Abstract
Attempts at eliciting neutralizing antibodies against human immunodeficiency virus (HIV)-1 have generally failed. Computationally designed epitope-scaffold platforms allow transplantation of structural epitopes to scaffold proteins. Human rhinovirus (HRV) allows such engrafting of HIV-1 epitopes on the surface scaffold proteins. However, since HRV infects only humans and great apes, the efficacy of chimeric HRV-based live viral vaccines is difficult to assess in animal models. Here, we used human ICAM-1 transgenic (hICAM-1 Tg) mice that support productive HRV infection to assess the efficacy of chimeric HRV expressing the HIV-1 membrane proximal external region (MPER) epitope, 4E10. Intranasal immunization with chimeric HRV in transgenic mice effectively induced antibodies that recognized 4E10 peptide as well as HIV-1 Env trimer. Importantly, the immunized mouse sera were able to neutralize HIV strains including those belonging to clades B and C. Moreover, intranasal immunization could bypass pre-existing immunity to HRV. Thus, chimeric HRV appears to provide a viable vaccine vehicle for HIV-1 immunization in humans.
Introduction
Given the continued failure of human immunodeficiency virus (HIV) vaccines in clinical trials, there is an urgent need for innovative strategies for designing an effective AIDS vaccine. The identification of numerous broadly neutralizing antibodies (bNAbs) in HIV-infected individuals inspired the enthusiasm for developing an antibody-based vaccine, since most neutralizing antibodies block the virus at the earliest steps in the viral replication cycle and confer long-term protection (reviewed in ref. 1). Furthermore, follow-up analysis for the only modestly successful HIV-1 vaccine clinical trial in Thailand revealed that the protection was correlated with V1V2 antibodies induced in the vaccinated individuals, highlighting the importance of antibodies for HIV-1 protection.2 However, attempts to generate NAbs by vaccination using envelope proteins have failed, in part, due to HIV virus escape mechanisms such as hypervariability of HIV-1 and conformational masking of the epitopes.3 In order to elicit potent neutralizing antibodies, it would be a wise strategy to focus specifically on conserved epitopes in the glycoprotein rather than using the entire viral spike (in which conformation masking usually occurs).
Computationally designed epitope-scaffold platforms allow transplantation of structural epitopes to scaffold proteins, and the approach was used successfully to elicit antibodies that neutralized human respiratory syncytial virus,4,5 as well as antibodies that recognize the natural structure of gp41 membrane-proximal external region (MPER) epitopes.6,7 The MPER is one of the most highly conserved sequences of Env. So far, there are four bNAb epitopes found to be located in the MPER, namely 2F5, 4E10, Z13e1, and 10E8. Among them, 2F5 epitope comprises MPER amino acids (aa) 662 to 667 (ELDKWA),8 and 4E10 epitope localizes to aa 671 to 676 (NWF(D/N)IT).9 2F5 MAb has greater potency, whereas 4E10 MAb has a broader neutralizing spectrum against various HIV-1 isolates.10 In our previous work, we grafted two conserved MPER epitopes of HIV gp41 (2F5 and 4E10 epitopes) on the surface of Human Rhinovirus (HRV) to develop a safe live virus vaccine.11,12 HRV, which causes mild forms of the common cold, is known to stimulate robust humoral and T-cell responses, including mucosal immune responses.13 HRV can accommodate a variety of foreign sequences including immunogenic HIV epitopes11,12,14 in the surface loop of the viral coat protein 2, designated the VP2 puff. This surface loop is, in fact, part of one of HRV's own immunogenic sites, constituting the largest of three loops forming the neutralizing immunogenic site II (NIm-II). We engineered segments from the 2F5 and 4E10 epitopes onto HRV14 via 0–3 linker residues to form a series of combinatorial libraries (leading to presentation of millions of conformation of the epitopes). By immunoselecting these HRV: 2F5/4E10 epitope libraries with the corresponding 2F5 or 4E10 bNAbs, we obtained chimeric viruses with greater antigenicity (which most likely present more natural conformations of the epitopes chosen) and used these to immunize guinea pigs. One HRV chimera carrying the 4E10 epitope was able to elicit neutralization of multiple HIV pseudoviruses (at IC40 level, it was able to neutralize 10 of 12 HIV-1 pseudotypes, including those with subtypes A, B, C, D, AE, and F, with CCR5 or CXCR4 co-receptor usage).12 These proof-of-concept results show that the HRV system displaying MPER epitopes can generate broadly neutralizing antibodies against HIV-1.
