Abstract
Being a transient carrier of genetic information, mRNA could be a versatile, flexible, and safe means for protein therapies. While recent findings highlight the enormous therapeutic potential of mRNA, evidence that mRNA-based protein therapies are feasible beyond small animals such as mice is still lacking. Previous studies imply that mRNA therapeutics require chemical nucleoside modifications to obtain sufficient protein expression and avoid activation of the innate immune system. Here we show that chemically unmodified mRNA can achieve those goals as well by applying sequence-engineered molecules. Using erythropoietin (EPO) driven production of red blood cells as the biological model, engineered Epo mRNA elicited meaningful physiological responses from mice to nonhuman primates. Even in pigs of about 20 kg in weight, a single adequate dose of engineered mRNA encapsulated in lipid nanoparticles (LNPs) induced high systemic Epo levels and strong physiological effects. Our results demonstrate that sequence-engineered mRNA has the potential to revolutionize human protein therapies.
Introduction
Messenger RNA is an intermediate carrier of genetic information that is used by organisms as template for protein expression. Thus, mRNA may also serve as a tool for the expression of proteins of interest by introducing exogenous molecules into target cells. This concept was first put to the test in the early 1970s by microinjecting RNA preparations into Xenopus oocytes, demonstrating the synthesis of RNA-encoded proteins.1,2 Meanwhile, loading of dendritic cells with antigen-encoding mRNA, described for the first time by Gilboa and colleagues, became a widely applied immunological approach.3
In the early 1990s, first studies demonstrated that exogenous mRNA can be used to direct protein expression in vivo. After local injection of various mRNAs into mouse muscle led to detectable protein levels, the treatment of vasopressin-deficient rats provided first evidence for mRNA as a potential therapeutic.4,5,6 However, several RNA structural features have been described as immunostimulatory due to interactions with various RNA sensors such as Toll-like receptors, RIG-I, and PKR.7,8,9,10,11,12,13,14 Activation of these receptors provides a danger signal which may interfere with the translational machinery. Whereas this feature could threaten the use of mRNA for protein therapies, immunostimulation may be beneficial for vaccines composed of antigen-encoding mRNA. Indeed, immunization with mRNAs coding for cancer antigens was successfully established and has already entered clinical trials.15,16,17,18,19
Analysis of various RNA preparations from bacteria and eukaryotic cells revealed significant differences as to their immunostimulatory properties.20 Obviously, cytokine secretion by RNA-transfected cells strongly correlated with the extent of nucleoside modifications. Transferring this concept to in vitro transcribed mRNA, for instance, pseudouridine-containing mRNAs reduced activation of known RNA sensors substantially.20,21,22 Although pseudouridine is primarily found in tRNA, rRNA, and small nuclear RNAs, pseudouridine-containing mRNAs were still translated and produced even more protein compared to unmodified mRNA.22,23 Accordingly, mRNA harboring modified nucleosides was suggested as means of choice for protein expression via mRNA.
Using enhanced green fluorescent protein mRNA, Rossi and colleagues confirmed that nucleoside modifications can strongly enhance protein expression and suppress cytokine secretion.24 In contrast to previous work, they applied a combination of pseudouridine and 5-methyl-cytidine which outperformed each single modification. As in earlier studies, unmodified nucleotides were completely replaced by their modified counterparts. With such mRNAs, the authors succeeded in reprogramming human cells to pluripotency. The same type of mRNA modification allowed vascular regeneration after myocardial infarction in mice by local expression of VEGF.25 However, different groups apparently favor different modified nucleosides.26 Moreover, according to the findings of Kormann et al.26, partially modified mRNA provides the best combination of high expression and low immunostimulation to achieve therapeutic effects in mice.
Two independent studies on erythropoietin-encoding mRNA found a superiority of nucleoside-modified compared to unmodified mRNA.26,27 Whereas unmodified Epo mRNA was immunostimulatory and failed to induce sufficient protein expression in mice, a few micrograms of nucleoside-modified mRNA were enough to achieve detectable protein expression and physiological effects upon direct administration to murine muscle.26 Complexation with TransIT, enabling intraperitoneal injections, reduced the dose threshold of nucleoside-modified mRNA to just a few nanogram in mice.27 Repeated treatments sustainably increased the hematocrit without any signs of immunostimulation, assessed via interferon-α secretion and anti-EPO antibody generation.27 In addition, a weight-adjusted mRNA dose enhanced serum levels of erythropoietin in Rhesus macaques.27 Unfortunately, physiological responses in monkeys were not reported in that study.
These data raised the question whether nucleoside-modification is an inevitable prerequisite for the development of mRNA-based protein therapies. Here, using sequence-engineered mRNA, we demonstrate that unmodified mRNA is fully suited for use in protein therapies. Engineered but chemically unmodified mRNA coding for erythropoietin achieved high systemic protein levels and strong physiological responses in vivo. Moreover, immunostimulation of that mRNA was at background levels. In combination with an appropriate formulation, e.g., LNPs designed for systemic administration,28 sequence-engineered Epo mRNA elicited meaningful physiological responses even in domestic pigs with a weight of 20 kg. Thus, strong evidence is provided that mRNA-based protein therapies are feasible in large animals, paving the way for the development of a novel class of human therapeutics.
Results
Potent protein expression in vitro with unmodified mRNA
Numerous studies from the early 1990s onwards advocate chemically unmodified mRNA as a suitable and potent means to induce antigen-specific immune responses,19,29,30,31,32,33,34 thereby indicating that such nucleic acids do give rise to expression of encoded proteins upon in vivo delivery. However, it is widely assumed and published that unmodified mRNA is improper for therapeutic purposes due to usually higher protein expression demands compared to vaccination and potentially detrimental immunostimulation. However, the finding that unmodified mRNA gives only very little protein expression contrasts with our experience with sequence-engineered nucleic acids.35 Hence, we set out to test the notion of unmodified mRNA being appropriate for the expression of therapeutic proteins.
