Abstract
Impaired systolic function, resulting from acute injury or congenital defects, leads to cardiac complications and heart failure. Current therapies slow disease progression but do not rescue cardiac function. We previously reported that elevating the cellular 2 deoxy-ATP (dATP) pool in transgenic mice via increased expression of ribonucleotide reductase (RNR), the enzyme that catalyzes deoxy-nucleotide production, increases myosin–actin interaction and enhances cardiac muscle contractility. For the current studies, we initially injected wild-type mice retro-orbitally with a mixture of adeno-associated virus serotype-6 (rAAV6) containing a miniaturized cardiac-specific regulatory cassette (cTnT455) composed of enhancer and promotor portions of the human cardiac troponin T gene (TNNT2) ligated to rat cDNAs encoding either the Rrm1 or Rrm2 subunit. Subsequent studies optimized the system by creating a tandem human RRM1-RRM2 cDNA with a P2A self-cleaving peptide site between the subunits. Both rat and human Rrm1/Rrm2 cDNAs resulted in RNR enzyme overexpression exclusively in the heart and led to a significant elevation of left ventricular (LV) function in normal mice and infarcted rats, measured by echocardiography or isolated heart perfusions, without adverse cardiac remodeling. Our study suggests that increasing RNR levels via rAAV-mediated cardiac-specific expression provide a novel gene therapy approach to potentially enhance cardiac systolic function in animal models and patients with heart failure.
Introduction
Heart failure, the inability of the heart to meet systemic demand for blood flow, is a growing burden on healthcare.1 A significant portion of these patients suffer from hypocontractility and low systolic function.2 However, there are few effective treatments that directly increase myocardial contraction. Currently available inotropic medications, such as dobutamine and millrinone, increase contractility via altering intracellular calcium but have adverse effects on heart failure survival.3,4 Targeted gene therapy for heart failure could potentially intervene directly in the diseased myocardium and enhance key molecular pathways. Several strategies including vector delivery of cDNAs encoding proteins involved in Ca2+ regulation and the β-adrenergic system have been pursued.5 The only completed clinical trial to date used a serotype-1 recombinant adeno-associated viral vector (rAAV1) to increase the expression of the sarcoplasmic/endoplasmic Ca2+ ATPase pump (SERCA2a). This trial demonstrated safety and efficacy, and importantly, provided benefits in clinical parameters that have extended beyond 3 years.6,7 Omecamtiv Mercarbol, a small molecule drug in clinical development, is a cardiac myosin activator that has been shown to increase left ventricular (LV) function independent of calcium and decrease filling pressure without increasing heart rate or oxygen consumption.8,9,10 However, Omecamtiv Mercarbol is delivered by repeat injections or chronic oral dosing and it increases systolic ejection time, leading to the shortening of diastole.10 While clearly beneficial, all current strategies except Omecamtiv Mercarbol target upstream effects rather than the myofilament where the contractile deficit often resides. We are pursuing a gene therapy approach that increases levels of the naturally occurring nucleotide, 2-deoxy adenosine triphosphate (dATP), and directly improves myocardial contraction at the level of the myofilament without impairing diastole.
Our previous work has shown that provision of dATP, instead of adenosine triphosphate (ATP), as a substrate for striated muscle myosin leads to enhanced contractile capacity. Replacing ATP with dATP in skinned cardiac preparations from mice, rats, rabbits, dogs, and humans increases the magnitude and rate of force development and shortening.11,12,13,14,15,16 We also found that raising cultured rat cardiomyocyte dATP levels to only ~1% of [ATP] by adeno-viral vector-mediated overexpression of ribonucleotide reductase (RNR; which converts ADP to dADP) under control of the constitutive cytomegalovirus promoter, significantly enhances the magnitude and rate of contraction, as well as the rate of relaxation, without altering intracellular Ca2+ transient amplitude.17,18 Cardiomyocytes from transgenic mice that over-express RNR (via the chicken beta-actin + cytomegalovirus (CAG) promotor)19 exhibit similarly enhanced contractile properties without altering Ca2+ transients, and have significantly increased LV function compared with wild-type littermates.18 Based on these results, overexpression of RNR and increased dATP content appear to have significant potential as a novel therapeutic strategy for treating systolic heart failure.
To explore the translational aspects of our gene therapy approach for treating heart failure, we have developed a series of rAAV6 vectors for the systemic delivery of cDNAs encoding the large (Rrm1) and small (Rrm2) subunits of RNR to intact myocardium. rAAV6 was chosen due to its highly efficient gene transfer to the myocardium.20,21,22,23,24 In cardiomyocytes, the therapeutic vector gene contained a human cardiac troponin-T regulatory cassette (cTnT455). For early proof-of-principle studies, we used separate rAAV6 vectors for cDNAs encoding the rat Rrm1 and Rrm2 subunits. We later evaluated a rAAV6 vector containing codon-optimized, tandem-arrayed human RRM1 and RRM2 cDNAs to test the hypothesis that elevating myocardial dATP with human RNR, via a rAAV gene therapy approach, improves cardiac function. For initial comparison studies of efficacy, we chose to study the effects in normal hearts, so as to limit any variability in basal cardiac function associated with damage or disease. We then tested the rAAV6 vector approach in a rodent model of myocardial infarction (MI).
In the current study, we demonstrate that both rat and human Rrm1 and Rrm2 cDNA-containing vectors produce significant elevation of LV function and this result is consistent when both coding subunits for RNR are contained in a single vector construct. Additionally, we demonstrate in a small trial study that LV function of infarcted rat hearts is improved, consistent with our recent report in an infarcted mini-pig model.25 Transduction, based on persistent rAAV genomes, occurred mostly in the heart and liver, and at a much lower level in skeletal muscle, but RNR enzyme overexpression was only detectable in the heart. Together, these encouraging results suggest that cardiomyocyte-specific gene therapy involving RNR-mediated increases in dATP levels is worth further testing in preclinical animal models of systolic heart failure.
Results
The experimental protocol to compare the effects of RNR overexpression in young adult C57BL/6J mice (3–5 months of age) is shown in Figure 1. Cardiac-specific overexpression of the RNR subunits was achieved via a miniaturized enhancer/promotor derived from the human cardiac troponin T gene (cTnT455; see Materials and Methods). Single or dual vector constructs (described in Supplementary Figure S1 and Table S1) were delivered 2 days following baseline echocardiography, and then mice were monitored either weekly or biweekly with echocardiography for 4 weeks. For vector comparison studies, mice were euthanized and hearts removed for Langendorff perfusion measurements at the 4-week endpoint. Tissues were processed for quantitative polymerase chain reaction (qPCR) analysis of vector genomes, immunohistochemistry, and western blots.
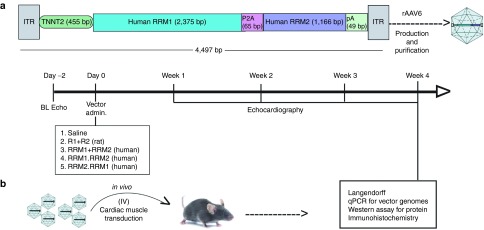
Figure 1.
