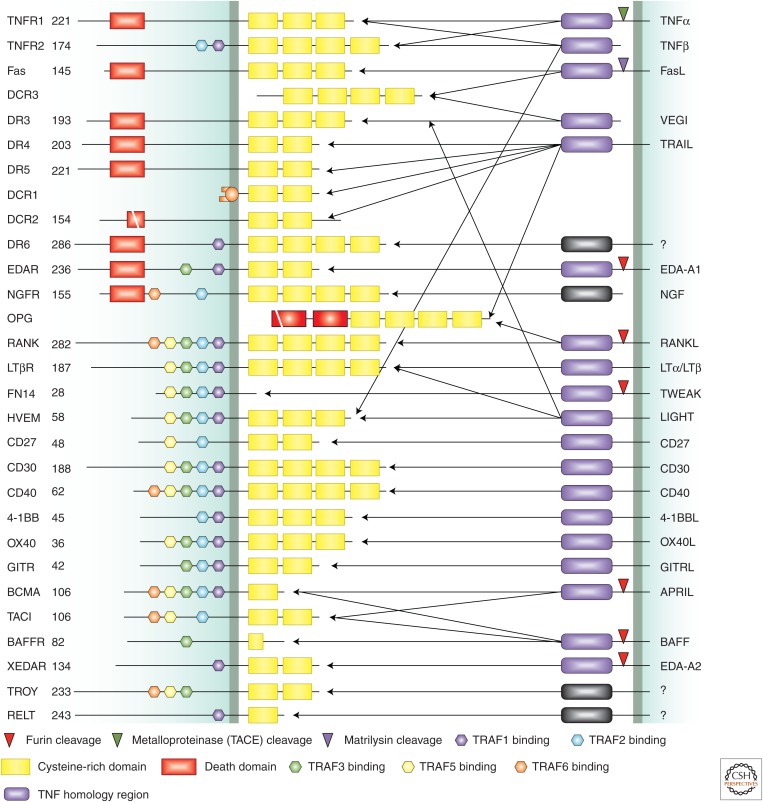
Members of the TNFR family have an amino-terminal extracellular region consisting of one to four cysteine-rich domains, each of which contains three conserved intrachain disulfide bridges. These are linked to a single transmembrane segment and a cytoplasmic region that lacks enzymatic activity (Locksley et al. 2001). Several receptors contain binding motifs for members of the TRAF family of ubiquitin ligases in their intracellular regions, and eight of them contain death domains (DDs), which are involved in apoptotic signaling (Fig. 7). Some instead lack intracellular and/or transmembrane regions and act as decoy receptors.
Figure 7.
The tumor necrosis factor (TNF) receptor family. The structural features of the members of the TNF receptor family (left) and their ligands (right) are shown. Cysteine-rich domains, death domains, and interaction motifs for various members of the TRAF family are indicated. Cleavage sites for various proteases involved in processing of the ligands are also shown. (From Aggarwal 2003; adapted, with permission.)
The TNF family ligands are also transmembrane proteins but have extracellular carboxyl termini. Intriguingly, these can be shed following cleavage by proteases, and some inhibit the effects of the membrane-bound ligand (Suda et al. 1997). The ligands are characterized by a conserved TNF homology domain that mediates receptor binding. An exception is nerve growth factor (NGF), which in addition to binding to an RTK also binds to p75, a member of the TNFR family. Several ligands of this family bind to more than one receptor (Fig. 7).
3.2. Activation of TNFRs
Most TNF family ligands are noncovalent trimers that form symmetric 3:3 complexes with their receptors. Homomeric as well as heteromeric receptor complexes have been described (Schneider et al. 1997). Preformed receptor oligomers exist and TNFR1 and TNFR2 have a pre-ligand-binding assembly domain (PLAD) that is required for the assembly of TNFR complexes (Chan et al. 2000). Whereas juxtaposition of the receptors is important for activation, trimerization itself appears not to be necessary because there are examples of agonistic bivalent monoclonal antibodies directed against the extracellular domain, and because one of the ligands, NGF, is a dimer.
3.3. Signaling via TNFRs
The receptors that have DDs, including TNFR1, Fas (also known as CD95), death receptor (DR) 3, DR4, DR5, DR6, EDAR, and the p75 NGF receptor, induce apoptosis and necrosis of cells (Moquin and Chan 2010; Newton and Dixit 2012; Green and Llambi 2014). Ligand-induced receptor activation leads to the formation of complexes referred to as death-inducing signaling complexes (DISCs), in which the receptor DDs interact with DDs in adaptor molecules such as TRADD and FADD. The protease procaspase 8 is also recruited to the DISC, triggering a caspase cascade that results in apoptosis.
In addition to apoptosis, TNFRs control survival and differentiation. Many of their effects are mediated by the TRAF family of ubiquitin ligases, which stimulate NF-κB, PI3K, and JNK and p38 MAPK pathways (Faustman and Davis 2010). Activation of NF-κB is of central importance and occurs downstream from all members of the TNFR family, except the decoy receptors. Moreover, certain members of the TNFR family stimulate cell proliferation, for example, by activation of the ERK1/2 MAPK.
3.4. TNFRs and Disease
Overactivity of members of the TNFR family is seen in immune-related diseases, and encouraging results have been obtained using inhibitory anti-TNF antibodies or ligand traps in the treatment of Crohn’s disease (Suenaert et al. 2002) and rheumatoid arthritis (Feldmann and Maini 2001). Similarly, blocking signaling via the receptors OX40, 4-1BB, CD27, and DR3 is effective in various immune diseases (Croft 2009). Promising attempts have also been made to induce apoptosis of cancer cells by treatment with agonists of TNFR1, Fas, and TRAIL receptors (Fox et al. 2010).
4. CELL ADHESION RECEPTORS AND MECHANOTRANSDUCERS
4.1. Mechanosensing Signaling through Integrins
Cells deploy multiple signaling mechanisms to sense the biophysical properties of their surroundings and communicate with their neighboring cells. Integrins are perhaps the best known cell-surface receptors involved in these essential processes. They function primarily in cell adhesion to the extracellular matrix (ECM) or to other cells, the latter through a repertoire of cell-surface ligands involved in cell–cell interactions. Integrins accumulate at cell–ECM and cell–cell contact points and orchestrate the local assembly of multimolecular structures known as adhesion complexes, often referred to as focal adhesions (FAs) or adhesomes (Hynes 2002; Geiger and Yamada 2011).
Integrins provide a link between the extracellular environment and the intracellular cytoskeleton, and their dynamic engagement at cell–ECM and cell–cell adhesions results in the rapid activation of multiple intracellular signaling circuits, a process known as outside-in signaling (Hynes 2002; Miranti and Brugge 2002). In turn, the adhesive properties of integrins and hence the strength of the interactions with the ECM and other cells are regulated by a variety of signaling pathways, which impinge on the interaction of integrins with a key cytoskeletal molecule, talin (see below); this process is known as inside-out signaling (Hynes 2002). Integrins can be regulated in this way by signals from growth factors acting on RTKs and GPCRs, as well as by inflammatory cytokines and bioactive lipids (Hynes 2002; Miranti and Brugge 2002). The latter is of particular importance for immune cells and platelets, in which integrins are maintained in an inactive state that enables cells to circulate in the bloodstream without interacting with endothelial cells in the blood vessel wall, but quickly deploy their adhesive properties in response to immune cell activation and the coagulation cascade (Moser et al. 2009). For example, cytokine-induced activation of the leukocyte integrin LFA1 (also known as αLβ2) leads to its rapid binding to intercellular adhesion molecule (ICAM)-1, an adhesion molecule expressed on the surface of activated endothelial cells, thereby stabilizing cell–cell interactions and facilitating the transmigration of circulating leukocytes through the endothelial cell layer into damaged tissues (Moser et al. 2009). The integrins also provide important survival signals, because many cells undergo anoikis, a form of programmed cell death, upon detaching from their surrounding ECM (Frisch and Screaton 2001). Overall, integrin-mediated cell adhesion controls cell migration, survival, growth, and differentiation, whereas at the organismal level, integrins play fundamental roles in tissue morphogenesis during development, and in the immune response, hemostasis, and tissue maintenance and repair (Hynes 2002; Miranti and Brugge 2002; Devreotes and Horwitz 2013).
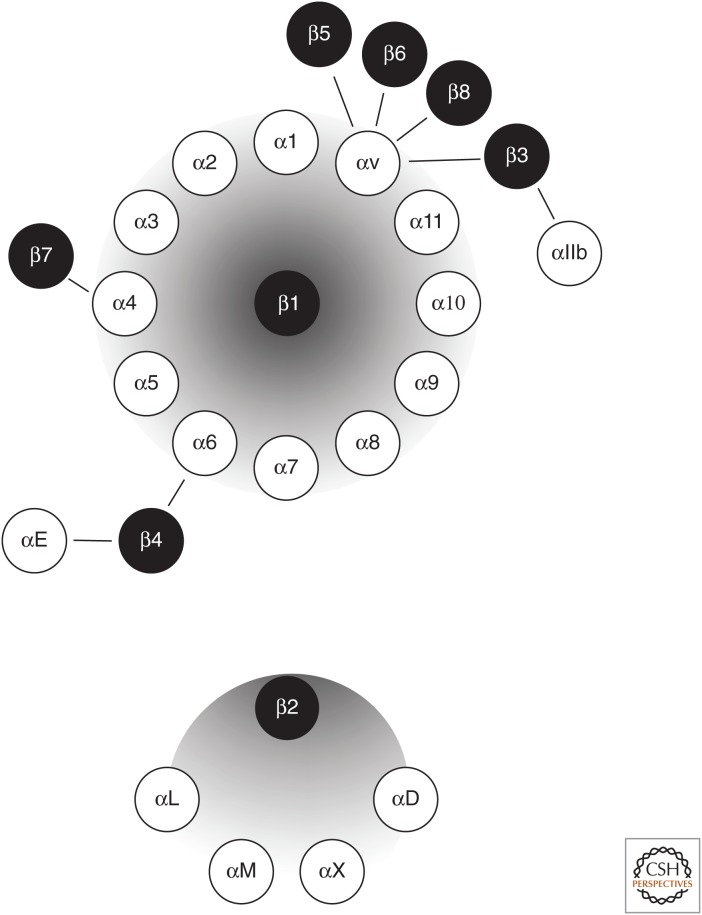
Each integrin is composed of a heterodimer of two transmembrane subunits (α and β). In humans, there are 18 α subunits (Fig. 8), which associate with eight different β subunits to form at least 24 heterodimeric complexes displaying distinct ligand-binding specificities and signaling capacities. Some of these dimers are widely expressed, whereas others show more restricted distributions and can therefore have more specific biological functions. Integrins typically have large extracellular regions, which interact with cell-surface ligands and with specific sequence motifs in ECM proteins, such as the tripeptide RGD motif originally described in fibronectin (Geiger et al. 2001; Hynes 2002). Some integrins are promiscuous and can bind to multiple ligands, for example, the ECM components vitronectin, fibrinogen, fibronectin, laminin, collagen, and tenascin. Other integrins show a more restricted binding pattern or bind to other cell adhesion receptors—for example, ICAMs, vascular cell adhesion molecule (VCAM), and E-cadherin (see Table 1). Most integrins possess a relatively short intracellular cytoplasmic domain (40–70 amino acids), with the exception of integrin β4, which has a long cytoplasmic domain.
