Abstract
Cerebrospinal fluid administration of recombinant adeno-associated viral (rAAV) vectors has been demonstrated to be effective in delivering therapeutic genes to the central nervous system (CNS) in different disease animal models. However, a quantitative and qualitative analysis of transduction patterns of the most promising rAAV serotypes for brain targeting in large animal models is missing. Here, we characterize distribution, transduction efficiency, and cellular targeting of rAAV serotypes 1, 2, 5, 7, 9, rh.10, rh.39, and rh.43 delivered into the cisterna magna of wild-type pigs. rAAV9 showed the highest transduction efficiency and the widest distribution capability among the vectors tested. Moreover, rAAV9 robustly transduced both glia and neurons, including the motor neurons of the spinal cord. Relevant cell transduction specificity of the glia was observed after rAAV1 and rAAV7 delivery. rAAV7 also displayed a specific tropism to Purkinje cells. Evaluation of biochemical and hematological markers suggested that all rAAV serotypes tested were well tolerated. This study provides a comprehensive CNS transduction map in a useful preclinical large animal model enabling the selection of potentially clinically transferable rAAV serotypes based on disease specificity. Therefore, our data are instrumental for the clinical evaluation of these rAAV vectors in human neurodegenerative diseases.
Introduction
Gene transfer of recombinant adeno-associated virus (rAAVs) holds promises to treat neurological disorders.1 Studies on different recombinant vector serotypes and modes of delivery to the central nervous system (CNS) showed that the combination of both rAAV serotype used and delivery routes play a key role in CNS transduction properties and thus in disease phenotype rescue outcome. However, one of the major hurdles to developing an effective clinical protocol for neurological disorders is the efficiency of vectors to reach the specific cell types in disease-specific CNS subdomains.
Attempts to treat CNS defects based on parenchymal delivery of rAAV vectors demonstrated efficacious proof-of-principle studies. Nonetheless, clinical trials using this approach showed limited benefit for Batten disease, Canavan disease, aromatic l-amino acid deficiency, and Parkinson's disease.2,3,4,5,6 One possible explanation for this limited success, in addition to potential local inflammatory responses, is that widespread distribution within the affected brain area from the injection site was inadequate and did not target relevant cell populations. The systemic delivery of rAAVs with the ability to efficiently cross the blood–brain barrier was recently shown as an attractive means to treat diseases with widespread CNS involvement.7,8,9,10 However, the vector doses required and exposure to visceral organs may raise concerns related to manufacturing costs and safety, respectively. An alternative route to efficiently transduce the CNS is based on delivery of viral vectors directly into the cerebrospinal fluid (CSF). CSF delivery can be achieved through ventricular, lumbar, and cisternal administration. The main advantage of CSF-mediated delivery is the exposure of the virus circulating in the CSF to a large CNS surface area resulting in a broad distribution of delivered viral particles within the CNS with a relatively limited amount of vector required.11 CSF-mediated delivery and spreading of molecules/virus within the CNS occurs in two steps: (i) pia mater fenestration allows access to superficial brain parenchyma (mainly composed of astroglia cells: glia limitans), which is in direct contact with the CSF. Access to deeper brain parenchyma areas is instead mainly mediated by perivascular CSF transport12 and (ii) uptake by brain cells, which depends on the specific tropism of rAAV serotypes.13
Therefore, the CSF delivery is an attractive administration route to develop CNS gene therapy–based approaches. Interestingly, proof-of-concept studies based on CSF delivery of rAAVs have been reported for various neurological disorders affecting CNS in different animal species.14,15,16,17 However, to date, little is known about how CSF delivery of potentially clinically transferable rAAV serotypes impacts transgene transduction patterns in specific CNS subdomains/cell types in large preclinical animal models. This information is crucial for the future development of clinical protocols for CNS diseases based on CSF delivery of rAAV vectors. Neurological disorders often exhibit an onset/presentation that affects discrete, disease-specific structures of the brain that then spread along the entire CNS with disease progression. Therefore, in principle, a viral vector that targets the CNS substructure initially affected in disease manifestation would result in an overall therapeutic benefit, and therapy via CSF delivery may mitigate the potential for systemic toxicity.
Here, we provide a detailed side-by-side analysis of CNS region/cell transduction specificity of eight rAAV serotypes (rAAV1, 2, 5, 7, 9, rh.10, rh.39, and rh.43) selected for their potential CNS tropism from the large portfolio of rAAV serotypes available.13,18 This transduction pattern has been evaluated together with an assessment of safety parameters CSF injection of the viral vectors in pigs, a large animal model that is useful for preclinical studies.19 These data will provide guidance and immediate impact for the design of future translational clinical studies.
Results
Pattern of GFP expression in the CNS upon intra-cisterna magna injection of rAAV1, 2, 5, 7, 9, rh.10, rh.39, and rh.43 in P30 pigs
Eight rAAV serotypes (rAAV1, 2, 5, 7, 9, rh.10, rh.39, and rh.43) carrying genes encoding for the green fluorescent protein (GFP) under the control of the CMV ubiquitous promoter were injected in the CSF at a dose of 1.5 × 1012 GC/Kg in wild-type (WT) pigs at age 30 days (P30) (Supplementary Table S1). CSF administration was based on an intra-cisterna magna (ICM) protocol previously described.20 Age-matched WT pigs ICM injected with phosphate-buffered saline (PBS) were used as controls.
In order to evaluate the transduction capability of the different rAAV serotypes, we first analyzed the GFP expression patterns in the CNS of injected animals. Injected pigs were sacrificed 1 month postinjection, and the brains were collected and sliced in eleven 0.5-cm-thick coronal sections covering the main regions of the brain and processed either for biochemical analysis or for GFP immunostaining (Supplementary Figure S1). The cervical region of the spinal cord was also collected (as slice S12) and analyzed in order to evaluate the transduction of rAAV vectors in the CNS regions localized posteriorly to the injection site including the neurodegenerative relevant areas of motor neurons (Supplementary Figure S1).
GFP immunohistochemical (IHC) analysis was performed on frozen sections cut from selected CNS coronal slices derived from both rAAV-injected and control PBS-injected pigs. Frontal and parietal cerebral cortex (from slices S4 and S5, respectively), hippocampus (from slice S8), cerebellum (from slice S11), basal ganglia (from slice, S4 and S5), and cervical tract of the spinal cord (from slice S12) were analyzed (Supplementary Figure S1). The GFP signal was distributed in several CNS regions in pigs that were injected with rAAV9, as seen in Figure 1. In the cerebral cortex, the GFP expression was evident in the layers I–IV, particularly in the glia limitans of layer I (Figure 1). Cerebellum also appeared transduced in rAAV9-injected pigs, with GFP signal observed in the Purkinje cells (Figure 1). rAAV9 vectors penetrated into the brain parenchyma also transducing the brain stem and basal ganglia, with GFP signal present in the accumbens and putamen (Supplementary Figure S2). Transduction was also evident in the dentate gyrus and CA3 areas of hippocampus (Figure 1).
Figure 1.
