Abstract
2H, 31P, and 1H-magic-angle-spinning (MAS) solid-state NMR spectroscopic methods were used to elucidate the interaction between sorbic acid, a widely used weak acid food preservative, and 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) bilayers under both acidic and neutral pH conditions. The linewidth broadening observed in the 31P NMR powder pattern spectra and the changes in the 31P longitudinal relaxation time (T1) indicate interaction with the phospholipid headgroup upon titration of sorbic acid or decanoic acid into DMPC bilayers over the pH range from 3.0 to 7.4. The peak intensities of sorbic acid decrease upon addition of paramagnetic Mn2+ ions in DMPC bilayers as recorded in the 1H MAS NMR spectra, suggesting that sorbic acid molecules are in close proximity with the membrane/aqueous surface. No significant 2H quadrupolar splitting (Δ γQ) changes are observed in the 2H NMR spectra of DMPC-d54 upon titration of sorbic acid, and the change of pH has a slight effect on Δ γQ, indicating that sorbic acid has weak influence on the orientation order of the DMPC acyl chains in the fluid phase over the pH range from 3.0 to 7.4. This finding is in contrast to the results of the decanoic acid/DMPC-d54 systems, where Δ γQ increases as the concentration of decanoic acid increases. Thus, in the membrane association process, sorbic acids are most likely interacting with the headgroups and shallowly embedded near the top of the phospholipid headgroups, rather than inserting deep into the acyl chains. Thus, antimicrobial mode of action for sorbic acid may be different from that of long-chain fatty acids.
Keywords: sorbic acid, fatty acid, antimicrobials, membrane, solid-state NMR
Introduction
Weak organic acids are widely used as preservatives in foods, cosmetics, and pharmaceuticals. Sorbic acid [(2E, 4E)-hexadienoic acid, Fig. 1A], for example, has been widely used in foods owing to its desirable physiological properties and its effectiveness against a wide range of microorganisms.[1] In spite their extensive use and studies, the working mechanism of weak acids is far from clarified. In conventional weak acid theory, the undissociated acid molecules are believed to cross the cell membrane freely, dissociate in the cytoplasm, and inhibit growth through acidification of the cytoplasm.[2,3] The concept that growth is inhibited only by undissociated acids was questioned and sorbic acid was proposed to be acting as a membrane-active agent.[4,5] More recently, the signal pathways set off by weak acids have been under investigation.[6] Overall, the antimicrobial activity of weak acids has been proposed to be due to a number of actions such as inhibition of enzyme activity,[7] accumulation of toxic anions,[8] inhibition of essential metabolic reactions as glycolysis,[9] membrane disruption,[5] induction of energetically unfavorable stress response,[10] or intracellular acidification.[11]
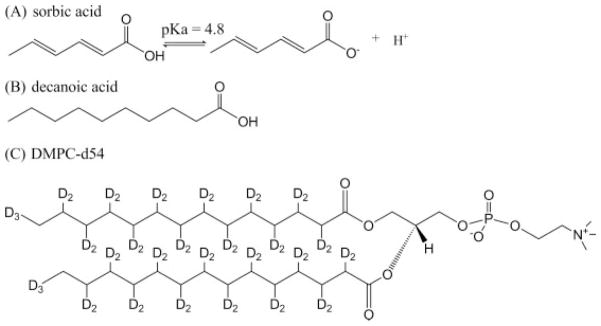
Figure 1.
The chemical structures of (A) sorbic acid, (B) decanoic acid, and (C) DMPC-d54.
The available data suggest the importance of membranes in the inhibition of sorbic acid and other weak organic acids. Microbial growth inhibition by sorbic acid is pH dependent, with greater inhibition at lower pH.[2] It was shown that sorbic acid is not effective at neutral pH where the dissociated form is the most populated state.[12] Similar to decanoic acid and octanoic acid, the undissociated form of sorbic acid is believed to be the active form and is thought to enter the cell by passive diffusion across the plasma membrane.[13,14] However, other studies have also shown antimicrobial activity of sorbic acid at higher pH levels (>6.5).[15] Transmission and scanning electron microscopy studies indicated outer membrane damage and increased hydrophobicity of the cell wall in sorbate-treated cells at pH 7.0, and dissociated sorbate was shown to have an inhibitory effect.[16] More generally speaking, the pH hypothesis which states that only the uncharged form of ionogeric molecules diffuse across a lipid bilayer[17] was recently questioned,[18] as well as the validity of Overton’s rule which states that the membrane permeability of a molecule correlates with its hydrophobicity.[19,20]
Solid-state NMR (SS-NMR) spectroscopy is an excellent tool to study biological membranes and the interactions of membranes with proteins as well as with small molecules.[21,22] The two distinct moieties of phospholipids, the phosphate headgroups, and the hydrocarbon acyl chain region, can be studied with different SS-NMR techniques. 2H NMR lineshapes are very sensitive to molecular motions and the quadrupolar splittings (ΔγQ) of selectively deuterated acyl chains of lipids directly reflect the motional order parameters within the acyl chain region.[23,24] 31P NMR spectroscopy provides another sensitive ‘reporter’ to monitoring the events occurring in the headgroup region and has been used widely to study the effects of various agents thought to act by perturbing the membranes.[25,26] 31P longitudinal relaxation time (T1) is very sensitive to fast lipid headgroup rotational motions and has been extensively described for phospholipids in biological membranes.[27] Magic-angle-spinning (MAS) experiments yield high-resolution 1H NMR spectra to study small molecules as well as peptides in the membranes. 1H NMR MAS studies coupled with the use of paramagnetic ions can reveal the location of the molecules with respect to the membrane.[28 – 30]
In our previous studies, fatty acids inserted into phospholipid bilayers were probed in model membrane systems using SS-NMR as well as electron paramagnetic resonance (EPR) spectroscopy.[31] In this study, the effects of sorbic acid on well-defined 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC, Fig. 1C) model membranes were investigated at different pH levels with 2H, 31P, and 1H-MAS NMR spectroscopic methods. Decanoic acid (Fig. 1B) was also studied to compare the membrane interaction modes between sorbic acid and fatty acids of longer chains to provide mechanistic insight of these important preservatives.