Figure 8.
The integrin family of cell adhesion receptors. Integrins are composed of a heterodimer of two transmembrane α and β subunits. The 18 α subunits and eight β subunits can form at least 24 heterodimeric complexes displaying distinct binding specificity and signaling capacity. (Adapted from Hynes 2002.)
Table 1.
Ligands for α–β integrin heterodimers
| Integrin | Representative ligands |
|---|---|
| α1β1 | Collagen, laminin |
| α2β1 | Collagen, laminin |
| α3β1 | Laminin, fibronectin |
| α4β1 | VCAM-1, fibronectin, thrombospondin |
| α4β7 | VCAM-1, fibronectin, MAdCAM-1 |
| α5β1 | Fibronectin, neural adhesion molecule L1 |
| α6β1 | Laminin |
| α6β4 | Laminin |
| α7β1 | Laminin |
| α8β1 | Osteopontin, fibronectin, vitronectin, tenascin |
| α9β1 | Tenascin, osteopontin, VCAM-1 |
| α10β1 | Collagen |
| α11β1 | Collagen |
| αEβ1 | E-cadherin |
| αLβ2 | ICAM-1, ICAM-2, ICAM-3 |
| αMβ2 | ICAM-1, ICAM-2, ICAM-4, fibrinogen |
| αXβ2 | ICAM-1, fibrinogen |
| αDβ2 | ICAM-3, VCAM-1 |
| αIIbβ3 | Fibrinogen, fibronectin, vitronectin, thrombospondin, von Willebrand factor |
| αVβ3 | Vitronectin, fibronectin, fibrinogen, osteopontin |
| αVβ5 | Vitronectin |
| αVβ6 | Vitronectin, fibronectin |
| αVβ8 | Fibronectin, laminin, collagen, vitronectin |
| α6β4 | Laminin |
| α4β7 | VCAM-1, fibronectin |
The conformation of the integrin extracellular domains changes dramatically upon cell–ECM and cell–cell adhesions (Shattil et al. 2010). This results in the separation of the intracellular cytoplasmic tails, which lack enzymatic activity but instead act as platforms for the assembly of multimeric protein complexes that are involved in signal transduction (Shattil et al. 2010; Wehrle-Haller 2012) and link integrins to the cytoskeleton. The direct binding of talin to the cytoplasmic tails of most activated integrin β subunits is one of the earliest events after integrins bind to the ECM. This causes the local accumulation of PIP2 upon recruitment of the lipid kinase phosphatidylinositol 4-phosphate 5-kinase via its interaction with talin and the subsequent recruitment of vinculin to the nascent adhesions (Legate and Fassler 2009; Moser et al. 2009; Shattil et al. 2010). This helps stabilize FAs, because integrin β subunits interact weakly with actin through talin in the absence of vinculin. Vinculin and talin also bind to α-actinin, which has a high affinity for actin and hence helps strengthen the interactions between β integrins and actin filaments. Other molecules linking the β subunits to actin include filamin, kindlins, migfilin, and integrin-linked kinase (ILK), a pseudokinase that bridges β integrins and parvin, another actin-binding protein (Hynes 2002; Legate et al. 2009; Geiger and Yamada 2011). ILK stabilizes the FA by retaining integrins in a clustered state and by reinforcing the connection with the cytoskeleton, whereas kindlins partner with talins in integrin inside-out signaling (Legate and Fassler 2009; Moser et al. 2009). Tensin is recruited to FAs and helps to establish additional connections between integrins and actin fibers. Ultimately, these multiple links between integrins and actin promote localized actin polymerization at sites of cell–ECM and cell–cell contact. This allows rapid assembly and disassembly of adhesion contacts and dynamic control of the cellular cytoskeleton during cell migration (Huttenlocher and Horwitz 2011; Devreotes and Horwitz 2013).
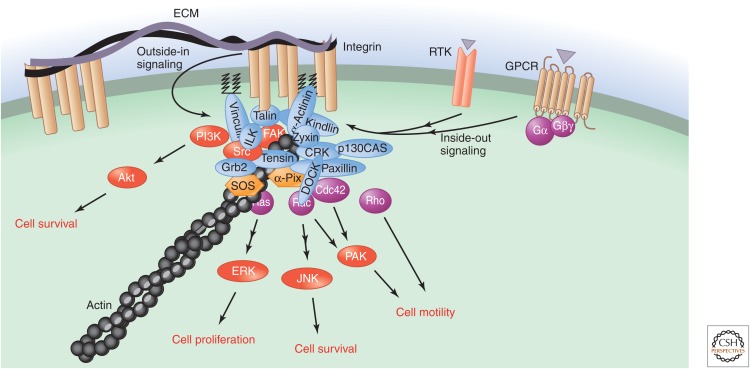
FAs (Geiger and Yamada 2011) contain many cytoskeletal, adaptor, and signaling proteins. Among them, multiple nonreceptor tyrosine kinases, including focal adhesion kinase (FAK), the related proline-rich tyrosine kinase 2 (Pyk2), Src, and Src-family kinases act both as signal transducers and as scaffolds (Fig. 9) (Miranti and Brugge 2002; Geiger and Yamada 2011). Activation of these tyrosine kinases upon integrin ligation results in their autophosphorylation and cross-phosphorylation at several tyrosine residues, creating binding sites for multiple signaling proteins. These include the adaptor protein Grb2, which leads to recruitment of SOS (a GEF for Ras) and the consequent activation of Ras and the MAPK ERK1/2. In the case of FAK, the phosphotyrosines recruit Src, Grb2, and the p85 subunit of PI3K via their SH2 domains (Miranti and Brugge 2002; Legate et al. 2009; Shattil et al. 2010). The latter generates PIP3 at the plasma membrane, which recruits ILK, Akt, and other proteins to the FA complex (Miranti and Brugge 2002; Shattil et al. 2010; Lee and Yaffe 2014). Akt relays prosurvival and growth-promoting signals. A direct substrate of FAK, paxillin, acts as a multidomain adaptor protein that forms a scaffold organizing a variety of signaling molecules, including Src, ILK, Crk, and vinculin (Miranti and Brugge 2002; Geiger and Yamada 2011). Crk is an adaptor protein that binds to tyrosine-phosphorylated paxillin or another adaptor protein, p130Cas, and can then use its SH3 domain to recruit multiple additional proteins. These include the GEF DOCK180, which activates the small GTPase Rac1 (Miranti and Brugge 2002; Legate et al. 2009). The Rac/Cdc42-GEF α-Pix is also activated upon integrin ligation by binding to phosphorylated paxillin and by the local accumulation of PIP3. Moreover, Src or FAK can phosphorylate and activate Vav, a Rac GEF that is recruited to the newly formed adhesive structures (Miranti and Brugge 2002; Legate et al. 2009). This results in the rapid remodeling of the actin cytoskeleton by Rho GTPases and their downstream effectors and the relay of the signals to the nucleus by JNK and p38 (Miyamoto et al. 1995; Miranti and Brugge 2002; Morrison 2012).
Figure 9.
Integrin-based cell adhesion and signaling. Integrin engagement at cell-matrix adhesions or interaction with a repertoire of cell-surface ligands results in the rapid assembly of a multifunctional protein network (Geiger and Yamada 2011) containing many cytoskeletal, adaptor, and signaling proteins. This contributes to cell adhesion and activates multiple signaling events. The adhesive properties of integrins are, in turn, regulated by a variety of signaling pathways; this is known as inside-out signaling.
The assembled protein network thus both contributes to adhesion to the ECM and other surrounding cells and orchestrates signaling events that enable the cells to respond appropriately to mechanical cues. In addition to outside-in and inside-out signaling, integrin activation can also induce the rapid clustering of multiple RTKs, such as EGFR, PDGFR, and FGFR (Miyamoto et al. 1996; Geiger and Yamada 2011). This enhances signaling in response to cell adhesion. Finally, note that rather than being rigid intracellular structures, most FAs are dynamic (Devreotes and Horwitz 2013), and regulation of their multiple components and adhesive properties by mitogens and chemoattractants, for example, is essential for the rapid dissolution of preexisting adhesions and the establishment of new contact sites during cell migration.
4.2. Signaling by Cell Adhesion Molecules (CAMs)
The formation of adhesive structures between adjacent cells, including adherens junctions and tight junctions, contributes to the establishment of cell polarity, differentiation, and survival and consequently key morphogenetic processes involved in embryonic development, control of tissue homeostasis, and tissue repair in adults. A large family of cell adhesion molecules (CAMs) provides mechanical adhesion among cells, while initiating signaling via networks that control cellular behavior in response to the microenvironment. In general, cadherin molecules mediate cell–cell adhesion at adherens junctions, claudins contribute to the formation of tight junctions, and immunoglobulin-like CAMs (Ig-CAMs) accumulate throughout the intercellular boundary (Cavallaro and Dejana 2011). Other CAMs, known as nectins, can participate in both adherens and tight junctions (Takai et al. 2008).
The cadherins are calcium-dependent, homophilic, cell–cell adhesion molecules expressed in nearly all cells in solid tissues. These molecules participate in cell–cell recognition, and only cells expressing the same type of cadherins may adhere to each other (Nose et al. 1988). The “classical” cadherins were originally named based on the tissue in which they are most prominently expressed, for example, E-cadherin in epithelial cells, VE-cadherin in vascular endothelial cells, and N-cadherin in nervous system and mesenchymal cells (Gumbiner 2005). These cadherins are single-pass transmembrane proteins that form a core adhesion complex in which a cadherin dimer binds through its extracellular region to another cadherin dimer on an adjacent cell in a calcium-dependent manner. The cadherin intracellular region is anchored in the plasma membrane and linked to the cytoskeleton through a family of proteins known as catenins (Gumbiner 2005). β-Catenin interacts with the distal part of the cadherin cytoplasmic tail, and p120 catenin interacts with a more proximal region (Gumbiner 2005). α-Catenin binds primarily to cadherin-associated β-catenin and provides a physical link to the actin cytoskeleton, by binding actin filaments either directly or indirectly through other actin-binding proteins, such as vinculin, α-actinin, and formins (Kobielak and Fuchs 2004; Mege et al. 2006). p120 regulates cell movement through its ability to control both cell adhesion by governing the availability of cadherins at the plasma membrane and actin cytoskeleton organization by regulating Rho GTPases (Anastasiadis and Reynolds 2001; Grosheva et al. 2001; Yanagisawa and Anastasiadis 2006). p120 can also directly affect gene expression by repressing transcription by scaffolding a nuclear complex including the gene silencer Kaiso (Daniel and Reynolds 1999).