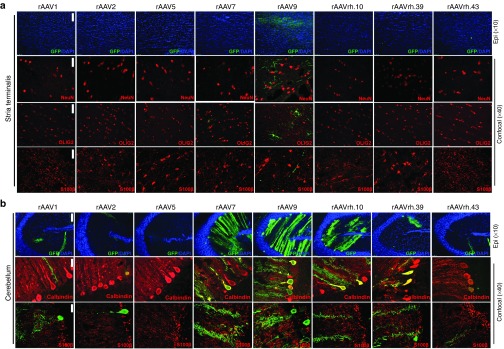
IHC analysis of GFP distribution pattern in main representative areas of pig brain upon ICM delivery of rAAV1, 2, 5, 7, 9, rh.10, rh.39, and rh.43. GFP expression in the brain of rAAV-injected pigs was evaluated 1 month after injection by IHC analysis in four representative 40-μm coronal cryosections (slice S4, S5, S8, and S11). The ×0.3/×0.4 pictures are scanned images (see Materials and Methods). Enlarged images (×4) from each slice showed transduction in layers I–IV of frontal cerebral cortex (slice S4), glia limitans (GL) of parietal cerebral cortex (slice S5), dentate gyrus (DG), and CA3 areas of hippocampus (slice S8) and cerebellum (Cb) (slice S11). Arrows indicated Purkinje cells. Bars for ×0.3 and ×0.4 images = 4 mm. Bar of enlarged images (×4) = 200 μm.
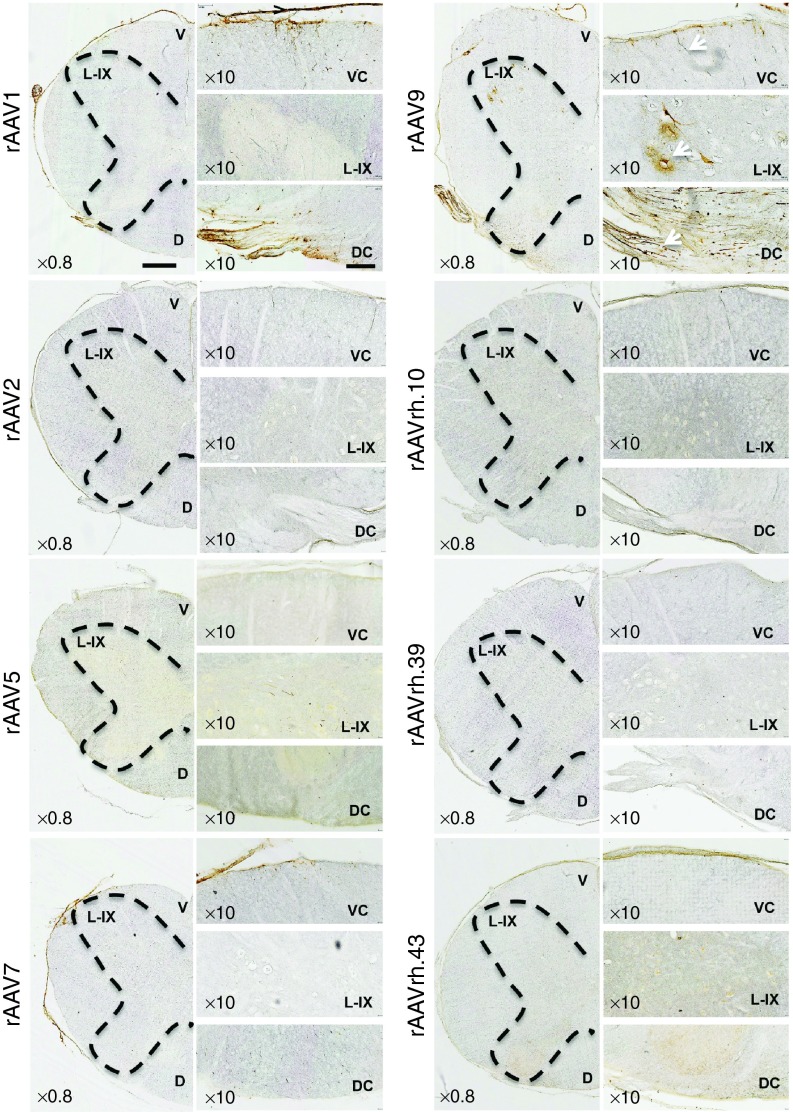
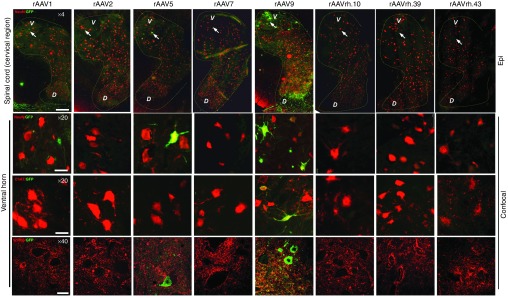
IHC analysis of rAAV1-injected pig brains showed that the first layer of the cerebral cortex was transduced with preferential localization of the GFP signal in the glia limitans (Figure 1 and Supplementary Figure S2). GFP expression was also observed in the Purkinje cell layer of the cerebellum and in the hippocampus (dentate gyrus and CA3; Figure 1). rAAV7 showed a distribution pattern similar to that observed for rAAV1, with higher transduction of Purkinje cell layer compared with the rAAV1 serotype (Figure 1 and Supplementary Figure S2). The glia limitans of the cerebral cortex and the Purkinje cells of the cerebellum appeared transduced also by the rAAV2 (Figure 1 and Supplementary Figure S2). The rAAV5 showed few GFP-positive cells in the layers I–IV of the cerebral cortex (Figure 1 and Supplementary Figure S2). GFP transduction was observed in the Purkinje cell layer of animals injected with rAAVrh.10, rAAVrh.39, and rAAVrh.43, while only a few GFP signal mainly localized to the layer I was detected in the cerebral cortex of these animals (Figure 1 and Supplementary Figure S2). rAAVrh.39-injected pigs also displayed a few GFP signal in dentate gyrus and CA3 areas of hippocampus (Figure 1). IHC analysis of the cervical region of the spinal cord showed that rAAV9 was able to transduce both neurons of lamina IX and fibers in the ventral column and fibers of gracile and cuneate fasciculi in the dorsal column (Figure 2). Both rAAV1 and rAAV7 displayed an astroglial transduction pattern in the spinal cord; however, the GFP signal was stronger in rAAV1-injected animals compared with rAAV7-injected animals (Figure 2). In the spinal cord, rAAV5 and rAAVrh.43 showed a weak GFP expression, which was localized in the gray matter of the ventral horn in rAAV5-injected animals and in the glia limitans of the ventral region in rAAVrh.43-injected animals (Figure 2). All remaining rAAV serotypes analyzed did not exhibit any detectable GFP signal in the spinal cord (Figure 2).
Figure 2.
IHC analysis of GFP distribution pattern in spinal cord of rAAV-injected pigs. IHC GFP staining on coronal cryosections (40 μm) of spinal cord cervical region from rAAV-injected pigs. The ×0.8 pictures are scanned images (see Materials and Methods). The black dashed lines indicate the butterfly shape of gray matter of the spinal cord. Magnification images (×10) show the ventral column (VC), the lamina IX (L-IX) of the ventral horn, and the dorsal column (DC) of the spinal cord. Arrows in the panels of rAAV9-injected pigs indicate fiber transduction in the ventral columns, motor neurons transduction in the L-IX, and transduction of gracile and cuneate fasciculi fibers in the dorsal column. Ventral (V) and dorsal (D) sides of the spinal cord are shown in the ×0.8 scanned images. Bar of ×0.80 images= 1 mm. Bar of ×10 images = 200 μm.