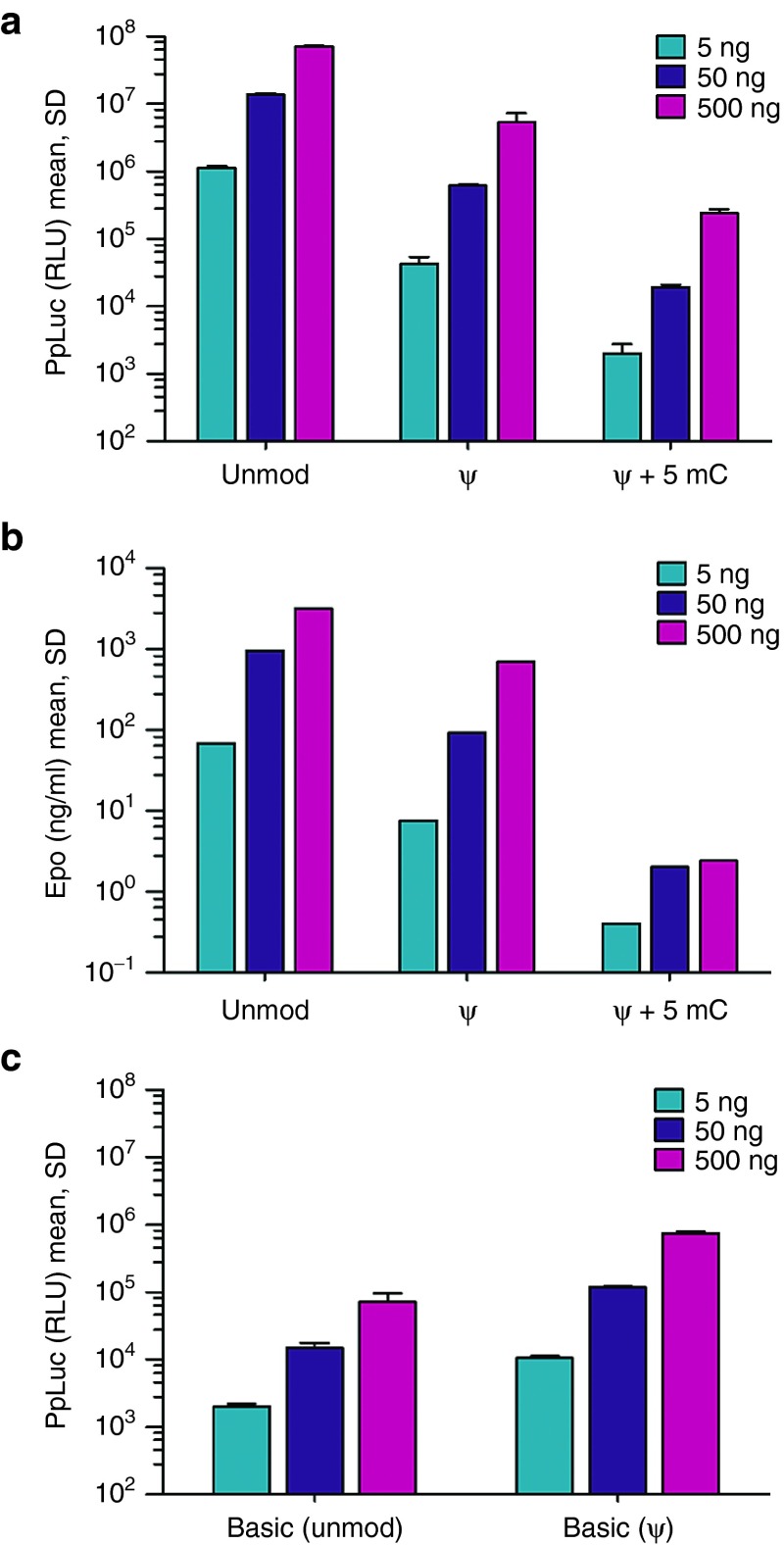
First, we designed a firefly luciferase-encoding mRNA applying a sequence optimization approach which adapts the codon usage and selects the most appropriate regulatory sequences such as 5' and 3' untranslated regions in a target and application specific manner. To test for potential additive effects, we also produced nucleoside-modified counterparts of the final sequence. Notably, in vitro protein expression revealed very high luciferase activity with the unmodified mRNA, while its nucleoside-modified counterparts gave rise to substantially lower protein levels (Figure 1a). This effect was not specific to luciferase as a similar result was obtained with a sequence-engineered mRNA coding for erythropoietin (Figure 1b). These observations appeared to be in contrast to previous reports demonstrating a superiority of modified mRNA. To exclude any general problems with our protocol for manufacturing nucleoside-modified mRNA and to test whether nucleoside modification interferes specifically with sequence-optimized mRNA, we also utilized a less advanced luciferase mRNA harboring widely used regulatory elements and standard codon optimization. Here, incorporation of pseudouridine showed the previously described superiority regarding protein expression compared to the corresponding unmodified mRNA, albeit at a low expression level (Figure 1c) (P < 0.0001 for all doses, student's t-test). Finally, comparison to a modified luciferase mRNA from a commercial supplier confirmed the effectiveness of sequence-engineering for which both adaptation of the open reading frame sequence as well as potent regulatory untranslated regions are essential parameters (Supplementary Figure S1).
Figure 1.
Sequence-engineered mRNA yields high protein expression in vitro. (a) Engineered, unmodified luciferase mRNA outperforms its nucleoside-modified counterparts. HeLa cells were lipofected in triplicate with the indicated amounts of a luciferase-encoding mRNA that was either unmodified or harbored the given nucleoside modifications. Luciferase expression was quantified 24 hours after transfection. (b) Engineered but unmodified mouse Epo mRNA is superior to nucleoside-modified variants. HeLa cells were lipofected in triplicate with different amounts of an erythropoietin-encoding mRNA that was either unmodified or modified by incorporation of the indicated nucleosides. Erythropoietin levels in the supernatant were quantified 24 hours after transfection. (c) Pseudouridine incorporation improves the expression of an unmodified basic luciferase mRNA. HeLa cells were transfected in triplicate with different amounts of a basic luciferase-encoding mRNA either being unmodified or harboring pseudouridine instead of the unmodified nucleotide. Protein expression was quantified 24 hours after transfection. unmod, engineered mRNA harboring the nucleotides A, U, G, and C; ψ, engineered mRNA in which pseudouridine replaces U; ψ+5mC, engineered mRNA in which U and C are replaced by pseudouridine and 5-methly-cytosine, respectively; basic, mRNA comprising a cap, a codon-optimized open reading frame, an α-globin 3'-UTR as well as a polyA.
Efficient protein expression and physiological effects in vivo with unmodified mRNA
The in vitro data already suggested that sequence-engineering can generate highly potent mRNA without any nucleoside modifications. However, in vitro expression data may not faithfully predict in vivo features. In order to facilitate a comparison to alternative approaches published in previous studies, we decided to use erythropoietin as our model protein. This model allows an evaluation on the basis of the level of protein expression as well as physiological parameters such as the number of reticulocytes and the hematocrit.