However, the neutralizing antibodies were not induced uniformly in all immunized guinea pigs and the neutralization titers were only modest, presumably because the animals are not permissive to HRV infection (lack of HRV receptor, human intercellular adhesion molecule-1 (ICAM-1), which is necessary for rhinovirus infection). Thus, HRV immunization in nonpermissive animals resembles vaccination with killed virus, essentially making it a suboptimal immunization strategy. Recently, human ICAM-1 transgenic (Tg) mice have been generated that support productive HRV infection.15 HRV infection in these mice leads to virus replication in the lungs that gets cleared within 4–5 days and results in the robust generation of antiviral antibodies. Therefore, it has the potential to serve as a suitable animal model to evaluate the chimeric HRV-HIV vaccine candidates.
In this study, hICAM-1 Tg mice were immunized with three promising chimeric viruses selected from the guinea pig model,12 and sera from the animals were evaluated for neutralizing ability against HIV-1 isolates. Further, we also tested whether pre-existing immunity to HRV could be overcome by intranasal administration as has been reported for adenovirus and influenza virus-based vaccines.16,17 Our data show that sera from the chimeric virus-immunized hICAM-1 Tg mice were able to recognize the 4E10 epitopic peptide, as well as HIV-1 gp140 trimer, with one of the chimeric viruses eliciting antibodies that could neutralize HIV strains belonging to Clades B and C.
Results
Immunogenicity of chimeric HRV-HIV viruses in hICAM-1 Tg mice
Among the 100 known rhinovirus serotypes, 90% employ hICAM-1 as their receptor for entering cells. Since hICAM-1 is only expressed by human cells, live HRV-based vaccines cannot be tested in commonly used animals such as mice and guinea pig that do not support HRV infection. Recently, human ICAM-1 Tg mice have become available that support human rhinovirus infection and generate robust anti-HRV neutralizing antibody response.15 Therefore, we aimed to test if hICAM-1 Tg mice could be used to evaluate HRV-HIV immunogenicity.
To elicit an antibody response, it is necessary for the epitopes to be displayed in the right conformation. Thus, we first assessed if intranasal infection of hICAM-1 Tg mice with HIV-HRV chimera leads to HIV-1 epitope expression in the lungs of infected mice. For this, mice were infected with two chimeric viruses, 12B1 and 22C1, from our previous study as well as with wild-type HRV-14 as negative control. Three days postinfection, sections of lungs were stained with 4E10 mAb to test for 4E10 epitope expression by fluorescence microscopy. Lung sections from wild-type HRV14 infected mice did not show staining (Figure 1a), while sections from both 12B1 and 22C1 HRV-HIV chimera infected mice showed strong 4E10 mAb staining in the cytoplasm of epithelial cells (Figure 1b,c), suggesting that the chimeric viruses displayed the 4E10 epitope in conformations that could be recognized by the mAb.
Figure 1.
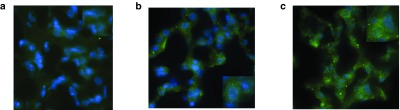
Expression of 4E10 epitope in the lungs of HRV-HIV-infected mice. Mice were infected with wild-type (WT) HRV (a) or HRV-HIV chimeras 12B1 (b) or 22C1 (c) and lung sections obtained 3 days later stained with 4E10 mAb followed by Alexa Fluor 488-labeled goat anti-human IgG. 4′,6-diamino-2-phenyl-indol was used to stain the nuclei. Section from a representative mouse of 3 tested is shown.
To test immunogenicity, we chose three representative chimeric HRV-HIV viruses 12B1, 22C1, and 13A3 from our previous work.12 hICAM-1 Tg mice were immunized intranasally with the selected chimeric viruses or with wild-type HRV-14 and keyhole limpet hemocyanin (KLH)-conjugated 4E10 peptide (intraperitoneally) as negative and positive controls respectively. In addition, wild-type C57BL mice (nontransgenic for hICAM-1) were also immunized with 12B1 as a control.