The formation of cadherin-dependent adhesions contributes to growth inhibition upon cell–cell contact. This has often been associated with inhibition of the canonical Wnt pathway by cadherins, which retain β-catenin at the plasma membrane and therefore limit the pool of free β-catenin available for nuclear signaling (Nelson and Nusse 2004). Recent evidence suggests that cadherin engagement can also activate the Hippo pathway, which results in the cytoplasmic retention of the transcriptional coactivator YAP that is necessary for cell growth (Kim et al. 2011). In epithelial cells, E-cadherin acts as a tumor and metastasis suppressor. Its expression and function are down-regulated or altered in many human cancers, and its reexpression decreases both the proliferative and invasive capacity of tumor cells (Vleminckx et al. 1991; Thiery and Sleeman 2006). However, the engagement of cadherins in newly formed cell contacts promotes cell proliferation and survival through the activation of MAPKs, PI3K, and Rho GTPases (Pece et al. 1999; Pece and Gutkind 2000; Braga and Yap 2005). This process often involves the engagement of growth factor receptors such as EGFR, VEGFR2, FGFR, and PDGFR, promoting their ligand-independent clustering and activation and prolonging the activation by ligands by enhancing receptor recycling and limiting their degradation (Carmeliet et al. 1999; Pece and Gutkind 2000; Suyama et al. 2002; Cavallaro and Dejana 2011).
Cadherins can also control multiple cellular processes by associating with Src-family kinases, G proteins of the Gα12/13 family, and phosphatases, such as density-enhanced phosphatase (RPTPη), the tyrosine phosphatase SHP2, and vascular endothelial protein tyrosine phosphatase (VE-PTP) (Cavallaro and Dejana 2011). Cadherins thus contribute to cell–cell recognition and adhesion while promoting cell survival and restricting cell motility/growth by regulating intracellular signaling at the plasma membrane. In some cases, the cadherin intracytoplasmic tail can translocate to the nucleus and regulate transcription after shedding of the ectodomain by cell-surface matrix and disintegrin family proteases and cleavage of the carboxy-terminal tail by intracellular proteases, such as γ-secretase (Cavallaro and Dejana 2011), which is reminiscent of Notch signaling.
Tight junctions involve numerous adhesion molecules, including occludin, junctional adhesion molecules (JAMs), and the claudin family of tetraspan transmembrane proteins, as well as intracellular adaptors, such as ZO1 and ZO2 (Tsukita et al. 2001). Claudins are the major adhesive proteins at tight junctions; whether they play a direct role in cell signaling is not clear yet.
Ig-CAMs are cell-surface glycoproteins that accumulate at the cell–cell boundary, and their homophilic interactions contribute to cell–cell recognition and adhesion in a calcium-independent fashion (Loers and Schachner 2007; Cavallaro and Dejana 2011). Although most Ig-CAMs have a transmembrane region and a cytoplasmic tail, some associate with the cell surface via a GPI anchor. In the former case, the cytoplasmic tail of Ig-CAMs can interact with cytoskeletal proteins such as actin, ankyrins, and spectrin and can also initiate signal transduction (Cavallaro and Dejana 2011). For example, the formation of NCAM-based adhesions results in the activation of a kinase cascade including CaMKIIα, the Src-family kinase Fyn, and FAK, and this promotes neurite outgrowth and neuronal survival (Bodrikov et al. 2008). NCAM also stimulates PKCβII by recruiting it to the membrane through the formation of a signalling complex involving a protein known as growth-associated protein 43 (GAP43) (Korshunova and Mosevitsky 2010). In common with cadherins, NCAM can interact with multiple growth factor receptors, including FGFR, regulating their signaling capacity.
Signaling by CAMs—either direct or indirect actions engaging and prolonging the activity of RTKs—is likely to play a key role during the formation of cell–cell contacts, particularly during embryonic development, morphogenesis, and tissue repair (Pece and Gutkind 2000; Dumstrei et al. 2002; Andl et al. 2003; Fedor-Chaiken et al. 2003). This may accelerate the growth rate without the need for elevated local levels of growth factors. The stabilization of cell–cell contacts may subsequently reduce signaling by ligand-activated RTKs by sequestering them in CAM-containing clusters, while favoring their ligand-independent activation of survival pathways, such as PI3K–Akt signaling (Pece and Gutkind 2000; Qian et al. 2004). Concomitantly, CAMs may restrict cell and organ overgrowth by limiting the availability of free β-catenin and YAP (Nelson and Nusse 2004; Kim et al. 2011). In these cases, CAMs may help prevent tumor formation while generating survival and differentiation signals. The microenvironment and RTK signaling networks can, in turn, modulate the localization, expression, and stability of CAMs and the proteins that they associate with, thus regulating their adhesive properties and signaling capacity. Conversely, CAMs can regulate RTK signaling, acting as rheostats governing the intensity and duration of their signals in response to environmental cues such as cell density, tissue architecture, and polarity.
5. NUCLEAR RECEPTORS
Nuclear receptors (NRs) comprise a large superfamily of intracellular transcription factors that can effect gene expression changes in response to a wide variety of lipophilic ligands (Sever and Glass 2013). In this respect, they differ from most other receptors in that they do not reside in the plasma membrane, which many of their ligands can cross. Typical NR ligands include steroids, vitamins, dietary lipids, cholesterol metabolites, and xenobiotics. Ligands for 24 NRs have been identified; the remaining family members are considered orphan receptors (Mangelsdorf et al. 1995). There are 48 NRs in humans (49 in mice and 18 in Drosophila). NRs are believed to be only one of two transcription factor families that are metazoan specific (King-Jones et al. 2005; Degnan et al. 2009). Because many are endocrine hormone receptors, this suggests a potentially critical role in the evolution of animal physiology. Hormonal NRs are typically classified by the type of ligands to which they bind, whereas orphans have various different abbreviations, indicating, for example, similarity to known receptors (e.g., estrogen-related receptor, ERR). Binding of their cognate ligands, in the most simplistic scenario, either changes the cellular localization of the NR or its interaction with repressive and activating cofactors in the nucleus. What distinguishes NRs from other genres of receptors is the ability to mediate transcription without intermediate signaling cascades. Instead, they directly bind to target genes. Nuclear–cytoplasmic cycling of some NRs allows them to have nongenomic effects and also to be targets of cytoplasmic signaling cascades (Wehling 1997).
5.1. Structure and Mechanisms of Receptor Activation
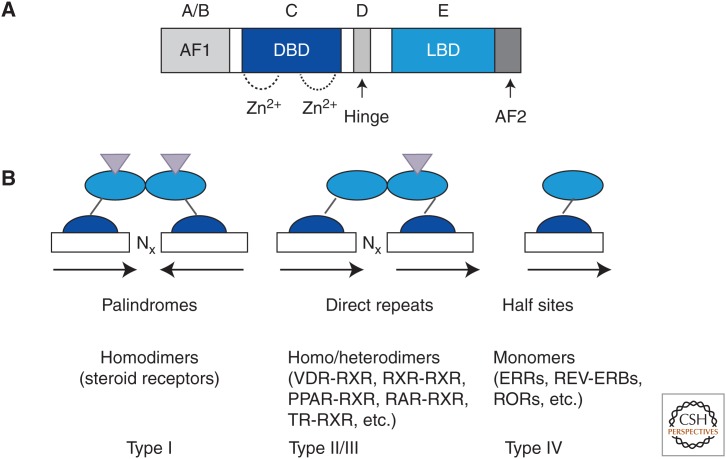
Nuclear receptors generally contain five functional domains. The A/B region at the amino terminus is divergent and in some NRs contains a ligand-independent transcriptional activation function domain (AF1). The highly conserved DNA-binding domain (DBD) is located in the central C domain. The carboxyl terminus is the E region, which contains the ligand-binding domain (LBD). A flexible hinge D region links the DBD and LBD (Fig. 10). The DBD contains tetracysteine (C4) zinc fingers that are unique to NRs and define membership in the superfamily. The DBD typically targets the NR to a specific DNA element termed a hormone-response element (HRE). The LBD is composed of 12 α helices and mediates ligand recognition, dimerization, interaction with coactivators/corepressors, and ligand-dependent transcriptional activation. The last helix, helix 12, within the LBD is the AF2 domain, which enables NRs to interact with short LxxLL motifs in coactivators or corepressors. These are termed the NR box or CoRNR box, respectively (Hu and Lazar 1999). Whereas AF1 domains vary greatly among all NRs and have a propensity to stay disordered, AF2 domains have similar structures (Warnmark et al. 2003).
Figure 10.
General structure and binding of nuclear receptors. (A) Domain organization of a typical nuclear receptor. (B) Three modes of signal transduction: as monomers, heterodimers, and homodimers. (From Sonoda et al. 2008: modified, with permission, © Elsevier.)
5.2. Nuclear Receptor Classification
NRs can be classified based on their DNA-binding mechanism (Fig. 10) or their ligand (Table 2). On the basis of their mode of actions, nuclear receptors are classified into four types. Type I and type III NRs are normally sequestered in the cytoplasm by heat shock proteins. Ligand binding to type I NRs dissociates heat shock proteins and results in nuclear enrichment. Type I NRs bind to DNA as homodimers, and the response elements they recognize are inverted repeats. Type II receptors, unlike type I receptors, are normally enriched in the nucleus and are bound to DNA even in the absence of ligand. They generally bind to DNA as heterodimers with the retinoid X receptor (RXR). In the absence of ligand, type II receptors repress transcription via association with corepressors. In the presence of ligand, corepressors are dissociated, and coactivators including histone-modifying enzymes are recruited by the type II receptors for gene activation. Examples of type II receptors include the thyroid hormone receptor (TR) and retinoic acid receptor (RAR). Type IV receptors bind as monomers and recognize half-site response elements.
Table 2.