Experimental protocol. (a) Schematic detailing the construct of the human cDNA containing vector. (b) Timeline of the administration of the cDNA containing vectors (either rat or human) to mice and the experimental protocols followed over the 4-week period. (R1+R2) = rAAV6 vectors containing either the rat R1 or R2 cDNAs driven by the human cTnT455 regulatory cassette (RC). (RRM1.RRM2), rAAV6-human (RRM1.RRM2)cTnT455 vector in which the RRM1 cDNA is 5' of the RRM2 cDNA and separated by a self-cleaving peptide sequence; RRM2.RRM1, treatment with an analogous vector with the RRM2 cDNA 5' of the RRM1 cDNA; (RRM1+RRM2), RRM1, and RRM2 cDNA containing vector.
rAAV6-ratR1cTnT455 and rAAV6-ratR2cTnT455 vectors enhance in-vivo and ex-vivo cardiac function
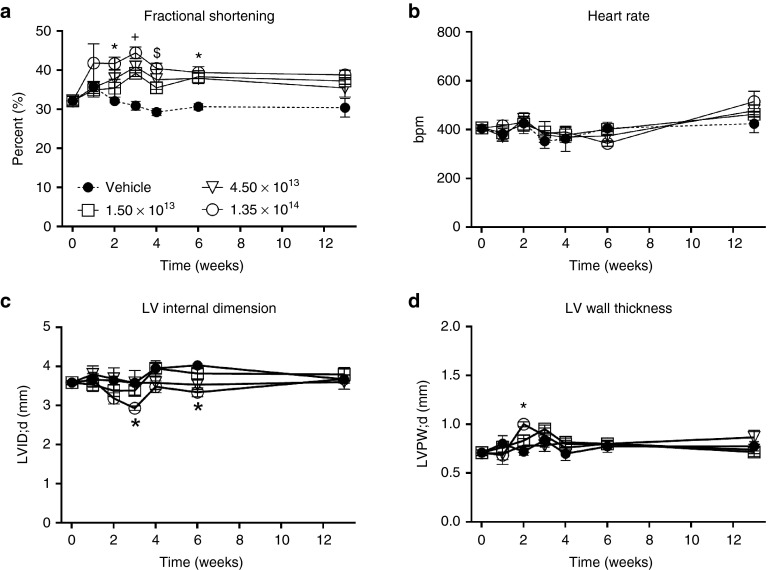
Prior to vector comparison studies, to obtain a vector dose-response measure of cardiac function, we performed a preliminary study with 3- to 5-month-old healthy mice systemically injected with 1.5 × 1013, 4.5 × 1013, or 1.35 × 1014 total vg/kg containing equal amounts of the rAAV6-ratR1cTnT455 (R1) and rAAV6-ratR2cTnT455 (R2) vectors. All three doses resulted in significantly elevated fractional shortening at 2, 3, 4, and 6 weeks postdelivery (Figure 2a), with no effect on heart rate (Figure 2b). At the end of 13 weeks, all three doses elicited an ~20–30% relative increase in fractional shortening as compared to vehicle treated hearts although this did not reach statistical significance (Figure 2a). During diastole, neither LV internal dimension (LVID;d; Figure 2c) nor LV posterior wall thickness (LVPDW;d; Figure 2d) was significantly altered in treated animals, particularly at the 4-week time point (Supplementary Table S2). These data suggest that all three doses of cTnT455-driven rat Rrm1 and Rrm2 expression result in a beat-to-beat elevation of cardiac performance in vivo. Importantly, there were no significant differences in diastolic dimensions, suggesting no evidence of LV dilation.
Figure 2.
Overexpression of rat R1 and R2 subunits enhances left ventricular systolic function in healthy adult mice. Serial measures of in-vivo cardiac function and morphometry obtained by echocardiography in mice receiving one of three doses (1.5, 4.5, and 1.35 × 1014 total vg/kg) containing equal amounts of each rAAV6-rat R1 and R2 vectors compared to saline-injected (control) hearts over a 13-week postinjection period. (a) Fractional shortening (FS); (b) heart rate; (c) left ventricular internal dimension in diastole (LVID;d); (d) Left ventricular poster wall thickness in diastole (LVPW;d). *P ≤ 0.05; 1.35 × 1013 vg/kg versus CON; ± P ≤ 0.05 All three doses versus CON; $P ≤ 0.05; 4.35 × 1013 and 1.35 × 1014 vg/kg versus CON (n = 4–6 per group per time point).
Langendorff isolated perfused hearts demonstrate significant improvements in ex vivo cardiac function
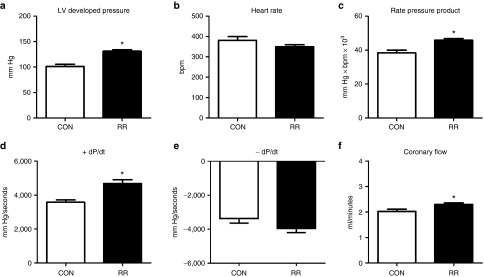
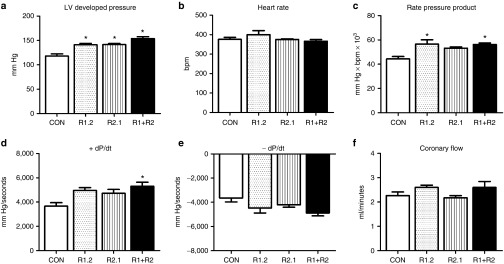
Next, 3- to 5-month-old healthy C57/B6J male mice (25–30 g) were treated with the R1 and R2 vectors at a dose of 7 × 1013 vg/kg, which comprises an intermediate (mid-range) dose to the 4.35 × 1013 vg/kg and 1.35 × 1014 vg/kg doses used in the 13-week study. After 4 weeks, the mice were euthanized and the hearts were subjected to Langendorff-isolated heart perfusions to determine the effect of RNR overexpression on ex-vivo cardiac function. Treatment significantly increased LV developed pressure (LVDevP) by ~30% without affecting spontaneous heart rate, leading to a significant elevation of the rate pressure product (Figure 3a–c). In addition, +dP/dt, an index of the rate of contractility, was ~30% higher in mouse hearts treated with the rat rAAV6 vectors (Figure 3d), while –dP/dt, a marker of ventricular relaxation, was ~15% faster but did not reach statistical significance (Figure 3e). Coronary flow was mildly, but significantly increased in treated hearts, consistent with perfusion–contraction matching due to increased myocardial workload (Figure 3f). These data demonstrate overexpression of rat RNR in adult, wild-type mice is effective in causing a significant elevation in measures of cardiac function.
Figure 3.
Isolated perfused hearts from adult mice with overexpressed R1 and R2 subunits exhibit increased cardiac function. Cardiac function was assessed in isolated perfused mouse hearts 4 weeks post-rAAV6 injection. (a) Left ventricular (LV) developed pressure (LV Dev P, the difference of LV systolic and LV diastolic pressures). (b) Heart rate (HR). (c) Rate pressure product (the product of LVDevP and HR). (d) Positive rate of pressure change calculated by the first derivative of the ascending LV pressure wave (+dP/dt), used as an index of the rate of pressure development. (e) Negative rate of pressure change calculated by the first derivative of the descending LV pressure wave (−dP/dt), used as an index of the rate of ventricular relaxation. (f) Coronary flow estimated by collecting the perfusate effluent over a 2-minute period. *P < 0.05 versus CON. CON, control; RR, heart from mouse treated with rAAV6-ratRrm1cTnT455 and rAAV6-ratRrm2cTnT455; n = 4 each group.
rAAV6 tissue transduction and endogenous and rAAV6-mediated RNR protein levels
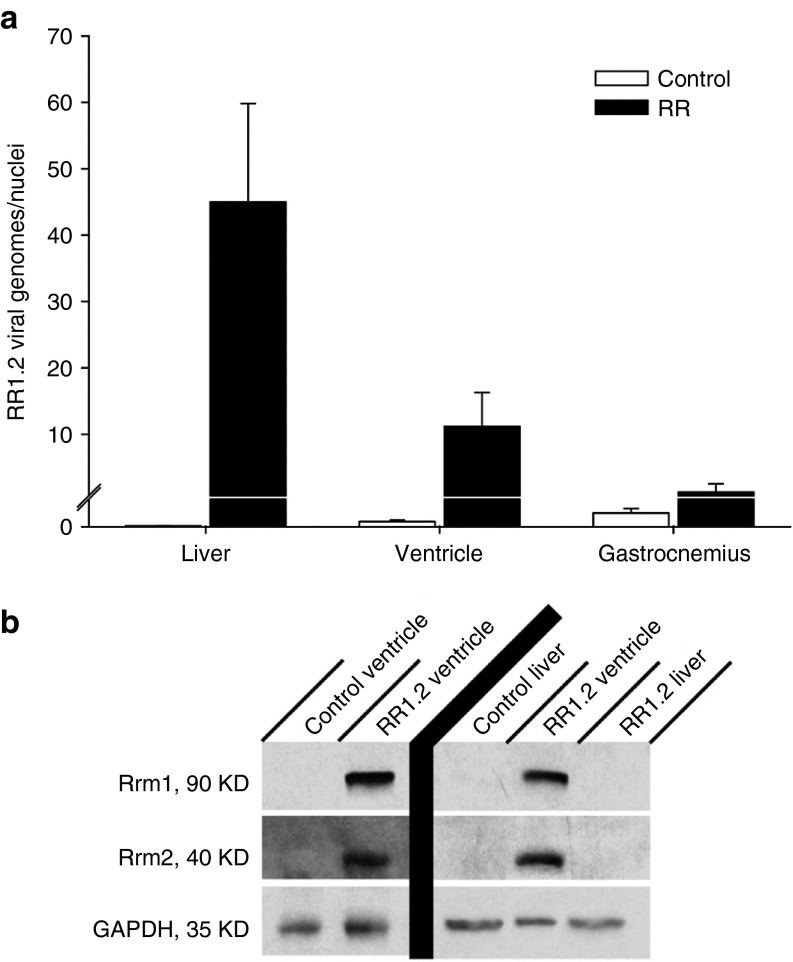
To assess the distribution of viral genomes 4 weeks following administration of the rAAV6 vectors, we performed qPCR on the liver, gastrocnemius, and left ventricle harvested from control and treated mice. qPCR analysis demonstrated significant viral genome (vg) uptake in the liver (45.0 ± 14.8 vg/nuclei) and ventricles (11.2 ± 5.1 vg/nuclei) of treated mice, with negligible uptake in the gastrocnemius (1.3 ± 1.3 vg/nuclei) (Figure 4a). Western blotting was used to assess endogenous and rAAV6-delivered Rrm1 and Rrm2 protein overexpression in ventricle and in liver tissue. As shown in Figure 4b, both Rrm1 and Rrm2 were significantly overexpressed by a similar amount in ventricles of treated mice, but not in ventricles of control mice. No Rrm1 or Rrm2 protein was detectible in the liver of control or treated mice, thus demonstrating the high specificity of the human cTnT455 regulatory cassette.