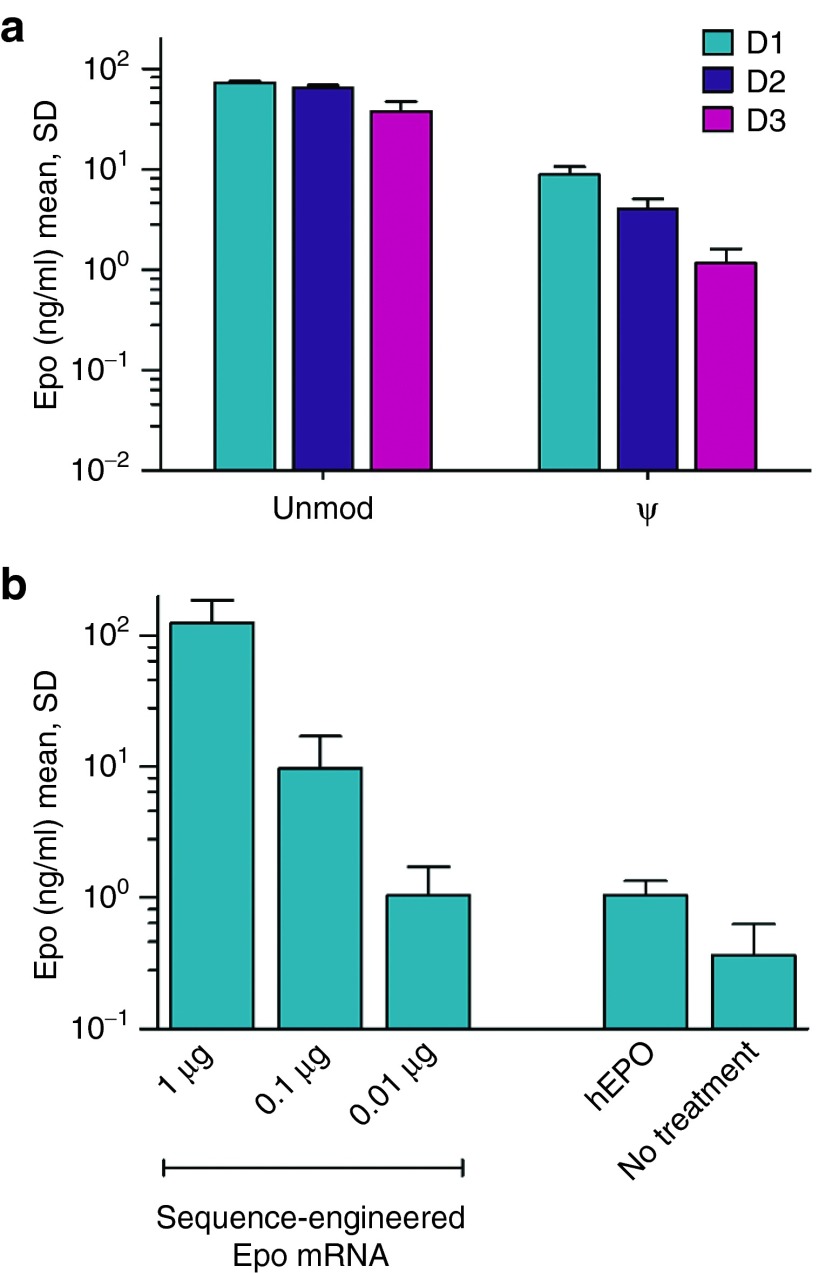
Intraperitoneal administration of TransIT-complexed sequence-engineered mEpo mRNA, either unmodified or harboring pseudouridine instead of uridine, into BALB/c mice confirmed our general in vitro observation of a reduced protein yield from nucleoside-modified mRNA (Figure 2a) (P < 0.0005 at all times, Student's t-test). A dose-titration revealed that substantial EPO levels could be obtained even with submicrogram quantities of mRNA (Figure 2b). Overall, protein levels were in the range of a previous study utilizing pseudouridine-modified mRNA formulated and administered in the same manner as here.27 However, our sequence-engineered mRNA gave rise to longer lasting protein expression.
Figure 2.
Sequence engineering of mRNA enables high level protein expression in vivo. (a) Engineered, unmodified mouse Epo mRNA yields higher protein levels in mice than the corresponding pseudouridine modified sequence. 1 μg of either unmodified or pseudouridine modified mRNA encoding erythropoietin was complexed with TransIT and intraperitoneally injected into mice. Serum erythropoietin (EPO) levels were determined at different times (1, 2, or 3 days) after administration. (b) Low nanogram doses of engineered, unmodified mouse Epo mRNA give rise to substantial protein levels in murine serum. The indicated amounts of TransIT-complexed unmodified Epo mRNA were administered intraperitoneally. As a control, 100 U of recombinant human EPO protein (hEPO) were given i.p. Serum EPO levels were determined 24 hours after treatment. unmod, engineered mRNA harboring the nucleotides A, U, G, and C; ψ, engineered mRNA in which pseudouridine replaces U. n = 4 for all groups.
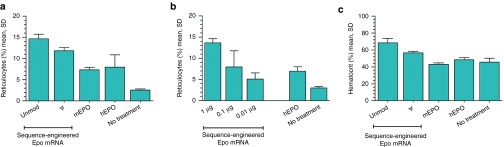
Administration of erythropoietin causes a burst release of reticulocytes representing precursors of mature erythrocytes into the blood stream. Accordingly, the administration of mEpo-encoding mRNA elicited an increase of reticulocyte numbers in mice (Figure 3a). As suggested by protein levels, the sequence-engineered mRNA outperformed its pseudouridine-modified counterpart (P < 0.01, Student's t-test). All doses down to 10 ng of unmodified mRNA significantly increased reticulocyte numbers (Figure 3b) (P < 0.05 for 10 ng versus untreated, Student's t-test). Taking into account the lower reticulocyte numbers obtained with 100 U of recombinant EPO protein as well as the lower background levels compared to a recent publication,27 the physiological effect of administering our engineered Epo mRNA is as strong as that observed with pseudouridine-modified mRNA. Likewise, the strong elevation of the hematocrit, the volume percentage of red blood cells in blood, is comparable with data from that study while effect sizes are largely corresponding to protein and reticulocyte levels (Figure 3c). In summary, these data demonstrate that sequence-engineered, unmodified mRNA is competitive with nucleoside-modified mRNA with respect to protein yield in vivo. As in vitro, the beneficial properties rely on optimizations of the open reading frame and regulatory sequences (Supplementary Figure S2).
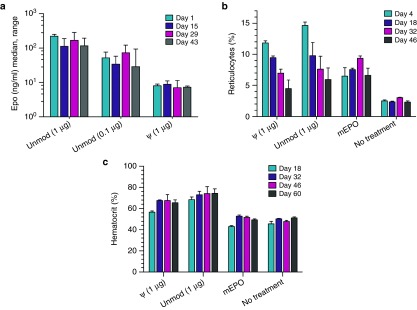
Figure 3.
Sequence engineered Epo mRNA elicits physiological effects in mice. (a) Sequence-engineered mouse Epo mRNA elicits strong reticulocyte responses in mice. Mice were intraperitoneally injected with either TransIT-complexed mRNA (1 μg) or recombinant erythropoietin (EPO) protein (hEPO: 100 U, mEPO: 800 ng). mRNA was either unmodified or harbored pseudouridine. The level of reticulocytes was determined 4 days after treatment. (b) Low nanogram doses of engineered but unmodified mouse Epo mRNA elicit substantial reticulocyte responses in mice. The indicated doses of TransIT-complexed Epo mRNA or 100 U of hEPO were intraperitoneally administered to mice. Reticulocytes were quantified 4 days after injection. (c) Sequence-engineered mouse Epo mRNA substantially increases the hematocrit in mice. Mice were treated with either TransIT-complexed mRNA (1 μg) or recombinant EPO protein (hEPO: 100 U, mEPO: 800 ng) on day 0 and 14. The hematocrit was measured on day 18. mRNA was either unmodified or harbored pseudouridine. unmod, engineered mRNA harboring the nucleotides A, U, G, and C; ψ, engineered mRNA in which pseudouridine replaces U; hEPO, recombinant human Epo protein; mEPO, recombinant murine Epo protein. n = 4 for all groups.