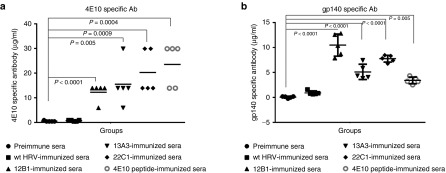
Groups of mice (five mice/group) were immunized by intranasal infection with 2.5 × 106 plaque forming unit (PFU) of the three HRV-HIV chimeras or wild-type HRV-14 for control after obtaining the preimmune serum samples. The infection was repeated twice at 3-week intervals and blood collected at weeks 0, 3, 6, and 9 postimmunization. To evaluate immunogenicity, first we tested the ability of the sera to bind to synthetic D7-N11 lactam-bridged 4E10 peptide (which has improved binding affinity for 4E10 mAb compared with linear 4E10 peptide12) by enzyme-linked immunosorbent assay (ELISA). The sera from the chimera-immunized hICAM-1 Tg mice were able to bind 4E10 peptide with an endpoint antibody titer ranging from an average of 12.4–20.4 µg/ml, similar to KLH-4E10 peptide immunized sera (average titer 23.6 µg/ml). In contrast, wild-type HRV-14 immunized and preimmune sera only had a background level of antibody (Figure 2a). Next, we tested if the sera could bind HIV-1 gp140 trimer (which has been shown to be stable18) by ELISA. The results showed that all of the individual chimeric virus-immunized mice sera could bind gp140 trimer corresponding to 5.2–10.5 µg/ml of 4E10 MAb, significantly higher than the preimmune sera (average 0.1 µg/ml) or wild-type HRV-14 immunized sera (average 0.9 µg/ml). Notably, the 12B1 immune sera showed the highest binding to the gp140 trimer with an average of 10.5 µg/ml (Figure 2b). These results indicated that HRV presenting the HIV-1 4E10 epitope could elicit antibodies in hICAM-1 Tg mice that recognize a constrained 4E10 peptide, as well as gp140 trimer.
Figure 2.
Sera from hICAM-1 Tg mice immunized with HRV-HIV chimera bind 4E10 peptide as well as HIV-1 SF162 gp140 trimer. (a) Serial dilutions of sera (week 9) were tested for binding 4E10 peptide (K7-D11 lactam-bridged Ac-NWFDITK7WLWD11KKK-NH2) by enzyme-linked immunosorbent assay (ELISA). Each serum sample was tested in triplicate and mean titers are shown for each mouse. The antibody titers were calculated by relating OD450 values of sera to that obtained with the ref 4E10 Mab and is shown as µg/ml. (b) Immunized mice sera (week 9) were tested for binding HIV-1 SF162 gp140 trimer at 1: 50 dilution by ELISA. Antibody titers were calculated as in (a). Unpaired Student t-test was used to calculate the significance between groups and P values are indicated.
Chimeric HRV-HIV virus immunized hICAM-1 Tg mice sera can neutralize HIVBaL
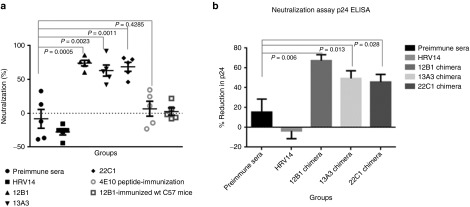
Next we tested the neutralizing ability of immune sera against a lab-generated HIV-1 strain, HIVBaL, using a Tat-regulated luciferase assay in TZM-bl cells. The result showed that all of the three chimeric virus-immunized mice sera were able to neutralize HIVBaL at a rate ranging from 59 to 78%, with 12B1 showing the highest neutralizing ability (78%). In contrast, the control preimmune sera (day 0) and sera from WT HRV-14 and 4E10 peptide immunized hICAM-1 Tg mice or 12B1 immunized WT C57BL mice did not show any neutralization (Figure 3a). To confirm neutralization, we also performed a multi-round replication assay. HIVBaL was incubated with 1: 10 dilution of sera and used to infect TZM-bl cells. Two days later, p24 antigen levels in the culture supernatant tested by p24 ELISA. All the three chimeric virus-immunized mice sera could decrease the viral p24 level from 48 to 68% compared to control, and the viral inhibition was significant when compared with either preimmune sera or wt HRV-immunized mice sera (Figure 3b). These results suggest that chimeric HRV-HIV could elicit anti-HIV neutralizing antibody in hICAM-1 Tg mice, but not in wild-type mice and thus, HRV replication in hICAM-1 Tg mice is important for enhancing the vaccine immunogenicity.
Figure 3.
Neutralizing ability of immunized mice sera against HIVBAL assessed by Tat-regulated luciferase assay and p24 enzyme-linked immunosorbent assay (ELISA). (a) HIV-BAL (200 pg of p24) was incubated with 1:50 dilution of sera for 2 hours at 37 °C before infecting TZM-bl cells and 24 hours after infection, the cell lysates were tested for luciferase activity. (b) HIV-BAL (200 pg of p24) was incubated with 1:10 dilution of sera for 2 hours at 37 °C before infecting TZM-bl cells and 48 hours after infection, the culture supernatants were tested for p24 antigen levels using p24 ELISA kit (from PerkinElmer). Preimmune sera and 12B1-immunized wild-type C57 mice were used as negative controls. Unpaired Student t-test was used to calculate the significance between preimmune sera group and sample groups. P values are indicated. Unpaired Student t-test was also used to compare between HRV14 immunized group and sample groups and the results were also significant, but have not been shown here.
