Summary
Regeneration starts with injury. Yet how injuries affect gene expression in different cell types, and how distinct injuries differ in gene expression remains unclear. We defined the transcriptomes of major cell types of planarians – flatworms that regenerate from nearly any injury – and identified 1,214 tissue-specific markers across 13 cell types. RNA sequencing on 619 single cells revealed that wound-induced genes were either expressed in nearly all cell types or specifically in one of three cell types (stem cells, muscle, or epidermis). Time-course experiments following different injuries indicated a generic wound response is activated with any injury regardless of the regenerative outcome. Only one gene, notum, was differentially expressed early between anterior- and posterior-facing wounds. Injury-specific transcriptional responses emerged 30 hours after injury, involving context-dependent patterning and stem-cell-specialization genes. The regenerative requirement of every injury is different; however, our work demonstrates that all injuries start with a common transcriptional response.
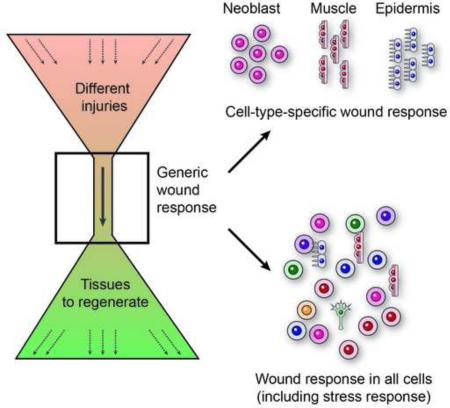
Graphical Abstract
Introduction
Wounding leads to a series of complex responses that are necessary for recovery (Gurtner et al., 2008). Recent studies in regenerative organisms, including planarians (Wenemoser et al., 2012), sea anemones (DuBuc et al., 2014), hydra (Lengfeld et al., 2009), and axolotls (Knapp et al., 2013) have demonstrated that wounding broadly impacts gene expression, including the activation of stress-response genes, tissue-patterning factors, matrix metalloproteinases, and growth factors. However, the functions of the vast majority of genes that are induced following injury remain unknown (DuBuc et al., 2014; Wenemoser et al., 2012).
Planarians are free-living flatworms with a remarkable regenerative capacity that is mediated by tissue-resident proliferative cells (neoblasts) that include pluripotent cells (Reddien and Sanchez Alvarado, 2004; Wagner et al., 2011). Following wounding, rapid gene expression changes are observed in both neoblasts and differentiated tissues (Wenemoser et al., 2012). A number of genes were shown to be activated at different wound types (Adell et al., 2009; Petersen and Reddien, 2009; Wenemoser et al., 2012), raising the possibility that a common transcriptional wound response precedes regeneration (Wenemoser et al., 2012). By contrast, it has been recently proposed that different injuries activate distinct transcriptional programs that subsequently converge to similar transcriptional programs later in regeneration (Kao et al., 2013). Determining whether wounds that will regenerate different anatomy begin with similar, identical, or very different transcriptional responses remains a central problem in understanding regeneration.
Some wound-induced genes, such as HSP90 and HSP70, are associated with general stress response; but others, such as follistatin, are critical for initiating regeneration (Gavino et al., 2013). By contrast, some wound-induced genes have known functions only in particular injuries. For example, wound-induced wnt1 expression has a known role in tail but not head regeneration (Adell et al., 2009; Petersen and Reddien, 2009), despite its induction at both wound types (Petersen and Reddien, 2009).
Multiple key questions about wound responses and how they associate with regeneration of different body parts remain unresolved. First, how does the transcriptional response to wounding map onto the different cell types at the site of injury? Second, how does the transcriptional response to injury differ depending on the injury type and the eventual regenerative outcome? Finally, which transcriptional changes are specific to the regeneration of particular anatomical structures and when do these changes appear?
We addressed these key questions by combining multiple experimental and computational approaches. We applied single-cell RNA sequencing (SCS) to 619 individual planarian cells and determined the transcriptomes of 13 distinct cell types, including all major planarian tissues, leading to the identification of 1,214 unique tissue markers. SCS from injured animals associated 49 wound-induced genes with the cell types that expressed them, revealing that major wound-induced gene classes were either expressed in nearly all cell types at the wound or specifically in one of three cell types (neoblast, muscle, and epidermis). Time-course experiments on bulk RNA from injuries leading to distinct regenerative outcomes determined that a single conserved transcriptional program was activated at essentially all wounds, except for the differential activation of a single gene, notum. Over 24 hours following the peak of this generic wound response, specialized transcriptional programs emerged, specific for the body parts requiring regeneration. Our results define a generic and conserved response to wounding, identify the cell types that drive it, and describe the subsequent transcriptional changes leading to regeneration.
Results
Single-cell sequencing of planarian cells
To dissect how different cell types transcriptionally respond to injuries, we used single-cell RNA sequencing (SCS), because it profiles the transcriptional responses of a cell and allows its cell type classification (Jaitin et al., 2014; Shalek et al., 2014). We isolated cells by fluorescent activated cell sorting (FACS; Fig 1A) from postpharyngeal wound sites that were collected from animals immediately following amputation or after a recovery period (4 or 12 hours post injury; hpi; Methods). In total, we sequenced RNA from 214 dividing neoblasts and 405 non-dividing cells (Table S1), and measured their gene expression by mapping the sequencing reads to the planarian transcriptome (Liu et al., 2013). On average, we detected the expression of 4,401 genes per cell (Fig S1A), with more than 91% of the cells expressing over 1,000 genes (Extended experimental procedures).
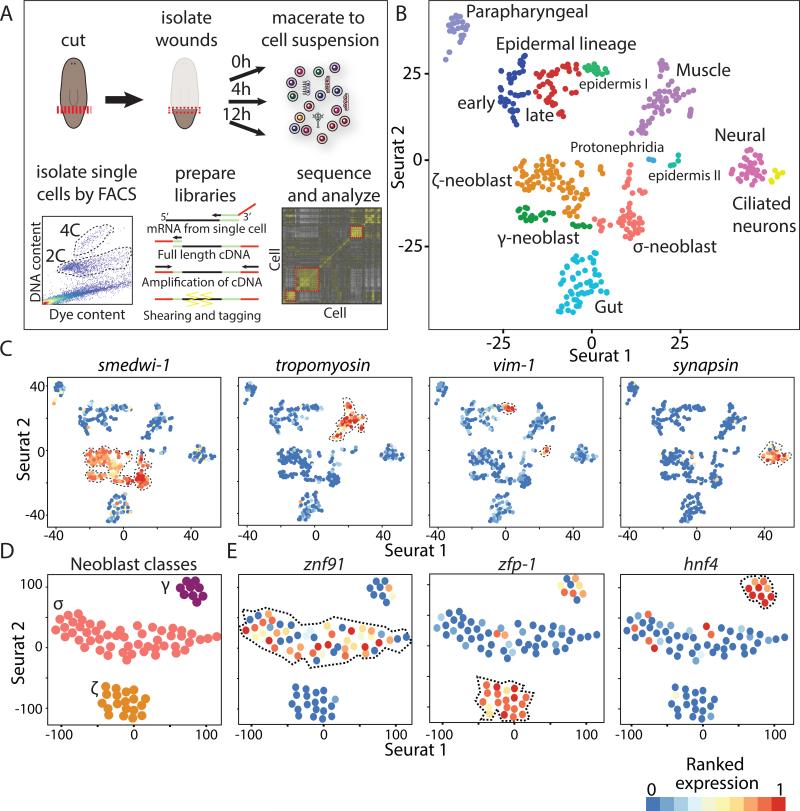
Figure 1. Unbiased detection of major planarian cell types by SCS.
(A) Illustration of SCS data generation and analysis. Animals were cut postpharyngeally (red line) and wound sites (red box) were isolated at 3 time points. Wound tissue was macerated, and dividing (4C) or non-dividing (2C) cells were isolated by FACS (Methods; dashed line shows gates). Sequencing libraries were prepared by cDNA-amplification and shearing, and libraries were sequenced and analyzed. (B) t-SNE plot of single cells. Cells (colored dots) are grouped by density clustering and labeled based on marker analysis. Cells shown are from the 2C (wounded and unwounded) and 4C (wounded) fractions. (C) Expression of canonical cell-type markers overlaid on t-SNE plots of the single-cells (dots); low and high ranked expression are colored by a gradient of blue, yellow, and red. (D) Analysis of the neoblast compartment. Shown are neoblasts (dots) from uninjured animals. Clusters are annotated based on multiple neoblast markers. (E) Expression of class-specific neoblast markers. See also Figure S1-2.