Unmodified mRNA allows repeated treatments
Beyond efficient protein expression, viable mRNA-based protein therapies should avoid stimulation of the innate immune system. In contrast to mRNA harboring nucleoside modifications, unmodified mRNA is considered to be immunostimulatory, giving rise to the secretion of various cytokines as reported in previous studies.24,25,26,27 This cytokine release may cause unwanted or even detrimental side effects such as a possibly fatal immune response against the encoded protein and, thus, should be minimized.
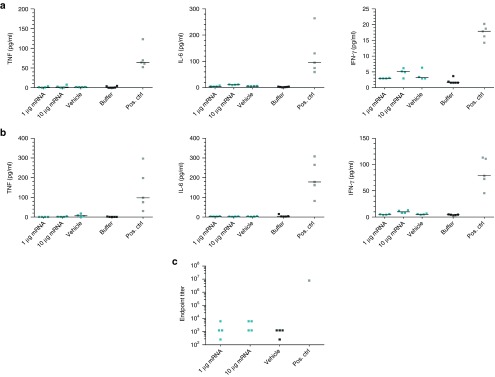
Since immunostimulatory unmodified mRNA was shown to harm cells upon consecutive transfections,24 we first investigated the impact of unmodified and pseudouridine-modified mEpo mRNA on primary human dermal fibroblasts after repeated transfections according to the protocol used by Warren et al. These analyses revealed that repeated transfections with mRNA do not require chemical modifications (Supplementary Figure S3). To examine to what extent the sequence-engineered, unmodified mRNA elicits any immunostimulation or even anti-EPO immune responses, mice were treated repeatedly with an Epo mRNA dose that gives rise to strong reticulocyte and hematocrit responses. In addition, to force the emergence of side effects, animals were challenged with a 10-fold higher mRNA dose as well. Six hours after the first mRNA treatment, blood samples were collected and the levels of proinflammatory cytokines such as TNF-α, IL-6, and IFN-γ were examined. None of the mice showed any substantial cytokine release upon mRNA administration; all measurements lay within the range observed for the formulation reagent only (which by itself could have some immunostimulatory potential) (Figure 4a). Similar results were obtained after six administrations within 3 weeks, i.e., after a more challenging treatment regime (Figure 4b). Histological and immunohistochemical analyses of the spleen, the main target organ of TransIT-complexed mRNA, did not show any disturbances of its integrity or immune status apart from a splenomegaly due to massive erythropoiesis, probably a direct consequence of mRNA-mediated splenic EPO expression (Supplementary Figure S4).
Figure 4.
Repeated treatments with engineered, unmodified Epo mRNA do not affect the immune status of mice inappropriately. (a) A single intraperitoneal injection of engineered, unmodified mouse Epo mRNA does not cause any substantial cytokine secretion. Mice were treated with different amounts of TransIT-complexed mouse Epo mRNA, vehicle only, or injection buffer. Six hours after injection, blood was collected and analyzed for the levels of various cytokines. As a positive control (pos. ctrl), mice were injected with an immunostimulatory RNA solution intramuscularly. (b) Repeated intraperitoneal injections of engineered but unmodified mouse Epo mRNA does not lead to any substantial cytokine secretion. Mice were treated twice a week for 3 weeks with different amounts of TransIT-complexed mRNA, vehicle only, or an injection buffer. Six hours after the last injection, blood was collected and analyzed for the levels of various cytokines. As a positive control, animals received a single dose of an immunostimulatory RNA solution intramuscularly. (c) Repeated intraperitoneal injections of engineered but unmodified mouse Epo mRNA did not induce an erythropoietin (EPO)-specific antibody response. Mice were treated twice a week for 3 weeks with different amounts of TransIT-complexed mRNA or vehicle only. Four weeks after the last administration, blood was collected and analyzed for the presence of EPO-specific antibodies. For comparison, an anti-EPO-antibody from rat was used as positive control. n = 5 for pos. ctrl for cytokine measurements, n = 4 for all other groups.
A potential consequence of an inappropriate immunostimulation could be the induction of antibodies specific to the encoded protein which may already occur upon a very weak or just locally restricted cytokine response. Hence, mice that received six intraperitoneal injections of a physiologically effective (0.05 mg/kg) as well as a 10-fold higher (0.5 mg/kg) dose of TransIT-complexed mRNA were analyzed for the emergence of EPO-specific antibodies 4 weeks after the last treatment. At that time, a potential antibody response should be fully developed. However, no anti-EPO antibodies could be detected using an assay with a detection limit of less than 0.1 ng/ml (Figure 4c).
To further corroborate the notion that engineered but unmodified mRNA can serve as a therapeutic, we also investigated EPO expression and corresponding physiological responses upon repeated treatments. To this end, mice received a highly effective dose of TransIT-complexed sequence-engineered mRNA, either unmodified or pseudouridine-modified, every other week. During the treatment period, doses did not lose efficacy with respect to serum EPO levels obtained 24 hours after administration (Figure 5a) (P = 0.235 for 1 μg mRNA, analysis of variance). However, as observed earlier, sequence-engineered mRNA elicited higher protein levels than its nucleoside-modified counterpart. Due to the very short half-life of EPO protein in serum and the transient nature of mRNA, all measurements reflect EPO expression in response to the latest treatment only.
Figure 5.
Engineered, unmodified Epo mRNA allows long-term/continued treatment of mice. (a) Protein yield from intraperitoneally administered mouse Epo mRNA is not affected by repeated treatments. Mice were repeatedly injected with 1 μg of either unmodified or pseudouridine-modified mRNA as well as 0.1 μg of unmodified mRNA at an interval of 2 weeks. Plasma erythropoietin (EPO) levels were determined 24 hours after each treatment. (b) Intraperitoneally injected mouse Epo mRNA increases reticulocyte counts even after multiple dosing. Mice received multiple doses of TransIT-complexed mRNA (1 μg, either unmodified or pseudouridine-modified) or mEPO (0.8 μg) at a biweekly interval starting on day 0 and were analyzed for reticulocytes 4 days after treatments. (c) Repeated intraperitoneal injections of Epo mRNA elicit a strong and sustained increase of the hematocrit. Mice received multiple doses of TransIT-complexed mRNA (1 μg, either unmodified or pseudouridine-modified) or mEPO (0.8 μg) at a biweekly interval starting on day 0 and were analyzed for the hematocrit at various times. n = 4 for all groups.