Nasal administration of chimeric HRV:HIV could bypass preimmunity to elicit HIV-1 neutralizing antibodies
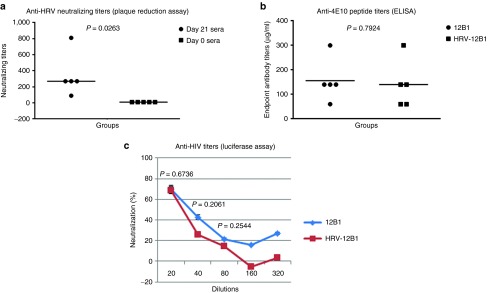
Since HRV is a common infection and there are many serotypes of HRV, pre-existing immunity to HRV is a concern for using HRV-based vaccines. However, for adenovirus, several reports suggest that nasal administration is able to bypass the preimmunity to the virus.16,17 Since hICAM-1 Tg mice allow productive infection of mice, we tested if nasal administration of HRV-HIV vaccine could similarly bypass preimmunity from an earlier HRV infection. For this, we first infected mice with wild-type HRV14. After confirming the generation of HRV neutralizing antibodies, we immunized the mice with 12B1 chimeric HRV-HIV. After boosting twice with 12B1, the sera at week 3 postimmunization were tested for their ability to neutralize HIVBaL. The plaque reduction neutralization assay results revealed that preimmunity to HRV14 was well established before immunization with 12B1, characterized by HRV neutralizing antibody titer (IC50) of 1: 270 (Figure 4a). The sera from these time points were unable to neutralize HIV-1. However, when these mice were subsequently immunized with 12B1 virus as described earlier, they developed anti-HIV-1 antibodies with 4E10 binding ability very similar to the mice that were not preimmunized with HRV14 (Figure 4b). HIV-1 neutralizing ability observed was also similar to that seen with HRV-naïve hICAM-1 Tg mice immunized with 12B1 (Figure 4c). We also confirmed that HRV-HIV infection in wt HRV preimmunized mice results in HIV antigen expression. For this, HRV immunized mice were intranasally infected with HRV-HIV, and 2 days later lung sections were stained with 4E10 mab as described in Figure 1. Indeed, the wt HRV preimmunized mice could express 4E10 epitope as efficiently as unimmunized mice (Supplementary Figure S1). We also tested if the wt HRV-immunized mice sera could neutralize HRV-HIV in vitro. The plaque reduction neutralization assay showed a dose-dependent neutralization of HRV-HIV (Supplementary Figure S2). Thus, the presence of neutralizing antibodies in the serum may not prevent at least a transient infection in the lungs. Taken together, these results show that pre-existing immunity against HRV can be bypassed by intranasal immunization, similar to the situation with adenovirus and influenza virus.
Figure 4.
Pre-existing immunity to human rhinovirus (HRV) does not compromise anti-HIV antibody response. (a) hICAM-1Tg mice were intranasally immunized with 2.5 × 106 wild-type HRV-14 and after 3 weeks, serial dilutions of sera, along with preimmune sera obtained before immunization (day 0) tested for neutralization of HRV-14 by plaque reduction assay. Each symbol represents an individual mouse. The neutralizing titers (PRNT 50) are shown as reciprocal of serum dilutions. (b) Prior HRV-14 immunized mice in (a) or naive hICAM-1 Tg mice were intranasally immunized with HRV:HIV chimera and sera obtained after 3 weeks titrated for binding 4E10 peptide by ELISA. Unpaired student T-test was used to test the significance between groups. (c) Dilutions of sera in (b) were tested for HIVBAL neutralization in TZM-bl cells as in Figure 2. % neutralization of HIVBAL was calculated by comparison of values with preimmune serum at each dilution.