Experimental
Chemicals
DMPC and DMPC-d54 were purchased from Avanti Polar Lipids Inc. (Alabaster, AL, USA) and used without further purification. All phospholipids were dissolved in chloroform and stored at −20 °C prior to use. Deuterium-depleted water and D2O were purchased from Isotec™ (Miamisburg, OH, USA). Sorbic acid, potassium sorbate, decanoic acid, tetrafluoroethanol (TFE), and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA).
NMR sample preparation
Multiamellar vesicles (MLVs) of DMPC/DMPC-d54 mixtures (in 3 : 1 ratio) containing or not containing the organic acid additives were prepared according to a previously reported procedure.[32] A total amount of ~70 mg of DMPC/DMPC-d54 lipids was dissolved in chloroform. Appropriate amounts of sorbic acid, potassium sorbate, or decanoic acid were dissolved in a minimal amount of TFE, respectively, and were mixed with the lipid solutions to obtain different additive : lipid ratios. The solvents (chloroform and TFE) were removed slowly under a steady stream of N2 gas and the samples were dried overnight in a vacuum desiccator. The resulting mixtures were hydrated with 190-μl HEPES buffer (30-mM HEPES, 20-mM NaCl, pH 6.0 or pH 7.4) or acetic acid buffer (20-mM NaCl, pH 3.0 or 4.4) respectively. The hydrated samples went through 5–10 freeze-thawing cycles to increase the homogeneity of the vesicles. The pellet (60–80 mg) was transferred to a 4-mm MAS rotor for 2H and 31P NMR spectroscopic studies.
For 1H MAS NMR experiments, the samples were prepared by following the same protocol except D2O buffers were used instead of deuterium-depleted water to hydrate the lipids vesicles.
NMR spectroscopy
All 2H and 31P NMR experiments were conducted on a Bruker Avance 500-MHz wide-bore NMR spectrometer (Bruker Biospin, Rheinstetten, Germany) equipped with a 4-mm triple resonance cross-polarization (CP)/MAS probe (Bruker Biospin). All experiments were conducted at 35 °C (more than 10° above 24 °C, the phase transition temperature of DMPC). Before NMR data acquisition, the sample was spun at 4 kHz in the MAS rotor at 35 °C for 15 min to allow for homogeneous sample distribution and temperature equilibration.
Powder pattern 2H NMR spectra were acquired at a 2H frequency of 76.773 MHz. A quadrupolar echo pulse sequence (90° − τ1 − 180° − τ2 acquire)[33] was used, with a recycle delay of 300 ms, echo delay τ1 of 40 μs, τ2 of 35 μs, 2H 90° pulse of 3.0 μs. The spectral width was set to 100 kHz. As many as 20 k scans were averaged and the free induction delays (FIDs) were processed with a linebroadening of 200 Hz with the Bruker software Topspin 2.0.
31P NMR spectra were collected at a 31P frequency of 202.456 MHz with the spin echo pulse sequence (90° − τ1 − 180° − τ2 − acquire) with 1H decoupling, using a 31P 90° pulse of 4.2 μs, a first echo delay τ1 of 20 μs, a second echo delay τ2 of 14μs, a decoupling power of 88 kHz, and a recycle delay of 5 s. The spectral width was set to 250 ppm. For the 31P NMR spectra, 256 scans were taken and the FIDs were processed using 100 Hz of line broadening with Topspin 2.0.
31P longitudinal relaxation times (T1) were measured using a standard one-dimensional direct phosphorous detection inversion-recovery pulse sequence (180° − T − 90°− acquire), with a pre-acquisition delay of 6 s, and 14 T values ranging from 0.1 ms to 6 s. The sample was spinning at a frequency of 4 kHz at the magic angle. T1 values were calculated by fitting the isotropic peak intensities into a single exponential function I(t) = I(0) + P × exp(−t/T1) by using Topspin 2.0.