In contrast to constant EPO responses, reticulocyte responses to sequential injections declined to some extent over time (Figure 5b). This effect was observed for both unmodified and modified mRNA but not with recombinant protein which, however, did not induce strong biological effects and was probably not delivered to the spleen due to the lack of a TransIT formulation. Whereas reticulocyte responses became more severely attenuated with more frequent administrations, temporary pausing of treatment led to a recovery of the reticulocyte response (Supplementary Figure S5). As opposed to mice receiving murine recombinant EPO protein, mRNA-injected animals were characterized by strongly elevated hematocrits (P < 0.01 for 1 μg versus untreated on all days, Student's t-test) as well as erythropoiesis induced splenomegaly in the absence of any other disturbances of the spleen (Figure 5c and Supplementary Figure S4). Moreover, more frequent administration of recombinant EPO appeared to also slightly suppress the extent of the reticulocyte response over time (Supplementary Figure S5). In summary, the attenuation of the reticulocyte response appears to be due to physiological control mechanisms or limitations of erythropoiesis upon excessive EPO availability rather than side effects of the mRNA-based protein therapy. Concerning this matter, no differences could be observed between sequence-engineered, unmodified mRNA and its pseudouridine-modified counterpart.
Engineered mRNA causes relevant effects in large animals
Having demonstrated that protein therapies on the basis of unmodified, sequence-engineered mRNA are in principle feasible in mice, we next asked whether such approaches can also offer therapeutic opportunities in large animals. Particularly for systemically acting proteins, the effective dose will be a function of animal size. It has yet to be shown that meaningful expression levels can be obtained in animals close to humans in weight with a feasible mRNA dose. Therefore, pigs with a weight of about 20 kg were treated with a weight-adjusted dose similar to that in mice. To this end, we applied a LNP formulation28 that, compared to TransIT complexes, offers a more clinically acceptable intravenous route of administration as well as the possibility of much higher mRNA doses if required. A single intravenous dose of sequence-engineered, unmodified mRNA (0.065 mg/kg) encapsulated in LNPs resulted in very high serum EPO levels (Figure 6a). As a consequence, reticulocyte numbers were strongly increased and animals showed a substantial and sustained elevation of the hematocrit (P < 0.05 for day 12 versus pretreatment, Student's t-test) (Figure 6a).
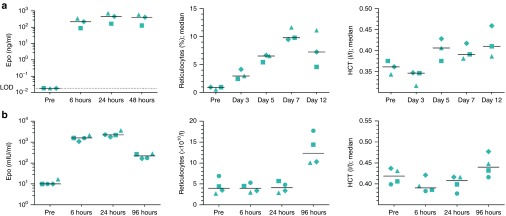
Figure 6.
Engineered, unmodified Epo mRNA can elicit systemic physiological responses in swine and nonhuman primates. (a) Intravenous mRNA injection into pigs gives rise to high serum erythropoietin (EPO) levels as well as to substantial physiological effects. Animals received 1.3 mg of lipid nanoparticle (LNP) encapsulated porcine Epo mRNA (0.065 mg/kg) on day 0 and were analyzed for EPO levels and hematological parameters at various times. Protein levels before treatment were below the limit of detection of the assay. (b) Intravenous mRNA injection into macaques increases EPO levels, reticulocyte numbers as well as the hematocrit. Animals received 100 μg of LNP encapsulated M. fascicularis Epo mRNA (0.037 mg/kg) and were analyzed for EPO levels and hematological parameters before and at various times after treatment. Distinct symbols are assigned to individual animals. Pre, prevalue before treatment, n = 3 for pig experiment, n = 4 for macaque study.
Finally, triggered by negative experiences of others with DNA-based approaches in humans and monkeys we analyzed the effect of unmodified Epo mRNA encapsulated in LNPs in cynomolgus monkeys. Animals received a single intravenous injection of a weight-adjusted dose (0.037 mg/kg) similar to that in mice. As observed in pigs, nonhuman primates revealed a considerable increase of serum EPO levels and reticulocyte counts as well as a meaningful raise of the hematocrit (increase for each individual from pretreatment to 96 hours after treatment; P < 0.05 for 96 hours versus pretreatment, Student's t-test) while there was no detectable cytokine release upon treatment (Figure 6b and Supplementary Figure S6).
Discussion
For the first time, the present study demonstrates that mRNA can give rise to therapeutically relevant protein levels in large animals. This advance was accomplished employing sequence-optimized, unmodified mRNA. Although high performance liquid chromatography (HPLC) purification depletes contaminants and, thus, contributes to reduced immunostimulation as well as improves mRNA expression to some extent,36,37,38 the lack of biological efficacy of HPLC-purified basic Epo mRNA highlights the importance and potency of sequence-engineering. Further, strong evidence is provided that such an mRNA does not cause inappropriate immunostimulation. This data together with recently published studies in mice substantiates the vision that mRNA approaches can revolutionize the field of protein-based therapies. Our work principally confirms that conventional unmodified mRNAs are inferior to molecules harboring specific nucleoside modifications as suggested by various recent reports,23,24,25,26,27 however, the picture changes dramatically with a sophisticated sequence-engineering.
Using a common model protein, erythropoietin, as well as a formulation reagent and an administration route that were previously applied to state-of-the-art pseudouridine mRNA, our data suggest that sequence-engineered mRNA is competitive with modified mRNA with respect to protein yield and physiological efficacy.27 Interestingly, as being attributed to nucleoside-modified mRNA, engineered mRNA neither induced substantial cytokine release nor elicited a protein-specific antibody response. Accordingly, the effects of a continued treatment of mice did not differ qualitatively between a sequence-engineered Epo mRNA and its pseudouridine-modified counterpart.
In contrast to findings by other groups, introducing nucleoside modifications was always detrimental to protein expression when applied to sequence-engineered mRNA. Part of the engineering process is the identification of optimal regulatory sequences outside of the open reading frame. Interestingly, choosing less effective sequences appears to dampen the negative effect of chemical modifications (Supplementary Figure S7a), suggesting an interference between chemical modifications and regulatory RNA elements. In addition, nucleoside modifications are well known to limit or even inhibit RNA sensor engagement.20,21,22 Combining these observations, it seems reasonable that modifications lead to a general weakening of protein-mRNA interactions. In line with this, we found that the incorporation of modified nucleosides such as pseudouridine impairs the function of internal ribosome entry site (IRES) elements (Supplementary Figure S7b). Collectively, this may imply that mRNAs harboring chemically modified nucleosides may be subject to much stronger limitations of maximum translation efficacy than solutions solely based on sequence-engineering due to a reduced activity or even inactivity of regulatory sequences in the presence of modifications.
As demonstrated, protein therapies based on sequence-engineered mRNA are characterized by a very good transferability between species. Simple body weight adjustments of the dose, i.e., similar doses per kg, were sufficient to gain substantial physiological effects in small to large animals. In healthy volunteers as well as anemic patients with chronic kidney disease, a dose of recombinant erythropoietin of about 600 IU/kg (which appears to be among the highest usually applied in clinical settings) led to maximum serum EPO levels of about 1,000 mIU/ml and an increase of the percentage of reticulocytes of about 2.39,40 These values are very much in line with those we obtained in nonhuman primates following intravenous administration of a reasonable dose of mRNA encapsulated in LNPs designed for hepatic delivery of nucleic acid therapeutics.41 Thus, the present study provides the first evidence that mRNA treatments can achieve meaningful biological effects in primates as well as the first proof of mRNA-based therapies being feasible in large animals, even for systemically acting proteins.