Breadth of neutralizing antibodies elicited in hICAM-1 Tg mice
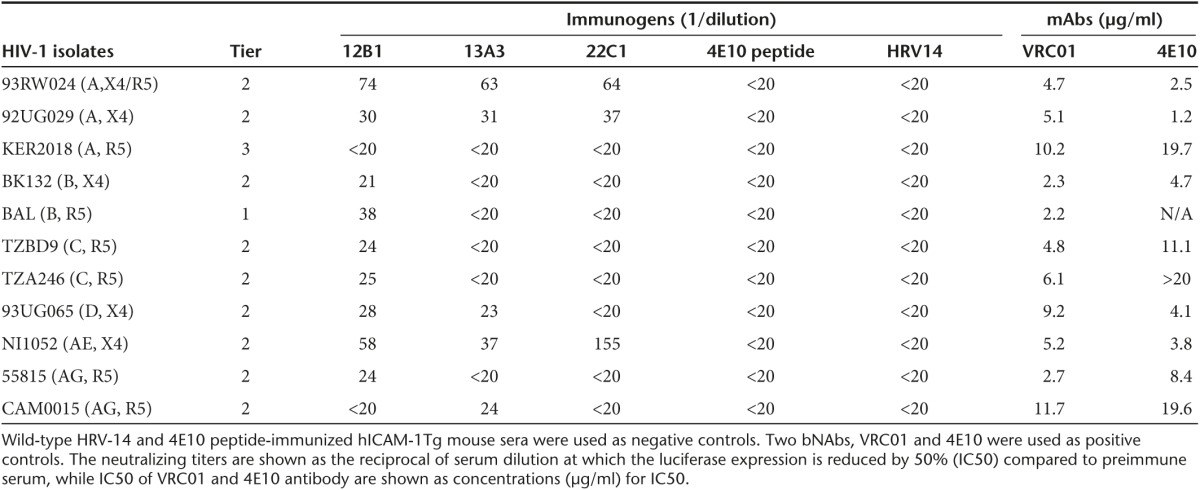
We next tested the potency and breadth of HIV-1 neutralization in sera from 12B1, 22C1 and 13A3 immunized mice. Sera from 4E10 peptide-immunized and wild-type HRV-14 immunized mice served as controls. Initially, we tested in house the neutralizing ability against 11 globally circulating HIV-1 primary isolates that include six HIV-1 subtypes (A, B, C, D, AE, and AG). Among the 11 chosen viruses, HIVBaL belongs to tier-1B category, Ker 2018 is a tier-3 virus and the rest belong to tier-2 viruses that are difficult to be neutralized. Neutralizing ability was assessed by Tat-regulated luciferase assay in TZM-bl cells infected with different isolates of HIV-1. The data plotted using the software Sigmaplot are depicted as neutralization curves, and the neutralization titers expressed as reciprocal titers at which 50% inhibition of virus infection was observed compared with preimmune serum (IC50). Supplementary Figure S3 shows the neutralization curves from five mice immunized with 12B1 chimeric virus. Average neutralization activity in the sera of five mice immunized with chimeric viruses 12B1, 13A3, and 22C1, as well as the wild-type HRV-14 and 4E10 peptide (KLH-conjugated NK15 peptide12) against 11 HIV-1 isolates are presented in Table 1. 12B1-immunized hICAM-1 Tg mice showed the broadest neutralization (9 of 11 HIV-1 isolates) compared to the other two chimeras (22C1 and 13A3).
Table 1. Neutralization of HIV-1 international primary isolates by sera from hICAM-1Tg mice immunized with chimeric viruses.
We also attempted to verify our result in Montefiori's reference lab at the Duke University using their stringent criteria (≥3× the signal in the preimmune serum against the same virus and/or ≥3× the signal of the same sample against the MLV negative control virus). However, we did not have sufficient amount of sera from the two 12B1-immunized mice that showed the best activity in our in house assay and we could only test three other mouse sera. In addition to a Clade B and a Clade C virus, neutralization was tested against 3 other tier 2 viruses. Although 3/3 and 2/3 sera showed neutralization against tier 1A Clade B and Clade C viruses respectively, statistically significant neutralization was not detected against the tier 1B and tier 2 strains tested (Supplementary Table S1). We therefore conclude that 12B1 chimera definitively induces antibodies that neutralize virus from Clades B and C and might have potential to neutralize other viruses.