1H MAS NMR experiments were conducted on a Bruker Avance 500 MHz standard bore NMR spectrometer (Bruker Biospin) equipped with a 4-mm double-resonance HR-MAS probe (Bruker Biospin) using a single-pulse sequence with a pre-acquisition delay of 5.0 s, a 90° pulse of 4.0 s. Sixty-four scans were averaged and the FIDs were processed using 0.3 Hz of line broadening with Topspin 2.0. The samples were spinning at a frequency of 4 kHz at the magic angle. The sample temperature was controlled at 26°C by using a BCU-05 unit (Bruker Biospin).
Results and Discussion
Interaction of sorbic acid with the acyl chain region by 2H NMR
To investigate whether the sorbic acid molecule can penetrate deeply into the hydrophobic acyl chain region of lipid bilayers, we first collected 2H NMR spectra of DMPC-d54, the lipids fully deuterated on the two acyl chains, in the presence or absence of weak acids at various pH levels.
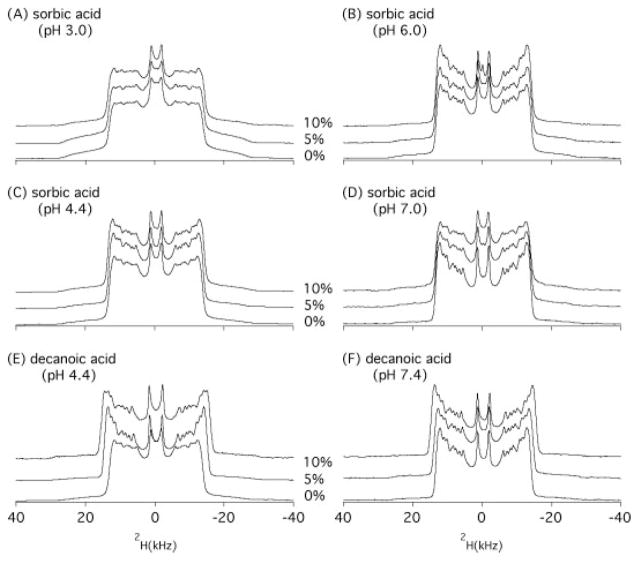
Figure 2 shows the 2H NMR spectra for sorbic acid/DMPC-d54 MLVs, along with the spectra of decanoic acid/DMPC-d54 vesicles at different pH levels. All spectra reveal typical ‘Pake patterns’ as anticipated for lipid MLVs. The two peaks with the smallest absolute separation are the signals originated from the terminal methyl group (−CD3) of the acyl chains of DMPC-d54. The maximal quadrupolar splitting (ΔνQ) is given by the largest distance between the two symmetric peaks in the powder-type spectra, which were generated from the deuterons on the first two methylenes nearby the carbonyl groups of the sn-1 and sn-2 acyl chains. The influence of organic acid additives is reflected in the size of the quadrupolar splittings, which depends on both the molecular conformation and the extent of molecular fluctuations of the labeled segments.
Figure 2.
2H NMR spectra of DMPC-d54 MLV samples as a function of concentration and pH: (A) DMPC-d54 bilayer with 0, 5, and 10 mol % sorbic acid at pH 3.0, (B) pH 4.4, (C) pH 6.0, (D) pH 7.4, (E) decanoic acid at pH 4.4, and (F) decanoic acid at pH 7.4, obtained using a quadrupolar echo pulse sequence under static condition at 35 °C.
The maximal quadrupolar splittings of the 2H NMR spectra in Fig. 2 are summarized in Table 1. In the absence of organic acids, the 2H quadrupolar splitting of DMPC-d54 is approximately 24.0–24.9 kHz. The quadrupolar splitting decreases very slightly due to the addition of sorbic acid at pH 3.0 or 4.4 (Figs 2A and C). At pH 6.0 and 7.4 (Figs 2B and D), no changes in the quadrupolar splittings were detected when adding 5 mol% or even 10 mol% of potassium sorbate to the DMPC-d54 MLVs. The data clearly indicate that sorbic acid does not penetrate deeply into the lipid hydrophobic core region within a pH range of 3.0–7.4.
Table 1.
Effect of sorbic acid or decanoic acid on the 2H maximal quadrupolar splittings (in kHz) of DMPC-d54 bilayers measured using static 2H solid-state NMR experiments at 35 °C
| DMPC | DMPC + 5%sorbic acid | DMPC + 10%sorbic acid | DMPC + 5%decanoic acid | DMPC + 10%decanoic acid | |
|---|---|---|---|---|---|
| pH 3.0 | 24.0 | 24.4 (+1.7%) | 24.8 (+3.3%) | – | – |
| pH 4.4 | 24.2 | 24.5 (+1.2%) | 24.9 (+2.9%) | 27.7 (+14.5%) | 29.6 (+22.3%) |
| pH 6.0 | 24.7 | 24.9 (+0.8%) | 25.0 (+1.2%) | – | – |
| pH 7.4 | 24.9 | 24.9 | 24.9 | 26.2 (+5.2%) | 27.6 (+10.8%) |
The values in parenthesis are the calculated percentage of increases of quadrupolar splittings upon addition of sorbic acid or decanoic acid.