Figure 4.
rAAV6 transduction and Rrm1 and Rrm2 protein levels in mouse liver, heart, and skeletal muscle 4 weeks postsystemic rAAV6 delivery. (a) Quantitative polymerase chain reaction of healthy 3–5-month-old control and rAAV6-R1.2 vector-treated mice demonstrating significant viral genome (vg) uptake in the liver and ventricles, with negligible uptake in gastrocnemius. (b) Western blot analysis of liver and ventricle tissue. Protein was extracted from heart and liver from control and vector-treated mice and western blots were performed. GAPDH was used as a loading control. Significant overexpression of both Rrm1 and Rrm2 was detected in ventricles but not in the livers of treated mice, and essentially no expression of either endogenous subunit was detected in control heart or liver.
RNR overexpression does not affect heart weight or cardiac tissue histology
In the study described above, both heart weight and body weight were similar for control and treated animals, such that the heart weight/body weight ratio did not differ at 1 month post-treatment (Supplementary Table S3). Additionally, histological assessment of LV wall tissue showed no observable pathology in either group (Figure 5a,b), consistent with our previous observations in adult transgenic mice that chronically overexpress Rrm1 and Rrm2 systemically.18 These findings, combined with the echocardiography data described above, demonstrate that there is little or no compensatory adverse cardiac remodeling at 1 month post-treatment with R1 and R2 vectors.
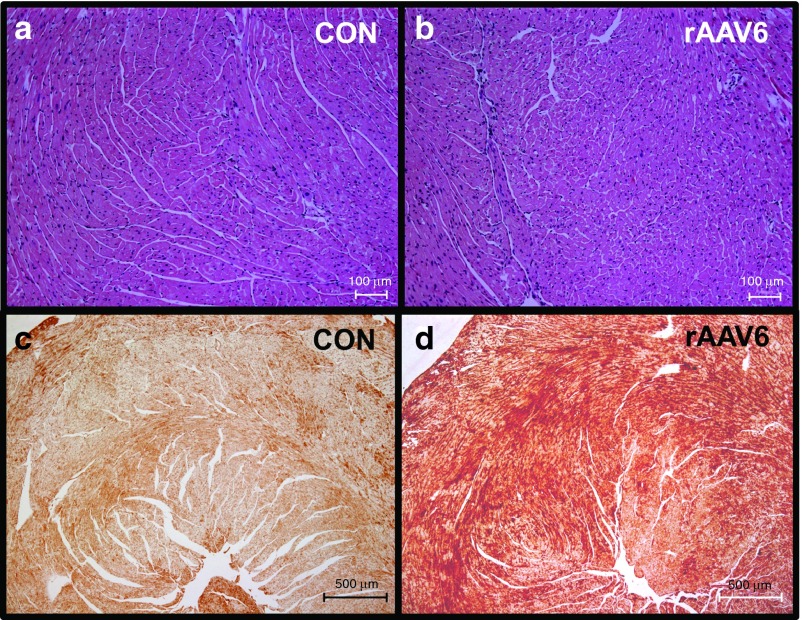
Figure 5.
Heart histology and Rrm1 distribution in rAAV6-rat(R1R2)cTnT455. At 1-month post-treatment, heart tissue was collected and sectioned for histological analysis of control (CON) and rAAV6-rat (R1R2)-treated mice. H & E staining (panels a and b) show morphology of ventricular tissue is not affected by overexpression of ribonucleotide reductase. Anti-Rrm1-stained sections taken at 4× (c and d) images show significant Rrm1 staining (brown) distributed evenly throughout the section, with much less Rrm1 staining in the CON.
Immunohistochemistry of ventricular tissue stained for Rrm1 showed significant overexpression and wide distribution of this protein throughout the ventricle in rAAV6-treated mice, with much lower levels in control tissue (Figure 5c,d). Together, these data demonstrate that systemic administration of rAAV6-R1R2cTnT455 results in cardiac-specific overexpression of Rrm1 and Rrm2. Importantly, although high levels of viral genomes were found in the liver, there was no detectable Rrm1 or Rrm2 protein expression in the livers of treated mice. Furthermore, liver mass did not differ between treated and control animals and there was no outward sign of disease as a result of viral genome presence at 1 month post-treatment (data not shown). These results validate the rAAV6-R1R2cTnT455 vector system and demonstrate that relatively low doses can be used as a tool to achieve cardiac specific RNR overexpression and enhanced ventricular function.
rAAV6-cTnT455 vectors for tandem overexpression of human RNR subunits
To test the effectiveness of human RNR and to assure that equivalent levels of each enzyme subunit would be produced in each transduced cardiomyocyte, we designed vectors carrying tandem RRM1 and RRM2 cDNAs separated by a 66 bp sequence encoding a self-cleaving P2A peptide. Since it was possible that retained portions of the P2A peptide at the N- and C-termini of RRM1 and RRM2 could adversely affect subunit functions, we designed tandem cDNAs in both the RRM1.RRM2 (R1.2) and RRM2.RRM1 (R2.1) 5′-to 3′ orders. When administered singly or in combination at a dose of 7 × 1013 vg/kg, vector treatment resulted in overexpression of both the RRM1 and RRM1 subunits (Supplementary Figure S2). As observed in mice treated with vector mixtures of AAV-carrying cTnT455-driven rat Rrm1 and Rrm2 cDNAs (Figure 3), all three human cDNA delivery strategies significantly increased LV DevP by ~20–30% without a significant effect on heart rate (Figure 6a,b). This translated into a similar increase in the rate pressure product that was significantly elevated in both the R1.R2 and R1+R2 treated hearts (Figure 6c). The +dP/dt was increased by ~30–45% in all treatment groups although the R1+R2 hearts were the sole group to reach statistical significance (Figure 6d). Similar to the overexpression of rat Rrm1 and Rrm2 subunits, the −dP/dt appeared faster in all treatment groups although no statistical significance was reached (Figure 6e). Despite increases in cardiac function, coronary flow remained similar in all groups (Figure 6f). Thus, the LV functional effects obtained with human RRM1 and RRM2 subunits were very similar to that obtained with rat subunits. Furthermore, overexpression of both human subunits from tandem cDNAs with an intervening self-cleaving peptide sequence was capable of eliciting increases in cardiac function.
Figure 6.