In silicon modeling of 12B1 in complex with 4E10 MAb
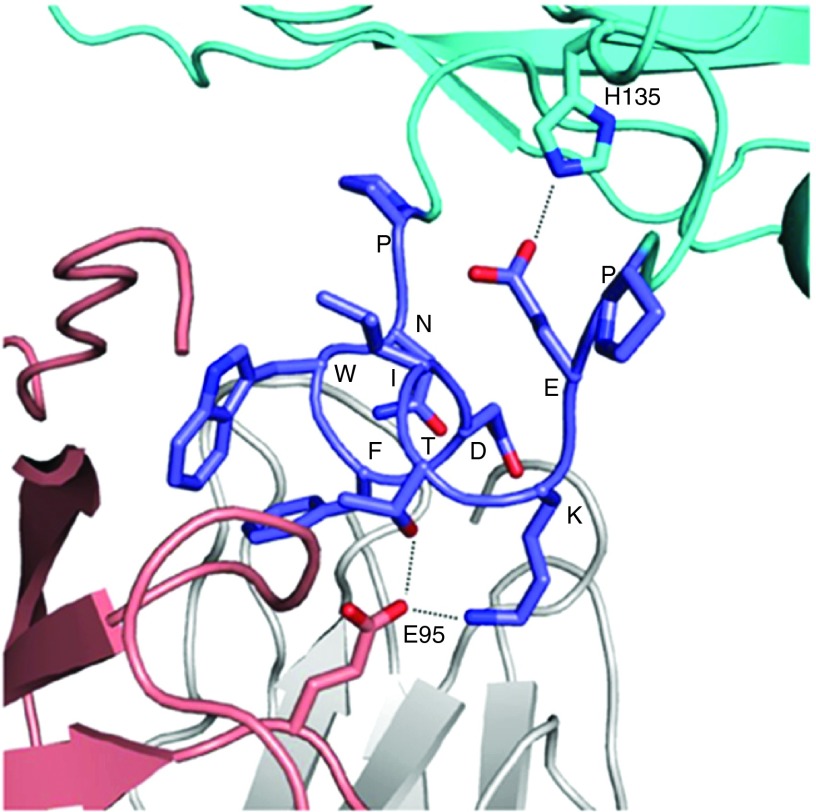
To elucidate why the chimeric virus 12B1 elicited the best neutralization, we performed a molecular modeling of 12B1 binding with mAb 4E10. We in silico mutated and truncated a 13-residue peptide (KGWNWFDITNWGK) bound to mAb 4E10 (PDB ID 1TZG19 into the PNWFDITKEP and adjusted the side chains of Lys and Glu for allowable conformations using the software COOT. We then inserted this the PNWFDITEP peptide into a position between residues 158 and 162 of VP2 of HRV-14 (PDB ID 4RHV) via adjusting main-chain conformation of the residues 158 and 162 in a manner that the antibody had no steric conflict with the virus capsid. Thereafter, we performed structural regularization for residues around the insertion sites using COOT followed by model energy minimization using software Crystallography & NMR system (CNS). The final assembled structural model containing the mAb 4E10 and the PNWFDITKEP-fused HRV14 capsid is represented using the software PyMOL (Figure 5). By examining potential interactions of PNWFDITKEP in this model, we found that the side chain of the introduced Lys next to Thr is likely able to promote the NWFDITK binding by creating an additional interaction with the E95 side chain of mAb 4E10 heavy chain, and the side chain of the introduced Glu is able to position the NWFDITK by interacting with the H135 side chain of virus V2 capsid (Figure 5). In addition, both terminal Pro residues are presumably capable of protruding this peptide from virus capsid surface and increasing its accessibility to the antibody.
Figure 5.
Computational modeling of chimeric human rhinovirus (HRV) 12B1 bound to 4E10 mAb. The PNWFDITKEP fused to HRV-14 colored in cyan is highlighted in blue, 4E10 mAb light and heavy chains are shown in silver and tint, respectively. The two terminal prolines of PNWFDITKEP protrude this peptide for accessing to 4E10 mAb. Potential interactions of Glu and Lys of the PNWFDITKEP are shown in dotted lines.
Discussion
It has been traditionally difficult to elicit NAbs against HIV-1. Here using HRV susceptible hICAM-1 Tg mice, we showed that HRV provides a convenient epitope-scaffold platform that allows transplantation of HIV-epitopes to elicit NAbs. We also showed that pre-existing HRV immunity is not a deterrent for intranasal immunization in mice. Thus, HRV appears to have potential for human vaccine applications.
The 4E10 antibody recognizing MPER region has been shown to exhibit broadly neutralizing activity against HIV-1. Although 4E10 antibody develops naturally in some HIV infected individuals, it has been difficult to generate 4E10-like antibodies by immunization with engineered immunogens. In an earlier study, we grafted the 4E10 epitope to HRV scaffold via 0–3 random linker residues, which provides the likelihood of forming millions of conformations. After immunoselection using 4E10 mAb, the chimeric viruses with highest binding to 4E10 mAb would theoretically present conformations more similar to those presented during natural infection. Although this approach has shown the potential of eliciting neutralizing antibodies, the rarity of success and very low titers obtained in the guinea pig model have raised questions regarding the applicability of this approach for human vaccination. Our results in this study using HRV-susceptible mice showed that the HRV-HIV chimera can induce antibodies that can neutralize viruses belonging to both Clades B and C. HRV-HIV infection here showed better results than in our previous study in HRV insusceptible guinea pigs using a HRV-HIV prime-peptide boost approach, suggesting that the HRV replication played a critical role for induction of neutralizing ability. However, it is possible that a peptide boost after HRV-HIV infection in hICAM-1 Tg mice might further improve the antibody titers. It must be cautioned, however, that there is always a possibility that mouse studies may not necessarily be reproducible in humans.