Conversely, the titration of decanoic acid at pH 4.4 (Fig. 2E) or pH 7.4 (Fig. 2F) increases the quadrupolar splittings significantly. Addition of 5 and 10 mol% of decanoic acid increases the quadrupolar splitting by 3.5 and 5.1 kHz at pH 4.4, and by 1.3 and 2.7 kHz atpH 7.4, respectively (Table 1). The increasing quadrupolar splittings directly reflect a decreasing efficiency of reorientational motion of the lipids upon interaction with decanoic acid. The results demonstrate that decanoic acid interacts with the lipid acyl chains at both pH 4.4 and pH 7.4. This result is consistent with our previous studies,[31] as well as the results of fatty acids in lipid membranes reported by others.[34]
Interaction of sorbic acid with the headgroup region by 31P NMR
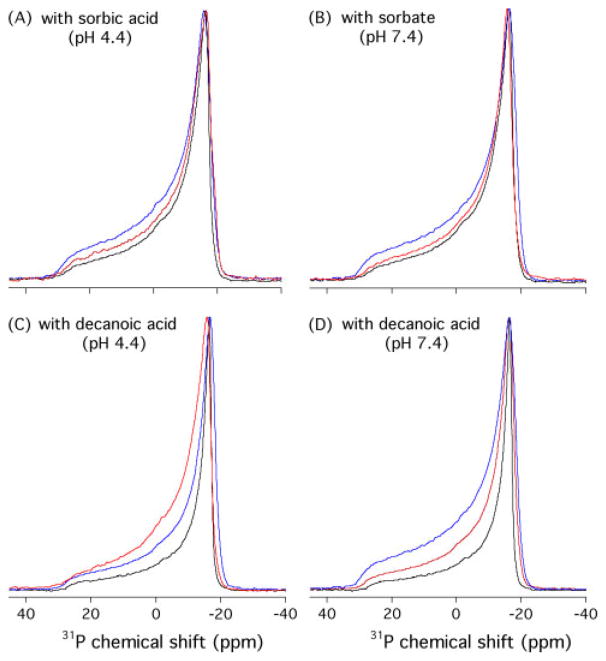
The interaction of sorbic acid and decanoic acid with the phospholipid headgroups of DMPC bilayers was studied with 31P NMR spectroscopy under both acidic and neutral conditions. Static 31P NMR spectra were collected on the same samples that used for the 2H NMR experiments. The 31P NMR spectra at pH 3.0 and 4.4 show very similar patterns, as well as those at pH 6.0 and 7.4. Only the 31P NMR spectra at pH 4.4 and 7.4 for DMPC MLVs containing varying amounts of sorbic acid or decanoic acid are presented.
As shown in Fig. 3, the 31P NMR spectra of DMPC MLVs are dominated by a pseudo-axially symmetric chemical shift tensor. The large chemical shift anisotropy (CSA) range exhibits characteristic lamellar lineshapes with a high field maximum (σ⊥) peak at ~?−17 ppm and a low field shoulder (σ||) at ~29 ppm. The obtained CSA Δσ( Δσ= σ|| − σ⊥) is ~46 ppm, typical of DMPC lipid bilayers in the fluid phase.[34] In all cases, the 31P NMR spectra of DMPC in the presence of sorbic acid or decanoic acid retain a similar motionally averaged powder pattern lineshape, suggesting that the DMPC bilayers remain in the liquid-crystalline Lα phase even after the addition of 10 mol% organic acids. 31P NMR spectra show considerable linewidth broadenings upon addition of sorbic acid or decanoic acid. This 31P NMR linewidth broadening indicates that sorbic acid or decanoic acid are interacting with the phosphorous headgroups of the lipids, either though a reduced local motion or a change in the orientation experienced by the phosphate headgroups.[26]
Figure 3.
1H-decoupled 31P NMR powder spectra of DMPC MLV samples as a function of concentration and pH: with sorbic acid at pH 4.4 (A) and 7.4 (B), with decanoic acid at pH 4.4 (C) and 7.4 (D), obtained using a Hahn-echo pulse sequence under static condition at 35 °C. The concentrations of the weak acids are 0 (in black), 5 (in red), and 10 (in blue) mol %, respectively.