The nuclear receptor superfamily
| Steroid receptors | |
| GR | Glucococorticoid |
| MR | Mineralocorticoid |
| AR | Androgens |
| PR | Progesterone |
| TRα, TRβ | Thyroid hormone |
| ERα, ERβ | Estrogen |
| Vitamin receptors | |
| VDR | Vitamin D |
| RARα, RARβ, RARγ | Retinoic acid |
| Fatty acids and derivatives | |
| PPARα, PPARβ/δ, PPARγ | Fatty acids |
| LXRα, LXRβ | Oxysterol |
| FXR | Bile acids |
| RXRα, RXRβ, RXRγ | Rexinoids |
| Xenobiotic receptors | |
| CAR | Androstane metabolites |
| PXR | Pregnane derivatives |
| Adopted orphan receptors | |
| HNF4α, HNF4γ | Fatty acids |
| REV-ERBα, REV-ERBβ | Heme |
| RORα, RORβ, RORγ | Cholesterol, retinoic acid |
| SF1, LRH1 | Phosphatidylinositols |
| ERRα, ERRβ, ERRγ | Estrogen |
| Orphan receptors | |
| SHP | |
| DAX1 | |
| TLX | |
| PNR | |
| GCNF | |
| TR2, 4 | |
| NR4A1, NR4A2, NR4A3 | |
| COUP-TFα, COUP-TFβ, COUP-TFγ |
5.3. Three Modes of Binding to Promoter Regions Influence Transcription
Hormone-responsive NRs are sequestered in the cytoplasm or are bound to HREs in repressive complexes that include chromatin modifiers such as histone deacetylases (HDACs), nuclear corepressors (N-CoRs), and SMRT proteins (for “silencing mediator of retinoid and thyroid receptors”) (Perissi et al. 2010). Ligand binding typically promotes nuclear translocation and/or recruitment of coactivators that displace corepressors to initiate transcription (Rosenfeld et al. 2006). Coactivators that mediate NR function include PGC1, SIRT1, p160, and p300 proteins (Sonoda et al. 2008). Mechanistically, NRs can mediate transcription (either repression or activation) in the following manner.
5.3.1. Homodimers
Type I and type III receptors bind to DNA as homodimers when bound to their cognate ligands. Type I receptors include all steroid receptors, and their response elements typically consist of two hexameric inverted (palindromic) repeat half-sites, for example, AGAACA (the glucocorticoid response element, GRE) or AGAACA (the estrogen response element, ERE). These sequences can mediate either activation or repression in response to hormone. The DBDs of steroid receptors are highly similar. Thus, the glucocorticoid receptor (GR), the mineralocorticoid receptor (MR), the androgen receptor (AR), and the progesterone receptor (PR) all bind to overlapping response elements. Type III receptors are similar to type I receptors except that they recognize direct repeats instead of inverted repeats.
5.3.2. Heterodimers
RXR forms heterodimers with various nonsteroidal members of the NR superfamily that do not bind to HREs efficiently by themselves. Depending on the type of NR, HREs have specific conformations. Unlike the type I receptors mentioned above, type II receptors form retinoid-acid-related-receptor (RXR)-containing heterodimers that recognize direct repeats of AGGTCA or similar sequences separated by specific spacing (Umesono et al. 1991). The RAR- and peroxisome proliferator-activated-receptor-γ (PPARγ)-containing dimers RAR–RXR and PPARγ-RXR bind to a direct repeat (DR-1), whereas the vitamin D3 receptor (VDR)-, TR-, and RAR-containing dimers VDR–RXR, TR–RXR, and RAR–RXR bind to DR-3, DR-4, and DR-5, respectively (Evans 2005).
5.3.3. Monomers
Type IV NRs bind to just one AGGTCA site as monomers in a ligand-independent manner. These receptors use an extended DBD. There are currently three known types of monomer-binding receptors, which are defined by unique sequences 5′ to the consensus site. REV-ERBs tend to bind to A(A/T)CTAGGTCA sequences, whereas ERRs bind to TNAAGGTCA sequences.
6. FUNCTIONS OF NUCLEAR RECEPTORS
6.1. Steroid and Thyroid Hormone Receptors
Steroid and thyroid receptors maintain homeostasis by acting as sensors for circulating hormones from the adrenals, ovaries, testes, and thyroid. GR, MR, AR, and PR are closely related and bind common DNA sequences. Following ligand treatment, these receptors translocate from the cytoplasm to the nucleus and bind to palindromes of the GR half-site AGAACA as homodimers. All other members of the superfamily, including ER and TR, bind to response elements that contain variations of the ER half-site AGGTCA.
6.2. Nonsteroidal Hormonal Receptors
Instead of acting as homodimers, all nonsteroidal hormone receptors, including TR, use RXR as a partner to form heterodimers. VDR and RAR are activated by calcitriol (vitamin D) and retinoic acid. RXR is conserved in invertebrates and may be an ancient member of the NR family. RXRs (α, β, and γ) are activated by the novel retinoid isomer 9-cis retinoic acid and other synthetic agents (rexinoids) (Mangelsdorf et al. 1992). RXR forms three different types of dimers. Other than the nonpermissive heterodimers mentioned above (e.g., RAR and VDR), which cannot be activated by the RXR ligand, RXR can form homodimers and permissive heterodimers (e.g., with PPAR and liver X receptor, LXR) that can be activated by RXR-ligand binding.
6.3. Receptors for Fatty Acids and Derivatives
The PPAR subfamily includes PPARα, PPARδ, and PPARγ. Each of the PPARs binds to common response elements as heterodimers with RXR, using fatty acid ligands, which function as metabolic sensors. PPARα is highly expressed in the liver and is the target of the fibrate class of drugs used to control hyperlipidemia (Issemann and Green 1990). PPARδ is universally expressed and promotes fatty-acid oxidation. PPARγ is required for adipogenesis, promotes fat storage, and has been widely studied for its role in the metabolic syndrome (Barak et al. 1999). LXRα and LXRβ and farnesoid X receptor (FXR) are sensors for cholesterol metabolites such as oxysterols and bile acids, respectively. Both LXRs and FXR form heterodimers with RXR. LXRs are important for maintaining cholesterol homeostasis, whereas FXR acts as the primary bile acid sensor by repressing expression of 7α-hydroxylase (CYP7A1), the rate-limiting enzyme in bile acid production, through up-regulation of two orphan NRs, SHP and LRH1, upon bile acid stimulation (Lu et al. 2001). SHP and LRH1 form inactivating heterodimers to repress transcription of CYP7A1, thus turning off bile acid production.
6.4. Xenobiotic Receptors
The xenobiotic receptors include the constitutive active/androstane receptor (CAR) and pregnane X receptor (PXR). Both CAR and PXR form heterodimers with RXR and bind promiscuously to different response elements (Lehmann et al. 1998; Sueyoshi et al. 1999). Their role is to induce expression of enzymes such as CYP3A that promote the metabolism of exogenous compounds (xenobiotics). Unlike typical NRs, PXR has a promiscuous LBD pocket that binds a wide variety of natural and synthetic compounds. Activation of xenobiotic receptors is not always beneficial to the host. For example, upon exposure to high doses of acetaminophen (a common component of pain killers and cocaine), CAR can mediate production of toxic by-products through up-regulation of three CYP enzymes that lead to acute hepatotoxicity (Zhang et al. 2002).
6.5. Adopted Orphan Receptors
Discovery of natural or synthetic ligands for several orphan NRs has led to their classification as “adopted” orphan receptors (e.g., REV-ERBα and REV-ERBβ). REV-ERBs are repressors that compete with retinoid-related orphan receptors (RORα, RORβ, and RORγ) for binding to (A/T)6RGGTCA HREs (Forman et al. 1994). The REV-ERB ligand is neither a lipid nor a hormone but, rather, a single heme molecule (Reinking et al. 2005). Many heme-containing proteins also bind to diatomic gases, and REV-ERBs are also responsive to NO and CO (see below). RORs are proposed to bind cholesterol and retinoid acid, although whether this is important in antagonizing REV-ERBs is not clear (Kallen et al. 2002; Stehlin-Gaon et al. 2003). NR5A subfamily members SF1 (NR5A1) and LRH1 (NR5A2) were originally thought to be constitutively active. Recent analysis, however, reveals that these receptors can bind to phosphatidylinositols, including PIP2 and PIP3 (Newton et al. 2014). They may therefore link transcription and phospholipid signaling (Krylova et al. 2005).
6.6. True Orphan Receptors
True orphans are constitutive activators or repressors that do not bind ligands. The NUR subfamily (NR4A1, NR4A2, and NR4A3) binds to DNA as monomers, homodimers, and heterodimers with RXR (or another NUR). NURs have no known ligands, and their activity is controlled by their expression levels. ERRα, ERRβ, and ERRγ interfere with ER and AR signaling. ERRs are activated by coactivators such as PGC1α, an important metabolic regulator (Hardie 2012). Germ-cell nuclear receptor (GCNF) is an unusual member of the NR superfamily that binds to a DR-0 motif and may function as a transcriptional repressor in stem cells (Fuhrmann et al. 2001).
NRs thus represent an important metazoan strategy for regulating gene expression programs by integrating different intracellular and extracellular signals. Moreover, crosstalk between NRs and other signal transduction pathways adds an additional layer of complexity to their already diverse functions.
7. GASES
The ability to sense and adapt to changes in levels of diatomic gases (O2, NO, and CO) is crucial for an organism’s survival. Like steroids, gases are able to cross the plasma membrane and bind to intracellular targets. Signal transduction via gases is predominantly mediated by heme-containing proteins. Binding of the gas to the sensor proteins via the heme moiety can affect their activity or stability. In the case of oxygen, the source of the gas is the environment. In contrast, nitric oxide and carbon monoxide can be generated in neighboring cells and function as bona fide paracrine signals (through covalent and noncovalent binding).
7.1. Signal Transduction via Oxygen
Levels of oxygen inside mammalian cells are mainly detected by hypoxia-inducible factor (HIF) (Wang et al. 1995). HIF is a heterodimer comprising a regulatory α subunit and a DNA-binding β subunit. At high oxygen levels, the HIFα subunit is hydroxylated by prolyl hydroxylase domain (PHD) proteins and targeted for proteosomal degradation by interacting with the von Hippel–Lindau (VHL) protein, which is the targeting subunit of a cullin RING ligase (CRL) 2 E3 ubiquitin ligase. The PHD proteins are dioxygenases that catalyze the incorporation of both atoms of molecular oxygen into their substrates. At low oxygen levels, HIFα degradation is inhibited and HIFα accumulates inside the cell. It associates with HIFβ, which binds to DNA elements termed hypoxia responsive elements (HREs) that have the core sequence RCGTG. HIF targets include erythropoietin, which stimulates red blood cell production; vascular endothelial factor (VEGF), which stimulates angiogenesis (growth of new blood vessels); and other enzymes in relevant pathways, including glycolytic enzymes.