Levels of GFP expression in CNS regions upon ICM injection of rAAV1, 2, 5, 7, 9, rh.10, rh.39, and rh.43 in P30 pigs
GFP expression levels in brain and spinal cord were quantified by western blotting experiments on 20 selected CNS areas of interest, which were dissected from coronal slices in both rAAV-injected and control PBS-injected pigs (Supplementary Figure S1). As seen in Figure 3, P30 injected pigs showed a GFP distribution pattern that overall correlated with the IHC data confirming the wide distribution of the rAAV9 serotype and underlining the specific distribution pattern of the other rAAV serotypes analyzed. However, the quantitative analysis allowed us to evaluate not only the distribution pattern of GFP but also the intensity of GFP expression in specific CNS regions of injected pigs.
Figure 3.
GFP expression levels in representative areas of the CNS of rAAV-injected pigs. Western blotting analysis with anti-GFP antibody performed on rAAV-injected pigs. Indicated areas were taken from the rostral to the caudal part of pig CNS (see Materials and Methods and Supplementary Figure S1 for details). The GFP protein levels were quantified in dissected areas by densitometric analysis of GFP signals. For the cerebral cortex, we analyzed frontal (from slice S1 and S2), parietal (from slice S5), and occipital (from slice S8–S10) regions. Values were normalized to both actin and purified GFP protein (70 ng loaded on sodium dodecyl sulfate gels) and expressed as arbitrary units (a.u., see Materials and Methods for details). Three animals for each serotype were analyzed. Bars represent means ± SEM from three samples (one sample for each animal).
Cerebral cortex. The cerebral cortex of rAAV9-injected pigs exhibited the highest intensity of GFP expression (Figure 3). rAAV1, rAAV5, and rAAV7 also showed relevant GFP protein levels in the cerebral cortex, with rAAV1 and rAAV7 displaying higher expression levels compared to rAAV5 (Figure 3).
Hippocampus. rAAV9 displayed the highest GFP expression levels in the hippocampus, followed by rAAV1, rAAV5, and rAAV7 serotypes (Figure 3). No detectable GFP expression was observed for all the other rAAV serotypes tested (Figure 3).
Midbrain and basal ganglia. The colliculi (superior and inferior) and the substantia nigra were transduced with the highest GFP expression intensity by the rAAV9 serotype (Figure 3). Quantitation of GFP protein levels in the substantia nigra showed higher GFP expression in rAAV5-injected animals compared to that observed in rAAV1- and rAAV7-injected animals (Figure 3). Low GFP protein levels were detected in the colliculi of pigs injected with rAAV1 serotype (Figure 3). rAAV9 exhibited the highest intensity of GFP expression in basal ganglia region followed by rAAV5, rAAV7, and rAAV1 (Figure 3). All other serotypes did not show detectable GFP expression in the midbrain and basal ganglia areas (Figure 3).
Cerebellum. The rAAV9 and rAAV7 serotypes showed similar levels of GFP expression in the cerebellum, and these protein levels were the highest among all rAAV serotypes analyzed (Figure 3). Lower levels of cerebellar GFP expression were displayed by the rAAVrh.39, rAAVrh.10, and rAAV1 serotypes (Figure 3). The GFP expression levels were very low or undetectable in the cerebellum of rAAV2-, rAAV5-, and rAAVrh.43-injected pigs (Figure 3).
Spinal cord. rAAV9 displayed the highest GFP protein levels in the spinal cord, followed by rAAV1, rAAV7, and rAAVrh.39 (Figure 3). All other serotypes did not show detectable GFP expression in the spinal cord region (Figure 3).