We also examined the effects of organic acids on the headgroup dynamics by using 31P MAS NMR longitudinal relaxation time of DMPC MLV samples. The changes in the 31P isotropic T1 values provide measurements of the amplitude of motion fluctuations on the fast (10−9 s) time scale, indicating changes in individual lipid dynamics.[35] Our 31P T1 values of pure DMPC MLV samples at 35 °C are in close agreement with a previously reported T1 value of 660 ms at the phosphorous frequency of 60.7 MHz and at 30 °C.[36] As shown in Fig. 4, at both pH 3.0 and 4.4, the 31P T1 values increase slightly upon adding 5 mol% sorbic acid, indicating that the presence of sorbic acid reduces the rate of rapid motion of the phosphate moiety. At pH 6.0 and 7.4, slight decreases in T1 were observed upon addition of sorbate. While the nature of interaction needs to be further investigated, all the 31P NMR results suggest that the phospholipid headgroups are undergoing perturbations upon association with sorbic acid or decanoic acid under both acidic and neutral conditions.
Figure 4.
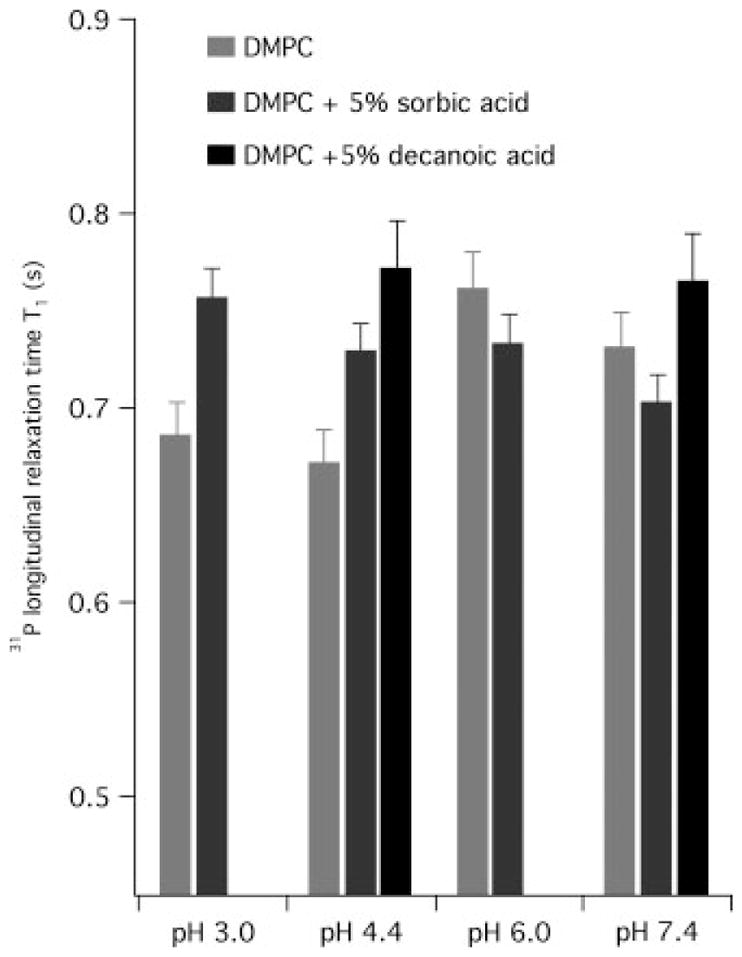
The comparison of 31P longitudinal relaxation times T1 at 35 °C between control MLV and MLVs with 5% mol % sorbic acid or decanoic acid measured at a 31P frequency of 202.456 MHz using inversion-recovery method, with 4 kHz magic-angle spinning.
The location of sorbic acid by 1H MAS NMR
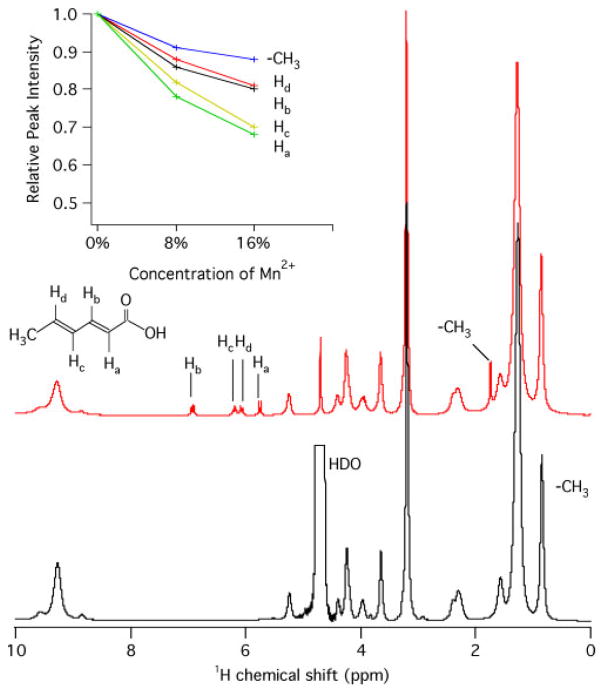
Because of the double bonds of sorbic acid, the proton signals of sorbic acid in 1H NMR spectra are easily distinguishable from that of the lipids; thus, providing a direct approach to track sorbic acid in the lipid membranes. The approximate location of sorbic acid in the DMPC bilayers was further investigated with 1H MAS NMR spectroscopy of DMPC/sorbate with and without Mn2+. Figure 5 shows the 1H MAS NMR spectra collected for DMPC in the presence of varying concentrations of sorbic acid and Mn2+. The peak assignment of sorbic acid was assisted by the previous report.[37] The intensities of 1H resonance peaks of sorbate protons decreased upon addition of Mn2+ions by about 10% after adding 5% Mn2+, as calculated from the peak intensities of sorbic acid, using the methyl group at the DMPC acyl chain end at 0.86 ppm as an internal standard. Higher concentrations of Mn2+reduce the peak intensities more significantly. The reduced peak intensities are consequences of paramagnetic T2 relaxation enhancement resulting from the addition of Mn2+ions, suggesting that the sorbate molecule is in the vicinity of Mn2+ ions, which are in the aqueous phase.[21] The 1H-MAS NMR data in the presence of paramagnetic ions again suggests that sorbate molecules are shallowly embedded on the surface of the DMPC bilayers.