We assessed the SCS data quality by comparing the expression of canonical neoblast markers (Guo et al., 2006; Reddien et al., 2005; Shibata et al., 1999) between sorted neoblasts and non-dividing cells. Neoblasts had a striking enrichment for these transcripts (Fig S1B; p < 1E-75). For example, smedwi-1 and bruli were overexpressed in neoblasts 217- and 140-fold, respectively, highlighting the expression data specificity.
Unbiased assignment of planarian cells to putative cell types
To define the cell types present at wounds, cells were clustered and analyzed according to their gene expression (Fig S1C). Initially, genes with high variance across cells were selected (Fig S1D-F; dispersion ≥ 1.5; Methods), because their expression levels can partition cells to groups (Jaitin et al., 2014; Shalek et al., 2013). Next, we used these genes as input for the recently published Seurat algorithm (Macosko et al., 2015; Satija et al., 2015) that extends the list of genes used for clustering by finding genes with significant expression structure across principal components (Extended experimental procedures; Fig S1G). Then, cells were embedded and visualized in a 2-dimensional space by applying t-Distributed Stochastic Neighbor Embedding on the genes selected by Seurat (t-SNE; Fig 1B; Methods). Finally, clusters were defined by applying density clustering (Ester et al., 1996) on the 2-dimensional embedded cells. Importantly, the time point at which cells were isolated did not affect cluster assignments (Table S1), indicating that the identity of a cell had a stronger impact on cluster assignment than did transcriptional responses to wounding. This process revealed 13 cell clusters (Fig 1B), which likely represented different major planarian cell types.
Detection of the major planarian cell types
Multiple approaches were used to assign cell type identity to the clusters, and to test whether cells in a cluster were of the same type. First, we plotted the expression of published cell-type-specific markers on the t-SNE plots (Fig 1C) and found that canonical tissue markers for major cell types were found exclusively in distinct clusters. This was highly suggestive of cluster identity for cell types, such as neoblast (Reddien et al., 2005), muscle (Witchley et al., 2013), neurons (Sanchez Alvarado et al., 2002), and epidermis (van Wolfswinkel et al., 2014).
Second, we identified cluster-specific genes by using a binary classifier (Sing et al., 2005) that quantified the ability of individual genes to partition cells assigned to one cluster from all other clusters by measuring the area under the curve (AUC) in a receiver operating characteristic curve (ROCC; Fig S1H; Methods). Similarly, we searched for markers that were expressed in multiple clusters displaying expression of the same canonical markers (e.g., smedwi-1 or synapsin; Fig 1C; Methods).
In total, 1,214 genes (false discovery rate; FDR < 0.1) were highly specific for a cluster or shared between cluster groups (Table S2). We used the multiple published anatomical markers found in this gene set to determine cluster identity for the following cell types: muscle (Witchley et al., 2013); gut (Forsthoefel et al., 2011); epidermis (van Wolfswinkel et al., 2014); early epidermal progenitors (prog-1) (Pearson and Sanchez Alvarado, 2010); late epidermal progenitors (agat-1) (Eisenhoffer et al., 2008; van Wolfswinkel et al., 2014); neoblasts including specialized neoblasts (Scimone et al., 2014; van Wolfswinkel et al., 2014), protonephridia (Scimone et al., 2011); and two neuronal types (Cowles et al., 2013; Sanchez Alvarado et al., 2002) (Fig 1B-E, S2; Table S2).
Finally, a single cluster was unique in lacking enriched expression of genes with published expression patterns. Whole-mount in situ hybridizations using RNA probes (WISH) on four of its top cluster-specific genes (Rab-11B, myoferlin, ESRP-1, and anoctamin) revealed strong parapharyngeal (pp) expression with a ventral anatomical bias (Fig S2A; Methods). Double fluorescent ISH (dFISH; Fig S2B) validated that single cells in the parapharyngeal region co-expressed these genes, indicating that this was indeed a cell type lacking prior molecular definition.
The clustering analysis we performed allowed detection of subpopulations of cells that appeared largely homogenous when examined only with canonical markers. For example, two adjacent clusters (Fig 1B) were determined to be neural based on specific expression of canonical neural markers, including synapsin, synaptotagmin, and prohormone convertase 2 (PC2; Fig 1C, S2D). However, one of these clusters co-expressed genes encoding known cilia components, such as bbs1, bbs9 (Fig S2D), ift88, and iguana (Glazer et al., 2010), suggesting that these might be neurons with sensory cilia (Louvi and Grove, 2011). The only other cell-type expressing these cilia genes was the epidermis (Fig S2D).
In the neoblast compartment, we detected three subpopulations representing the recently described σ-, ζ-, and γ- type neoblasts (van Wolfswinkel et al., 2014) (Fig 1D-E), and revealing multiple putative markers unique to each subpopulation (Table S2; Fig 1E), such as znf91, a previously undescribed gene encoding a zinc finger protein showing the highest specificity to the σ-Neoblasts (AUC=0.81, FDR=2.6E-5; Fig 1E; Table S2).
Importantly, the dissection of planarian cell types and their associated gene expression generated an extensive repository of cell-type-specific markers for every major cell type, including signaling molecules, receptors and transcription factors (TFs), as well as profiles of their co-expression (available at https://radiant.wi.mit.edu/app/).
Identification of cell-type-specific wound-induced genes
Knowing which cell types express particular wound-induced genes is important for understanding how the wound response differs across injuries with different anatomy. However, the cell-type specificity of only a small number of wound-induced genes is known (Wenemoser et al., 2012; Witchley et al., 2013).
Since SCS data is often noisy and incomplete (Jaitin et al., 2014), we first defined a comprehensive list of wound-induced genes by RNA sequencing of bulk samples from two different injury types. We profiled the expression of anterior-facing (head removal) and posterior-facing (trunk and tail removal) wounds in the prepharyngeal region (Fig 2A) by isolating RNA, in triplicate, at four time-points (0, 3, 6, and 12 hpi; Fig 2A; Methods).
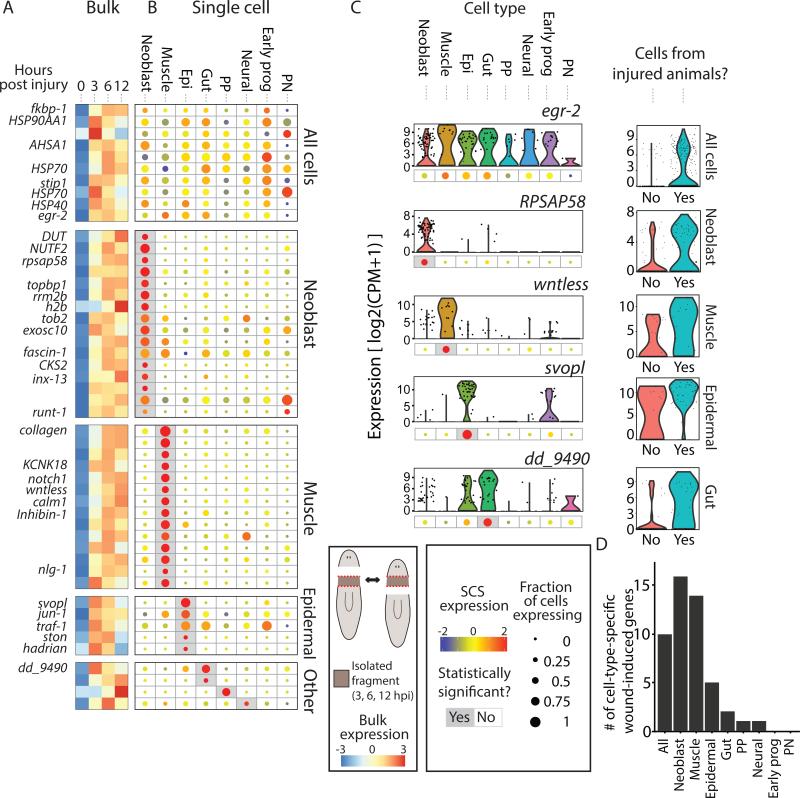
Figure 2. Cell-type-specific expression of wound-induced genes.