It is generally thought that the induction of MPER bNAbs is likely to be hampered due to autoreactivity of the epitopes resulting in immune tolerance. In fact, autoreactivity of 2F5 and 4E10 epitopes have been demonstrated in humans20,21 and in knockin mice.22 However, a systematic analysis of binding affinity using an autoantigen microarray showed that 4E10 binding to HIV epitopes is 2–3 log higher than that for autoantigens, which is vastly different from pathogenic autoantibodies like the sera of patients with SLE.23 Moreover, 4E10 and 10E8-like antibodies have been induced by various experimental approaches.7,24 It is also thought that HIV-1 bNAbs require a long CDR3 loop, which is absent in mice, rendering it a more difficult model to elicit bNAbs. Further, bNAbs during natural HIV infection requires prolonged maturation of the antibodies by accumulation of somatic hypermutation. These may be the reasons for the limited breadth of neutralization observed here. However, further dissection of the B-cell response after vaccination and elucidation of the structure of anti-HIV antibody in complex with chimeric HRV-HIV will be necessary to reveal the exact mechanism/s involved.
Human rhinovirus as a potential HIV-1 vaccine vehicle has some unique advantages: the vector can display 60 copies of the vaccine epitopes on the surface of each particle, and being a live virus infection, further amplifies the epitope presentation. This key advantage of amplification during productive infection was missing in our earlier studies in guinea pigs that showed rather modest titers of NAb production following vaccination. On the other hand, our present study in HRV susceptible mice clearly shows the potential of chimeric HRV as a viable vaccine option. However, since HRV infection in humans is common and over a hundred serotypes of HRV exists, there is always a concern that prior infection with HRV would dampen the vaccine efficacy. Our results showing that intranasal infection can bypass pre-existing immunity would argue against this possibility. In fact, in AdHu5-vaccinated nonhuman primates, anti-AdHu5 neutralizaing antibody was undetectable in nasal cavity even though seum samples showed a high neutralizing antibody level.17,25 Our results also show that the presence of neutralizing antibodies in the serum may not prevent at least a transient infection in the lungs. These results are also consistent with similar studies in adenovirus and influenza infections. Despite this, if pre-existing immunity in humans still remains a problem, this can be overcome by changing from HRV14 to other serotypes.
In summary, our study showed that neutralizing antibodies against HIV-1 could be elicited by immunization of HRV displaying a single 4E10 epitope. However, 4E10 is one of the first generation of broadly neutralizing antibodies and compared with other recently identified HIV-1 bNAbs, both potency and breadth of 4E10 are relatively limited. Therefore, displaying broader and more potent NAb epitopes on HRV will be likely improve immunogenicity against HIV-1. bNAbs recognize regions critical for virus life cycle (e.g., entry pathways such as CD4/CCR5 binding) and thus, escape results in fitness cost to the virus.26 Nevertheless, the virus can escape and expression of the epitope on HRV is unlikely to influence this process. That is why it is preferable to elicit multiple bNAbs targeting different regions. In fact, studies of SIV infection in rhesus macaque as well of HIV-1 in humanized mice have shown a combination of three to four bNAbs might be needed for complete protection.27,28 Hence, a mixture of chimeric HRV displaying different NAb epitopes or engineering chimeric viruses presenting multiple neutralizing epitopes (especially displaying more accessible, more potent epitopes without poly-reactivity) as well as adopting optimal prime-boost regimen will likely be needed to further improve HRV-based HIV-1 vaccine efficacy.
Materials and Methods
Ethics statement. All work including animal studies were conducted following the guidelines for the Care and Use of Laboratory Animals of National Research Council, USA. The authors received approval from the Institutional Animal Care and Use Committee (IACUC) of the Texas Tech University Health Sciences Center (protocol number 8007).
Cells culture. HeLa cells and TZM-bl cells (purchased from ATCC, Manassas, VA) were cultured in complete Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mmol/l L-glutamine (PSG). Hela cells were used to produce, propagate, and titer the chimeric HRV:HIV viruses, as well as wild-type HRV-14.
Chimeric rhinoviruses. Propagation of chimeric HRV-HIV viruses and their titration have been described previously.11,12 Briefly, 1 × 109 of Hela cell suspension culture was infected with chimeric viruses (multiplicity of infection (MOI) 0.1) at 34.5 °C. After 10–11 hours of infection, the cells were centrifuged and the pellet was subjected to three cycles of freeze thaw followed by treatment with DNase I. The cell lysate was then ultracentrifuged with a 30% sucrose cushion at 25,000 rpm for 2.5 hours at 4 °C in a Beckman SW 28 Ti rotor. The virus pellet was resuspended in 20 mmol/l Tris-HCl, pH 7.4.