In order to further characterize the GFP transduction pattern, we quantified delivered rAAV genomes in different CNS areas of injected pigs by quantitative polymerase chain reaction. High copy numbers of rAAV genomes were observed in the CNS regions in which strong GFP expression was observed, while negligible values of genome copy numbers were observed in the CNS regions with low/absent GFP expression, thus supporting the GFP expression profiles obtained by both IHC and western blotting experiments (Supplementary Figure S3).
Cell type transduction upon ICM injection of rAAV 1, 2, 5, 7, 9, rh.10, rh.39, and rh.43 in P30 pigs
In order to precisely evaluate the specific cell types transduced by the different rAAV serotypes analyzed, we co-labeled the CNS coronal sections from rAAV-injected pigs with anti-GFP and antibodies for cell-specific markers of glial and neuronal cells.
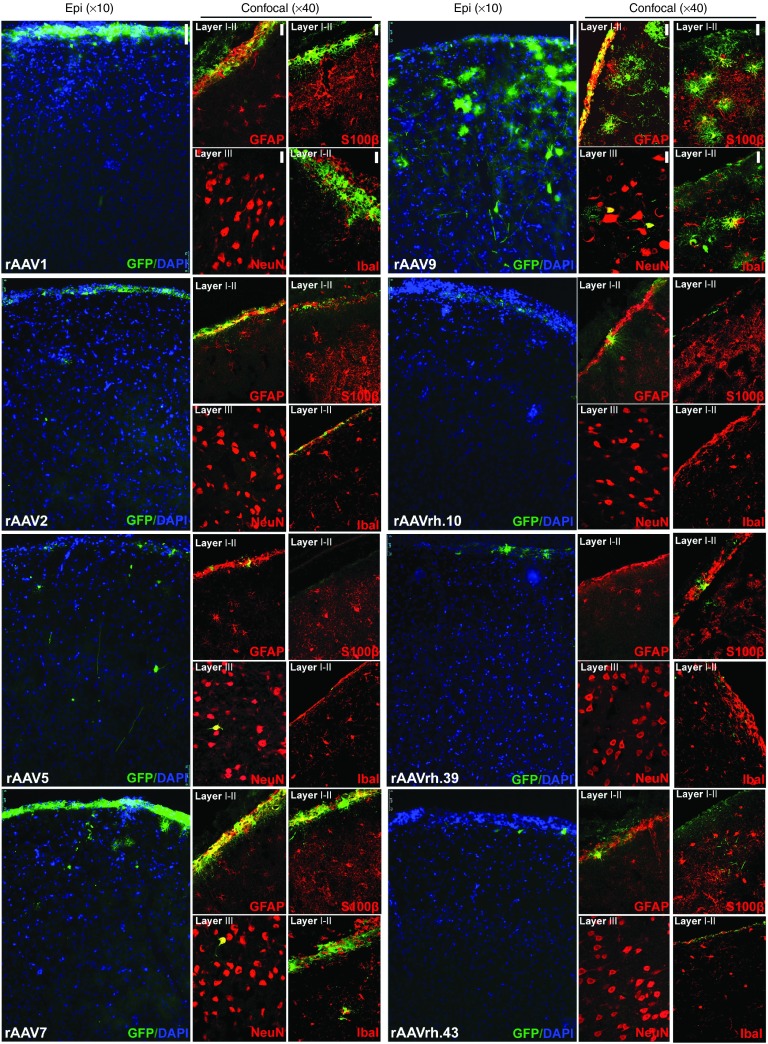
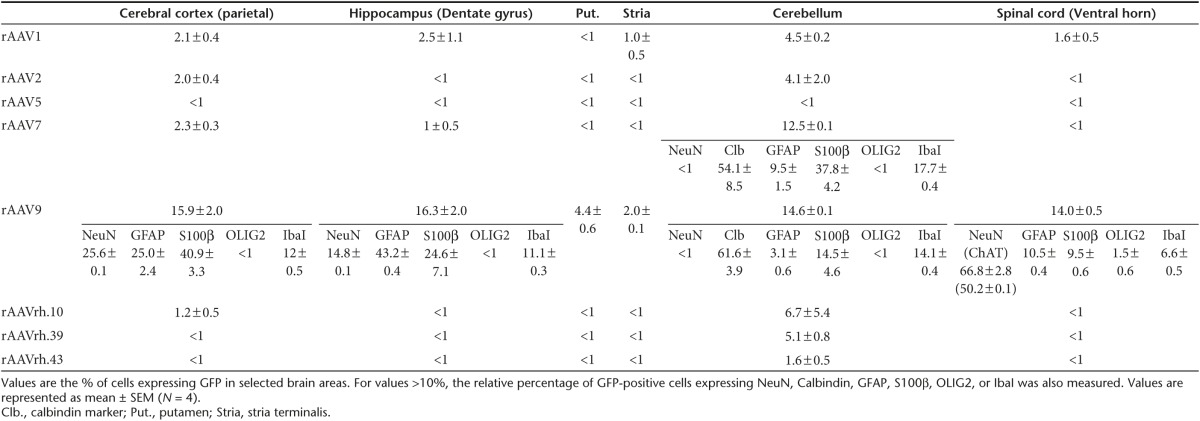
Cerebral cortex. Analysis of parietal cerebral cortex (layers I–IV from slice S6) revealed that ~16% of cells were GFP-positive in rAAV9-injected pigs, ~2–3% of cells were GFP-positive in rAAV1-, rAAV2-, and rAAV7-injected pigs, while less than 1% of GFP-positive cells were found for all other serotypes (Figure 4 and Table 1). In rAAV1-, rAAV2-, and rAAV7-injected animals GFP-expressing cells almost completely co-localized with either GFAP or S100β astroglial markers in the layer I of cerebral cortex (GFAP and S100β stain, respectively, fibrous and protoplasmic astrocytes), while few/no GFP-positive cells co-localizing with the microglial marker Iba1 or the neuronal marker NeuN were found, thus indicating a predominant transduction of cerebral cortical astrocytes for rAAV1-, rAAV2-, and rAAV7 serotypes (Figure 4). In rAAV9-injected pigs, ~26% of GFP-containing cells were GFAP-positive (mostly in the glia limitans), ~41% were S100β-positive, ~12% were Iba1-positive, and ~25% were NeuN-positive, thus indicating the capability of this serotype to efficiently transduce both neurons and different types of glial cells in this region (Figure 4 and Table 1). In the cerebral cortex of pigs injected with all other rAAV serotypes, the few GFP-positive cells found mostly co-localized with astroglial markers in the layer I (Figure 4).
Figure 4.
Cell type tropism of rAAV serotypes in the cerebral cortex of injected pigs. Epi-fluorescent images (×10) showed GFP-expressing cells co-stained with DAPI in the parietal cerebral cortex (layers I–IV from slice 6) of rAAV-injected pigs. Confocal images (×40) showed GFP co-localization with neuronal (NeuN), astroglial (GFAP, S100β), and microglial (Iba1) markers in the same region. Bar for epifluorescent images = 75 μm. Bar for confocal images = 50 μm.
Table 1. Percentage of GFP transduction.
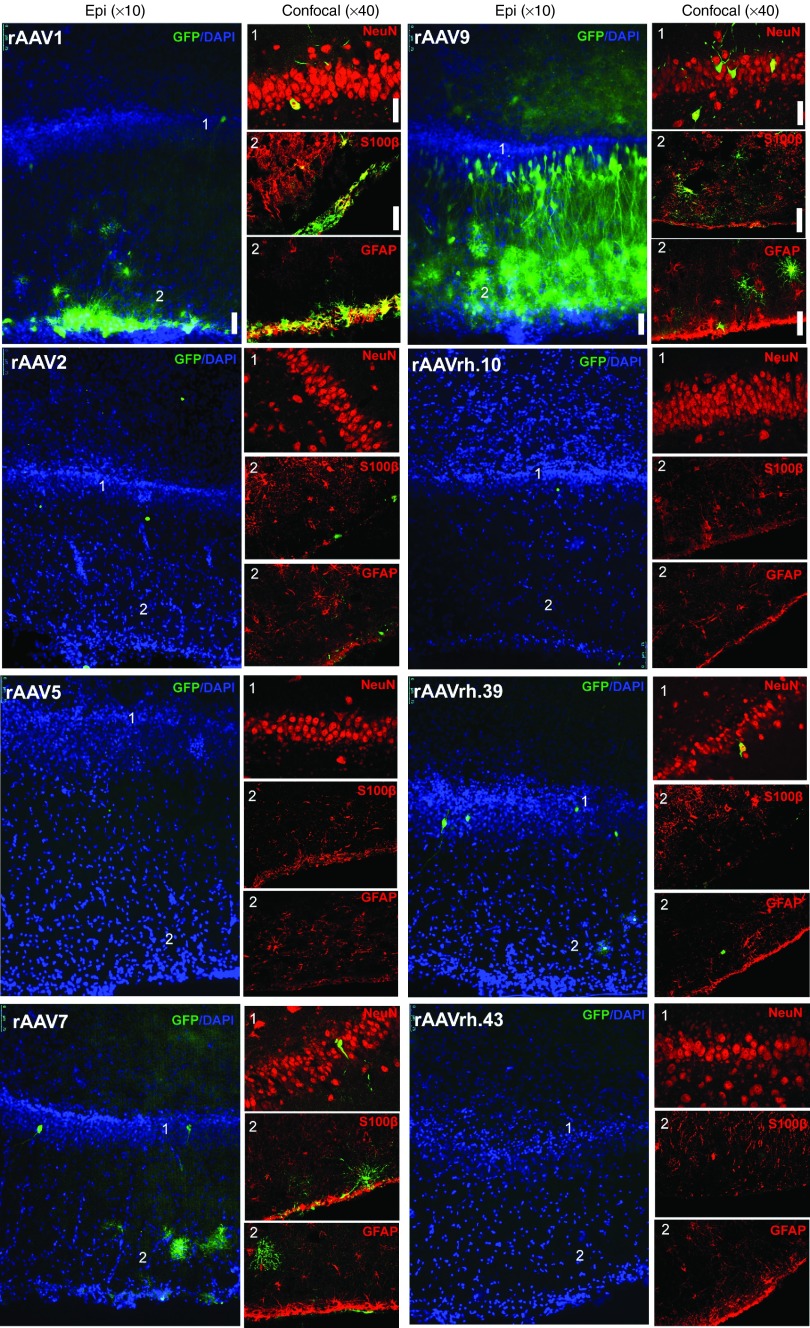
Hippocampus. Analysis of the dentate gyrus and CA3 areas of hippocampus in rAAV9-injected pigs revealed the presence of several GFP-expressing cells, which, as in the cerebral cortex, reflected both neuronal and astroglial cells transduction (Figure 5, Supplementary Figure S4, and Table 1). In the dentate gyrus area, GFP-positive cells co-localized with NeuN (~15%), GFAP (~43%), S100β (~45%), and Iba1 (~11%) markers (Figure 5 and Table 1). GFP-positive cells co-localizing with NeuN, GFAP, and S100β markers were also observed in the hippocampus (dentate gyrus and CA3) of rAAV1-injected pigs (Figure 5, Supplementary Figure S4, and Table 1). Few GFP-positive cells co-localizing with astroglial and neuronal markers were observed in rAAV7- and rAAVrh.39-injcted pigs, while negligible GFP-positive cells were found in pigs injected with all other serotypes (Figure 5, Supplementary Figure S4, and Table 1).