7.2. Signal Transduction via Nitric Oxide
Nitric oxide (NO) was first discovered as an endothelial-derived relaxing factor (EDRF) and subsequently found to be a powerful mediator in other biological processes, including platelet aggregation, synaptic transmission, and inflammatory responses (Moncada et al. 1991). Vascular NO is a key regulator of endothelial function and stimulates blood vessel dilation, prevents platelet aggregation and adhesion, and inhibits proliferation of vascular smooth muscle cells. NO is produced by NO synthase (NOS) from the amino acid l-arginine and detected by the heme-containing enzyme soluble guanylyl cyclase (sGC). Synthesis of NO from arginine involves two mono-oxygenation steps and requires one O2, one NADPH, and tetrahydrobiopterin (BH4).
The electronic structure of NO makes it an excellent ligand for heme, allowing it to bind to sGC heme at a low and nontoxic concentration. sGC is a heterodimer composed of α1 and β1 subunits. H105 in the β1 amino terminus is the heme ligand (Wedel et al. 1994). In a two-step process, NO first binds to the sGC heme moiety, forming a 6-coordinate complex. In the second step, the heme-H105 bound breaks forming a 5-coordinate complex. The second step triggers a conformational change in the sGC catalytic domain, leading to its activation. When activated by NO, sGC converts GTP to cGMP, which then acts as a second messenger (Newton et al. 2014). Besides sGC, NO can also interact with other heme-containing enzymes, including methionine synthase, cytochrome c oxidase, cytochrome P450, hemoglobin, and ferritin (Moncada and Palmer 1991; Brown and Cooper 1994; Danishpajooh et al. 2001). REV-ERBα and REV-ERBβ have also been shown to be able to bind NO through their heme-containing LBDs, which results in inhibition of the ability of REV-ERB to repress transcription (Pardee et al. 2009).
In S-nitrosylation, NO covalently links to the thiol side chain of cysteine residues in target proteins and modifies their activities. An example of NO-mediated S-nitrosylation is the modification of β-catenin in the adherens junction to increase endothelial permeability (Thibeault et al. 2010).
7.3. Signal Transduction via Carbon Monoxide
Endogenous carbon monoxide is generated by heme oxygenase (HO), an enzyme that degrades heme into iron, biliverdin, and CO in aging red blood cells and is induced by many cellular stressors and toxins, including UV, hydrogen peroxide, heavy metals, and arsinite. Like NO, CO has affinity for heme-containing proteins and activates sGC by binding to its heme motif, which stimulates similar biological processes, including intestinal smooth muscle relaxation and neural transmission (Rodgers 1999). In olfactory neurons, CO acts as a neurotransmitter via sGC to produce cGMPs (Verma et al. 1993). It can also bind to REV-ERBs, although at a lower affinity. In addition, CO can bind to other circadian-rhythm-regulating transcription factors, such as neuronal PAS domain protein 2 (NPAS2), which controls circadian gene expression. Upon exposure to CO, NPAS2 cannot form heterodimers with its partner Bmal (Dioum et al. 2002).
Gases are thus not only cell stress indicators, but also important endogenous signaling molecules. Their levels are also crucial and often disrupted in pathological processes, including cardiovascular disease, cancers, and CNS diseases. Future work on diatomic gas signaling thus has the potential lead to the development of entirely new classes of therapeutic agents.
8. CONCLUDING REMARKS
Signaling mechanisms controlling cell growth, migration, survival, and differentiation involve a plethora of different types of ligands, including proteins, peptides, and certain lipids, that bind to receptors at the cell surface, and steroids and gases that can pass across the plasma membrane and bind to intracellular receptors. Recent work has provided immense insights into how receptors are activated after ligand binding, how they initiate signaling inside the cell, and how signaling is terminated. A striking feature is that regulated dimerization or oligomerization is involved in the control of many plasma membrane receptors and intracellular signaling molecules. Moreover, signal transduction often occurs by the reversible formation of complexes and specific translocations of signaling molecules, rather than by random diffusion events. Another striking feature is that many different types of posttranslational modifications of proteins are involved in the regulation of the activity, stability, and subcellular localization of signaling molecules. Furthermore, different signaling inputs converge on a few well-conserved intracellular pathways, several of which link to transcriptional regulation of gene programs.
Whereas the initial phase of signal transduction often involves amplification mechanisms, such as inhibition of phosphatases in conjunction with activation of tyrosine kinases, subsequent phases contain different negative feedback and termination mechanisms, acting at many different levels. There is also extensive crosstalk between different signaling pathways. An important question that remains to be answered is exactly how specificity of signaling is achieved. Moreover, most of our knowledge about receptor signal transduction mechanisms comes from studies of cultured cells; much more work is needed before we understand signal transduction at the organismal level.
The Ryk and Ror families of RTK, for example, bind members of the Wnt family. Ryk has a Wnt inhibitory factor-1 (WIF1) domain, and the two Ror receptors have cysteine-rich domains related to a domain in the Frizzled family of serpentine receptors to which ligands of the Wnt family bind (van Amerongen et al. 2008).
Editors: Lewis Cantley, Tony Hunter, Richard Sever, and Jeremy Thorner
Additional Perspectives on Signal Transduction available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection
- Aggarwal BB. 2003. Signalling pathways of the TNF superfamily: A double-edged sword. Nat Rev Immunol 3: 745–756. [DOI] [PubMed] [Google Scholar]
- Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, Tarcic G, Siwak D, Lahad J, Jacob-Hirsch J, et al. 2007. A module of negative feedback regulators defines growth factor signaling. Nat Genet 39: 503–512. [DOI] [PubMed] [Google Scholar]
- Anastasiadis PZ, Reynolds AB. 2001. Regulation of Rho GTPases by p120-catenin. Curr Opin Cell Biol 13: 604. [DOI] [PubMed] [Google Scholar]
- Ancot F, Foveau B, Lefebvre J, Leroy C, Tulasne D. 2009. Proteolytic cleavages give receptor tyrosine kinases the gift of ubiquity. Oncogene 28: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Andl CD, Mizushima T, Nakagawa H, Oyama K, Harada H, Chruma K, Herlyn M, Rustgi AK. 2003. Epidermal growth factor receptor mediates increased cell proliferation, migration, and aggregation in esophageal keratinocytes in vitro and in vivo. J Biol Chem 278: 1824–1830. [DOI] [PubMed] [Google Scholar]
- Andreeva AV, Kutuzov MA, Voyno-Yasenetskaya TA. 2007. Scaffolding proteins in G-protein signaling. J Mol Signal 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkhipov A, Shan Y, Das R, Endres NF, Eastwood MP, Wemmer DE, Kuriyan J, Shaw DE. 2013. Architecture and membrane interactions of the EGF receptor. Cell 152: 557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballon DR, Flanary PL, Gladue DP, Konopka JB, Dohlman HG, Thorner J. 2006. DEP-domain-mediated regulation of GPCR signaling responses. Cell 126: 1079–1093. [DOI] [PubMed] [Google Scholar]
- Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM. 1999. PPARγ is required for placental, cardiac, and adipose tissue development. Mol Cell 4: 585–595. [DOI] [PubMed] [Google Scholar]
- Benovic JL, Pike LJ, Cerione RA, Staniszewski C, Yoshimasa T, Codina J, Caron MG, Lefkowitz RJ. 1985. Phosphorylation of the mammalian β-adrenergic receptor by cyclic AMP-dependent protein kinase. Regulation of the rate of receptor phosphorylation and dephosphorylation by agonist occupancy and effects on coupling of the receptor to the stimulatory guanine nucleotide regulatory protein. J Biol Chem 260: 7094–7101. [PubMed] [Google Scholar]
- Berman DM, Gilman AG. 1998. Mammalian RGS proteins: Barbarians at the gate. J Biol Chem 273: 1269–1272. [DOI] [PubMed] [Google Scholar]
- Blumer JB, Smrcka AV, Lanier SM. 2007. Mechanistic pathways and biological roles for receptor-independent activators of G-protein signaling. Pharmacol Ther 113: 488–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockaert J, Pin JP. 1999. Molecular tinkering of G protein-coupled receptors: An evolutionary success. EMBO J 18: 1723–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodrikov V, Sytnyk V, Leshchyns’ka I, den Hertog J, Schachner M. 2008. NCAM induces CaMKIIα-mediated RPTPα phosphorylation to enhance its catalytic activity and neurite outgrowth. J Cell Biol 182: 1185–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouschet T, Martin S, Henley JM. 2005. Receptor-activity-modifying proteins are required for forward trafficking of the calcium-sensing receptor to the plasma membrane. J Cell Sci 118: 4709–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga VMM, Yap AS. 2005. The challenges of abundance: Epithelial junctions and small GTPase signalling. Curr Opin Cell Biol 17: 466. [DOI] [PubMed] [Google Scholar]
- Brown GC, Cooper CE. 1994. Nanomolar concentrations of nitric oxide reversibly inhibit synaptosomal respiration by competing with oxygen at cytochrome oxidase. FEBS Lett 356: 295–298. [DOI] [PubMed] [Google Scholar]
- Brzostowski JA, Kimmel AR. 2001. Signaling at zero G: G-protein-independent functions for 7-TM receptors. Trends Biochem Sci 26: 291–297. [DOI] [PubMed] [Google Scholar]
- Burgess AW, Cho HS, Eigenbrot C, Ferguson KM, Garrett TP, Leahy DJ, Lemmon MA, Sliwkowski MX, Ward CW, Yokoyama S. 2003. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol Cell 12: 541–552. [DOI] [PubMed] [Google Scholar]
- Cabrera-Vera TM, Vanhauwe J, Thomas TO, Medkova M, Preininger A, Mazzoni MR, Hamm HE. 2003. Insights into G protein structure, function, and regulation. Endocr Rev 24: 765–781. [DOI] [PubMed] [Google Scholar]