Overexpression of human RRM1 and RRM2 proteins increases cardiac function in isolated mouse hearts. Cardiac function was assessed in isolated mouse hearts perfused on a Langendorff apparatus. (a) Left ventricular (LV) developed pressure (LV Dev P, the difference of LV systolic and LV diastolic pressures). (b) Heart rate (HR). (c) Rate pressure product (the product of LVDevP and HR). (d) Positive rate of pressure change calculated by the first derivative of the ascending LV pressure wave (+dP/dt), used as an index of the rate of pressure development. (e) Negative rate of pressure change calculated by the first derivative of the descending LV pressure wave (−dP/dt), used as an index of the rate of ventricular relaxation. (f) Coronary flow estimated by collecting the perfusate effluent over a 2-minute period. *P < 0.05 versus CON. CON, control; R1.2, heart from mice treated with a rAAV6-human (RRM1.RRM2)cTnT455 vector in which the RRM1 cDNA is 5' of the RRM2 cDNA and separated by a self-cleaving peptide sequence; R2.1, treatment with an analogous vector with the RRM2 cDNA 5' of the RRM1 cDNA; (R1+R2), mouse heart with RRM1 and RRM2 cDNA containing vector; n = 4 each group.
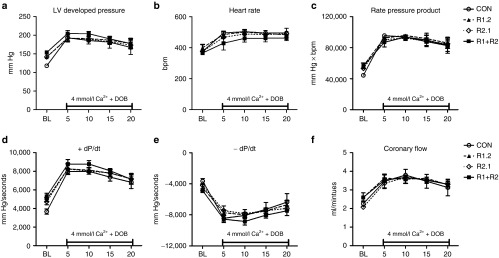
Although baseline cardiac function was elevated in the R1+R2 treated mouse hearts (Figure 6 and Supplementary Figure S3), it was possible that the response to an acute increase in cardiac workload could be impaired. Therefore, the response of the vector treated hearts to a high workload challenge was tested by administering a perfusate with a high calcium concentration combined with infusing a low dose of dobutamine. This pharmacological strategy increases the contractile force of the mouse heart with a simultaneous increase in heart rate. As shown in Figure 7, all hearts demonstrated a significant increase in cardiac functional parameters during the 20-minute high workload challenge. Importantly, all treated hearts (R1.2, R2.1, and R1+R2) exhibited responses comparable to the control group in all parameters measured (Figure 7a–f). These findings indicate that viral-directed overexpression of the RRM1 and RRM2 subunit in mouse hearts does not impair the response to an acute increase in cardiac workload despite elevated function at baseline.
Figure 7.
Overexpression of human RRM1 and RRM2 proteins does not impair the response to increased workload in isolated mouse hearts. Cardiac function in response to an acute increase in workload was examined in isolated mouse hearts using a Langendorff apparatus. The increase in workload was achieved by perfusing the heart with 4 mmol/l calcium and 50 nmol/l dobutamine for 20 minutes. (a) LV developed pressure (LV Dev P, the difference of LV systolic and LV diastolic pressures). (b) Heart rate (HR). (c) Rate pressure product (the product of LVDevP and HR). (d) Positive rate of pressure change calculated by the first derivative of the ascending LV pressure wave (+dP/dt), used as an index of the rate of pressure development. (e) Negative rate of pressure change calculated by the first derivative of the descending LV pressure wave (−dP/dt), used as an index of the rate of ventricular relaxation. (f) Coronary flow estimated by collecting the perfusate effluent over a 2-minute period. n = 4 each group. CON, control; R1.2, heart from mice treated with a rAAV6-human (RRM1.RRM2)cTnT455 vector in which the RRM1 cDNA is 5' of the RRM2 cDNA and separated by a self-cleaving peptide sequence; R2.1, treatment with an analogous vector with the RRM2 cDNA 5' of the RRM1 cDNA; (R1+R2), mouse heart with RRM1 and RRM2 cDNA containing vector; n = 4 each group.
Injection of rAAV6-rat(R1R2)cTnT455 in rescues in vivo cardiac function in rats subjected to MI
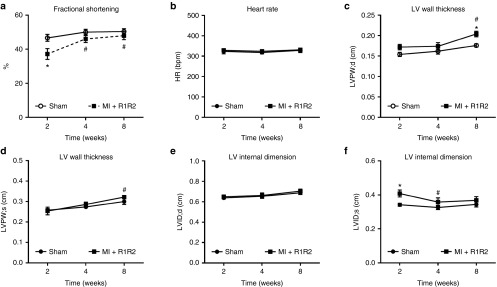
To explore the possibility of vector-driven overexpression of RNR to positively affect in vivo cardiac function in a disease model, we subjected rats to coronary artery ligation to induce MI. Five days post-MI, rats received an injection of rAAV6-R1R2cTnT455 (2.5 × 1013 vg/kg) into the noninfarcted myocardium and cardiac function was monitored with serial echocardiography. At 2 weeks post-MI, treated rats demonstrated a significantly decreased fractional shortening, but this recovered and remained at values comparable to sham surgery rats at 4 and 8 weeks (Figure 8a). LV poster wall thickness during both systole and diastole was significantly increased at 8 weeks in the treatment group (Figure 8c,d). Although no significant differences were noted in LVID in diastole (Figure 8e), LVID during systole significantly decreased at 4 weeks in the treatment group, which remained at 8 weeks (Figure 8f). These data indicate the potential for RNR vector delivery to improve and sustain in-vivo cardiac function in an animal model of MI.
Figure 8.
Injection of rAAV6-rat(R1R2)cTnT455 improves in-vivo cardiac function in rats subjected to myocardial infarction (MI). Direct injection of rAAV6-rat(R1R2) into the noninfarcted myocardium was made 5 days after ligation of the left anterior descending artery (MI ± R1R2) or sham surgery. Serial echocardiography measures were performed at 2, 4, and 8 weeks postinfarction. (a) Fractional shortening (FS); (b) heart rate; (c) Left ventricular (LV) poster wall thickness in diastole (LVPW;d); (d) Left ventricular poster wall thickness in systole (LVPW;s); (e) Left ventricular internal dimension in diastole (LVID;d); (f) LV internal dimension in systole (LVID;s). *P ≤ 0.05 versus Sham. #P ≤ 0.05 versus MI ± R1R2 (n = 5 each group).
Discussion
The purpose of this study was to develop a safe and effective rAAV6 that selectively overexpresses RNR in cardiomyocytes, resulting in enhanced contraction by elevating intracellular dATP levels. We first generated two rAAV6 vectors expressing the rat cDNA sequences of Rrm1 and Rrm2 under control of a human cardiac troponin-T regulatory cassette (cTnT455) and showed elevated cardiac function after systemic injection of mixtures containing an equal number of the separate vectors in healthy 3- to 5-month-old mice. Importantly, these results were consistent with the values obtained with both echocardiography and isolated perfused hearts in our previous report of transgenic RNR overexpressing mice.18 Next, we synthesized codon-optimized sequences of the human RRM1 and RRM2. To facilitate the delivery of both human RRM1 and RRM2 by a single rAAV6 vector, we then inserted a sequence encoding the self-cleaving P2A peptide between the two tandem gene sequences so that both RNR subunits would be expressed at similar levels. We also generated individual rAAV6 vectors containing human RRM1 and RRM2 genes for comparison to the separate rAAVs carrying rat R1 and R2 cDNAs, and to the single human vector containing both enzymatic subunits.
Systemic injection of rAAV6 carrying the tandem human cDNAs resulted in similar elevations of cardiac function, when measured by Langendorff perfusion, as observed following systemic injections of two separate vectors carrying either the human or rat cDNAs. The two-vector R1R2 delivery strategy is similar in its effectiveness to previous studies in which the delivery of cDNAs encoding the N- and C-terminal portions of mini-dystrophin with two separate rAAV vectors was beneficial to dystrophic mice.26,27,28 One potential consequence of elevated basal cardiac function is an increased energetic demand. In our previous study using transgenic mice, we observed an approximate 10% decrease in baseline myocardial energetic status as determined by the phosphocreatine to ATP ratio (PCr:ATP).18 When challenged with dobutamine, the maximum cardiac function achieved and the decline in PCr:ATP ratio was similar in both control and RR transgenic mice. Since the response to an acute increase in workload of hearts treated with human cDNAs was similar to control, it is likely that the energetic status is not comprised in our present study.