Figure 5.
Cell type tropism of rAAV serotypes in the hippocampus of injected pigs. Epi-fluorescent images (×10) showed GFP-expressing cells co-stained with DAPI in the dentate gyrus of hippocampus of rAAV-injected pigs. Confocal images (×40) showed GFP co-localization with NeuN, S100β, and GFAP markers in the same region. Bar for epifluorescent images: 75 μm. Bar for confocal images: 50 μm.
Basal ganglia. In the stria terminalis and putamen areas of basal ganglia, only rAAV9-injected animals displayed detectable GFP-positive cells (Figure 6a and Table 1). Confocal images of stria terminalis showed that these GFP-expressing cells co-localized with the oligodendroglial marker OLIG2 (Figure 6a). Remarkably, analysis of this region also showed the presence of some GFP-positive signal, which, although did not co-localize with astroglial or neuronal markers, narrowed several projections, thus suggesting transduction of neuronal bodies resident in other brain regions that circuit with stria terminalis (Figure 6a).
Figure 6.
Cell type tropism of rAAV serotypes in the stria terminalis and cerebellum of injected pigs. Epi-fluorescent images (×10) showed GFP-expressing cells co-stained with DAPI in the (a) stria terminalis and in the (b) cerebellum of rAAV-injected pigs. Confocal images (×40) showed GFP co-localization with NeuN, OLIG2, and S100β markers in the (a) stria terminalis and with (b) Calbindin and S100β in the cerebellum of rAAV-injected pigs. Bar for epifluorescent images = 75 μm. Bar for confocal images = 50 μm.
Cerebellum. In the cerebellum, several GFP-positive cells were observed in rAAV7- and rAAV9-injected pigs (respectively ~12 and ~15%), lower numbers of GFP-expressing cells were found in rAAVrh.10, rAAVrh.39-, rAAV2-, and rAAV1-injected pigs, while few/negligible GFP-positive cells were found for all other serotypes (Figure 6b and Table 1). Co-localization analysis with Calbindin marker revealed that these values reflected an efficient transduction of Purkinje cells (Figure 6b and Table 1). GFP co-localization with either Iba1 or astroglial markers were also found in both rAAV9- and rAAV7-injected pigs (Figure 6b and Table 1).
Spinal cord. In the spinal cord, several GFP-expressing cells were observed in rAAV9-injected pigs (Figure 7 and Table 1). In the ventral horn, GFP-positive cells displayed a predominant co-localization with NeuN marker (Figure 7 and Table 1). A few GFP signal co-localizing with NeuN marker was observed in both rAAV1- and rAAV5-injected pigs, while no other serotype showed detectable GFP-positive cells in this region (Figure 7 and Table 1). The use of choline acetyltransferase (ChAT) marker revealed that NeuN-positive GFP-expressing cells were mostly motor neurons in rAAV9-injected animals (Figure 7 and Table 1). The other serotypes tested were negative for ChAT co-staining, including rAAV5, thus indicating that the NeuN-positive GFP-expressing cells in the spinal cord of rAAV5-injected pigs represent a different neuronal subtype (Figure 7 and Table 1).
Figure 7.
Cell type tropism of rAAV serotypes in the spinal cord of injected pigs. Epi-fluorescent scanned images (×4) showed GFP-expressing cells co-stained with NeuN in the spinal cord (cervical region) of rAAV-injected pigs. Confocal images (×20 and ×40) showed GFP co-localization with either NeuN or ChAT (marker of motor neurons) in the ventral horn of the spinal cord (cervical region). Arrows in epifluorescent ×4 images indicate the lamina IX area containing the motor neurons. D, dorsal horn; V, ventral horn. Bar for ×4 images = 400 μm. Bar for ×20 images = 100 μm. Bar for ×40 images = 150 μm.
Evaluation of toxicity and immune-responses after ICM injection of rAAV 1, 2, 5, 7, 9, rh.10, rh.39, and rh.43 in P30 pigs
We then evaluated the presence of a humoral immune response in treated animals. Serum and CSF were collected and screened before and 1 month after injection in order to determine the presence of neutralizing antibodies (NAbs) against the rAAV capsids. None of pigs analyzed showed preexisting NAbs either in serum or in CSF (Supplementary Table S2). At the time of sacrifice, no NAbs were observed in the serum or CSF of rAAV9-, rAAVrh39-, and rAAVrh43-injected pigs (Supplementary Table S2). In contrast, all animals injected with rAAV2 exhibited NAbs in both serum and CSF. Results for the remaining rAAV serotypes were variable with some, but not all, animals displaying NAbs in serum and CSF (Supplementary Table S2).
In order to assess early safety signals, panels of biochemical and hematological markers potentially indicative of renal or hepatic damage/failure, as well as inflammation, were evaluated, and piglet weight was determined on the day of the injection and at sacrifice to evaluate normal physiological development. No significant differences were detected between rAAV-injected animals and controls for markers tested before injection and at the end of the experimental protocol (Supplementary Table S3). The change in the variables that were observed in pigs (either controls or rAAV-injected) when comparing pre- and post-injection time is easily related to the physiological developmental stages of the pig.21 Moreover, weight increase in rAAV-injected pigs was about 75%, similar to that observed in control PBS-injected pigs (Supplementary Table S1). No differences in behavioral patterns were observed between rAAV- and PBS-injected pigs. None of the animals showed pain-related postures or signs. A small number of animals (six pigs injected with rAAV5 and rAAV9) exhibited a very mild ataxia that resolved within 12 hours post-injection. In the following days, they were able to eat, drink, and interact normally, without any signs of stress.
Overall, these data suggest that the ICM procedure and rAAV administration were safe and well tolerated over a 1-month period without a significant impact on a number of key physiological parameters.
Discussion
In this study, we generated a comprehensive and detailed CNS transduction map for eight recombinant rAAV viral vector serotypes administered via CSF delivery in a large animal model, Sus scrofa. We evaluated the transduction efficiency in 20 regions covering the entire brain from the prefrontal to the occipital region, including the spinal cord. This analysis encompassed the quantification of rAAV-delivered GFP protein levels and the characterization of both the CNS distribution and cellular-specific transduction profile for each of the serotypes tested (summarized in Supplementary Table S4). Importantly, since CSF-mediated transport provides CSF-circulating molecules access to both superficial and deeper regions of the brain parenchyma,12 the observed CNS distribution of rAAV vectors reflected the intrinsic capability of rAAV serotypes to transduce specific brain regions when taken up by different cells (i.e., tropism).
Overall, our data showed a significant differential tropism of the rAAV serotypes tested, as illustrated by the GFP expression patterns observed. Moreover, the present study provides important information on both regional and cellular specificity of the transduction patterns that suggest potential therapeutic advantages/disadvantages when considering rAAV-based treatments for both cell-autonomous and non–cell-autonomous CNS disorders. Injection of rAAV9 resulted in the widest GFP expression along the entire CNS, showing efficient and widespread transduction of neurons and glial cells of different layers of cerebral cortex, basal ganglia, midbrain, and brain stem areas including motor neurons of the spinal cord. These data are consistent with the CNS transduction pattern of rAAV9 described by us and other groups.14,15,16,22,23,24 In contrast to the wide-transduction spectrum of rAAV9, the other rAAV serotypes tested displayed more prominent cell-specific transduction profiles. In particular, rAAV1 and rAAV7 efficiently transduce the glia limitans, with rAAV7 also showing a specific Purkinje cell tropism. rAAVrh.10 also displayed a tropism to Purkinje cells, as previously reported in rats.17 Transduction of microglial cells was observed for several rAAV serotypes tested including rAAV1, rAAV2, rAAV7, and rAAV9.