While local administrations may be possible without the need for an mRNA delivery system,26,42 a protective formulation will be mandatory for systemic delivery due to the ubiquity and abundance of RNases. The present data suggest that an effective unmodified mRNA is not dependent on a specific formulation. However, formulations differ with respect to their characteristics and have probably to be selected on a case-by-case basis. Regarding erythropoietin expression in pigs, the intravenously administered LNP-formulation of engineered mRNA appeared to be more effective than intraperitoneally injected TransIT-complexes. Nevertheless, even a weight-adjusted dose of TransIT-complexed porcine Epo mRNA elicited a statistically significant and meaningful physiological response in pigs with a body weight of about 20 kg (Supplementary Figure S8). Other crucial parameters of the formulation are certainly a minimization of side effects, which is particularly important in scenarios where long-term treatment is required, as well as the convenience of the administration route. Collectively, there is now profound evidence that mRNA, not least sequence-engineered but otherwise unmodified mRNA, can revolutionize protein therapy. Thus, the approach currently awaits its first clinical proof of principle.
Materials and Methods
mRNA sequence engineering. The codons of the open reading frame are adapted in order to improve translation and half-life of the mRNA. To this end, only the most GC-rich codons were used for each amino acid. To provide the optimized open reading frame sequence with an optimal combination of untranslated sequences, it is subjected to a screening process applying preselected sequences. For this preselection of efficacious regulatory sequences, various biological sources were screened for potent enhancer and stabilizer elements. Unless otherwise stated, the sequence-engineered mRNAs harbored a cap, an optimized open reading frame, a 5'-UTR from HSD17B4 (hydroxysteroid (17-β) dehydrogenase 4), a 3'-UTR from ALB (albumin), a polyA, and a histone stem loop. Detailed sequence information on those engineered mRNAs is given in the Supplementary Material. Further sequence information is available on request.
mRNA synthesis. In brief, linearized plasmid harboring the sequence of interest downstream of a T7 promoter was transcribed using T7 RNA polymerase (Thermo Scientific, Braunschweig, Germany). For capping of the RNA, m7G capping and 2'-O-methyltransferase kits (CellScript, Madison, WI) were used. 100% replacement was used for mRNAs that harbored chemically-modified nucleosides. All mRNAs lacking modified nucleosides (sequence-engineered or not) as well as all nucleoside-modified mRNAs were purified according to the same protocol by reversed-phase chromatography using a PLRP-S stationary phase and an acetonitrile gradient in a triethylammonium acetate buffer. A detailed protocol has been described elsewhere.38
mRNA formulation for in vivo application. For intraperitoneal administration, mRNAs were formulated with TransIT-mRNA (Mirus Bio, Madison, WI) according to a recently described protocol.27 Intravenous administration of mRNA to macaques or pigs was conducted using mRNA encapsulated in LNPs prepared at Acuitas Therapeutics (Vancouver, Canada). The Acuitas mRNA delivery platform has been developed from technology shown previously to enable siRNA-dependent hepatic gene silencing across species in rodents, nonhuman primates and humans.28,43,44 LNPs are prepared using a self-assembly process in which an aqueous solution of mRNA at pH 4.0 is rapidly mixed with a solution of lipids dissolved in ethanol.43 LNPs used in this study are similar in composition to those described previously, which contain an ionizable cationic lipid/phosphatidylcholine/cholesterol/PEG-lipid (50:10:38.5:1.5 mol/mol), encapsulated RNA-to-total lipid ratio of ~0.05 (wt/wt) and a diameter of ~80 nm.28,43 At blood pH, LNPs exhibit a net neutral surface charge but become positively charged in acidified endosomes following ApoE-mediated endocytosis by hepatocytes in vivo, resulting in endosome disruption and release of mRNA into the cytoplasm.28,41,45 Acuitas will make the LNPs available on request.
Cell transfection. For in vitro transfection of HeLa cells, mRNAs were complexed with Lipofectamine 2000 (Life Technologies, Darmstadt, Germany). In brief, cells were seeded into 96-well plates (104 cells/well) 1 day before transfection. On the next day, mRNA was complexed according to the manufacturer's instruction using 1.5 μl transfection reagent per μg of mRNA. Lipofection was conducted in Opti-MEM in the absence of serum for 90 minutes. Afterwards, medium was replaced by serum-complemented Roswell Park Memorial Institute 1640 medium.
Luciferase measurement. Cells transfected with mRNA encoding Photinus pyralis luciferase (PpLuc) were lysed 24 hours after transfection. To this end, the medium was removed, cells were covered with Passive Lysis Buffer (Promega, Mannheim, Germany), incubated for 15 minutes at RT, frozen at −80 °C and thawed. To determine luciferase expression, 20 μl lysate were analyzed in a Hidex Chameleon plate reader using Beetle-Juice (PJK GmbH, Kleinblittersdorf, Germany).
Animal experiments. All experiments were approved by the responsible authorities (Regional Council Tuebingen) and carried out in accordance with national laws. BALB/c female mice 7–9 weeks of age were purchased from Janvier Labs (Le Genest-Saint-Isle, France). For intraperitoneal injections, TransIT-formulated mRNA encoding murine EPO was administered in a total volume of 100 μl. Cynomolgus monkeys (2.5–2.8 kg) were housed and experiments were performed at Huntingdon Life Science (Huntingdon, UK). Animals received a single dose of LNP-formulated mRNA (100 μg) encoding EPO from Macaca fascicularis in phosphate buffered saline pH 7.4. The total volume for intravenous injections was 2 ml. Hungarian large white, domestic pigs (female, approximately 20 kg) were housed and experiments were conducted at ATRC Aurigon Toxi-Coop Research Center (Dunakeszi, Hungary). For intraperitoneal injection, 360 μg of TransIT-formulated mRNA encoding porcine EPO was administered in a total volume of 25 ml. Animals received two injections on consecutive days. For intravenous administration, pigs received 1.3 mg of porcine Epo mRNA formulated with LNPs in phosphate-buffered saline pH 7.4. The total volume per animal was 26 ml.
Quantification of EPO levels. Upon transfection of HeLa cells with mRNA coding for murine EPO, mouse EPO levels in the culture medium were measured 24 hours later using a mouse EPO ELISA kit (R&D Systems, Wiesbaden, Germany). This kit was also used to determine EPO levels in the plasma of treated mice. For plasma preparation, a few microliters of blood were collected, heparinized, and centrifuged. For pigs, blood samples were collected into serum vials, kept at room temperature for at least 20 minutes, and centrifuged. Porcine EPO levels in the supernatant were measured using a mouse EPO ELISA kit (R&D Systems) with cross reactivity to pig EPO and recombinant pig EPO protein (Cusabio, Wuhan, China) as standard. Serum of cynomolgus monkeys was analyzed with a human EPO ELISA kit (R&D Systems) that cross reacts with macaque EPO.