Although systemic delivery of AAV6 leads to vector uptake by both cardiac and noncardiac tissue, primarily liver and skeletal muscle, inclusion of the human cardiac-specific cTnt455 regulatory cassette in the rAAV6 constructs restricts the RNR subunit expression to cardiac muscle. This could enhance safety in conjunction with potential human trials, because overexpression of RNR in proliferating tissues could perturb growth controls.19,29 Although we detected rAAV6-delivered RRM1 throughout the ventricle, its expression, together with that of RRM2 exclusively to postmitotic cardiomyocytes, would be unlikely to activate their proliferation. Additionally, we observed no overt signs of toxicity in treated mice after 13 weeks. This is of importance since global overexpression of RNR in the transgenic mice has been associated with the development of lung neoplasms19 and the current study demonstrates the tight cardiac-specific transcriptional activity of the cTnT455 regulatory cassette. The elevation of basal cardiac function at 1 month postdelivery of rAAV6 vectors was very similar to results from our previous study of young adult transgenic mice that overexpress Rrm1 and Rrm2.18 In addition, the response to the calcium plus dobutamine administration was not elevated above controls, likely due to the achievement of maximum myocardial workload, which was consistent with our observations in transgenic mice.18 The transgenic mouse utilized a chicken β-actin promoter with a cytomegalovirus (CAG) enhancer, thereby overexpressing the enzyme subunits at high levels in all cell types, compared to the restricted cardiac-specific overexpression in the current study. Thus, the increased cardiac function in both studies is likely due to a cardiac-specific effect of elevated dATP. This hypothesis is supported by in vitro, reductionist studies in which we have demonstrated that similar levels of Rrm1 and Rrm2 overexpression in cultured adult rat cardiomyocytes (via adenoviral transduction of Rrm1 and Rrm2 using a cytomegalovirus promoter) leads to significant increases in the magnitude and rate of shortening, and an acceleration of relaxation.17 The level of dATP in these cells was increased ~10-fold, becoming ~1% of the total adenosine triphosphate nucleotide pool. Obtaining higher levels of dATP in cardiomyocytes might be even more effective in improving cardiac function, but may be difficult to achieve by overexpression of Rrm1 and Rrm2 due to the autoregulation of RNR (i.e., RNR is stimulated by ATP and inhibited by dATP binding to the allosteric site with a feedback inhibition (Ki) of ~50 µmol/l30,31). Since ATP levels are 5–8 mmol/l in cardiomyocytes, this Ki may effectively limit cellular levels to ~1–2% of the total adenosine triphosphate pool. While methods such as mutation of the Rrm1 A-site can elevate cellular dATP levels even further,30,32,33 the levels achieved by our approach appear to be sufficient to have powerful positive effects on cardiac muscle contraction.
The mechanisms underlying dATP-mediated enhancement of cardiac function are myofilament specific and have been investigated in our previous studies.11,12,13,14,15,17,34,35 Replacing ATP with dATP increases cross-bridge binding and enhances both the magnitude and rate of force development.11,12,13,14 Our recent molecular dynamics simulations of myosin suggest that with dADP.Pi in the nucleotide binding pocket there are local (pocket) structural changes associated with loss of the Phe 129 contact from the missing oxygen at the 2' position of the ribose ring. This results in translated structural changes on the actin binding surface that increase myosin affinity for actin.36 We (ref. 14) and others37,38 have shown that contractile activation is very dependent on initial cross-bridge binding and is highly cooperative. Thus, enhanced myosin binding affinity with dATP provides a mechanism whereby large increases in contractile activation occur with comparatively small, early increases in cross-bridge binding, especially in the range of [Ca2+]i concentrations that occur during a cardiac twitch. Importantly, this highly cooperative process means that small amounts of dATP (e.g., 1% of the ATP pool) are sufficient to achieve substantial increases in contractility. While other cellular ATPases may be able to utilize dATP, this low level in cells likely precludes any significant effect on their activity. After dATP is hydrolyzed by myosin, it can be rephosphorylated many times by creatine kinase and other enzymes, so increases in dATP can confer long-lived effects on contractility.12
Implications for translational research
We recently reported that dATP enhances contraction in chemically-demembranated multicellular ventricular wall preparations and in isolated myofibrils from human subjects with end-stage heart failure.16 Additionally, we have recently reported that administration of the human AAV6-RRM1.RRM2cTnt455 vector via coronary catheter (a limited delivery approach) restores much of the lost function when delivered 2 weeks following MI in Yucatan mini-pigs, as measured by echocardiographic and hemodynamic parameters, at 1 and 2 months post-treatment.25 Also noted in this study was the potential of the treatment to impact measures of diastolic function, a notable effect that could have additional therapeutic potential. Importantly, no safety or toxicity issues were observed. This study, together with our current study, is an important step in moving this therapeutic approach toward the clinic.
It is important to note that the present study has several limitations that need to be evaluated further in future studies of large animal models as our therapeutic strategy moves toward clinical testing. First, we elected not to use a control viral vector in this study as the goal was to assess the resultant effect of the treatment and this would not be the standard in clinical trials. In addition, rAAV6 has been previously used in rodents without the observation of significantly altered cardiac function.39 Second, the fractional shortening values obtained in the studies with mice were relatively low compared with commonly reported values and is most likely due to an anesthesia effect.40,41 However, since heart rates are similar between groups throughout the time course, the increase in fractional shortening in the treatment groups are valid and consistent with the data obtained in the isolated heart perfusion experiments. Third, this study's primary focus was to identify the optimal dose that could elicit changes in cardiac function in small animal models under both healthy and infarcted hearts. Our recent paper has extended these findings in a large animal model.25 However, studies examining the long-term efficacy on our viral delivery strategy are definitely required and are forthcoming.
Despite the limitations presented above, there are several features that make this gene therapy approach unique, including: (i) the overexpressed RRM1 and RRM2 proteins are not replacing a defective or missing protein but instead changes transcriptional regulation by the tandem expression of RRM1 and RRM2 through the TnT regulatory cassette result in the elevated production of RNR and subsequently, dATP, which is the therapeutic agent. It should be noted that the post-transcriptional regulatory mechanisms that regulate the levels of dATP are still in place and prevent the accumulation of abnormally high levels of dATP; (ii) the therapeutic elevation of endogenous RRM1 and RRM2 enzyme levels would not be anticipated to cause any immunological issues as they are self-antigens; (iii) this is the first proposed use of a cellular nucleotide manipulation to improve in vivo cardiac function. dATP is unique in that it is a natural cellular nucleotide that can be used by myosin to augment contraction; (iv) both contraction and relaxation of cardiac muscle are enhanced by dATP, thus potentially improving systolic and diastolic function; and (v) the dATP produced by transduced cells can diffuse through gap junctions to surrounding cells, making transduced cells resident drug (nucleotide) delivery devices.42 Thus, transduction of only a minority of cardiomyocytes may be sufficiently effective in eliciting robust functional changes. Our previous report that even relatively small RNR-expressing grafts of human stem cell-derived cardiomyocytes can substantially enhance ventricular function when transplanted into normal adult rat hearts supports this idea.42
Materials and Methods
Cardiac-specific regulatory cassette. cTnT455 was derived from the human cardiac troponin T (TNNT2) gene. TNNT2's cardiac-specific transcriptional activity and the approximate 5'- and 3'-boundaries of its promoter were originally described in the chicken and rat.43,44 We subsequently aligned the 5'-sequences from human and other mammalian species, and carried out an extensive deletion analysis within the (−447 to +48) portion of the human sequence to identify conserved and nonconserved subregions that could be eliminated without substantially reducing its transcriptional activity in transiently transfected neonatal rat cardiomyocytes. These assays also led to delineation of “separate” enhancer and proximal promoter regions of human TNNT2, and disclosed that inserting two copies of the 130 bp miniaturized enhancer 5' of the miniaturized promoter provided significantly higher transcriptional activity than a single copy. The resulting 455 base pair “cTnT455” regulatory cassette was then ligated into the pAAV-hPLAP plasmid and used to generate rAAV6 vectors by the Chamberlain and Odom lab. These were tested for transcriptional activity in normal C57BL/6 mice following retro-orbital injections and were expressed at high levels in ventricular cardiac muscle and at only background levels (0.06–2%) in all other tissues, including skeletal muscle. cTnT455 is available upon request, and a complete description of its stepwise derivation, final sequence, and transcriptional characterization will be published elsewhere.