An important aspect of translational studies is the demonstration of safety. While the experiments herein described were not designed to fully investigate the potential for treatment-related toxicities, an effort was made to look for early safety signals. Beside transient ataxia post-surgery in some animals, no overt signs of distress were observed. Moreover, consistent with previous studies,25 we did not observe preexisting NAbs against any of the rAAV capsids tested while NAbs were present only for some serotypes (rAAV1, rAAV2, rAAV5, rAAV7, and rAAVrh.10) in a few injected animals (Supplementary Table S2). Nevertheless, it has been described that low levels of circulating NAbs do not interfere with CNS gene transfer after CSF delivery.14,15,26 Recently, Samaranch et al.27 described antigen-presenting cell–mediated neuroinflammation against non-self protein (GFP). In the present study, we did not evaluate the presence of such an inflammatory response; however, its potential toxicity could be overcome by the use of a self-protein as reported.27 Moreover, although we observed a lack of immunotoxicity at 1 month after injection, we do not exclude that tardive immune responses (e.g., cell-mediated) might be present in treated animals after long-term analysis (>8 weeks).
The data described herein demonstrate the value of rAAV delivery via CSF as a mode to efficiently transduce the CNS, compare, contrast, and map the tropism, transduction efficiency, and cell specificity of eight rAAV serotypes in a large animal model. Therefore, although translating these results to human should take into account potential differences in the tropism features of the AAV vectors tested, our work provides a guide to investigators to better match rAAV serotypes with the needs of specific neurodegenerative diseases that could be potentially treated with a gene-therapy approach.
Materials and Methods
Viral vectors. rAAV vectors were produced by the TIGEM rAAV Vector Core by triple-plasmid transfection of HEK293 cells and were purified by two rounds of CsCl2 gradient centrifugation.28 Viral vector titers (genome copies/ml) were determined by real-time PCR quantification using TaqMan (Applied Biosystems, Foster City, CA) and dot-blot analysis.28
The final titer of each preparation was calculated as the average between the PCR quantification and dot-blot results.
Animals, rAAV administration, and tissue collection. The study was conducted in accordance with the provisions of European Economic Community Council Directive 86/609 adopted by the Italian Government (DL 27/01/1992 No. 116) under the local approval of the Ethical Committee of the University of Bologna and under the approval of Italian Ministry of Health.
All animal studies have been approved by the authors.
The animals enrolled were WT large White × Duroc hybrids, and the sex ratio between females and castrated males was ~1:1.
These animals were transferred to our facility on the day of weaning (28th day after birth) and housed in multiple stalls with infrared heating lamps. They were strictly monitored in order to rule out any pathology that may have affected the entire experiment.
AVV administration. All of the activities performed on the day of the injection have been thoroughly described by Romagnoli et al.20 In brief, animal received an i.m. bolus of tiletamine-zolazepam (5 mg/kg) 10 minutes before induction; general anesthesia was achieved using sevoflurane with an induction mask. After orotracheal intubation and stabilization, venous access for fluid therapy was achieved from an auricular vein. Blood samples (6 ml) were collected through the femoral artery.
The dorsal area of the neck was trimmed and surgically prepared, and the puncture of the cisterna magna was performed as previously described.20 One ml of CSF was collected before the injection in order to analyze it as a preinjection physiological standard. The dose of 1.5 × 1012 GC/Kg of viral vector in the volume range from 0.5 to 2.8 ml was injected slowly to avoid a sudden increase in intracranial pressure. Piglets were then placed in Trendelenburg position for 2 minutes in order to help the injected compound to spread toward the more rostral parts of the CNS. Animals were then monitored until complete recovery.
During the following days, all animals were strictly monitored in order to rule out any possible side effect of the procedure and to evaluate any changes in behavior and consequentially in welfare.
Sacrifice and tissue collection. Procedures for anesthesia, venous access, and blood sampling were exactly the same as performed on the day of the injection. CSF samples were collected by puncturing the lumbar spinal space rather than the cisterna magna. Percutaneous cystocentesis was performed to obtain sterile urine samples (3 ml). Animals were then sacrificed with a single bolus (0.3 ml/kg) of Tanax and total body perfusion with Dulbecco's phosphate-buffered saline was started. After median sternotomy, the right atrium was opened and the left ventricle was infused with 500 ml of warm Dulbecco's phosphate-buffered saline (+38 °C) and 1,000 ml of cold Dulbecco's phosphate-buffered saline (+4 °C); blood ejected from the right atrium was drained using a surgical aspirator.
As far as CNS samples, collected tissues were the whole brain and cervical region of spinal cord. Dissection was performed using the technique described by Wischnitzer29 modified by Prof C. Bombardi. Blood samples were collected using a vacuum system, and tubes with K3ethylenediaminetetraacetic acid anticoagulant, citrate, and clot activator were used. Samples were processed within 1 hour from collection and analyzed or stored at −80 °C until analysis.
All animals had a complete blood work (ADVIA 2120, Siemens Healthcare Diagnostics, Tarrytown, NY) including complete blood count with hematocrit value, hemoglobin concentration, erythrocyte indices, platelet count, white blood cell with differential white blood cell counts and blood smear examination, and a chemistry profile including aspartate transaminase, alanine transaminase, creatinine, urea, total protein, albumin, albumin to globulin ratio. All chemistry analyses were carried out on an automated chemistry analyser (Olympus AU 400, Beckman Coulter/Olympus, Brea, CA).
Evaluation of AAV vector copy number in the CNS. Genomic DNA was extracted from five selected regions of CNS using a DNeasy Blood and Tissue Extraction kit (Qiagen, Valencia, CA). We selected CNS regions, which were efficiently transduced at least by three viral vectors. DNA concentration was determined by using a Nanodrop. Real-time PCR was performed on 100 ng of genomic DNA using a LightCycler SYBR green I system (Roche, Almere, The Netherlands). For the amplification, the EGFP fwd (5′ AGC AGC ACG ACT TCT TCA ACT CC 3′) and EGFP rev (5′ CCA TGA TAT AGA CGT TGT GG 3′) were used. Amplification was run on a LightCycler 96 device (Roche) with standard cycles. A standard curve was generated, using the corresponding AAV vector plasmid pAAV2.1CMV-EGFP.
GFP immunoblotting. To quantify the GFP protein, 20 main regions covering the entire CNS of injected pigs were dissected and then snap-frozen in liquid nitrogen (see Supplementary Figure S1). Approximately 70 mg of each dissected region were homogenized with TissueLyser using 10 volumes (700 µl) of 3× Flag lysis buffer (50 mmol/l Tris–HCl pH8, 200 mmol/l NaCl, 1% Triton X100, 1 mmol/l ethylenediaminetetraacetic acid, and 50 mmol/l 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) and a protease inhibitor cocktail (Sigma-Aldrich, St Louis, MO). The lysates were incubated in ice for 1 hour and cleared by centrifugation. Protein concentration was determined using the Bio-Rad colorimetric assay (Bio-Rad, Hercules, CA). Thirty micrograms of homogenate protein were separated on a 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred to polyvinyl difluoride membrane at 30 V over night. As control, 70 ng of purified GFP was loaded on sodium dodecyl sulfate gels. Anti-GFP rabbit polyclonal (1:1,000, NB600-308; Novus BIO, Littleton, CO) and anti-β-actin mouse monoclonal antibodies (1:2,000, A5441, Sigma-Aldrich) were, respectively, used for the detection of GFP and actin proteins. The membranes were incubated with secondary antirabbit and antimouse antibodies (1:5,000, CALBIOCHEM, San Diego, CA), and protein bands were visualized using UVP-ChemiDoc-It (Life Science Software, Hopkinton, MA). Relative band intensity was quantified by densitometric analysis using ImageJ software (National Institutes of Health, Bethesda, MD).