Figure 5.
1H HR-MAS NMR spectra measured for pure DMPC (bottom), and DMPC/10 mol % sorbate (middle), at pH 7.4 obtained at 35 °C using a single-pulse sequence with 4 kHz sample spinning at the magic angle. The relative resonance intensities of sorbic acid were plot against the Mn2+ concentration (up).
DMPC is a zwitterionic lipid, carrying a positive charge at the N atom and a negative charge at the P–O bond. The DMPC MLVs used in this study were in fully hydrated aqueous dispersions. Electrostatic interactions with headgroups contribute to the initial binding of weak acids with the membranes. The association of the hydrophobic portion of weak acids with the hydrophobic acyl chains of membranes may drive the weak acids’ insertion into the core region of membranes. The exact location or distribution of weak acids in the membrane can be described by a balanced interplay between the hydrophobic and electrostatic interactions. Under acidic conditions, further stabilization may be achieved if hydrogen bonds can be formed between weak acids (H-bond donor) and the phosphate group (H-bond acceptor). The pKa value of sorbic acid is 4.8.[2] About 98.5% of sorbic acid would be in the undissociated forms at pH 3.0, as calculated from the Henderson–Hasselbalch equation.[38] Similarly, about 71.5, 5.9, and 0.3% of sorbic acid would be the undissociated form at pH 4.4, 6.0, and 7.4, respectively. The pKa of decanoic acid is 6.8,[39] suggesting about 99.6 and 20.1% in the undissociated form at pH 4.4 and 7.4, respectively. The octanol–water partition coefficients (log Poctanol/water) of sorbic acid and decanoic acid are 1.33 and 4.09, respectively. The observed changes in quadrupolar splittings correlate well with the length of the chains of weak acids and the pH values, suggesting a key role of the hydrophobic acyl portion in the weak monocarboxylate acids in their membrane insertion. From the 2H NMR spectra at pH 3.0 and 4.4, it can be seen that only a very small percentage of the undissociated forms of sorbic acid may insert into the membrane, resulting in a weak effect on the quadrupolar splittings. The deuterium quadrupolar splittings offer direct evidence that sorbic acid does not penetrate deeply into the hydrocarbon core region of the membranes in the pH range from 3.0 to 7.4.
The 31P NMR and 1H MAS NMR spectra, on the other hand, clearly indicate the interfacial interactions of the sorbic acid with the headgroups and sorbic acid is shallowly embedded in the lipid bilayers. The 2H NMR studies also indicate two distinct types of membrane interactions: (i) sorbic acid binds to the polar lipid headgroups near the bilayer surface (adsorption) and (ii) decanoic acid penetrates into the bilayer interface (absorption). This is consistent with an early study, which suggests that decanoic acid has a mode of action distinct from weak acid preservatives, where inhibition of yeast growth by decanoic acid was attributed to a consequence of rapid membrane rupture and loss of cytoplasm.[4]
The relative molecular size of the weak acids and the lipids can also help rationalizing the observed difference in membrane interaction of sorbic acid and decanoic acid. Simple molecular mechanics calculations using MM2[40] show that the length of sorbic acid molecule (the distance between C1 and C6) is about 6.3 Å, while the length of decanoic acid is ~11.5 Å in extended conformation. The headgroup region of lipid bilayers is about 8 Å long and the half hydrophobic thickness of ~12.5 Å.[41] The polar carboxylate group of sorbic acid is energetically favored to be located in the vicinity of the charged headgroups of lipids; thus, the medium length carbon chain of sorbic acid is unable to reach the hydrophobic core of the lipid bilayers. The two carbon–carbon double bonds at α, β, and γ , and δ positions of sorbic acid molecule make it a very rigid molecule. This, along with the medium length of the carbon chain, may make the sorbic acid molecule difficult to insert deeply into the membrane and induce significant disturbing effects on the hydrophobic regions of lipid membranes, which can be detected with the 2H NMR spectroscopy.