Generation of rAAV6-R1R2cTnT455 vectors. The pAAV-cTnT455-hPLAP plasmid was prepared in the Vector Core of the Seattle Wellstone Muscular Dystrophy Research Center (and kindly provided by Center Director, Jeff Chamberlain, University of Washington, Seattle, WA) and used as backbone to subclone PCR fragments containing the rat RNR cDNAs Rrm1 and Rrm2. Briefly, the Rrm1 cDNA was PCR amplified using the following primer sequences: (forward: AGTCGACatgcatgtgatcaagcgagat; reverse: gatatctcaggatccacacatcagacactc). This amplicon was subsequently ligated into the pAAV-cTnT455 vector using the restriction enzymes SalI and EcoRV while the pAAV-cTnT455-hPLAP was digested with HindIII, treated with Klenow enzyme to blunt-end the DNA fragments followed by purification and a second digestion with SalI and subsequently sequence verified. To generate the pAAV-cTnT455-RR2, we performed a SalI and HindIII double digestion of the pAAV-cTnT455-hPLAP. The PCR product generated the RR2 coding sequence containing the SalI/HindIII flanking 5' and 3', respectively. Following a standard ligation reaction and transformation into Sure2 Escherichia coli, a single clone containing the RR2 insert was confirmed by restriction analysis (SalI and HindIII) followed by sequence verification. The primer sequences utilized to generate the Rrm2 amplicon were: forward: GTCGACatgctctcggtccgcgccccgct; reverse: aagcttttagaagtcagcatccaaggtgaa. For studies with rat RNR, the two subunits were carried in separate AAV6 vectors, each with the cTnT455 regulatory cassette; AAV6-ratR1cTnT455 (R1) + AAV6-ratR2cTnT455 (R2). For ease of reading, we have designated this dual vector system rAAV6-ratR1R2cTnT455 (R1R2).
Human vector constructs. The final construct was synthesized by Genscript (Piscataway, NJ) and contained the following elements: human cTnT455 RRM1-P2A-RRM2-polyA signal flanked by NotI sites. The P2A self-cleaving peptide sequence was encoded by a 66 bp sequence45 and was inserted between the subunit cDNAs in two different tandem arrays: RRM1-P2A-RRM2 and RRM2-P2A-RRM1. Human RRM1 (NM_001033.3) and RRM2 (NM_001034) sequences were codon optimized for expression in human using Genscript's OptimumGene Codon Optimization Analysis (includes analysis of codon usage bias, GC content, CpG dinucleotide content, mRNA secondary structure, cryptic splicing sites, premature polyA sites, internal chi sites and ribosomal binding sites, negative CpG islands, RNA instability motifs, repeat sequences, and restriction sites that may interfere with cloning). The synthesized construct was sequenced and cloned into the pAAVhr-green fluorescent protein (GFP) recipient vector (Stratagene, San Diego, CA) in place of GFP using NotI/NotI sites.
Virus production. Recombinant AAV6 vector was produced and prepared as previously described.46,47 Briefly, rAAV6 vectors were produced by CaPO4 cotransfection of HEK 293D cells with plasmids containing rAAV-cTnT455-RR1.2-SpA vector genomes flanked by AAV2 ITR's and pDGM6 packaging/helper genes.48 Cells were harvested and processed through a microfluidizer (Microfluidics, Newton, MA), filtered through a 0.22-μm filter, and vector particles were purified by affinity chromatography on a HiTrap heparin column (GE Healthcare, Chalfont St. Giles, UK). The rAAV6 eluate was then layered onto a 40% sucrose gradient and concentrated by ultracentrifugation at 27,000 RPM for 18 hours at 4 °C, followed by solubilization in Hanks buffered saline solution (Invitrogen, Carlsbad, CA). Vector genome titer was determined by Southern blot analysis with a DNA standard of known quantity using a 32P-labeled oligonucleotide probe to the SV40 polyadenylation signal (5′-CACTGCATTCTAGTTGTGGTTTGTC-3′) or the rabbit β-globin poly-adenylation signal (5′-TGAATAAAAGATCCTTA-3′) when titering rAAV6-cTnT455-human RRM1 and RRM2 vectors.
Viral delivery. Three- to five-month-old C57BL/6 mice were used for intravenous delivery of the virus. All animal experiments were approved by the University of Washington (UW) Animal Care Committee and were carried out in accordance with federal guidelines. Animals were housed in the Department of Comparative Medicine at the UW and were cared for in accordance with the US NIH Policy on Humane Care and Use of Laboratory Animals.
Treated mice received injections of the rAAV6 vector diluted in Hanks buffered saline solution while control mice received injections of Hanks buffered saline solution alone. All injections were administered in a volume of 0.2 ml via the right retro-orbital sinus route while the mice were under anesthesia with isoflurane. For the dose–response experiments, mice (25–30 g) were injected with a dose of either 1.5 × 1013, 4.5 × 1013, or 1.35 × 1014 total vg/kg containing equal amounts of the rAAV6-ratR1cTnT455 (R1) and rAAV6-ratR2cTnT455 (R2) vectors. A second cohort of mice received an injection of 7 × 1013 vg/kg of each rAAV6-ratR1cTnT455 (R1) and rAAV6-ratR2cTnT455 (R2) vector. A third cohort received injections of the human cDNAs RRM1.RRM2 (R1.2), RRM2.RRM1 (R2.1), or RRM1 and RRM2 (R1 ± R2) at 7 × 1013 vg/kg for each vector.
Myocardial infarction model. Adult male rats (F344/NHsd) received permanent ligation of the left anterior descending artery to induce MI. An additional cohort of rats undergoing sham surgery went through a similar procedure but without ligation of the left anterior descending. Five days post-MI, rats received an injection of 2.5 × 1013 vg/kg of each rAAV6-ratR1cTnT455 (R1) and rAAV6-ratR2cTnT455 (R2) into the noninfarcted myocardium.
Protein and DNA analysis. Total DNA was isolated from frozen heart tissue using the DNeasy Mini Kit (Qiagen, Hilden, Germany) according to manufacturer's guidelines. qPCR was performed using iTaq Universal Probes Supermix (Bio-Rad, Hercules, CA) with the following primer/probe sequences: cTnT455: forward: CCCAGTCCCCGCTGAGA; 5′ UTR RRM1l; reverse: AGGTTCCAGGCGCTGCT; probe: (5FAM) TGAGCAGACGCCTCCAGGATCTGTC (BHQ1a-5FAM). Pax7: forward: CAAGGCCGGGTCAATCAG, reverse: AGATGACA CAGGGCCGGA; probe: (5HEX) CGACCCCTGCCTAACCACATCCG (BHQ1a-5HEX).
All samples were run in triplicates, and average quantities are reported unless otherwise indicated. A standard curve was run on each plate using a plasmid that contained a mouse Pax7 gene as well as the plasmid vector used for virus production. Vector genomes per nuclei were calculated by normalizing to Pax7 genomes in each sample.
For protein analysis, frozen heart tissue was prepared for western blot analysis as previously described.17 The following primary antibodies were used: anti-Rrm1 (t-16) Goat polyclonal IgG (Santa Cruz Biotechnology, Dallas, TX); anti-Rrm2 (E-16) Goat polyclonal IgG (Santa Cruz Biotechnology), and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) Rabbit polyclonal IgG (Rockland Immunochemicals, Limerick, PA). Protein band density was quantified using open access software, Image J (National Institutes of Health, Bethesda, MD). Each band was normalized to the protein standard, GAPDH.
Echocardiography. The animals were anesthetized using 1% isofluorane. Images were collected with a GE Vivid 7 system using 13MHz linear transducer (General Electric Healthcare, Chalfont, United Kingdom). LV end-diastolic (LVEDD) and LV end-systolic (LVESD) dimensions were determined using M-mode measurements obtained by short-axis views at the mid-papillary level. All data were averaged from at least three cardiac cycles. Fractional shortening was calculated from these data using the formula: (LVEDD-LVESD)/LVEDD × 100. All echocardiography was carried out by a single reader who was blinded to the treatment of the animals.