(A) The expression of wound-induced genes, as detected by bulk RNA-sequencing, is shown at different time points (0, 3, 6, and 12 hpi). Shown is the average expression of the anterior- and posterior-facing time courses. Rows and columns represent genes and time points, respectively. Gene expression is colored according to the z-transformed expression (z-score range is −3 to 3). Shown are wound-induced genes for which cell-type specificity was determined. (B) The corresponding cell-type-specific gene expression is shown in a dot-plot map. Dot size represents the proportion of cells expressing the gene (see key; 0-1), and the color represents normalized expression in cells expressing the gene (blue-to-red, low-to-high expression). Gray background represents statistically significant enrichment in a cell type (FDR ≤ 0.01; Extended experimental procedures). Genes are ordered according to their controlled enrichment p-value. Genes assigned to the All cells were overexpressed following wounding in multiple cell types (Methods). Cell-type acronym labels: NB, Neoblasts; Epi, epidermal lineage; Early prog, early epidermal progenitors; PP, para- pharyngeal; PN, protonephridia). (C) Left panels: Representative genes with wound-induced expression in different cell types. Expression across cell types is shown in violin plots with corresponding dot-plots beneath. Right panels: violin plots comparing the expression in cells of the cell type the gene was found to be enriched in between uninjured and injured animals. (D) Summary of the detected cell-type-specific wound-induced genes.
The bulk sequencing data revealed that 128 genes were overexpressed in at least one time point compared to the 0 hpi (uninjured) samples, in at least one of the two wound types (fold-change ≥ 2; FC; FDR ≤ 0.05; Fig 2A; Table S3; Methods). To determine what cell types participated in the wound response, we compared the SCS expression of the 128 wound-induced genes (i) between cells isolated from uninjured animals and injured animals; and (ii) between different cell types using only cells isolated following wounding (Fig 2B-C; Methods). In total, we detected the cell-type-specificity of 49 of the 128 genes (38%). Ten of these genes were wound-induced in nearly all cell types (Fig 2), with six of them annotated as general stress response factors, including heat-shock protein 90 (HSP90), HSP70, and HSP40 (Methods). Only one of the genes encoded a transcription factor, egr-2 (Fig 2B-C).
Strikingly, most of the cell-type-specific genes (35/49; 71%; Fig 2D) were wound-induced in one of three cell types. 16 genes were enriched in neoblasts, including genes related to proliferation (e.g., H2B, topbp1, rrm2b) and neural regeneration (runt-1, known to be induced in neoblasts) (Sandmann et al., 2011; Wenemoser et al., 2012). In muscle cells, we found enrichment for 14 wound-induced genes, including five genes that were implicated in major signaling pathways, including Wnt, BMP, and TGF-β, which are essential for proper patterning of planarian tissues (Reddien, 2011; Witchley et al., 2013). Importantly, as the number of muscle cells sequenced was smaller than many other cell types (e.g., the number of gut cells was almost twice the number of muscle cells), these results cannot be explained by an increased statistical power resulting from larger sample size. Finally, five genes were enriched in epidermal lineage cells, including Smed-jun-1 (Wenemoser et al., 2012). In addition, a small number of genes (1-2) were wound-induced in three other cell types: gut, parapharyngeal (Fig S3A), and neural cells.
Our results are supported by two recent studies that examined the co-expression of several wound-induced genes with cell-type-specific markers. nlg1, inhibin-1, and wntless were found to be specifically wound-induced in muscle cells of injured animals (Witchley et al., 2013), whereas jun-1, TRAF-1, ston, and hadrian were found to be localized to the epidermis (Wenemoser et al., 2012).
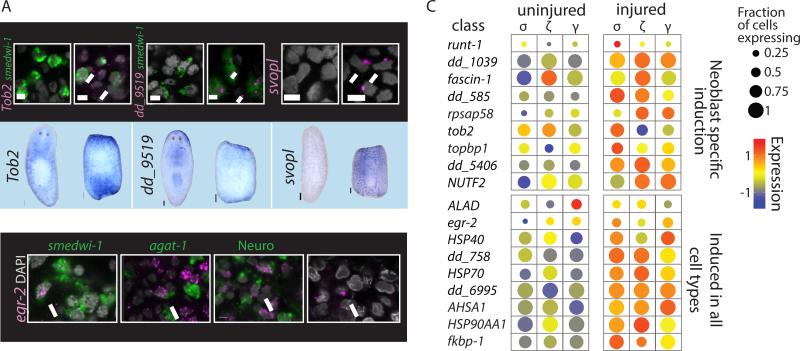
We used multiple approaches to validate our results. First, we examined the co-localization of three candidates (svopl, dd_9519, Tob2) with published cell-type markers. dFISH analysis found, in all cases, high specificity of expression to the identified cell type in the single-cell analysis (Fig 3A). Furthermore, we tested whether egr-2 was indeed wound-induced in multiple cell types (Fig 2), and found that it was co-localized with markers for neoblasts (smedwi-1), epidermal progenitors (agat-1), neural cells (PC2, synapsin, and synaptotagmin), and with differentiated epidermis (outermost epidermal layer; Fig 3B).
Figure 3. Analysis of Cell-Type-Specific Expression After Injury.
(A) Validations of tissue-specific wound-induced genes. Upper panel: dFISH analysis (scale = 5μm) of cell-type-specific wound-induced gene (magenta) and a cell-type marker (green), or imaging of the outermost layer (epidermis). Nuclei labeled with DAPI (gray). White arrows point to co-expressing cells. Lower panel: WISH analysis comparing gene expression in intact and amputated animals (scale = 100μm). (B) dFISH analysis of egr-2 (magenta) with markers of multiple tissues in animals 12 hpi (green; smedwi-1 – neoblasts; agat-1 – epidermal progenitors; Neuro (pooled RNA probes for PC2, synapsin, synaptotagmin) – Neural tissue; epidermal cells were imaged by the outermost layer of the animals). WISH/FISH analysis was done on at least 15 fragments for each gene. (C) Gene expression comparison of uninjured and injured neoblasts. Shown are dot plots of neoblast-specific wound-induced genes (top panel) and genes found to be wound-induced in most or all cell types (bottom panel) in the different neoblast classes. Dot size represents the fraction of expressing cells (0-1); color represents the expression levels (z-score) in the fraction of expressing cells.
Next, we tested whether different neoblast subpopulations (van Wolfswinkel et al., 2014) responded differently to wounding (Fig 3C). We compared the gene expression of neoblasts representing the general neoblast pool (σ), the epidermal progenitors (ζ), and the putative gut progenitors (γ) between uninjured and injured animals. Interestingly, while some wound-induced genes were overexpressed in specific populations (e.g., runt-1 in the σNeoblasts), most genes changed similarly across neoblast subtypes (Fig 3C).
This analysis demonstrates that the cell-type architecture of the wound response involves: (i) genes induced broadly in most or all cell types; (ii) multiple genes induced in a cell-type-specific manner in one of three types of cells: neoblast, muscle, or epidermis; and (iii) rare individual genes expressed in a specific cell type (gut, parapharyngeal, or neural cells).
A single gene, notum, detectably differentiates between anterior and posterior wound responses
How similar are the transcriptional responses to distinct injuries? The cell types that express wound-induced genes are widespread across the planarian body, and in principle, could mount a similar transcriptional response at injuries requiring regeneration of distinct tissues.
However, the extent of similarity in wound responses between distinct injuries is yet to be resolved. To address this question we searched for wound-induced genes that were enriched at anterior- over posterior-facing wounds, or vice versa, at any of the three time-points (3, 6, and 12 hpi; Methods; Fig 2A, 4A; Table S3). Importantly, these two wound types had very similar tissue composition, but required distinct regenerative outcomes (Fig 4A).
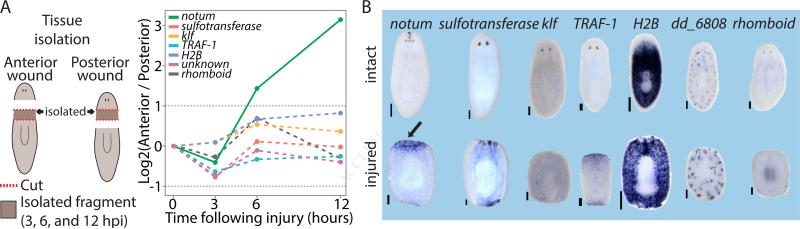
Figure 4. notum is the only gene detectably induced asymmetrically at wounds.
(A) The gene expression profiles of injuries with different wound orientation (anterior and posterior; left panel) are compared in time-course experiments of tissues isolated from the same location. Plotted is the log2 ratio of differentially expressed genes between the two wound types (FDR ≤ 0.05; fold-change ≥ 1.5). Dashed lines represent genes that could not be validated by WISH, and that are likely false-positives. (B) WISH validations of wound-induced genes shown in panel A (performed on at least 10 animals). Top panel shows gene expression in intact animals compared to the expression in amputated trunks (bottom panel). Amputated animals were fixed at the time point showing peak asymmetry in expression. Only notum showed asymmetrical expression following wounding (black arrow). Scale=100 μm. See also Figure S3.