Determination of reticulocyte counts. For mice, a small volume of blood was drawn from animals, mixed with an appropriate amount of heparin, and analyzed using Retic-COUNT (BD Biosciences, Heidelberg, Germany) according to the manufacturer's instructions. Stained cells were analyzed on a FACS Canto (BD Biosciences). Reticulocyte levels are given as percentage of total red blood cells. For pigs, blood was drawn from the cranial vein and collected into ethylenediaminetetraacetate (EDTA)-coated vials. Reticulocytes were measured using a Sysmex XT-2000iV automated hematology analyzer at Aurigon. Reticulocyte levels are given as percentage of total red blood cells. For cynomolgus monkeys, blood was collected from a suitable vein, EDTA was added as anticoagulant, and reticulocytes were counted with an Advia 120 hematology system at Huntingdon.
Determination of the hematocrit. For mice, the volume ratio of blood cells was determined using hematocrit capillaries (KABE Labortechnik, Nümbrecht-Elsenroth, Germany). In brief, capillaries were filled with heparinized blood, sealed on one end with wax (Hirschmann Laborgeräte, Eberstadt, Germany), centrifuged, and analyzed according to the manufacturer's instructions. For pigs and macaques, hematocrits were determined with a Sysmex XT-2000iV and Advia 120 hematology analyzer, respectively.
Measurement of cytokine secretion. Mice received six injections of TransIT-complexed mRNA or control solutions within 3 weeks. Six hours after the first and last treatment, blood was collected, heparinized, and plasma preparations were analyzed for various cytokines. TNF-α, IL-6, and IFN-γ were measured by Cytometric Bead Array analysis (BD Biosciences).
Detection of EPO-specific antibody responses. The induction of EPO-specific antibodies in response to repeated treatments with Epo mRNA was analyzed by enzyme-linked immunosorbent assay (ELISA). To this end, plates were coated with mouse EPO protein and incubated with plasma from mice 4 weeks after they received six injections of mRNA or control solution within 3 weeks. As positive control, a rat anti-mouse EPO antibody (R&D Systems) was applied in the ELISA. EPO-specific antibodies were detected with goat anti-mouse and anti-rat IgG antisera labelled with peroxidase. The ELISA allowed detection of a concentration of anti-EPO antibody as low as about 100 pg/ml.
Histological spleen analysis. Spleens of euthanized mice were embedded in paraffin and sections subjected to hematoxylin and eosin staining as well as to immunohistochemical analyses according to standard protocols by the Mouse Facility at the Institute of Pathology (Tübingen, Germany).
SUPPLEMENTARY MATERIAL Figure S1. mRNA sequence-engineering improves protein expression in vitro. Figure S2. mRNA sequence-engineering improves protein expression in vivo. Figure S3. Repeated transfections of primary cells with mRNA do not require chemical modifications. Figure S4. TransIT-complexed Epo-mRNA induces massive erythropoiesis in the spleen without causing histological or immunological disturbances. Figure S5. Desensitization by high and/or frequent EPO expression transiently reduces the physiological response to the hormone. Figure S6. LNP-encapsulated engineered, unmodified Epo mRNA does not show inappropriate stimulation of the innate immune system. Figure S7. Incorporation of pseudouridine shows negative interference with functional RNA elements. Figure S8. TransIT-complexed engineered Epo mRNA can elicit systemic physiological responses in swine.
Acknowledgments
We are very grateful to A. Urbschat, I. Biermann, N. Stäbler, and T. Dam for their excellent technical help throughout this project. We thank I. Hoerr, A. Buck, U. Kruse, and N. Horscroft for critically reading the manuscript. A.T., S.G., P.B., M.F.-M., and T.S. are employees of CureVac GmbH developing therapeutics based on sequence-engineered mRNA. B.L.M. and M.J.H. are employees of Acuitas Therapeutics, a company providing delivery solutions for molecular therapeutics using lipid nanoparticles.
Supplementary Material
References
- Gurdon, JB, Lane, CD, Woodland, HR and Marbaix, G (1971). Use of frog eggs and oocytes for the study of messenger RNA and its translation in living cells. Nature 233: 177–182. [DOI] [PubMed] [Google Scholar]
- Laskey, RA, Gurdon, JB and Crawford, LV (1972). Translation of encephalomyocarditis viral RNA in oocytes of Xenopus laevis. Proc Natl Acad Sci USA 69: 3665–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boczkowski, D, Nair, SK, Snyder, D and Gilboa, E (1996). Dendritic cells pulsed with RNA are potent antigen-presenting cells in vitro and in vivo. J Exp Med 184: 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, JA, Malone, RW, Williams, P, Chong, W, Acsadi, G, Jani, A et al. (1990). Direct gene transfer into mouse muscle in vivo. Science 247(4949 Pt 1): 1465–1468. [DOI] [PubMed] [Google Scholar]
- Conry, RM, LoBuglio, AF, Wright, M, Sumerel, L, Pike, MJ, Johanning, F et al. (1995). Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res 55: 1397–1400. [PubMed] [Google Scholar]
- Jirikowski, GF, Sanna, PP, Maciejewski-Lenoir, D and Bloom, FE (1992). Reversal of diabetes insipidus in Brattleboro rats: intrahypothalamic injection of vasopressin mRNA. Science 255: 996–998. [DOI] [PubMed] [Google Scholar]
- Alexopoulou, L, Holt, AC, Medzhitov, R and Flavell, RA (2001). Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413: 732–738. [DOI] [PubMed] [Google Scholar]
- Diebold, SS, Kaisho, T, Hemmi, H, Akira, S and Reis e Sousa, C (2004). Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303: 1529–1531. [DOI] [PubMed] [Google Scholar]
- Heil, F, Hemmi, H, Hochrein, H, Ampenberger, F, Kirschning, C, Akira, S et al. (2004). Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303: 1526–1529. [DOI] [PubMed] [Google Scholar]
- Hornung, V, Ellegast, J, Kim, S, Brzózka, K, Jung, A, Kato, H et al. (2006). 5'-Triphosphate RNA is the ligand for RIG-I. Science 314: 994–997. [DOI] [PubMed] [Google Scholar]
- Kato, H, Takeuchi, O, Sato, S, Yoneyama, M, Yamamoto, M, Matsui, K et al. (2006). Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441: 101–105. [DOI] [PubMed] [Google Scholar]
- Pichlmair, A, Schulz, O, Tan, CP, Näslund, TI, Liljeström, P, Weber, F et al. (2006). RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science 314: 997–1001. [DOI] [PubMed] [Google Scholar]
- Rehwinkel, J, Tan, CP, Goubau, D, Schulz, O, Pichlmair, A, Bier, K et al. (2010). RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140: 397–408. [DOI] [PubMed] [Google Scholar]
- Nallagatla, SR, Hwang, J, Toroney, R, Zheng, X, Cameron, CE and Bevilacqua, PC (2007). 5'-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science 318: 1455–1458. [DOI] [PubMed] [Google Scholar]