Plasma membranes have long been believed to be the targets of weak acid antimicrobial agents.[2] It was hypothesized that membranes act as an allosteric regulator or a ‘receptor’ with which a substrate can interact with different binding sites in a pH-dependent manner.[42] It has also been shown that perturbing the permeability of the cytoplasmic membranes might not immediately be responsible for the biological activity of several surfactants.[43] The present study indicates that inserting deep into the membranes may not be a prerequisite for sorbic acid to act as an antimicrobial agent. Considering the weak effects of sorbic acid exerting on the membrane at such high concentration (3/70 mg lipids) in this study and the low concentration (4 mM, 0.6 mg/ml) where potassium sorbate shows genotoxic action in human lymphocytes,[44] membrane disruption is not sufficient to justify, alone, the enhanced antimicrobial activity of sorbic acid at low pH, and the membrane-disruption mechanism is unlikely to be predominant mode of action of sorbic acid at these concentrations. As more evidence comes out, we may speculate that rather than acting as a nonspecific antimicrobial agent, sorbic acid might act on specific bacterial targets.
The data available so far do not exclude the possibility that sorbic acid might induce membrane damage in a way dependent on the composition of the membranes. The interaction of sorbic acid with cholesterol or other components in the membranes, for instance, might amplify the weak effects sorbic acid exerted on the membrane. Thus, further experiments are in progress to investigate the effects of sorbic acids on other lipid bilayers, which may be good models representative of bacterial membranes.
Acknowledgments
This work was partly supported by NIH grant (GM60259-01) and NSF award (CHE-0645709) to G.A.L. The Bruker 500-MHz wide-bore NMR spectrometer was obtained from a National Science Foundation grant given to G.A.L. (10116333).
References
- 1.Lück E, Jager M, Laichena SF. Antimicrobial Food Additives: Characteristics, Uses, Effects. 2. Springer-Verlag; Berlin Heidelberg: 1997. [Google Scholar]
- 2.Sofos JN. Sorbate Food Preservatives. CRC-Press Inc; Boca Raton: 1989. [Google Scholar]
- 3.Brul S, Coote P. Int J Food Microbiol. 1999;50:1. doi: 10.1016/s0168-1605(99)00072-0. [DOI] [PubMed] [Google Scholar]
- 4.Stratford M, Anslow PA. FEMS Microbiol Lett. 1996;142:53. doi: 10.1111/j.1574-6968.1996.tb08407.x. [DOI] [PubMed] [Google Scholar]
- 5.Stratford M, Anslow PA. Lett Appl Microbiol. 1998;27:203. doi: 10.1046/j.1472-765x.1998.00424.x. [DOI] [PubMed] [Google Scholar]
- 6.Brul S, Coote P, Oomes S, Mensonides F, Hellingwerf K, Klis F. Int J Food Microbiol. 2002;79:55. doi: 10.1016/s0168-1605(02)00179-4. [DOI] [PubMed] [Google Scholar]
- 7.Freese E, Sheu CW, Galliers E. Nature. 1973;241:321. doi: 10.1038/241321a0. [DOI] [PubMed] [Google Scholar]
- 8.Eklund T. J Gen Microbiol. 1985;313:73. doi: 10.1099/00221287-131-1-73. [DOI] [PubMed] [Google Scholar]
- 9.Krebs HA, Wiggins D, Stubbs M, Sols A, Bedoya F. Biochem J. 1983;214:657. doi: 10.1042/bj2140657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bracey D, Holyoak CD, Coote PJ. J Appl Microbiol. 1998;85:1056. doi: 10.1111/j.1365-2672.1998.tb05271.x. [DOI] [PubMed] [Google Scholar]
- 11.Plumridge A, Hesse SJA, Watson AJ, Lowe KC, Stratford M, Archer DB. Appl Environ Microbiol. 2004;70:3506. doi: 10.1128/AEM.70.6.3506-3511.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marín S, Abellana M, Rubinat M, Sanchis V, Ramos AJ. Int J Food Microbiol. 2003;87:251. doi: 10.1016/s0168-1605(03)00068-0. [DOI] [PubMed] [Google Scholar]
- 13.Viegas CA, Rosa MF, Sá-Correia I, Novais JM. Appl Environ Microbiol. 1989;55:21. doi: 10.1128/aem.55.1.21-28.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warth AD. J Gen Microbiol. 1977;43:215. [Google Scholar]
- 15.Skirdal I, Eklund T. J Appl Bacteriol. 1993;74:191. doi: 10.1111/j.1365-2672.1993.tb03014.x. [DOI] [PubMed] [Google Scholar]