In vitro function. LV function was measured in Langendorff isolated heart preparations as previously described.18,49 In brief, excised mouse hearts were perfused at a constant pressure of 80 mmHg with a modified Krebs Henseleit buffer consisting of (mmol/l): 118 sodium chloride (NaCl), 25 sodium carbonate (NaHCO3), 5.3 potassium chloride (KCl), 2.0 calcium chloride (CaCl2), 1.2 magnesium sulfate (MgSO4), 0.5 ethylenediaminetetraacetic acid (EDTA), 5.5 glucose, and 0.5 pyruvate, equilibrated with 95% O2 and 5% CO2 (pH 7.4). Temperature was maintained at 37.5 °C throughout the protocol. After 20 minutes of equilibration, baseline function was monitored for 10–20 minutes at a fixed end diastolic pressure of 8–10 mmHg by way of a water-filled balloon inserted into the LV. After baseline measurements, the calcium concentration in the perfusate was changed from 2 to 4 mmol/l and dobutamine was infused at 5% of the coronary flow at a final concentration of 50 nmol/l for 20 minutes to simulate an acute increase in cardiac work (high workload challenge). During the entire experimental protocol, a continuous recording of LV function (via the balloon connected to a pressure transducer) was monitored with the use of a data acquisition system (PowerLab, ADInstruments, Colorado Springs, CO).
Histology. Hearts from anesthetized mice were rapidly excised, rinsed in phosphate-buffered saline, and sliced into 2-mm-thick short-axis sections. A minimum of three sections were taken from each heart, starting at the apex. All sections were fixed in formalin, processed, and paraffin-embedded. Paraffin blocks were section at 4 µm. To assess tissue morphometry, staining with hematoxylin and eosin (H&E) was performed using a standard protocol, dehydrated through an ethanol gradient, and coverslipped using mounting media. In adjacent sections, immunostaining was performed with primary antibodies against Rrm1 (Goat polyclonal anti-R1 (1:50), Santa Cruz Biotechnology). Samples were then labeled with a biotinylated rabbit anti-Goat (1:500) secondary antibody (Thermo Scientific Pierce Antibodies, Grand Island, NY) and developed with 3,3′-diaminobenzidine (Sigma-Aldrich, St Louis, MO) and mounted.
Statistical analysis. A two-tailed Student's t-test was used for comparisons involving two groups. One-way analysis of variance followed by Bonferroni's multiple comparison test was used for multiple group comparisons. Analysis of variance with repeated measures followed by Bonferroni's multiple comparison tests was used for time-course comparisons. Significance was tested at the P < 0.05 level using GraphPad Prism 5.0 (GraphPad Software, San Diego, CA). Data are presented as mean ± standard error of the mean.
SUPPLEMENTARY MATERIAL Table S1. Single and dual tandem R1 and R2 cDNA constructs of recombinant AAV vectors. Table S2. In-vivo echocardiography data from mice treated with graded vector doses containing equal concentrations of AAV6-rat R1cTnt455 and AAV6-rat R1cTnt455. Table S3. Heart weight and body weight measurements of control and rAAV6 R1-R2 treated mice. Figure S1. Plasmid map for rAAV production enabling cardiac specific overexpression of human R1R2. Figure S2. Rrm1 and Rrm2 protein expression in hearts from mice treated with human vectors. Figure S3. Ejection fraction of mice treated with human vectors.
Acknowledgments
This work was supported by grants from the National Institutes of Health (NIH): R01 HL111197 (to M.R. and S.D.H.), R01 HD048895 (to M.R.), R01 HL084642, P01HL094374, P01 GM81619, and U01 HL100405 (to C.E.M.), and 1U54 AR065139-01A1 (to S.D.H. and G.L.O.). S.C.K. is funded by the American Heart Association 14SDG18590020. G.L.O. and S.D.H. are also funded by the Muscular Dystrophy Association. S.G.N. is funded by NIH T32 EB001650. F.M.-H. is funded by T32 HL07828 and an American Heart Association post-doctoral fellowship. M.R. is an Established Investigator of the American Heart Association. Viral vectors were developed and produced in the University of Washington Senator Paul D. Wellstone Muscular Dystrophy Cooperative Research Center, with partial support from BEAT Biotherapeutics, Inc. M.R. and C.E.M. are co-founders and equity holders in BEAT Biotherapeutics, Inc. G.G.M. is a consultant for BEAT Biotherapeutics, Inc.
Supplementary Material
References
- Go, AS, Mozaffarian, D, Roger, VL, Benjamin, EJ, Berry, JD, Borden, WB et al.; American Heart Association Statistics Committee and Stroke Statistics Subcommittee. (2013). Heart disease and stroke statistics–2013 update: a report from the American Heart Association. Circulation 127: e6–e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray, JJ (2010). Clinical practice. Systolic heart failure. N Engl J Med 362: 228–238. [DOI] [PubMed] [Google Scholar]
- Abraham, WT, Adams, KF, Fonarow, GC, Costanzo, MR, Berkowitz, RL, LeJemtel, TH et al.; ADHERE Scientific Advisory Committee and Investigators; ADHERE Study Group. (2005). In-hospital mortality in patients with acute decompensated heart failure requiring intravenous vasoactive medications: an analysis from the Acute Decompensated Heart Failure National Registry (ADHERE). J Am Coll Cardiol 46: 57–64. [DOI] [PubMed] [Google Scholar]
- Teerlink, JR, Metra, M, Zacà, V, Sabbah, HN, Cotter, G, Gheorghiade, M et al. (2009). Agents with inotropic properties for the management of acute heart failure syndromes. Traditional agents and beyond. Heart Fail Rev 14: 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjar, RJ (2013). Potential of gene therapy as a treatment for heart failure. J Clin Invest 123: 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessup, M, Greenberg, B, Mancini, D, Cappola, T, Pauly, DF, Jaski, B et al.; Calcium Upregulation by Percutaneous Administration of Gene Therapy in Cardiac Disease (CUPID) Investigators. (2011). Calcium Upregulation by Percutaneous Administration of Gene Therapy in Cardiac Disease (CUPID): a phase 2 trial of intracoronary gene therapy of sarcoplasmic reticulum Ca2+-ATPase in patients with advanced heart failure. Circulation 124: 304–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsebo, K, Yaroshinsky, A, Rudy, JJ, Wagner, K, Greenberg, B, Jessup, M et al. (2014). Long-term effects of AAV1/SERCA2a gene transfer in patients with severe heart failure: analysis of recurrent cardiovascular events and mortality. Circ Res 114: 101–108. [DOI] [PubMed] [Google Scholar]
- Morgan, BP, Muci, A, Lu, PP, Qian, X, Tochimoto, T, Smith, WW et al. (2010). Discovery of omecamtiv mecarbil the first, selective, small molecule activator of cardiac Myosin. ACS Med Chem Lett 1: 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teerlink, JR (2009). A novel approach to improve cardiac performance: cardiac myosin activators. Heart Fail Rev 14: 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teerlink, JR, Clarke, CP, Saikali, KG, Lee, JH, Chen, MM, Escandon, RD et al. (2011). Dose-dependent augmentation of cardiac systolic function with the selective cardiac myosin activator, omecamtiv mecarbil: a first-in-man study. Lancet 378: 667–675. [DOI] [PubMed] [Google Scholar]
- Regnier, M and Homsher, E (1998). The effect of ATP analogs on posthydrolytic and force development steps in skinned skeletal muscle fibers. Biophys J 74: 3059–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier, M, Lee, DM and Homsher, E (1998). ATP analogs and muscle contraction: mechanics and kinetics of nucleoside triphosphate binding and hydrolysis. Biophys J 74: 3044–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier, M, Rivera, AJ, Chen, Y and Chase, PB (2000). 2-deoxy-ATP enhances contractility of rat cardiac muscle. Circ Res 86: 1211–1217. [DOI] [PubMed] [Google Scholar]