The intensity of bands corresponding to the GFP expressed in brain lysates was quantified by densitometric analysis. Each of these values was divided for both the value resulting from densitometric quantitation of the band corresponding to the purified GFP (which was loaded on the same sodium dodecyl sulfate gel in which the brain samples were loaded) and the value resulting from densitometric quantitation of actin. The resulting ratio was expressed as arbitrary units (a.u.)
Immunolabeling. IHC staining of GFP was performed on 40 μm floating cryosections derived from brain and spinal cord of injected pigs.
Sections were washed in PBS/Triton X-100 0.1% (wash buffer), incubated for 30 minutes at room temperature in 1% hydrogen peroxidase diluted in PBS 1× and incubated overnight at 4 °C in primary antibody solution: PBS 1×/ Triton X-100 0.1%, 2% horse serum, rabbit anti-GFP (1:1,500, NB600-308; Novus BIO). Anti-GFP was detected with MACH4 Universal HRP-Polymer Biotin-Free Detection Polymer Detection Kit (BIOCARE Medical, Concord, CA). After 1 hour, we washed three times the sections and developed the signal with diaminobenzidine (Vectorstain Kit; Vector Laboratories, Burlingame, CA). Sections were then mounted with CV Ultra Mounting Media (Leica, Wetzlar, Germany).
To identify the cell types, we performed immunofluorescence experiments on 30-μm cryosections. Samples were blocked in TBS1X-Triton 0.3% and Donkey Serum 5% for 1 hour at room temperature and incubated with anti-GFP either rabbit polyclonal (1:1,000, NB600-308; Novus BIO) or chicken polyclonal 9 (1:800, AB13970; Abcam, Cambridge, UK), anti-S100β mouse monoclonal (1:800, AB66028; Abcam), anti-IBA1 rabbit polyclonal (1:500, 234003; Synaptic System, Gottingen, Germany), anti-NeuN mouse monoclonal (1:400, MAB377; Millipore), anti-GFAP mouse monoclonal (1:200, MAB3402; Millipore, Amsterdam, the Netherlands), anti-Olig2 mouse monoclonal (1:200, MABN50; Millipore), and anti-CHAT goat poyclonal (1:100, AB144P; Millipore) to, respectively, identify GFP protein, neurons, astrocytes, oligodendrocytes, and motor neurons. After overnight incubation with primary antibody, sections were washed in PBS 1× and incubated with Alexa Fluor 488-conjugated donkey antirabbit (1:500; Invitrogen, Carlsbad, CA), Alexa Fluor 594-conjugated donkey anti-mouse (1:500; Invitrogen), and Alexa Fluor 594-conjugated donkey anti-goat (1:500; Invitrogen). Rinsed sections were mounted with VECTASHIELD mounting medium with 4',6-diamidino-2-phenylindole, dihydrochloride (DAPI) (Vinci-Biochem, Florence, Italy) and analyzed by either epi-fluorescent and confocal microscopy.
GFP quantitation (% of GFP-positive cells) in immunofluorescence experiments. We quantified the GFP immunofluorescence signal in the CNS of injected pigs in five main CNS regions (parietal cerebral cortex from slice S6, dentate gyrus of hippocampus from slice S8, putamen from slice S5, cerebellum from slice S11, and lamina IX of the cervical tract of spinal cord (slice S12). One square millimeter area of four different sections (30 μm) stained with DAPI and co-labeled with GFP, and the specific cell type markers (NeuN, Calbindin, ChAT, GFAP, S100β, Iba1, or OLIG2) were scanned in z-stack using Zeiss LSM 710 microscope equipped with a Zeiss confocal-scanning laser using a 40× objective (Zeiss, Oberkochen, Germany). The exposure time used to acquire all images was identical. The co-localization analysis was performed by the microscope ZEISS 2008 program. Total number of DAPI- and GFP-positive signals were counted using the cell-counter program (ImageJ software) with a fixed threshold. Total GFP cell counts present in the 1 mm2 area analyzed were then expressed as percentage of total cells (DAPI positive) expressing the GFP (%GFP-positive cells). For CNS regions in which the %GFP-positive cells was >10%, NeuN-, Calbindin-, ChAT-, GFAP-, S100β-, Iba1-, or OLIG2-positive cells were also counted and expressed as percentage of GFP-positive cells expressing the specific cell type marker.
Microscopy. 3,3'-Diaminobenzidine-processed brain and spinal cord sections were digitized using a Scan-Scope slide scanner (Leica scn400). Virtual slides were viewed using Leica digital image hub, and images were generated using the same software. Immunofluorescence images of the entire cervical region of the spinal cord were digitized using a Scan-Scope slide scanner (Leica scn400). Fluorescence images at ×10 magnification were visualized by epi-fluorescence microscope. Confocal microscopy was performed with a Zeiss LSM 710 confocal microscope equipped with a Zeiss confocal-scanning laser using 40× objective.
Neutralizing antibody assay. The presence of NAbs to rAAV capsid was assessed, on serum and CSF collected at the day of injection and 1 month after injection, as previously described.30
Data analysis. Biochemical and hematological results among groups were compared using nonparametric statistics (Mann–Whitney U-test, Kruskal–Wallis ANOVA, and Friedman test for paired data). Data are expressed as mean ± 1 SEM (n = 3). A P value of <0.05 was considered to be statistically significant.
SUPPLEMENTARY MATERIAL Figure S1. Scheme of pig brain and spinal cord dissection. Figure S2. IHC analysis of GFP distribution pattern in basal ganglia of pig brain upon ICM delivery of rAAV1, 2, 5, 7, 9, rh.10, rh.39, and rh.43. Figure S3. Distribution of rAAV vector genomes following ICM injection in pig. Figure S4. Cell type tropism of rAAV serotypes in the CA3 area of the hippocampus of injected pigs. Table S1. Summary of injections in P30 pigs. Table S2. Neutralizing anti-AAV antibody levels. Table S3. Biochemical and hematological parameters. Table S4. Summary of GFP transduction.
Acknowledgments
The authors thank Graciana Diez-Roux, Shipeng Yuan, and Vivian Choi for critical reading of the manuscript. Edoardo Nusco and Rossella Venditti for technical help. We acknowledge the rAAV vector core (TIGEM) for rAAV production and neutralizing antibody assay, the Advanced Microscopy and Imaging core (TIGEM), Donatella Montanaro and the Pathology core (CEINGE) for IHC image scanning. Shire funded this work as part of a project within Discovery Biology and Translational Research.
A.-R.G., Y.H., and S.L.K. are employees and stockholders of Shire. The other authors declare that they have no conflict of interest.