- 16.Statham JA, McMeekin TA. J Appl Microbiol. 1988;65:469. [Google Scholar]
- 17.Shore PA, Brodie BB, Hogben CAM. J Pharmacol Exp Ther. 1957;119:361. [PubMed] [Google Scholar]
- 18.Thomae AV, Wunderli-Allenspach H, Krämer SD. Biophys J. 2005;89:1802. doi: 10.1529/biophysj.105.060871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grime JM, Edwards MA, Rudd NC, Unwin PR. Proc Natl Acad Sci USA. 2008;105:14277. doi: 10.1073/pnas.0803720105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Missner A, Kugler P, Antonenko YN, Pohl P. Proc Natl Acad Sci USA. 2008;105:E123. doi: 10.1073/pnas.0809606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seydel JK, Wiese M. Drug-Membrane Interactions: Analysis, Drug Distribution, Modeling. WILEY-VCH Verlag GmbH; Weinheim: 2002. p. 87. [Google Scholar]
- 22.Lorigan GA. In: NMR Spectroscopy of Biological Solids. Ramamoorthy A, editor. CRC/Taylor & Francis Group; Boca Raton: 2005. p. 237. [Google Scholar]
- 23.Seelig J. Q Rev Biophys. 1977;10:353. doi: 10.1017/s0033583500002948. [DOI] [PubMed] [Google Scholar]
- 24.Minto RE, Adhikari PR, Lorigan GA. Chem Phys Lipids. 2004;132:55. doi: 10.1016/j.chemphyslip.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Roberts JE, Griffin RG. In: Phosphorus NMR in Biology. Burt CT, editor. CRC Press; Boca Raton: 1987. p. 63. [Google Scholar]
- 26.Bechinger B, Seelig J. Biochemistry. 1991;30:3923. doi: 10.1021/bi00230a017. [DOI] [PubMed] [Google Scholar]
- 27.Dufourc EJ, Mayer C, Stohrer J, Althoff G, Kothe G. Biophys J. 1992;61:42. doi: 10.1016/S0006-3495(92)81814-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ader C, Schneider R, Seidel K, Etzkorn M, Baldus M. Biochem Soc Trans. 2007;35:991. doi: 10.1042/BST0350991. [DOI] [PubMed] [Google Scholar]
- 29.Buffy JJ, Hong T, Yamaguchi S, Waring AJ, Lehrer RI, Hong M. Biophys J. 2003;85:2363. doi: 10.1016/s0006-3495(03)74660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boland MP, Middleton DA. Phys Chem Chem Phys. 2008;10:178. doi: 10.1039/b712892d. [DOI] [PubMed] [Google Scholar]
- 31.Nusair NA, Tiburu EK, Dave PC, Lorigan GA. J Magn Reson. 2004;168:228. doi: 10.1016/j.jmr.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 32.Lu J, Blazyk J, Lorigan GA. Biochim Biophys Acta2006. 1758;1303 doi: 10.1016/j.bbamem.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Davis JH, Jeffrey KR, Bloom M, Valic MI, Higgs TP. Chem Phys Lett. 1976;42:390. [Google Scholar]
- 34.Lindström F, Thurnhofer S, Vetter W, Gröbner G. Phys Chem Chem Phys. 2006;8:4792. doi: 10.1039/b607460j. [DOI] [PubMed] [Google Scholar]
- 35.Gehman JD, Luc F, Hall K, Lee TH, Boland MP, Pukala TL, Bowie JH, Aguilar MI, Separovic F. Biochemistry. 2008;47:8557. doi: 10.1021/bi800320v. [DOI] [PubMed] [Google Scholar]
- 36.Rajan S, Kang SY, Gutowsky HS, Oldfield E. J Biol Chem. 1981;256:1160. [PubMed] [Google Scholar]
- 37.Cigić IK, Plavec J, Mozina SS, Zupancic-Kralj L. J Chromatogr A. 2001;905:359. doi: 10.1016/s0021-9673(00)00996-1. [DOI] [PubMed] [Google Scholar]
- 38.Arroyo-López FN, Bautista-Gallego J, Duran-Quintana MC, Garrido-Fernández A. Food Microbiol. 2008;25:566. doi: 10.1016/j.fm.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 39.Small DM, Cabral DJ, Cistola DP, Parks JS, Hamilton JA. Hepatology. 1984;4:77S. doi: 10.1002/hep.1840040814. [DOI] [PubMed] [Google Scholar]
- 40.Allinger NL, Yuh YH, Lii JH. J Am Chem Soc. 1989;111:8551. [Google Scholar]
- 41.Sengupta D, Smith JC, Ullmann GM. Biochim Biophys Acta2008. 1778;2234 doi: 10.1016/j.bbamem.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 42.Avdeef A, Box KJ, Comer JEA, Hibbert C, Tam KY. Pharm Res. 1998;15:209. doi: 10.1023/a:1011954332221. [DOI] [PubMed] [Google Scholar]
- 43.Glover RE, Smith RR, Jones MV, Jacksonc SK, Rowlands CC. FEMS Microbiol Lett. 1999;177:57. doi: 10.1111/j.1574-6968.1999.tb13713.x. [DOI] [PubMed] [Google Scholar]
- 44.Mpountoukas P, Vantarakis A, Sivridis E, Lialiaris T. Food Chem Toxicol. 2008;46:2390. doi: 10.1016/j.fct.2008.03.021. [DOI] [PubMed] [Google Scholar]