- *.Cantrell D. 2014. Signaling in lymphocyte activation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, Lampugnani MG, Moons L, Breviario F, Compernolle V, Bono F, Balconi G, Spagnuolo R, Oostuyse B, Dewerchin M, et al. 1999. Targeted deficiency or cytosolic truncation of the VE-cadherin gene in mice impairs VEGF-mediated endothelial survival and angiogenesis. Cell 98: 147–157. [DOI] [PubMed] [Google Scholar]
- Cavallaro U, Dejana E. 2011. Adhesion molecule signalling: Not always a sticky business. Nat Rev Mol Cell Biol 12: 189–197. [DOI] [PubMed] [Google Scholar]
- Chan FK, Chun HJ, Zheng L, Siegel RM, Bui KL, Lenardo MJ. 2000. A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science 288: 2351–2354. [DOI] [PubMed] [Google Scholar]
- Chuang TT, Iacovelli L, Sallese M, De Blasi A. 1996. G protein-coupled receptors: Heterologous regulation of homologous desensitization and its implications. Trends Pharmacol Sci 17: 416–421. [DOI] [PubMed] [Google Scholar]
- Ciraolo E, Iezzi M, Marone R, Marengo S, Curcio C, Costa C, Azzolino O, Gonella C, Rubinetto C, Wu H, et al. 2008. Phosphoinositide 3-kinase p110β activity: Key role in metabolism and mammary gland cancer but not development. Sci Signal 1: ra3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coso OA, Chiariello M, Yu J-C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind JS. 1995. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell 81: 1137–1146. [DOI] [PubMed] [Google Scholar]
- Costa C, Martin-Conte EL, Hirsch E. 2011. Phosphoinositide 3-kinase p110γ in immunity. IUBMB Life 63: 707–713. [DOI] [PubMed] [Google Scholar]
- Croft M. 2009. The role of TNF superfamily members in T-cell function and diseases. Nat Rev Immunol 9: 271–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JM, Reynolds AB. 1999. The catenin p120ctn interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol Cell Biol 19: 3614–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danishpajooh IO, Gudi T, Chen Y, Kharitonov VG, Sharma VS, Boss GR. 2001. Nitric oxide inhibits methionine synthase activity in vivo and disrupts carbon flow through the folate pathway. J Biol Chem 276: 27296–27303. [DOI] [PubMed] [Google Scholar]
- Degnan BM, Vervoort M, Larroux C, Richards GS. 2009. Early evolution of metazoan transcription factors. Curr Opin Genet Dev 19: 591–599. [DOI] [PubMed] [Google Scholar]
- De Vos AM, Ultsch M, Kossiakoff AA. 1992. Human growth hormone and extracellular domain of its receptor: Crystal structure of the complex. Science 255: 306–312. [DOI] [PubMed] [Google Scholar]
- *.Devreotes P, Horwitz AR. 2013. Signaling networks that regulate cell migration. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA, McKnight SL. 2002. NPAS2: A gas-responsive transcription factor. Science 298: 2385–2387. [DOI] [PubMed] [Google Scholar]
- Dorsam RT, Gutkind JS. 2007. G-Protein-coupled receptors and cancer. Nat Rev Cancer 7: 79–94. [DOI] [PubMed] [Google Scholar]
- Dumstrei K, Wang F, Shy D, Tepass U, Hartenstein V. 2002. Interaction between EGFR signaling and DE-cadherin during nervous system morphogenesis. Development 129: 3983–3994. [DOI] [PubMed] [Google Scholar]
- Endres NF, Das R, Smith AW, Arkhipov A, Kovacs E, Huang Y, Pelton JG, Shan Y, Shaw DE, Wemmer DE, et al. 2013. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell 152: 543–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RM. 2005. The nuclear receptor superfamily: A rosetta stone for physiology. Mol Endocrinol 19: 1429–1438. [DOI] [PubMed] [Google Scholar]
- Faustman D, Davis M. 2010. TNF receptor 2 pathway: Drug target for autoimmune diseases. Nat Rev Drug Discov 9: 482–493. [DOI] [PubMed] [Google Scholar]
- Fedor-Chaiken M, Hein PW, Stewart JC, Brackenbury R, Kinch MS. 2003. E-cadherin binding modulates EGF receptor activation. Cell Commun Adhes 10: 105–118. [PubMed] [Google Scholar]
- Feldmann M, Maini RN. 2001. Anti-TNFα therapy of rheumatoid arthritis: What have we learned? Annu Rev Immunol 19: 163–196. [DOI] [PubMed] [Google Scholar]
- Flower DR. 1999. Modelling G-protein-coupled receptors for drug design. Biochim Biophys Acta 1422: 207–234. [DOI] [PubMed] [Google Scholar]
- Forman BM, Chen J, Blumberg B, Kliewer SA, Henshaw R, Ong ES, Evans RM. 1994. Cross-talk among RORα1 and the Rev-erb family of orphan nuclear receptors. Mol Endocrinol 8: 1253–1261. [DOI] [PubMed] [Google Scholar]
- Fox NL, Humphreys R, Luster TA, Klein J, Gallant G. 2010. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) receptor-1 and receptor-2 agonists for cancer therapy. Expert Opin Biol Ther 10: 1–18. [DOI] [PubMed] [Google Scholar]
- Frisch SM, Screaton RA. 2001. Anoikis mechanisms. Curr Opin Cell Biol 13: 555–562. [DOI] [PubMed] [Google Scholar]
- Fuhrmann G, Chung AC, Jackson KJ, Hummelke G, Baniahmad A, Sutter J, Sylvester I, Scholer HR, Cooney AJ. 2001. Mouse germline restriction of Oct4 expression by germ cell nuclear factor. Dev Cell 1: 377–387. [DOI] [PubMed] [Google Scholar]
- Fukuhara S, Chikumi H, Gutkind JS. 2001. RGS-containing RhoGEFs: The missing link between transforming G proteins and Rho? Oncogene 20: 1661–1668. [DOI] [PubMed] [Google Scholar]
- Geiger B, Yamada KM. 2011. Molecular architecture and function of matrix adhesions. Cold Spring Harb Perspect Biol 3: a005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B, Bershadsky A, Pankov R, Yamada KM. 2001. Transmembrane crosstalk between the extracellular matrix and the cytoskeleton. Nat Rev Mol Cell Biol 2: 793–805. [DOI] [PubMed] [Google Scholar]
- *.Green DR, Llambi F. 2014. Cell death signaling. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosheva I, Shtutman M, Elbaum M, Bershadsky AD. 2001. p120 catenin affects cell motility via modulation of activity of Rho-family GTPases: A link between cell–cell contact formation and regulation of cell locomotion. J Cell Sci 114: 695–707. [DOI] [PubMed] [Google Scholar]
- Gumbiner BM. 2005. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol 6: 622–634. [DOI] [PubMed] [Google Scholar]
- Gutkind JS. 1998. The pathways connecting G protein-coupled receptors to the nucleus through divergent mitogen-activated protein kinase cascades. J Biol Chem 273: 1839–1842. [DOI] [PubMed] [Google Scholar]
- Gutkind JS. 2000. Regulation of mitogen-activated protein kinase signaling networks by G protein-coupled receptors. Sci STKE 2000: re1. [DOI] [PubMed] [Google Scholar]
- Hanyaloglu AC, von Zastrow M. 2008. Regulation of GPCRs by endocytic membrane trafficking and its potential implications. Annu Rev Pharmacol Toxicol 48: 537–568. [DOI] [PubMed] [Google Scholar]
- *.Hardie DG. 2012. Organismal carbohydrate and lipid homeostasis. Cold Spring Harb Perspect Biol 4: a006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Harrison DA. 2012. The JAK/STAT pathway. Cold Spring Harb Perspect Biol 4: a011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart MJ, Jiang X, Kozasa T, Roscoe W, Singer WD, Gilman AG, Sternweis PC, Bollag G. 1998. Direct stimulation of the guanine nucleotide exchange activity of p115 RhoGEF by Gα13. Science 280: 2112–2114. [DOI] [PubMed] [Google Scholar]
- Heldin C-H. 1995. Dimerization of cell surface receptors in signal transduction. Cell 80: 213–223. [DOI] [PubMed] [Google Scholar]
- *.Hemmings BA, Restuccia DF. 2012. PI3K-PKB/Akt pathway. Cold Spring Harb Perspect Biol 4: a011189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SY, Kudo M, Chen T, Nakabayashi K, Bhalla A, van der Spek PJ, van Duin M, Hsueh AJ. 2000. The three subfamilies of leucine-rich repeat-containing G protein-coupled receptors (LGR): Identification of LGR6 and LGR7 and the signaling mechanism for LGR7. Mol Endocrinol 14: 1257–1271. [DOI] [PubMed] [Google Scholar]
- Hu X, Lazar MA. 1999. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature 402: 93–96. [DOI] [PubMed] [Google Scholar]
- Hubbard KB, Hepler JR. 2006. Cell signalling diversity of the Gqα family of heterotrimeric G proteins. Cell Signal 18: 135–150. [DOI] [PubMed] [Google Scholar]
- Huttenlocher A, Horwitz AR. 2011. Integrins in cell migration. Cold Spring Harb Perspect Biol 3: a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO. 2002. Integrins: Bidirectional, allosteric signaling machines. Cell 110: 673–687. [DOI] [PubMed] [Google Scholar]
- *.Ingham PW. 2012. Hedgehog signaling. Cold Spring Harb Perspect Biol 4: a011221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issemann I, Green S. 1990. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature 347: 645–650. [DOI] [PubMed] [Google Scholar]
- Jiang G, Hunter T. 1999. Receptor signaling: When dimerization is not enough. Curr Biol 9: R568–R571. [DOI] [PubMed] [Google Scholar]
- *.Julius D, Nathans J. 2012. Signaling by sensory receptors. Cold Spring Harb Perspect Biol 4: a005991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jura N, Endres NF, Engel K, Deindl S, Das R, Lamers MH, Wemmer DE, Zhang X, Kuriyan J. 2009. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell 137: 1293–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen JA, Schlaeppi JM, Bitsch F, Geisse S, Geiser M, Delhon I, Fournier B. 2002. X-ray structure of the hRORα LBD at 1.63 Å: Structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORα. Structure 10: 1697–1707. [DOI] [PubMed] [Google Scholar]
- Kang JS, Liu C, Derynck R. 2009. New regulatory mechanisms of TGFβ receptor function. Trends Cell Biol 19: 385–394. [DOI] [PubMed] [Google Scholar]
- Kim N, Stiegler AL, Cameron TO, Hallock PT, Gomez AM, Huang JH, Hubbard SR, Dustin ML, Burden SJ. 2008. Lrp4 is a receptor for Agrin and forms a complex with MuSK. Cell 135: 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NG, Koh E, Chen X, Gumbiner BM. 2011. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc Natl Acad Sci 108: 11930–11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Jones K, Charles JP, Lam G, Thummel CS. 2005. The ecdysone-induced DHR4 orphan nuclear receptor coordinates growth and maturation in Drosophila. Cell 121: 773–784. [DOI] [PubMed] [Google Scholar]
- Kniazeff J, Prezeau L, Rondard P, Pin JP, Goudet C. 2011. Dimers and beyond: The functional puzzles of class C GPCRs. Pharmacol Ther 130: 9–25. [DOI] [PubMed] [Google Scholar]