Supplementary Material
References
- Simonato, M, Bennett, J, Boulis, NM, Castro, MG, Fink, DJ, Goins, WF et al. (2013). Progress in gene therapy for neurological disorders. Nat Rev Neurol 9: 277–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worgall, S, Sondhi, D, Hackett, NR, Kosofsky, B, Kekatpure, MV, Neyzi, N et al. (2008). Treatment of late infantile neuronal ceroid lipofuscinosis by CNS administration of a serotype 2 adeno-associated virus expressing CLN2 cDNA. Hum Gene Ther 19: 463–474. [DOI] [PubMed] [Google Scholar]
- Leone, P, Shera, D, McPhee, SW, Francis, JS, Kolodny, EH, Bilaniuk, LT et al. (2012). Long-term follow-up after gene therapy for canavan disease. Sci Transl Med 4: 165ra163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwu, WL, Muramatsu, S, Tseng, SH, Tzen, KY, Lee, NC, Chien, YH et al. (2012). Gene therapy for aromatic L-amino acid decarboxylase deficiency. Sci Transl Med 4: 134ra61. [DOI] [PubMed] [Google Scholar]
- Marks, WJ Jr, Bartus, RT, Siffert, J, Davis, CS, Lozano, A, Boulis, N et al. (2010). Gene delivery of AAV2-neurturin for Parkinson's disease: a double-blind, randomised, controlled trial. Lancet Neurol 9: 1164–1172. [DOI] [PubMed] [Google Scholar]
- LeWitt, PA, Rezai, AR, Leehey, MA, Ojemann, SG, Flaherty, AW, Eskandar, EN et al. (2011). AAV2-GAD gene therapy for advanced Parkinson's disease: a double-blind, sham-surgery controlled, randomised trial. Lancet Neurol 10: 309–319. [DOI] [PubMed] [Google Scholar]
- Foust, KD, Nurre, E, Montgomery, CL, Hernandez, A, Chan, CM and Kaspar, BK (2009). Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol 27: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, B, Li, S, Wang, H, Guo, Y, Gessler, DJ, Cao, C et al. (2014). Global CNS transduction of adult mice by intravenously delivered rAAVrh.8 and rAAVrh.10 and nonhuman primates by rAAVrh.10. Mol Ther 22: 1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, FJ, Naughton, BJ, Zaraspe, K, Murrey, DA, Meadows, AS, Clark, KR et al. (2015). Broad functional correction of molecular impairments by systemic delivery of scAAVrh74-hSGSH gene delivery in MPS IIIA mice. Mol Ther 23: 638–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, YH, Chang, M and Davidson, BL (2009). Molecular signatures of disease brain endothelia provide new sites for CNS-directed enzyme therapy. Nat Med 15: 1215–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen, MK, Bjornsson, CS, Dymecki, SM, Gilbertson, RJ, Holtzman, DM and Monuki, ES (2013). The choroid plexus and cerebrospinal fluid: emerging roles in development, disease, and therapy. J Neurosci 33: 17553–17559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliff, JJ, Wang, M, Liao, Y, Plogg, BA, Peng, W, Gundersen, GA et al. (2012). A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid β. Sci Transl Med 4: 147ra111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murlidharan, G, Samulski, RJ and Asokan, A (2014). Biology of adeno-associated viral vectors in the central nervous system. Front Mol Neurosci 7: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinderer, C, Bell, P, Gurda, BL, Wang, Q, Louboutin, JP, Zhu, Y et al. (2014). Intrathecal gene therapy corrects CNS pathology in a feline model of mucopolysaccharidosis I. Mol Ther 22: 2018–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurigot, V, Marcó, S, Ribera, A, Garcia, M, Ruzo, A, Villacampa, P et al. (2013). Whole body correction of mucopolysaccharidosis IIIA by intracerebrospinal fluid gene therapy. J Clin Invest (epub ahead of print). [DOI] [PMC free article] [PubMed]
- Samaranch, L, Salegio, EA, San Sebastian, W, Kells, AP, Bringas, JR, Forsayeth, J et al. (2013). Strong cortical and spinal cord transduction after AAV7 and AAV9 delivery into the cerebrospinal fluid of nonhuman primates. Hum Gene Ther 24: 526–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hordeaux, J, Dubreil, L, Deniaud, J, Iacobelli, F, Moreau, S, Ledevin, M et al. (2015). Efficient central nervous system AAVrh10-mediated intrathecal gene transfer in adult and neonate rats. Gene Ther 22: 316–324. [DOI] [PubMed] [Google Scholar]
- Weinberg, MS, Samulski, RJ and McCown, TJ (2013). Adeno-associated virus (AAV) gene therapy for neurological disease. Neuropharmacology 69: 82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind, NM, Moustgaard, A, Jelsing, J, Vajta, G, Cumming, P and Hansen, AK (2007). The use of pigs in neuroscience: modeling brain disorders. Neurosci Biobehav Rev 31: 728–751. [DOI] [PubMed] [Google Scholar]
- Romagnoli, N, Ventrella, D, Giunti, M, Dondi, F, Sorrentino, NC, Fraldi, A et al. (2014). Access to cerebrospinal fluid in piglets via the cisterna magna: optimization and description of the technique. Lab Anim 48: 345–348. [DOI] [PubMed] [Google Scholar]
- Egeli, AK, Framstad, T and Morberg, H (1998). Clinical biochemistry, haematology and body weight in piglets. Acta Vet Scand 39: 381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spampanato, C, De Leonibus, E, Dama, P, Gargiulo, A, Fraldi, A, Sorrentino, NC et al. (2011). Efficacy of a combined intracerebral and systemic gene delivery approach for the treatment of a severe lysosomal storage disorder. Mol Ther 19: 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, SJ, Nagabhushan Kalburgi, S, McCown, TJ and Jude Samulski, R (2013). Global CNS gene delivery and evasion of anti-AAV-neutralizing antibodies by intrathecal AAV administration in non-human primates. Gene Ther 20: 450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federici, T, Taub, JS, Baum, GR, Gray, SJ, Grieger, JC, Matthews, KA et al. (2012). Robust spinal motor neuron transduction following intrathecal delivery of AAV9 in pigs. Gene Ther 19: 852–859. [DOI] [PubMed] [Google Scholar]
- Calcedo, R and Wilson, JM (2013). Humoral Immune Response to AAV. Front Immunol 4: 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini, MA, Bu, J, Richards, AM, Treleaven, CM, Sullivan, JA, O'Riordan, CR et al. (2014). Translational fidelity of intrathecal delivery of self-complementary AAV9-survival motor neuron 1 for spinal muscular atrophy. Hum Gene Ther 25: 619–630. [DOI] [PubMed] [Google Scholar]
- Samaranch, L, San Sebastian, W, Kells, AP, Salegio, EA, Heller, G, Bringas, JR et al. (2014). AAV9-mediated expression of a non-self protein in nonhuman primate central nervous system triggers widespread neuroinflammation driven by antigen-presenting cell transduction. Mol Ther 22: 329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria, M, Ferrara, A and Auricchio, A (2013). AAV2/8 vectors purified from culture medium with a simple and rapid protocol transduce murine liver, muscle, and retina efficiently. Hum Gene Ther Methods 24: 392–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wischnitzer, S (1972). Atlas and Dissection Guide for Comparative Anatomy. 2nd edn. W H Freeman and Company: San Francisco. [Google Scholar]
- Mussolino, C, della Corte, M, Rossi, S, Viola, F, Di Vicino, U, Marrocco, E et al. (2011). AAV-mediated photoreceptor transduction of the pig cone-enriched retina. Gene Ther 18: 637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.