Out of the 128 wound-induced genes, only one gene (notum) had a biased expression of more than two-fold in one of the amputations compared to the other, in at least one time point (Fig 4A). Even with relaxed thresholds (FC ≥ 1.5; FDR ≤0.1) we found that only seven genes were overexpressed at one of the injuries compared to the other (Fig 4A). We tested the expression data predictions by WISH, and strikingly, only notum displayed asymmetric expression, with the six other genes having no robust differential expression in anterior and posterior wound sites (Fig 4B). The one true-positive gene, notum, is known to be activated at all wounds but to have stronger expression at anterior-facing compared to posterior-facing wounds (Petersen and Reddien, 2011). Importantly, notum is essential for establishing correct head-tail regeneration in planarians (Petersen and Reddien, 2011).
We extended this analysis by screening 218 additional genes by WISH; these genes represented a diversity of fold changes for wound induction and genes that were below threshold for significant difference between wound types. All wound-induced genes had similar expression at anterior and posterior-facing injuries (Fig S3B; Table S3-4). These data strongly indicate that following anterior or posterior amputations, the same transcriptional response to wounding is immediately activated, except for higher expression of a single gene, notum, at anterior-facing wounds.
Comparison of responses to diverse injuries through extended time-course experiments
The striking similarity in the wound response following two amputations types is consistent with the possibility that a generic wound response would be activated following any injury, even when regeneration is not required (Wenemoser et al., 2012). To test this hypothesis, we studied distinct injuries requiring regeneration of different body parts in time courses that span the wound response and extended to subsequent regenerative phases (0-120 hpi; Fig 5A, S4A; Table S5).
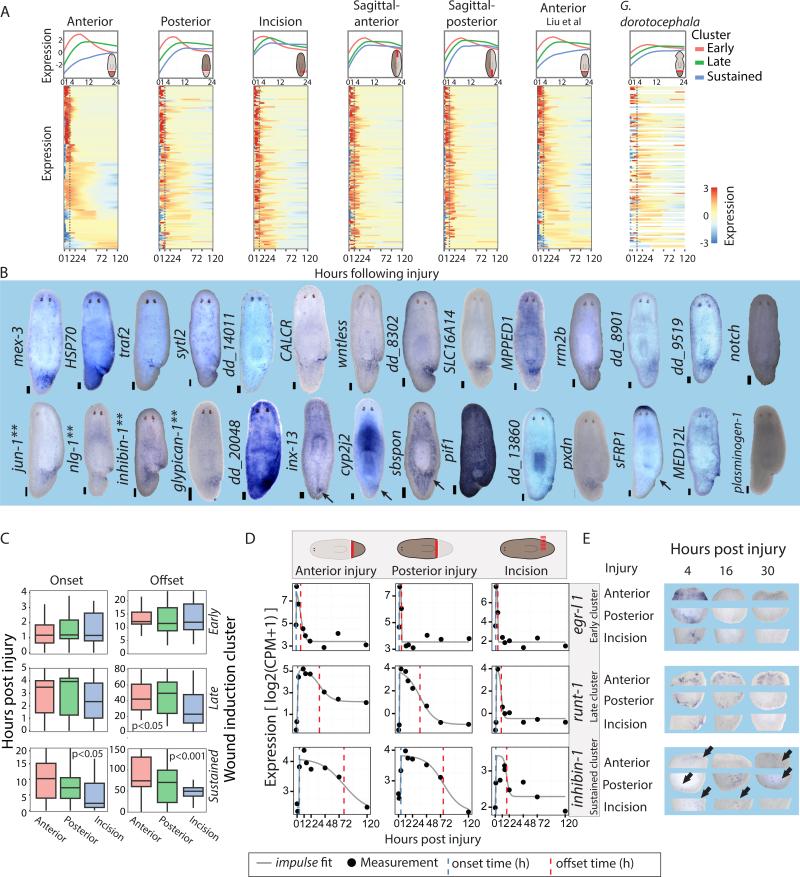
Figure 5. Time-course analysis reveals a generic response to wounding.
(A) Expression of wound-induced genes at different planarian injuries. A core set of 128 wound-induced genes is plotted in different extended time courses. Worm illustrations show the injury site and isolated tissue location (red block line). Top panel: the expression of different wound-induced clusters from 0 to 24 hpi (lines are loess fit of wound-induced gene expression in each cluster; the same genes were used in all panels). Bottom panel: The expression of the wound-induced genes from 0 to 120 hpi is shown according to fitting of individual genes to a constrained impulse model (Chechik and Koller, 2009) (shown is row z-score; blue-to-red, low-to-high expression, respectively). Right-most column: Conservation of the wound response in anteriorly regenerating G. dorotocephala. Gene order follows orthology assignment between G. dorotocephala and S. mediterranea (Methods; white lines represent genes with no ortholog assigned). (B) WISH analysis of wound-induced genes. Shown are representative animals 4 or 12 hours following incision (scale = 100 μm; ** denote genes for which WISH analysis of incision was previously published). (C) Analysis of onset and offset times in different wound-induced genes clusters and injuries, as computed using the impulse model (ks-test). (D) Expression of representative genes from the early (egr-l 1), late (runt-1), and sustained (inhibin-1) clusters (0-120 hpi) is shown in time course data. Gene expression data points (black dots) are plotted with the impulse fit function (gray line). Onset and offset times, blue and red dashed lines, respectively. (E) WISH validation of onset and decay times for the genes shown in panel D. Gene expression is shown for the three types of injuries tested (anterior, posterior, and incision). Scale = 100 μm. See also Figure S4.
At every time point we isolated wound sites from the following injuries: (1) postpharyngeal anterior-facing; (2) postpharyngeal posterior-facing; (3) sagittal-anterior; (4) sagittal-posterior; and (5) a lateral incision, which did not require regeneration (Fig 5A, S4A; Methods). Gene expression was measured by RNA-seq and compared to uninjured equivalent anatomical regions. In addition, a recently published head regeneration RNA-seq data set was incorporated (Liu et al., 2013).
To test if the same transcriptional response was activated in every injury, a comprehensive collection of wound-induced genes was required. We therefore determined whether the 128 gene list (described above) included the majority of wound-induced genes without detecting an abundance of false positives. WISH was performed on 225 genes (Table S4), which covered a wide range of fold changes and FDR following wounding. We found that a threshold of FC > 2 balanced sensitivity (57%) with precision (88%). This analysis estimates that the total number of wound-induced genes, detectable with utilized methods, is approximately 224 (s.d.=27), an appreciably small (~1%) fraction of all planarian genes (Fig S4B-E; Table S4; Methods).
A common response to wounding activated following diverse injuries
To test whether a generic transcriptional program is activated at every injury, we evaluated how many of the 128 wound-induced genes were induced within 16 hours following the injuries described above. 85% of the genes were overexpressed in at least 5 time-courses (FC > 1.5; Table S5; Methods); fold-changes in time courses that did not meet this threshold were often (43%) just below it. We tested by WISH whether the wound-induced genes that did not appear to be overexpressed by RNA-seq in a given time course were indeed not induced by that injury type. In all cases, the genes were actually expressed at the tested injury site (9/9 incisions; Table S5). Furthermore, we tested 10 additional of the 128 wound-induced genes that appeared to be lowly induced in incisions (2 > FC > 1.5) and eight genes that appeared to be lowly induced in posterior amputations (2 > FC > 1.5), and found that they were in fact induced in all cases (Table S5). By contrast, tissues far from the injury (Online Methods) showed upregulation of a fraction of the wound-induced genes (15%; Fig S4G), with many of these genes (9/23) associated with stress responses.
To further validate that tissue removal was not required for activating the wound-response program, we compared the expression of 35 randomly selected wound-induced genes by WISH in intact, amputated, or incised animals at their time of peak expression (Fig 5B; Table S5; Methods). All 35 genes were induced following amputations, and strikingly, 34/35 (97%) of the genes were detectably overexpressed following incisions, corroborating the time-course experiments (Fig 5A; Table S5). sulfotransferase, which was not detectably overexpressed by WISH, was at least two-fold overexpressed in all RNA-seq time courses. Collectively, these results strongly suggest that a single generic transcriptional program was activated at every injury. This response might include genes that are insignificant for many types of injuries, but essential for the recovery from others. Consistent with this possibility, RNAi of only 8 of 62 wound-induced genes displayed a detectable phenotype (Table S3), further suggesting that many wound-induced genes are not essential for survival and recovery after injury.