- Kobielak A, Fuchs E. 2004. α-Catenin: At the junction of intercellular adhesion and actin dynamics. Nat Rev Mol Cell Biol 5: 614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobilka BK. 2011. Structural insights into adrenergic receptor function and pharmacology. Trends Pharmacol Sci 32: 213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Kopan R. 2012. Notch signaling. Cold Spring Harb Perspect Biol 4: a011213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MXG. 2009. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 137: 216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Kornbluth S, Fissore R. 2014. Vertebrate reproduction. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a006064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunova I, Mosevitsky M. 2010. Role of the growth-associated protein GAP-43 in NCAM-mediated neurite outgrowth. Adv Exp Med Biol 663: 169–182. [DOI] [PubMed] [Google Scholar]
- Krylova IN, Sablin EP, Moore J, Xu RX, Waitt GM, MacKay JA, Juzumiene D, Bynum JM, Madauss K, Montana V, et al. 2005. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell 120: 343–355. [DOI] [PubMed] [Google Scholar]
- *.Laplante M, Sabatini DM. 2012. mTOR signaling. Cold Spring Harb Perspect Biol 4: a011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Lee MJ, Yaffe MB. 2014. Protein regulation in signal transduction. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a005918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legate KR, Fassler R. 2009. Mechanisms that regulate adaptor binding to β-integrin cytoplasmic tails. J Cell Sci 122: 187–198. [DOI] [PubMed] [Google Scholar]
- Legate KR, Wickstrom SA, Fassler R. 2009. Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev 23: 397–418. [DOI] [PubMed] [Google Scholar]
- Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. 1998. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest 102: 1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. 2010. Cell signaling by receptor tyrosine kinases. Cell 141: 1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengyel E, Sawada K, Salgia R. 2007. Tyrosine kinase mutations in human cancer. Curr Mol Med 7: 77–84. [DOI] [PubMed] [Google Scholar]
- Leushacke M, Barker N. 2011. Lgr5 and Lgr6 as markers to study adult stem cell roles in self-renewal and cancer. Oncogene 31: 3009–3022. [DOI] [PubMed] [Google Scholar]
- Lim X, Nusse R. 2013. Wnt signaling in skin development, homeostasis, and disease. Cold Spring Harb Perspect Biol 5: a008029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S-Y, Makino K, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung M-C. 2001. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol 3: 802–808. [DOI] [PubMed] [Google Scholar]
- Liongue C, Ward AC. 2007. Evolution of Class I cytokine receptors. BMC Evol Biol 7: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locksley RM, Killeen N, Lenardo MJ. 2001. The TNF and TNF receptor superfamilies: Integrating mammalian biology. Cell 104: 487–501. [DOI] [PubMed] [Google Scholar]
- Loers G, Schachner M. 2007. Recognition molecules and neural repair. J Neurochem 101: 865–882. [DOI] [PubMed] [Google Scholar]
- Lopez-Ilasaca M, Li W, Uren A, Yu J-C, Kazlauskas A, Gutkind JS, Heidaran MA. 1997. Requirement of phosphatidylinositol-3 kinase for activation of JNK/SAPKs by PDGF. Biochem Biophys Res Commun 232: 273–277. [DOI] [PubMed] [Google Scholar]
- Lu TT, Repa JJ, Mangelsdorf DJ. 2001. Orphan nuclear receptors as eLiXiRs and FiXeRs of sterol metabolism. J Biol Chem 276: 37735–37738. [DOI] [PubMed] [Google Scholar]
- Luttrell LM, Gesty-Palmer D. 2010. Beyond desensitization: Physiological relevance of arrestin-dependent signaling. Pharmacol Rev 62: 305–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz S, Shankaranarayanan A, Coco C, Ridilla M, Nance MR, Vettel C, Baltus D, Evelyn CR, Neubig RR, Wieland T, et al. 2007. Structure of Gαq-p63RhoGEF-RhoA complex reveals a pathway for the activation of RhoA by GPCRs. Science 318: 1923–1927. [DOI] [PubMed] [Google Scholar]
- Magalhaes AC, Dunn H, Ferguson SS. 2012. Regulation of GPCR activity, trafficking and localization by GPCR-interacting proteins. Br J Pharmacol 165: 1717–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Borgmeyer U, Heyman RA, Zhou JY, Ong ES, Oro AE, Kakizuka A, Evans RM. 1992. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes Dev 6: 329–344. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. 1995. The nuclear receptor superfamily: The second decade. Cell 83: 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May LT, Leach K, Sexton PM, Christopoulos A. 2007. Allosteric modulation of G protein-coupled receptors. Annu Rev Pharmacol Toxicol 47: 1–51. [DOI] [PubMed] [Google Scholar]
- *.McCaffrey LM, Macara IG. 2012. Signaling pathways in cell polarity. Cold Spring Harb Perspect Biol 4: a009654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mege RM, Gavard J, Lambert M. 2006. Regulation of cell–cell junctions by the cytoskeleton. Curr Opin Cell Biol 18: 541–548. [DOI] [PubMed] [Google Scholar]
- Miaczynska M, Pelkmans L, Zerial M. 2004. Not just a sink: Endosomes in control of signal transduction. Curr Opin Cell Biol 16: 400–406. [DOI] [PubMed] [Google Scholar]
- Miranti CK, Brugge JS. 2002. Sensing the environment: A historical perspective on integrin signal transduction. Nat Cell Biol 4: E83–E90. [DOI] [PubMed] [Google Scholar]
- Miyamoto S, Teramoto H, Coso OA, Gutkind JS, Burbelo PD, Akiyama SK, Yamada KM. 1995. Integrin function: Molecular hierarchies of cytoskeletal and signaling molecules. J Cell Biol 131: 791–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S, Teramoto H, Gutkind JS, Yamada KM. 1996. Integrins can collaborate with growth factors for phosphorylation of receptor tyrosine kinases and MAP kinase activation: Roles of integrin aggregation and occupancy of receptors. J Cell Biol 135: 1633–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncada S, Palmer RM. 1991. Inhibition of the induction of nitric oxide synthase by glucocorticoids: Yet another explanation for their anti-inflammatory effects? Trends Pharmacol Sci 12: 130–131. [DOI] [PubMed] [Google Scholar]
- Moncada S, Palmer RM, Higgs EA. 1991. Nitric oxide: Physiology, pathophysiology, and pharmacology. Pharmacol Rev 43: 109–142. [PubMed] [Google Scholar]
- Moquin D, Chan FK-M. 2010. The molecular regulation of programmed necrotic cell injury. Trends Biochem Sci 35: 434–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Morrison DK. 2012. MAP kinase pathways. Cold Spring Harb Perspect Biol 4: a011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M, Legate KR, Zent R, Fassler R. 2009. The tail of integrins, talin, and kindlins. Science 324: 895–899. [DOI] [PubMed] [Google Scholar]
- Moustakas A, Heldin C-H. 2009. The regulation of TGFβ signal transduction. Development 136: 3699–3714. [DOI] [PubMed] [Google Scholar]
- Mu Y, Sundar R, Thakur N, Ekman M, Gudey SK, Yakymovych M, Hermansson A, Dimitriou H, Bengoechea-Alonso MT, Ericsson J, et al. 2011. TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer. Nat Commun 2: 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan JF, Pan H, Lu Q. 2010. Arrestin domain-containing protein 3 recruits the NEDD4 E3 ligase to mediate ubiquitination of the β2-adrenergic receptor. EMBO Rep 11: 605–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WJ, Nusse R. 2004. Convergence of Wnt, β-catenin, and cadherin pathways. Science 303: 1483–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Newton K, Dixit VM. 2012. Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol 4: a006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Newton AC, Bootman M, Scott JD. 2014. Second messengers. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a005926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni C-Y, Murphy MP, Golde TE, Carpenter G. 2001. γ-Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science 294: 2179–2181. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn F, Ravetch JV. 2008. Fcγ receptors as regulators of immune responses. Nat Rev Immunol 8: 34–47. [DOI] [PubMed] [Google Scholar]
- Nose A, Nagafuchi A, Takeichi M. 1988. Expressed recombinant cadherins mediate cell sorting in model systems. Cell 54: 993–1001. [DOI] [PubMed] [Google Scholar]
- O’Hayre M, Vázquez-Prado J, Kufareva I, Stawiski EW, Handel TM, Seshagiri S, Gutkind JS. 2013. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nat Rev Cancer 13: 412–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL. 2005. Regulation of the polarity protein Par6 by TGFβ receptors controls epithelial cell plasticity. Science 307: 1603–1609. [DOI] [PubMed] [Google Scholar]
- Pardee KI, Xu X, Reinking J, Schuetz A, Dong A, Liu S, Zhang R, Tiefenbach J, Lajoie G, Plotnikov AN, et al. 2009. The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBβ. PLoS Biol 7: e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T. 2004. Specificity in signal transduction: From phosphotyrosine–SH2 domain interactions to complex cellular systems. Cell 116: 191–203. [DOI] [PubMed] [Google Scholar]
- Pece S, Gutkind JS. 2000. Signaling from E-cadherins to the MAPK pathway by the recruitment and activation of epidermal growth factor receptors upon cell–cell contact formation. J Biol Chem 275: 41227–41233. [DOI] [PubMed] [Google Scholar]
- Pece S, Chiariello M, Murga C, Gutkind JS. 1999. Activation of the protein kinase Akt/PKB by the formation of E-cadherin-mediated cell–cell junctions. Evidence for the association of phosphatidylinositol 3-kinase with the E-cadherin adhesion complex. J Biol Chem 274: 19347–19351. [DOI] [PubMed] [Google Scholar]
- Perissi V, Jepsen K, Glass CK, Rosenfeld MG. 2010. Deconstructing repression: Evolving models of co-repressor action. Nat Rev Genet 11: 109–123. [DOI] [PubMed] [Google Scholar]
- Pierce KL, Premont RT, Lefkowitz RJ. 2002. Seven-transmembrane receptors. Nat Rev Mol Cell Biol 3: 639–650. [DOI] [PubMed] [Google Scholar]