- Regnier, M, Martin, H, Barsotti, RJ, Rivera, AJ, Martyn, DA and Clemmens, E (2004). Cross-bridge versus thin filament contributions to the level and rate of force development in cardiac muscle. Biophys J 87: 1815–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari, BB, Regnier, M, Rivera, AJ, Kreutziger, KL and Martyn, DA (2004). Cardiac length dependence of force and force redevelopment kinetics with altered cross-bridge cycling. Biophys J 87: 1784–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussavi-Harami, F, Razumova, MV, Racca, AW, Cheng, Y, Stempien-Otero, A and Regnier, M (2015). 2-Deoxy adenosine triphosphate improves contraction in human end-stage heart failure. J Mol Cell Cardiol 79: 256–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte, FS, Dai, J, Buckley, K, Feest, ER, Adamek, N, Geeves, MA et al. (2011). Upregulation of cardiomyocyte ribonucleotide reductase increases intracellular 2 deoxy-ATP, contractility, and relaxation. J Mol Cell Cardiol 51: 894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski, SG, Kolwicz, SC, Korte, FS, Luo, Z, Robinson-Hamm, JN, Page, JL et al. (2013). Transgenic overexpression of ribonucleotide reductase improves cardiac performance. Proc Natl Acad Sci USA 110: 6187–6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X, Page, JL, Surtees, JA, Liu, H, Lagedrost, S, Lu, Y et al. (2008). Broad overexpression of ribonucleotide reductase genes in mice specifically induces lung neoplasms. Cancer Res 68: 2652–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasano, T, Kikuchi, K, McDonald, AD, Lai, S and Donahue, JK (2007). Targeted high-efficiency, homogeneous myocardial gene transfer. J Mol Cell Cardiol 42: 954–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, G, Bish, LT, Sleeper, MM, Mu, X, Sun, L, Lou, Y et al. (2011). Transendocardial delivery of AAV6 results in highly efficient and global cardiac gene transfer in rhesus macaques. Hum Gene Ther 22: 979–984. [DOI] [PubMed] [Google Scholar]
- Katz, MG, Fargnoli, AS, Swain, JD, Tomasulo, CE, Ciccarelli, M, Huang, ZM et al. (2012). AAV6-βARKct gene delivery mediated by molecular cardiac surgery with recirculating delivery (MCARD) in sheep results in robust gene expression and increased adrenergic reserve. J Thorac Cardiovasc Surg 143: 720–726.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, JM, Navarro, S, Doki, T, Stewart, JM, Mitsuhashi, N and Kearns-Jonker, M (2012). Gene transfer of heme oxygenase-1 using an adeno-associated virus serotype 6 vector prolongs cardiac allograft survival. J Transplant 2012: 740653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raake, PW, Schlegel, P, Ksienzyk, J, Reinkober, J, Barthelmes, J, Schinkel, S et al. (2013). AAV6.βARKct cardiac gene therapy ameliorates cardiac function and normalizes the catecholaminergic axis in a clinically relevant large animal heart failure model. Eur Heart J 34: 1437–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota, S, Carey, J, Reinecke, H, Leggett, J, Teichman, S, Laflamme, MA et al. (2015). Ribonucleotide reductase-mediated increase in dATP improves cardiac performance via myosin activation in a large animal model of heart failure. Eur J Heart Fail 17: 772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, A, Yue, Y, Lai, Y and Duan, D (2008). A hybrid vector system expands adeno-associated viral vector packaging capacity in a transgene-independent manner. Mol Ther 16: 124–130. [DOI] [PubMed] [Google Scholar]
- Ghosh, A, Yue, Y, Shin, JH and Duan, D (2009). Systemic Trans-splicing adeno-associated viral delivery efficiently transduces the heart of adult mdx mouse, a model for duchenne muscular dystrophy. Hum Gene Ther 20: 1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom, GL, Gregorevic, P, Allen, JM and Chamberlain, JS (2011). Gene therapy of mdx mice with large truncated dystrophins generated by recombination using rAAV6. Mol Ther 19: 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aye, Y, Li, M, Long, MJ and Weiss, RS (2015). Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies. Oncogene 34: 2011–2021. [DOI] [PubMed] [Google Scholar]
- Caras, IW and Martin, DW Jr (1988). Molecular cloning of the cDNA for a mutant mouse ribonucleotide reductase M1 that produces a dominant mutator phenotype in mammalian cells. Mol Cell Biol 8: 2698–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, A and Reichard, P (1998). Ribonucleotide reductases. Annu Rev Biochem 67: 71–98. [DOI] [PubMed] [Google Scholar]
- Reichard, P, Eliasson, R, Ingemarson, R and Thelander, L (2000). Cross-talk between the allosteric effector-binding sites in mouse ribonucleotide reductase. J Biol Chem 275: 33021–33026. [DOI] [PubMed] [Google Scholar]
- Fairman, JW, Wijerathna, SR, Ahmad, MF, Xu, H, Nakano, R, Jha, S et al. (2011). Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization. Nat Struct Mol Biol 18: 316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreutziger, KL, Piroddi, N, McMichael, JT, Tesi, C, Poggesi, C and Regnier, M (2011). Calcium binding kinetics of troponin C strongly modulate cooperative activation and tension kinetics in cardiac muscle. J Mol Cell Cardiol 50: 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier, M, Rivera, AJ, Chase, PB, Smillie, LB and Sorenson, MM (1999). Regulation of skeletal muscle tension redevelopment by troponin C constructs with different Ca2+ affinities. Biophys J 76: 2664–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski, SG, Adamek, N, Geeves, MA, Gay, E, Kolwicz Jr, SC, Murry, CE, et al. (2013). 2-deoxy-ATP alters myosin structure to enhance cross-bridge cycling and improve cardiac function. Biophys J 104:17a. [Google Scholar]
- McDonald, KS and Moss, RL (2000). Strongly binding myosin crossbridges regulate loaded shortening and power output in cardiac myocytes. Circ Res 87: 768–773. [DOI] [PubMed] [Google Scholar]
- Dobesh, DP, Konhilas, JP and de Tombe, PP (2002). Cooperative activation in cardiac muscle: impact of sarcomere length. Am J Physiol Heart Circ Physiol 282: H1055–H1062. [DOI] [PubMed] [Google Scholar]
- Masson, R, Nicklin, SA, Craig, MA, McBride, M, Gilday, K, Gregorevic, P et al. (2009). Onset of experimental severe cardiac fibrosis is mediated by overexpression of Angiotensin-converting enzyme 2. Hypertension 53: 694–700. [DOI] [PubMed] [Google Scholar]
- Gao, S, Ho, D, Vatner, DE and Vatner, SF (2011). Echocardiography in Mice. Curr Protoc Mouse Biol 1: 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachon, RE, Scharf, BA, Vatner, DE and Vatner, SF (2015). Best anesthetics for assessing left ventricular systolic function by echocardiography in mice. Am J Physiol Heart Circ Physiol 308: H1525–H1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundy, SD, Murphy, SA, Dupras, SK, Dai, J, Murry, CE, Laflamme, MA et al. (2014). Cell-based delivery of dATP via gap junctions enhances cardiac contractility. J Mol Cell Cardiol 72: 350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mar, JH, Antin, PB, Cooper, TA and Ordahl, CP (1988). Analysis of the upstream regions governing expression of the chicken cardiac troponin T gene in embryonic cardiac and skeletal muscle cells. J Cell Biol 107: 573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G, Yeh, HI and Lin, JJ (1994). Characterization of cis-regulating elements and trans-activating factors of the rat cardiac troponin T gene. J Biol Chem 269: 30595–30603. [PubMed] [Google Scholar]
- Kim, JH, Lee, SR, Li, LH, Park, HJ, Park, JH, Lee, KY et al. (2011). High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6: e18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbert, CL, Allen, JM and Miller, AD (2001). Adeno-associated virus type 6 (AAV6) vectors mediate efficient transduction of airway epithelial cells in mouse lungs compared to that of AAV2 vectors. J Virol 75: 6615–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankinship, MJ, Gregorevic, P, Allen, JM, Harper, SQ, Harper, H, Halbert, CL et al. (2004). Efficient transduction of skeletal muscle using vectors based on adeno-associated virus serotype 6. Mol Ther 10: 671–678. [DOI] [PubMed] [Google Scholar]
- Gregorevic, P, Blankinship, MJ, Allen, JM, Crawford, RW, Meuse, L, Miller, DG et al. (2004). Systemic delivery of genes to striated muscles using adeno-associated viral vectors. Nat Med 10: 828–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolwicz, SC, Jr, Tian, R (2010). Assessment of cardiac function and energetics in isolated mouse hearts using 31P NMR spectroscopy. J Vis Exp 42:e2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.