The response to wounding terminates earlier when regeneration is not required
Whereas different injuries activated essentially the same genes, the dynamics of their expression across injuries could be different. We therefore fit the gene expression data to a quantitative model (impulse) (Chechik and Koller, 2009; Sivriver et al., 2011) that extracted transcriptional parameters for every wound-induced gene (Fig 5A, C; Methods), including their onset and offset times (time to reach half maximal expression and time to return to half baseline expression, respectively; Methods). Wound-induced genes were then clustered based on their fitted expression into three groups with significantly different onset and offset parameters (Figure 5A, B-D). Based on these parameters, wound-induced clusters were labeled as: early (n=44), late (n=53), and sustained (n=31). Most of the wound-induced stress-response genes, such as HSP70, HSP90, and HSP40, were part of the early cluster, rapidly induced and fast to decay (Table S5), and our SCS data showed that they are induced in nearly all cell types (Fig 2A-B).
The late cluster included many cell-type-specific wound-induced genes, such as patterning factors overexpressed selectively in muscle cells following wounding (Fig 2A-D; Table S5) (Witchley et al., 2013). Strikingly, in every injury, patterning factors were overexpressed with a median onset of less than 4h even without any tissue loss. Such a rapid induction for these genes is remarkable considering that the time-scale of regeneration and its associated patterning is days to over a week (Reddien and Sanchez Alvarado, 2004).
Next, we compared the onset and offset times of wound-induced gene clusters across injuries (Fig 5C). The onset (~1 hpi) and offset (~12 hpi) of the early cluster did not differ significantly between injuries (ks-test p > 0.05; following Bonferroni correction). Similarly, the late cluster was already induced at ~3 hpi in each injury; however, the offset time, following an incision, was almost 20 hours earlier compared to anterior and posterior regeneration (Fig 5C; p < 0.05). Finally, the onset and offset of the sustained cluster were significantly earlier in the incision (p < 0.05), suggesting that lack of tissue was required for the response to sustain, or alternatively, that tissue fusion was sufficient to terminate it.
We tested these results by selecting candidates from each wound-induced gene cluster and performing WISH time courses (Fig 5D-E) on animals that suffered different injuries. Comparison between the fitted data (Fig 5D) and the in situ gene expression (Fig 5E, S4F) further validated that (i) early cluster genes (e.g. egr-l 1) displayed similar onset and offset times across injuries; and that (ii) late and sustained cluster genes (e.g. runt-1 and inhibin-1) had similar expression across injuries in early time points but their expression returned to baseline earlier at incisions. Together, these results indicated that while the same set of genes is activated at every injury, the duration of their activation is shorter when regeneration is not required.
The generic wound response is conserved in a related planarian species
To assess if the generic wound-response program described above in Schmidtea mediterranea is conserved in other species, we used a second planarian model, Girardia dorotocephala (Flickinger and Coward, 1962). We sequenced and assembled its transcriptome and found high-confidence orthologs for 95/128 (74%) of the wound-induced genes (Extended experimental methods; File S1; Table S6). RNA sequencing on anterior-facing wounds revealed strong and significant correlation between the fold changes of wound-induced genes in both organisms (Pearson r=0.56; p = 5.1e-09), with genes from all three clusters of wound induction (i.e, early, late, and sustained) being up-regulated. The overexpressed genes included cell-type-specific wound-induced S. mediterranea genes expressed in muscle (wntless, notum), neoblasts (runt-1, Tob2, inx-13), and epidermis (jun-1, ston). Furthermore, both a gut- and a parapharyngeal-specific gene were induced following injury. In total, 61% (58/95) of the S. mediterranea wound-induced genes were detectably overexpressed following wounding in G. dorotocephala (Table S6). The activation of orthologous stress-response, patterning, and proliferation-related genes, further highlights key conserved components of the generic wound response.
The generic wound response is followed by a specific regenerative response
The response to wounding was nearly identical in different injuries, despite preceding regeneration of very different anatomy. We therefore used our extended time course data to search for the onset of injury-specific gene expression. We compared the expression of known head-enriched genes (n=43) (Gurley et al., 2010; Reddien, 2011; Scimone et al., 2014; van Wolfswinkel et al., 2014; Vogg et al., 2014) between tail fragments that regrew heads and incisions that did not require regeneration (Fig 6A-B). Fitting the gene expression of regenerating animals (Fig 6; Methods) revealed that they had a wide range (>90 hours) of onset values, which was significantly later than the wound-induced genes (ks-test p=9.2E-11).
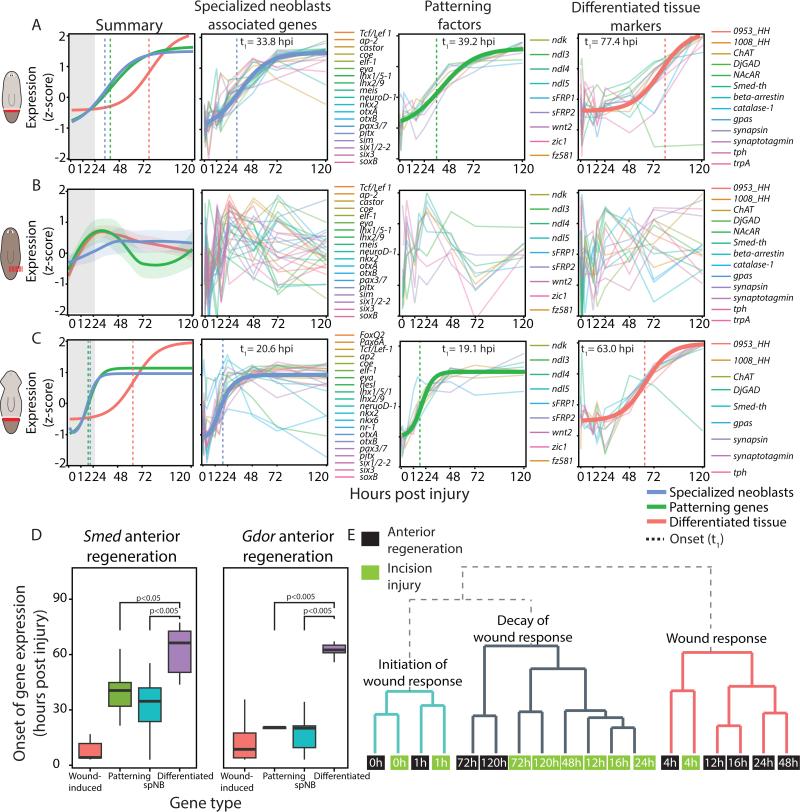
Figure 6. Injury-specific regeneration occurs in a temporally defined order.
(A) Summary panel: shown is a fit of the normalized median expression of neoblast specialization-associated genes, injury-specific patterning factors, and terminally differentiated tissue markers (blue, green and red, respectively). Matching colored vertical lines mark the onset times of the corresponding group of genes. Gray box highlights the wound-response phase. Other panels: bold lines represent impulse model fit of the genes used for modeling the dynamics of the group; thin lines represent individual genes. Onset time is marked by a vertical dashed line. (B) The genes used for panel A were plotted with the incision time-course data in which there was no missing tissue. Shown is a loess fit (bold lines) and confidence interval of the z-scores for each class of genes (lightly colored area) as the data could not be fit to the impulse model. Individual panels show a non-specific response following wounding. (C) A similar analysis performed on anteriorly regenerating G. dorotocephala revealed a similar order of events to amputation in S. mediterranea. (D) Box plot showing the onset time of different groups of genes following amputation. Boxes represent the interquartile range, thick lines are the median. Statistical significance was tested by a ks-test. (E) Dendrogram illustrating the similarity of gene expression of wound-induced genes in samples from the anterior regeneration and the incision time courses. Each node represents a sample (0-120 hpi; green and black nodes, incision and anterior samples, respectively). Annotations on the tree represent the interpretation of samples in clade.
Genes were categorized based on previously suggested functions to three groups: (i) tissue patterning factors, which were previously associated with expression in muscle (Witchley et al., 2013); (ii) genes associated with specialized neoblasts (Scimone et al., 2014; van Wolfswinkel et al., 2014); and (iii) markers of differentiated anterior tissues. All three groups were highly upregulated during anterior regeneration, but they were separable into two distinct phases (Fig 6A). During the first phase, genes enriched in specializing neoblasts (34 hpi) and anteriorly expressed patterning genes (39 hpi) were upregulated. Subsequently, almost 40 hours later, genes enriched in differentiated head cell types were upregulated (Fig 6A; ks-test p = 4.4E-4; 77 hpi). Similar phases were found for orthologous genes in G. dorotocephala (Fig 6C). Importantly, both regenerative phases were separable from the generic wound-response onset by over 24 hours (Fig 6D; ks-test p=9.2E-11). By contrast, in animals suffering incisions we could not detect significant expression changes in any of the genes associated with regeneration (Fig 6B), which prohibited fitting to the impulse model, indicating that these were indeed part of a specific regenerative response.