- Qian X, Karpova T, Sheppard AM, McNally J, Lowy DR. 2004. E-cadherin-mediated adhesion inhibits ligand-dependent activation of diverse receptor tyrosine kinases. EMBO J 23: 1739–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C, Stenmark H. 2009. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458: 445–452. [DOI] [PubMed] [Google Scholar]
- Rajagopal S, Rajagopal K, Lefkowitz RJ. 2010. Teaching old receptors new tricks: Biasing seven-transmembrane receptors. Nat Rev Drug Discov 9: 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Red Brewer M, Choi SH, Alvarado D, Moravcevic K, Pozzi A, Lemmon MA, Carpenter G. 2009. The juxtamembrane region of the EGF receptor functions as an activation domain. Mol Cell 34: 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinking J, Lam MM, Pardee K, Sampson HM, Liu S, Yang P, Williams S, White W, Lajoie G, Edwards A, et al. 2005. The Drosophila nuclear receptor e75 contains heme and is gas responsive. Cell 122: 195–207. [DOI] [PubMed] [Google Scholar]
- Renauld J-C. 2003. Class II cytokine receptors and their ligands: Key antiviral and inflammatory modulators. Nat Rev Immunol 3: 667–676. [DOI] [PubMed] [Google Scholar]
- Rodgers KR. 1999. Heme-based sensors in biological systems. Curr Opin Chem Biol 3: 158–167. [DOI] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK. 2006. Sensors and signals: A coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev 20: 1405–1428. [DOI] [PubMed] [Google Scholar]
- Rosenfeldt H, Vazquez-Prado J, Gutkind JS. 2004. P-REX2, a novel PI-3-kinase sensitive Rac exchange factor. FEBS Lett 572: 167–171. [DOI] [PubMed] [Google Scholar]
- *.Samelson LE. 2011. Immunoreceptor signaling. Cold Spring Harb Perspect Biol 3: a011510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Sassone-Corsi P. 2012. The cyclic AMP pathway. Cold Spring Harb Perspect Biol 4: a011148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee S, Carmillo P, Whitty A. 2006. Quantitative analysis of the activation mechanism of the multicomponent growth-factor receptor Ret. Nat Chem Biol 2: 636–644. [DOI] [PubMed] [Google Scholar]
- Schneider P, Thome M, Burns K, Bodmer JL, Hofmann K, Kataoka T, Holler N, Tschopp J. 1997. TRAIL receptors 1 (DR4) and 2 (DR5) signal FADD-dependent apoptosis and activate NF-κB. Immunity 7: 831–836. [DOI] [PubMed] [Google Scholar]
- *.Sever R, Brugge JS. 2014. Cancer. Cold Spring Harb Perspect Med 10.1101/cshperspect.a006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Sever R, Glass CK. 2013. Signaling by nuclear receptors. Cold Spring Harb Perspect Biol 5: a016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattil SJ, Kim C, Ginsberg MH. 2010. The final steps of integrin activation: The end game. Nat Rev Mol Cell Biol 11: 288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy SK, Lefkowitz RJ. 2011. β-Arrestin-mediated receptor trafficking and signal transduction. Trends Pharmacol Sci 32: 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Pei L, Evans RM. 2008. Nuclear receptors: Decoding metabolic disease. FEBS Lett 582: 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehlin-Gaon C, Willmann D, Zeyer D, Sanglier S, Van Dorsselaer A, Renaud JP, Moras D, Schule R. 2003. All-trans retinoic acid is a ligand for the orphan nuclear receptor RORβ. Nat Struct Biol 10: 820–825. [DOI] [PubMed] [Google Scholar]
- Suda T, Hashimoto H, Tanaka M, Ochi T, Nagata S. 1997. Membrane Fas ligand kills human peripheral blood T lymphocytes, and soluble Fas ligand blocks the killing. J Exp Med 186: 2045–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suenaert P, Bulteel V, Lemmens L, Noman M, Geypens B, Van Assche G, Geboes K, Ceuppens JL, Rutgeerts P. 2002. Anti-tumor necrosis factor treatment restores the gut barrier in Crohn’s disease. Am J Gastroenterol 97: 2000–2004. [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. 1999. The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J Biol Chem 274: 6043–6046. [DOI] [PubMed] [Google Scholar]
- Suyama K, Shapiro I, Guttman M, Hazan RB. 2002. A signaling pathway leading to metastasis is controlled by N-cadherin and the FGF receptor. Cancer Cell 2: 301–314. [DOI] [PubMed] [Google Scholar]
- Takai Y, Miyoshi J, Ikeda W, Ogita H. 2008. Nectins and nectin-like molecules: Roles in contact inhibition of cell movement and proliferation. Nat Rev Mol Cell Biol 9: 603–615. [DOI] [PubMed] [Google Scholar]
- Taussig R, Gilman AG. 1995. Mammalian membrane-bound adenylyl cyclases. J Biol Chem 270: 1–4. [DOI] [PubMed] [Google Scholar]
- Terrillon S, Bouvier M. 2004. Roles of G-protein-coupled receptor dimerization. EMBO Rep 5: 30–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibeault S, Rautureau Y, Oubaha M, Faubert D, Wilkes BC, Delisle C, Gratton JP. 2010. S-Nitrosylation of β-catenin by eNOS-derived NO promotes VEGF-induced endothelial cell permeability. Mol Cell 39: 468–476. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. 2006. Complex networks orchestrate epithelial–mesenchymal transitions. Nat Rev Mol Cell Biol 7: 131. [DOI] [PubMed] [Google Scholar]
- Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL. 1998. SARA, a FYVE domain protein that recruits Smad2 to the TGFβ receptor. Cell 95: 779–791. [DOI] [PubMed] [Google Scholar]
- Tsukita S, Furuse M, Itoh M. 2001. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol 2: 285–293. [DOI] [PubMed] [Google Scholar]
- Umesono K, Murakami KK, Thompson CC, Evans RM. 1991. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell 65: 1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen R, Mikels A, Nusse R. 2008. Alternative wnt signaling is initiated by distinct receptors. Sci Signal 1: re9. [DOI] [PubMed] [Google Scholar]
- Vaqué JP, Dorsam RT, Feng X, Iglesias-Bartolome R, Forsthoefel DJ, Chen Q, Debant A, Seeger MA, Ksander BR, Teramoto H, et al. 2013. A genome-wide RNAi screen reveals a Trio-regulated Rho GTPase circuitry transducing mitogenic signals initiated by G protein-coupled receptors. Mol Cell 49: 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma A, Hirsch DJ, Glatt CE, Ronnett GV, Snyder SH. 1993. Carbon monoxide: A putative neural messenger. Science 259: 381–384. [DOI] [PubMed] [Google Scholar]
- Vleminckx K, Vakaet L Jr, Mareel M, Fiers W, van Roy F. 1991. Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell 66: 107–119. [DOI] [PubMed] [Google Scholar]
- Wang GL, Jiang BH, Rue EA, Semenza GL. 1995. Hypoxia-inducible factor 1 is a basic-helix–loop–helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci 92: 5510–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lupardus P, Laporte SL, Garcia KC. 2009. Structural biology of shared cytokine receptors. Annu Rev Immunol 27: 29–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnmark A, Treuter E, Wright AP, Gustafsson JA. 2003. Activation functions 1 and 2 of nuclear receptors: Molecular strategies for transcriptional activation. Mol Endocrinol 17: 1901–1909. [DOI] [PubMed] [Google Scholar]
- Wedel B, Humbert P, Harteneck C, Foerster J, Malkewitz J, Bohme E, Schultz G, Koesling D. 1994. Mutation of His-105 in the β1 subunit yields a nitric oxide–insensitive form of soluble guanylyl cyclase. Proc Natl Acad Sci 91: 2592–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehling M. 1997. Specific, nongenomic actions of steroid hormones. Annu Rev Physiol 59: 365–393. [DOI] [PubMed] [Google Scholar]
- Wehrle-Haller B. 2012. Assembly and disassembly of cell matrix adhesions. Curr Opin Cell Biol 24: 569–581. [DOI] [PubMed] [Google Scholar]
- Wehrman T, He X, Raab B, Dukipatti A, Blau H, Garcia KC. 2007. Structural and mechanistic insights into nerve growth factor interactions with the TrkA and p75 receptors. Neuron 53: 25–38. [DOI] [PubMed] [Google Scholar]
- Welch HC, Coadwell WJ, Ellson CD, Ferguson GJ, Andrews SR, Erdjument-Bromage H, Tempst P, Hawkins PT, Stephens LR. 2002. P-Rex1, a PtdIns(3,4,5)P3- and Gβγ-regulated guanine-nucleotide exchange factor for Rac. Cell 108: 809–821. [DOI] [PubMed] [Google Scholar]
- Whistler JL, Enquist J, Marley A, Fong J, Gladher F, Tsuruda P, Murray SR, Von Zastrow M. 2002. Modulation of postendocytic sorting of G protein-coupled receptors. Science 297: 615–620. [DOI] [PubMed] [Google Scholar]
- Willard FS, Kimple RJ, Siderovski DP. 2004. Return of the GDI: The GoLoco motif in cell division. Annu Rev Biochem 73: 925–951. [DOI] [PubMed] [Google Scholar]
- *.Wrana JL. 2013. Signaling by the TGFβ superfamily. Cold Spring Harb Perspect Biol 5: a011197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi J, Kawano T, Nagao M, Kaziro Y, Itoh H. 2000. Gi-dependent activation of c-Jun N-terminal kinase in human embryonal kidney 293 cells. J Biol Chem 275: 7633–7640. [DOI] [PubMed] [Google Scholar]
- Yanagisawa M, Anastasiadis PZ. 2006. p120 catenin is essential for mesenchymal cadherin-mediated regulation of cell motility and invasiveness. J Cell Biol 174: 1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. 2001. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2: 127–137. [DOI] [PubMed] [Google Scholar]
- Yoshimura A, Mori H, Ohishi M, Aki D, Hanada T. 2003. Negative regulation of cytokine signaling influences inflammation. Curr Opin Immunol 15: 704–708. [DOI] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al. 2012. Regulation of the Hippo–YAP pathway by G-protein-coupled receptor signaling. Cell 150: 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzawa S, Opatowsky Y, Zhang Z, Mandiyan V, Lax I, Schlessinger J. 2007. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell 130: 323–334. [DOI] [PubMed] [Google Scholar]
- Zhang J, Huang W, Chua SS, Wei P, Moore D.D. 2002. Modulation of acetaminophen-induced hepatotoxicity by the xenobiotic receptor CAR. Science 298: 422–424. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. 2006. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125: 1137–1149. [DOI] [PubMed] [Google Scholar]
- Zikherman J, Weiss A. 2009. Antigen receptor signaling in the rheumatic diseases. Arthritis Res Ther 11: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. 2011. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12: 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwang Y, Yarden Y. 2009. Systems biology of growth factor-induced receptor endocytosis. Traffic 10: 349–363. [DOI] [PubMed] [Google Scholar]