- Rittig, SM, Haentschel, M, Weimer, KJ, Heine, A, Muller, MR, Brugger, W et al. (2011). Intradermal vaccinations with RNA coding for TAA generate CD8+ and CD4+ immune responses and induce clinical benefit in vaccinated patients. Mol Ther 19: 990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel, B, Aulwurm, S, Probst, J, Stitz, L, Hoerr, I, Rammensee, HG et al. (2006). Therapeutic anti-tumor immunity triggered by injections of immunostimulating single-stranded RNA. Eur J Immunol 36: 2807–2816. [DOI] [PubMed] [Google Scholar]
- Weide, B, Carralot, JP, Reese, A, Scheel, B, Eigentler, TK, Hoerr, I et al. (2008). Results of the first phase I/II clinical vaccination trial with direct injection of mRNA. J Immunother 31: 180–188. [DOI] [PubMed] [Google Scholar]
- Weide, B, Pascolo, S, Scheel, B, Derhovanessian, E, Pflugfelder, A, Eigentler, TK et al. (2009). Direct injection of protamine-protected mRNA: results of a phase ½ vaccination trial in metastatic melanoma patients. J Immunother 32: 498–507. [DOI] [PubMed] [Google Scholar]
- Fotin-Mleczek, M, Duchardt, KM, Lorenz, C, Pfeiffer, R, Ojkić-Zrna, S, Probst, J et al. (2011). Messenger RNA-based vaccines with dual activity induce balanced TLR-7 dependent adaptive immune responses and provide antitumor activity. J Immunother 34: 1–15. [DOI] [PubMed] [Google Scholar]
- Karikó, K, Buckstein, M, Ni, H and Weissman, D (2005). Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23: 165–175. [DOI] [PubMed] [Google Scholar]
- Anderson, BR, Muramatsu, H, Jha, BK, Silverman, RH, Weissman, D and Karikó, K (2011). Nucleoside modifications in RNA limit activation of 2'-5'-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res 39: 9329–9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, BR, Muramatsu, H, Nallagatla, SR, Bevilacqua, PC, Sansing, LH, Weissman, D et al. (2010). Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res 38: 5884–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó, K, Muramatsu, H, Welsh, FA, Ludwig, J, Kato, H, Akira, S et al. (2008). Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther 16: 1833–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, L, Manos, PD, Ahfeldt, T, Loh, YH, Li, H, Lau, F et al. (2010). Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7: 618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangi, L, Lui, KO, von Gise, A, Ma, Q, Ebina, W, Ptaszek, LM et al. (2013). Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat Biotechnol 31: 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kormann, MS, Hasenpusch, G, Aneja, MK, Nica, G, Flemmer, AW, Herber-Jonat, S et al. (2011). Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat Biotechnol 29: 154–157. [DOI] [PubMed] [Google Scholar]
- Karikó, K, Muramatsu, H, Keller, JM and Weissman, D (2012). Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol Ther 20: 948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman, M, Ansell, SM, Mui, BL, Tam, YK, Chen, J, Du, X et al. (2012). Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew Chem Int Ed Engl 51: 8529–8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon, F, Krishnan, S, Lenzen, G, Magné, R, Gomard, E, Guillet, JG et al. (1993). Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur J Immunol 23: 1719–1722. [DOI] [PubMed] [Google Scholar]
- Hoerr, I, Obst, R, Rammensee, HG and Jung, G (2000). In vivo application of RNA leads to induction of specific cytotoxic T lymphocytes and antibodies. Eur J Immunol 30: 1–7. [DOI] [PubMed] [Google Scholar]
- Petsch, B, Schnee, M, Vogel, AB, Lange, E, Hoffmann, B, Voss, D et al. (2012). Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat Biotechnol 30: 1210–1216. [DOI] [PubMed] [Google Scholar]
- Diken, M, Kreiter, S, Vascotto, F, Selmi, A, Attig, S, Diekmann, J et al. (2013). mTOR inhibition improves antitumor effects of vaccination with antigen-encoding RNA. Cancer Immunol Res 1: 386–392. [DOI] [PubMed] [Google Scholar]
- Kreiter, S, Diken, M, Selmi, A, Diekmann, J, Attig, S, Hüsemann, Y et al. (2011). FLT3 ligand enhances the cancer therapeutic potency of naked RNA vaccines. Cancer Res 71: 6132–6142. [DOI] [PubMed] [Google Scholar]
- Kreiter, S, Selmi, A, Diken, M, Koslowski, M, Britten, CM, Huber, C et al. (2010). Intranodal vaccination with naked antigen-encoding RNA elicits potent prophylactic and therapeutic antitumoral immunity. Cancer Res 70: 9031–9040. [DOI] [PubMed] [Google Scholar]
- Schlake, T, Thess, A, Fotin-Mleczek, M and Kallen, KJ (2012). Developing mRNA-vaccine technologies. RNA Biol 9: 1319–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascolo, S (2004). Messenger RNA-based vaccines. Expert Opin Biol Ther 4: 1285–1294. [DOI] [PubMed] [Google Scholar]
- Probst, J, Weide, B, Scheel, B, Pichler, BJ, Hoerr, I, Rammensee, HG et al. (2007). Spontaneous cellular uptake of exogenous messenger RNA in vivo is nucleic acid-specific, saturable and ion dependent. Gene Ther 14: 1175–1180. [DOI] [PubMed] [Google Scholar]
- Karikó, K, Muramatsu, H, Ludwig, J and Weissman, D (2011). Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res 39: e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, WK, Goon, BL, Guilfoyle, MC and Wacholtz, MC (1998). Pharmacokinetics and pharmacodynamics of recombinant human erythropoietin after single and multiple subcutaneous doses to healthy subjects. Clin Pharmacol Ther 64: 412–423. [DOI] [PubMed] [Google Scholar]
- McGowan, T, Vaccaro, NM, Beaver, JS, Massarella, J and Wolfson, M (2008). Pharmacokinetic and pharmacodynamic profiles of extended dosing of epoetin alfa in anemic patients who have chronic kidney disease and are not on dialysis. Clin J Am Soc Nephrol 3: 1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope, MJ (2014). Enhancing siRNA delivery by employing lipid nanoparticles. Therapeutic Delivery 5: 663–673. [DOI] [PubMed] [Google Scholar]
- Mays, LE, Ammon-Treiber, S, Mothes, B, Alkhaled, M, Rottenberger, J, Müller-Hermelink, ES et al. (2013). Modified Foxp3 mRNA protects against asthma through an IL-10-dependent mechanism. J Clin Invest 123: 1216–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, MA, Jayaraman, M, Matsuda, S, Liu, J, Barros, S, Querbes, W et al. (2013). Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol Ther 21: 1570–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho, T, Adams, D, Silva, A, Lozeron, P, Hawkins, PN, Mant, T et al. (2013). Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N Engl J Med 369: 819–829. [DOI] [PubMed] [Google Scholar]
- Akinc, A, Querbes, W, De, S, Qin, J, Frank-Kamenetsky, M, Jayaprakash, KN et al. (2010). Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol Ther 18: 1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.