Hierarchal clustering of samples from the anterior regeneration and incision time courses, using wound-induced gene expression, further supported the conclusion that gene expression changes are sustained only when tissue is missing (Fig 6E). Samples from early time points (0, 1, and 4 hpi) from incisions and anterior amputations formed a cluster, because of similarities in early wound response. However, starting at 12 hpi, the wound-induced gene expression at incisions was largely eliminated (Fig 5A-C) and these samples clustered with 72 and 120 hpi samples from anterior-regenerating fragments.
Our results support a model of a sequentially activated regenerative program starting with the generic wound-response (0-24 hpi), followed by expression of injury-specific patterning factors and specialized neoblast genes (~30 hpi), and finally with the appearance of differentiated tissues (~70 hpi).
Discussion
The ability of planarians to regenerate from almost any injury, combined with the wide array of methods established for their study, make them a unique system for studying regeneration initiation. Here, we took a single cell RNA sequencing approach, combined with bulk tissue sequencing from multiple distinct wound types to characterize the transcriptional responses associated with planarian regeneration initiation. Our data supports a model in which a generic transcriptional program is activated by wounding to accommodate the regeneration of diverse tissue types depending on the nature of the injury (7A). How can a generically activated transcriptional program be activated, if every injury involves different combinations of cell types at unpredictable wound sites? We found that the generic wound response includes stress-related responses in all cell types and cell-specific responses in neoblasts, muscle, and epidermis that are distributed throughout the planarian body (7B). Finally, following the generic wound response, injury-specific transcription is activated, including patterning and stem cell specialization genes, that precedes the appearance of differentiated tissue markers by ~40 hr (7C). Together, these results link a common transcriptional wound response with divergent regenerative outcomes.
Wound-response polarity is likely determined by a single gene, notum
To find genes activated at wounds associated with different regenerative outcomes, we performed RNA sequencing on two wound types that regenerate different tissues, heads or tails. Strikingly, only one gene, notum, a Wnt-pathway inhibitor (Gerlitz and Basler, 2002), demonstrated a strong bias in expression (over two fold) to one of the two injuries. notum was previously shown to be preferentially expressed at anterior-facing wounds over posterior-facing wounds and to be required for the head-versus-tail regeneration decision (Petersen and Reddien, 2011). However, whether other genes showed similar expression asymmetry was unknown. We tested over 200 additional genes that appeared to show any expression bias to one of the two injuries, but found none that were clearly preferentially induced at one wound type over the other. Other subtle transcriptional differences could exist between these wounds, but were undetectable by RNA-seq and WISH. Therefore, our analyses suggest that notum is the only gene with a transcriptional response distinguishing anterior and posterior-facing wounds up to 24 hpi, which is striking given these wounds will initiate completely different regenerative programs.
A generic, conserved, response to wounding precedes regeneration
Several planarian genes were previously shown to be induced following wounding, even without tissue loss, suggesting that they are generically induced by the injury (Petersen and Reddien, 2011; Wenemoser et al., 2012). Interestingly, a few of these genes, such as wnt1, are important planarian patterning genes (Petersen and Reddien, 2009). Through the usage of time-course experiments from different anatomical positions, we rigorously tested the hypothesis that a common transcriptional program is activated at every type of wound. We found that indeed all wound responses start the same, regardless of the eventual regenerative outcome. We estimated that the generic response involves the upregulation of 224 genes in the first 12 hours following injury. When there was no missing tissue to regenerate, the wound-response initiated largely normally, but decayed earlier.
We propose that the generic wound response acts as a funnel between the varied injuries an organism might suffer and subsequent varied regenerative outcomes (Fig 7). As such, the generic response includes all the necessary components for promoting survival and allowing regeneration of any tissue. The generic response is modified with time to achieve the necessary regenerative outcome. In parallel to the transcriptional wound-response, massive neoblast proliferation (Wenemoser and Reddien, 2010) and apoptosis (Pellettieri et al., 2010) take place following any injury – even at injuries that will not require substantial regeneration, such as following needle puncture. Strikingly, these processes appear to be interconnected: following the initial generic wound response a sequence of events involving the activation of context-dependent transcriptional programs (Lapan and Reddien, 2012; Scimone et al., 2011), mitosis (Wenemoser and Reddien, 2010), and apoptotic (Pellettieri et al., 2010) responses are observed only when the injury requires regeneration.
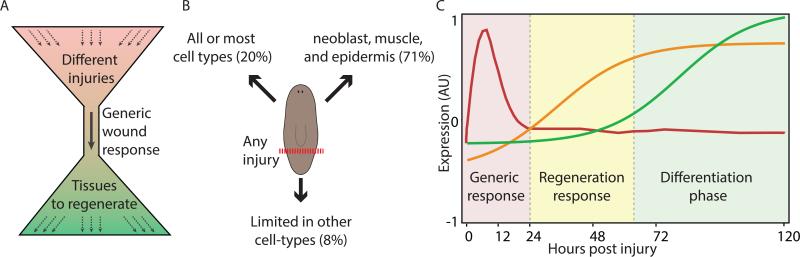
Figure 7. Model for planarian wound-response and initiation of regeneration.
(A) Planarians regenerate from almost any injury through a single transcriptional response. (B) Transcriptional changes following the wound response are divided into three cellular components. (C) A temporal model of planarian regeneration. Every injury triggers a prototypical generic response (red box; red line). If regeneration is not required following the injury, the response will decline. Otherwise, the expression of an injury-specific response emerges (yellow box; yellow line). These responses involve patterning molecules and neoblast-associated fate specialization genes. About three days following the injury, expression of differentiated tissue markers appears in association with the emergence of the newly regenerated structures (green box; green line).
Cell-type specific wound-response genes
How could activation of the same transcriptional program be accommodated by diverse wound locations (injuries through the brain versus tail, for instance), where different cell types juxtapose the wound?
Analysis of some genes activated by wounding showed that multiple tissues are involved, including the epidermis (Wenemoser et al., 2012) and muscle (Witchley et al., 2013), although it remained unclear, to what extent these results are generalizable. We compiled a list of wound-induced genes through time-course experiments, and assessed their expression in single cells from wounds. Our results demonstrated that the response to wounding has three components (Fig 7B): (i) A non-specific component, with genes expressed in nearly all cell types following wounding, including multiple stress-response genes. (ii) A specific component, including 71% of the cell-type-specific genes, with preferential expression in one of three cell types: neoblast, muscle, or epidermis. This component included multiple patterning factors (Witchley et al., 2013), transcription factors, and genes associated with proliferation. (iii) Finally, individual wound-induced genes were expressed in gut, parapharyngeal cells, and neurons, reflecting unique physiological responses in these tissues following wounding. The architecture of the wound response - comprised of genes activated in any cell type at the wound and cell-type specific genes activated in cells widespread in the body – enables the same genes to be activated at essentially all wounds.
Several lines of evidence support the accuracy of wound-induced expression cell type assignments: First, wound-induced expression was much lower before injuries (RNA-seq and WISH); therefore, cells with strongest SCS expression are the best candidates to explain wound-induced expression. Second, in most cases, SCS expression was mostly limited to a single cell type. Third, dFISH validated cell type assignments for a set of tested genes. Finally, direct comparison of neoblasts isolated from intact and injured animals was in agreement with the SCS analysis.
The onset of regeneration and the pruning of the wound response
Through extended time-course experiments, we found that 24 hours following the peak of wound response, patterning genes associated with regeneration (Reddien, 2011; Witchley et al., 2013) were overexpressed, hand-in-hand, with transcription factors associated with neoblast specialization (Scimone et al., 2014). Upregulation of these genes emerged almost 40 hours before the upregulation of differentiated tissue markers. We therefore suggest that regeneration can be modeled by three components of gene-expression changes (Fig 7C): (i) activation of a generic wound-response (~224 genes), which allows the animal to mount a regenerative response to essentially any injury (0-16 hpi). (ii) Expression of patterning factors and neoblast specialization genes, specific to the identity of tissues being regenerated (~36 hpi). (iii) Expression of differentiated tissue markers associated with functional new tissue (Fig 7C; 72 hpi).
A unique repository of cell-type specific expression
This work presents the first application of SCS to planarians. Therefore, many of the profiled cell types were not previously studied at the molecular level in detail. This analysis therefore generated a unique repository, including 1,214 unique cell-type-specific markers, including signaling molecules, receptors, and transcription factors. We developed an online resource that allows accessing the transcriptome of every cell from all identified cell types, available at https://radiant.wi.mit.edu/app/.
Previous studies profiled the gene expression of several planarian cell types through the application of specially developed cell-isolation techniques (Forsthoefel et al., 2012). While successful in studying the targeted tissue, such approaches are not readily applicable to every cell type. Furthermore, as these methods are applied to cell populations, they do not reveal cell-to-cell heterogeneity or gene co-expression in individual cells (Shalek et al., 2013). By contrast, the single-cell expression data allowed us to generate comprehensive co-expression profiles in every profiled cell type, as well as their cell-type expression heterogeneity (online resource).
Concluding remarks
Our analysis suggests a simple and unifying model for the planarian wound response. SCS data indicate that a large component of this response is driven specifically by three abundant tissues (Fig 7B) that allow the response to take place regardless of the anatomy and location of the wound site. Instead of tailoring the response for the desired outcome, the response logic operates in an “act-first” mechanism: activating a program that is sufficient for recovery from any injury. This program is subsequently replaced with an injury-specific response appropriate for regeneration from a specific injury (Fig 7).
Experimental procedures
Planarian culture
Clonal lines of asexual Schmidtea mediterranea (CIW4) and G. dorotocephala were maintained as previously described (van Wolfswinkel et al., 2014).
Single cell library construction
Libraries were prepared using the SmartSeq2 method, as previously described (Picelli et al., 2013; Picelli et al., 2014). Briefly, RNA from single-cells was reverse transcribed with a poly-dT anchored oligo and a template-switching oligo. cDNA was then amplified. Sequencing libraries were prepared using the Nextera XT kit (Illumina).
Sequencing reads mapping
Sequencing reads were mapped to the S. mediterranea dd_Smed_v4 assembly (http://planmine.mpi-cbg.de/; (Liu et al., 2013)) using Novoalign v2.08.02 with parameters [-o SAM -r Random] and were converted to BAM using samtools v1.1 (Li et al., 2009). Read count, for every sample, was calculated with bedtools v2.20.1 (Quinlan and Hall, 2010). Read counts were normalized by edgeR (Robinson et al., 2010). G. dorotocephala libraries were similarly mapped to a de novo transcriptome assembly (File S1).
Single-cell data clustering
An expression matrix for all cells was prepared for analysis in R v3.1.1. Samples expressing less than 1000 or more than 9000 genes were discarded from further analysis. Genes that were used for t-SNE representation and density-based clustering (Ester et al., 1996) were selected by identifying principal components that contribute to the variance using the Seurat method (Macosko et al., 2015; Satija et al., 2015) (Extended experimental procedures).
Detection of cluster-specific genes
Cluster-specific genes were detected by enrichment analysis (McDavid et al., 2013) on genes displaying at least 2-fold enrichment in a cluster compared to all other clusters. Controlled p-value, for each gene, were calculated using the Seurat package (Satija et al., 2015). Then, a binary classifier was used on every cell-type-specific gene (FDR < 0.1; (Sing et al., 2005)). The classifier quantified, for each of the genes tested, its ability to partition the cells it was enriched in from all other cells. For every gene the true positive rate (TPR; sensitivity) and false positive rate (FPR; 1 - specificity) were calculated, and a receiver operating characteristic curve (ROCC) was generated (Fig S1H).
WISH using RNA probes
WISH was performed as previously described (Pearson et al., 2009).
Gene cloning
Genes were amplified from planarian cDNA using gene-specific primers (Extended experimental procedures) and cloned into a pGEM vector (Promega).
Gene annotation
Previously undescribed genes were annotated by best-BLAST hit [e<1E-5) against a sequence database including planarian, human, mouse, fly, and C. elegans sequences. If BLAST hits were not found, the contig id from the transcriptome assembly (Liu et al., 2013) was used. See extended experimental methods for a list of all annotations used in the figures and their corresponding contig id in the assembly.
Double-stranded RNA synthesis
Double stranded RNA (dsRNA) was synthesized as previously described (Petersen and Reddien, 2008). RNA was quantified by Nanodrop (Thermo Fisher Scientific Inc.) to have at least 5 ug/ul.
Illumina library preparations for anterior and posterior time courses
Prepharyngeal fragments were isolated in biological triplicates and placed in TRIzol Reagent (0 hpi). Anterior-facing or posterior-facing wounds were amputated as prepharyngeal fragments at 3, 6, and 12 hpi in biological triplicates. RNA was purified according to manufacturer's instructions (Life Technologies) and sequencing libraries were prepared with a TruSeq RNA sample prep kit V2 (Illumina).
Illumina library preparations for extended time courses
Wound tissues were isolated and put in TRIzol. Tissues were lysed with Qiagen TissueLyser II, and RNA was extracted according to the manufacturer's instructions. Libraries were prepared as previously described (Engreitz et al., 2014; Schwartz et al., 2014) (extended experimental procedures).
Detection of differentially expressed genes and genes and with putative asymmetric wound expression
Wound-induced genes were called using triplicate time course experiments by using the edgeR exactTest function to compare expression at every wounding time point to 0 h. Genes called as wound induced met the following thresholds in at least one time point (FDR ≤ 0.05; fold-change ≥ 2; minimal expression of RPKM 6 in at least 2 of 21 libraries). Putative asymmetric expression was detected by comparing anterior and posterior wound-induced gene expression from matched time points using exactTest. All genes with FDR ≤ 0.05 and fold-change ≥ 1.5 were tested by WISH analysis, as well as 218 other genes not meeting these thresholds (Table S4).
Single cell isolation and FACS
Cells from postpharyngeal wound-sites were isolated and sorted (Hayashi et al., 2006) into 96-well microplates containing 5 ul Buffer TCL (Qiagen) + 1% 2-mercaptoethanol.
Detection of onset and offset of wound-induction
To extract onset and offset parameters of genes, expression data from each time course were used for fitting by the impulse model (Chechik and Koller, 2009; Chechik et al., 2008) using a Matlab implementation (Sivriver et al., 2011) with constraint parameters [retries = 100; t1 ≥ 0; t2 ≥ 0; h0 ≥ 0; h1 ≥ 0; h2 ≥ 0; β1 ≥ 0; β2 ≤ 0].
Supplementary Material
Acknowledgements
We thank Travis R. Rogers for experimental support, David Gennert for SCS protocol help, and Naomi Habib for Matlab implementation of the impulse algorithm. We thank Ben Kleaveland, Schraga Schwartz, Eran Mick, Sarit Edelheit, Guy Bushkin, and Ophir Shalem for critical reading, and the Reddien lab members for manuscript comments. OW was supported by an EMBO long-term fellowship, and is the Howard Hughes Medical Institute Fellow of The Helen Hay Whitney Foundation. We acknowledge NIH (R01GM080639) support. PWR is a Howard Hughes Medical Institute Investigator and an associate member of the Broad Institute of Harvard and MIT. AR acknowledges the Klarman Cell Observatory. RS was supported by NIH F32 HD075541. AR is an SAB member at ThermoFisher Scientific and Syros Pharmaceuticals and a consultant for Driver Genomics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
OW and PWR conceived and designed the overall study. OW, LEC, and AP designed and performed experiments. OW analyzed sequencing data with feedback from RS and AR. OW and PWR wrote the manuscript with comments from all authors.
Accession numbers
Sequencing data were deposited into the Short Read Archive (SRA) under project id PRJNA276084.
References
- Adell T, Salo E, Boutros M, Bartscherer K. Smed-Evi/Wntless is required for beta-catenin-dependent and -independent processes during planarian regeneration. Development. 2009;136:905–910. doi: 10.1242/dev.033761. [DOI] [PubMed] [Google Scholar]
- Chechik G, Koller D. Timing of gene expression responses to environmental changes. Journal of Computational Biology. 2009;16:279–290. doi: 10.1089/cmb.2008.13TT. [DOI] [PubMed] [Google Scholar]
- Chechik G, Oh E, Rando O, Weissman J, Regev A, Koller D. Activity motifs reveal principles of timing in transcriptional control of the yeast metabolic network. Nature Biotechnology. 2008;26:1251–1259. doi: 10.1038/nbt.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles MW, Brown DD, Nisperos SV, Stanley BN, Pearson BJ, Zayas RM. Genome-wide analysis of the bHLH gene family in planarians identifies factors required for adult neurogenesis and neuronal regeneration. Development. 2013;140:4691–4702. doi: 10.1242/dev.098616. [DOI] [PubMed] [Google Scholar]
- DuBuc TQ, Traylor-Knowles N, Martindale MQ. Initiating a regenerative response; cellular and molecular features of wound healing in the cnidarian Nematostella vectensis. BMC Biology. 2014;12:24. doi: 10.1186/1741-7007-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhoffer GT, Kang H, Sánchez Alvarado A. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell. 2008;3:327–339. doi: 10.1016/j.stem.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159:188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ester M, Kriegel H-P, Sander J, Xu X. A density-based algorithm for discovering clusters in large spatial databases with noise; Paper presented at: Kdd..1996. [Google Scholar]
- Flickinger RA, Coward SJ. The induction of cephalic differentiation in regenerating Dugesia dorotocephala in the presence of the normal head and in unwounded tails. Developmental Biology. 1962;5:179–204. doi: 10.1016/0012-1606(62)90009-x. [DOI] [PubMed] [Google Scholar]
- Forsthoefel DJ, James NP, Escobar DJ, Stary JM, Vieira AP, Waters FA, Newmark PA. An RNAi screen reveals intestinal regulators of branching morphogenesis, differentiation, and stem cell proliferation in planarians. Developmental cell. 2012;23:691–704. doi: 10.1016/j.devcel.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsthoefel DJ, Park AE, Newmark PA. Stem cell-based growth, regeneration, and remodeling of the planarian intestine. Developmental Biology. 2011;356:445–459. doi: 10.1016/j.ydbio.2011.05.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavino MA, Wenemoser D, Wang IE, Reddien PW. Tissue absence initiates regeneration through Follistatin-mediated inhibition of Activin signaling. Elife. 2013;2:e00247. doi: 10.7554/eLife.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlitz O, Basler K. Wingful, an extracellular feedback inhibitor of Wingless. Genes & Development. 2002;16:1055–1059. doi: 10.1101/gad.991802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer AM, Wilkinson AW, Backer CB, Lapan SW, Gutzman JH, Cheeseman IM, Reddien PW. The Zn finger protein Iguana impacts Hedgehog signaling by promoting ciliogenesis. Developmental Biology. 2010;337:148–156. doi: 10.1016/j.ydbio.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Peters AH, Newmark PA. A Bruno-like gene is required for stem cell maintenance in planarians. Developmental Cell. 2006;11:159–169. doi: 10.1016/j.devcel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Gurley KA, Elliott SA, Simakov O, Schmidt HA, Holstein TW, Sánchez Alvarado A. Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Developmental Biology. 2010;347:24–39. doi: 10.1016/j.ydbio.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature. 2008;453:314–321. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Asami M, Higuchi S, Shibata N, Agata K. Isolation of planarian X-ray- sensitive stem cells by fluorescence-activated cell sorting. Development, Growth & Differentiation. 2006;48:371–380. doi: 10.1111/j.1440-169X.2006.00876.x. [DOI] [PubMed] [Google Scholar]
- Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao D, Felix D, Aboobaker A. The planarian regeneration transcriptome reveals a shared but temporally shifted regulatory program between opposing head and tail scenarios. BMC genomics. 2013;14:797. doi: 10.1186/1471-2164-14-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp D, Schulz H, Rascon CA, Volkmer M, Scholz J, Nacu E, Le M, Novozhilov S, Tazaki A, Protze S, et al. Comparative transcriptional profiling of the axolotl limb identifies a tripartite regeneration-specific gene program. PLoS One. 2013;8:e61352. doi: 10.1371/journal.pone.0061352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapan SW, Reddien PW. Transcriptome analysis of the planarian eye identifies ovo as a specific regulator of eye regeneration. Cell reports. 2012;2:294–307. doi: 10.1016/j.celrep.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengfeld T, Watanabe H, Simakov O, Lindgens D, Gee L, Law L, Schmidt HA, Ozbek S, Bode H, Holstein TW. Multiple Wnts are involved in Hydra organizer formation and regeneration. Developmental Biology. 2009;330:186–199. doi: 10.1016/j.ydbio.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SY, Selck C, Friedrich B, Lutz R, Vila-Farre M, Dahl A, Brandl H, Lakshmanaperumal N, Henry I, Rink JC. Reactivating head regrowth in a regeneration-deficient planarian species. Nature. 2013;500:81–84. doi: 10.1038/nature12414. [DOI] [PubMed] [Google Scholar]
- Louvi A, Grove EA. Cilia in the CNS: the quiet organelle claims center stage. Neuron. 2011;69:1046–1060. doi: 10.1016/j.neuron.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDavid A, Finak G, Chattopadyay PK, Dominguez M, Lamoreaux L, Ma SS, Roederer M, Gottardo R. Data exploration, quality control and testing in single-cell qPCR- based gene expression experiments. Bioinformatics. 2013;29:461–467. doi: 10.1093/bioinformatics/bts714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Sánchez Alvarado A. Formaldehyde-based whole-mount in situ hybridization method for planarians. Developmental Dynamics. 2009;238:443–450. doi: 10.1002/dvdy.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BJ, Sánchez Alvarado A. A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development. 2010;137:213–221. doi: 10.1242/dev.044297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellettieri J, Fitzgerald P, Watanabe S, Mancuso J, Green DR, Sánchez Alvarado A. Cell death and tissue remodeling in planarian regeneration. Developmental Biology. 2010;338:76–85. doi: 10.1016/j.ydbio.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW. Smed-betacatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science. 2008;319:327–330. doi: 10.1126/science.1149943. [DOI] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW. A wound-induced Wnt expression program controls planarian regeneration polarity. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17061–17066. doi: 10.1073/pnas.0906823106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW. Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science. 2011;332:852–855. doi: 10.1126/science.1202143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nature methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R. Full- length RNA-seq from single cells using Smart-seq2. Nature protocols. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW. Constitutive gene expression and the specification of tissue identity in adult planarian biology. Trends in genetics : TIG. 2011;27:277–285. doi: 10.1016/j.tig.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Annual review of cell and developmental biology. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Alvarado A, Newmark PA, Robb SM, Juste R. The Schmidtea mediterranea database as a molecular resource for studying platyhelminthes, stem cells and regeneration. Development. 2002;129:5659–5665. doi: 10.1242/dev.00167. [DOI] [PubMed] [Google Scholar]
- Sandmann T, Vogg MC, Owlarn S, Boutros M, Bartscherer K. The head- regeneration transcriptome of the planarian Schmidtea mediterranea. Genome biology. 2011;12:R76. doi: 10.1186/gb-2011-12-8-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nature Biotechnology. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Kravarik KM, Lapan SW, Reddien PW. Neoblast specialization in regeneration of the planarian Schmidtea mediterranea. Stem Cell Reports. 2014;3:339–352. doi: 10.1016/j.stemcr.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Srivastava M, Bell GW, Reddien PW. A regulatory program for excretory system regeneration in planarians. Development. 2011;138:4387–4398. doi: 10.1242/dev.068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Shuga J, Trombetta JJ, Gennert D, Lu D, Chen P, Gertner RS, Gaublomme JT, Yosef N, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N, Umesono Y, Orii H, Sakurai T, Watanabe K, Agata K. Expression of vasa(vas)-related genes in germline cells and totipotent somatic stem cells of planarians. Developmental Biology. 1999;206:73–87. doi: 10.1006/dbio.1998.9130. [DOI] [PubMed] [Google Scholar]
- Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- Sivriver J, Habib N, Friedman N. An integrative clustering and modeling algorithm for dynamical gene expression data. Bioinformatics. 2011;27:i392–400. doi: 10.1093/bioinformatics/btr250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Wagner DE, Reddien PW. Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell. 2014;15:326–339. doi: 10.1016/j.stem.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogg MC, Owlarn S, Perez Rico YA, Xie J, Suzuki Y, Gentile L, Wu W, Bartscherer K. Stem cell-dependent formation of a functional anterior regeneration pole in planarians requires Zic and Forkhead transcription factors. Developmental Biology. 2014;390:136–148. doi: 10.1016/j.ydbio.2014.03.016. [DOI] [PubMed] [Google Scholar]
- Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 2011;332:811–816. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D, Lapan SW, Wilkinson AW, Bell GW, Reddien PW. A molecular wound response program associated with regeneration initiation in planarians. Genes & Development. 2012;26:988–1002. doi: 10.1101/gad.187377.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D, Reddien PW. Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Developmental Biology. 2010;344:979–991. doi: 10.1016/j.ydbio.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witchley JN, Mayer M, Wagner DE, Owen JH, Reddien PW. Muscle cells provide instructions for planarian regeneration. Cell Reports. 2013;4:633–641. doi: 10.1016/j.celrep.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.