Recombinant proteins, peptides, antibodies, and HIV-1 isolates. Recombinant HIV-1 SF162 gp140 trimer was obtained from NIH AIDS reagent program (donated by Dr. L Stamatatos, Seattle Biomedical Research Institute, Seattle, WA). 4E10 peptide (K7-D11 lactam-bridged Ac-NWFDITK7WLWD11KKK-NH2 (NK-15)) and KLH-conjugated 4E10 peptide (KLH-C-NWFDITK7WLWD11KKK-NH2 (KLH-NK-15))12 were purchased from Chempep (Wellington, FL). 4E10 mAb was purchased from PolyMun, Vienna, Austria. VRC01 antibody encoding vector was obtained from NIH AIDS reagent program (donated by Mascola J. et al.), expressed and purified in the laboratory. Internationally circulating HIV-1 isolates were obtained from NIH AIDS reagent program, propagated in peripheral blood mononuclear cells (PBMCs) and stocks titrated for p24 levels by ELISA.
hICAM-1 Tg mice and immunization. All animal work was approved by TTUHSC IACUC. hICAM-1 Tg mice were obtained from Dr. Johnston L laboratory, Imperial College London, bred at TTUHSC animal facility and used at 6–8 weeks of age. Groups of hICAM-1 Tg mice (five mice/group) were infected intranasally with different clones of chimeric HRV (2.5 × 106 PFU) as described in ref. 12. The mice were boosted twice with the same chimeric viruses at week 3 and week 6. The blood obtained at weeks 0, 3, 6, and 9 were used for evaluating the antibody response. Wild-type HRV14 (intranasally) and KLH-conjugated peptide (KLH-C-NWFDITKWLWDKKKK-NH2, 40 µg with complete Freund's adjuvants for priming, and incomplete Freund's adjuvants for boosting twice, intraperitoneally) immunized mice were used as controls.
ELISA. Indirect ELISA was performed in triplicate to assess the immunized mice sera binding to 4E10 peptide and gp140 trimer. Briefly, 10 µg/ml of 4E10 peptide or gp140 trimer was coated on to 96-well Maxisorp plates (Nunc, Denmark) in 50 mmol/l sodium borate, pH 8.5 at 4 °C overnight, and then blocked with 5% skimmed milk in phosphate-buffered solution (PBS) for 2 hours at 37 °C. After washing the plates five times with PBS/0.2% Tween 20 (PBS-T), serially diluted mouse sera (4E10 MAb was used as positive control) were added to the wells and incubated for 2 hours at 37 °C. Plates were washed and 1.0 µg/ml of horse radish peroxidase (HRP)-labeled sheep anti-mouse IgG (Fc specific, Cat# A170, Sigma, MO, for mouse sera detection) or HRP-labeled sheep anti-human IgG (Fc specific, Cat# A168, Sigma, for 4E10 MAb detection) was added, incubated for 1 hour at 37 °C. After washing, peroxidase substrate (0.3 mg/ml tetramethylbenzidine dissolved in 10% dimethyl sulfoxide in 0.18 mol/l sodium citrate, pH 3.95) was added. The reaction was catalyzed by the addition of H2O2 to 0.009% to develop color for 20 minutes, and then stopped by the addition of an equal volume of 1 mol/l H2SO4. OD450 measurements were then recorded. Antibody titers are expressed as reciprocal of serum dilutions, and the positive titer was defined as > 3 × mean of background (PBS-coated well). All data are reported as the average of three measurements.
Immunofluorescence staining. Lung tissues were collected immediately after euthanasia, rinsed 5–10 minutes in ice-cold PBS and fixed in 4% fresh paraformaldehyde in PBS (PH = 7.4) for 6 hours at 4 °C. Fixed lung tissues were embedded within optimal cutting temperature compound at −80 °C. Frozen sections (4 µm thick) underwent an antigen retrieval procedure using Trilogy pretreatment solution (Cell Marque, Rocklin, CA, Cat# 920P-04) at 40 °C for 10 minutes. Then all sections were blocked in 3% BSA/PBS at room temperature for 1 hour and stained with 4E10 antibody at 1:500 dilution in 3% bovine serum albumin (BSA)/PBS at 4 °C overnight. After washing three times, the sections were incubated with Alexa Fluor 488-labeled goat anti-human IgG (Life Technologies, Carlsbad, CA, Cat.# A-11013) secondary antibody (1:500 dilution in 3% BSA/PBS) at room temperature for 1 hour. Nuclei were stained with 4′,6-diamino-2-phenyl-indol and sections examined with a Nikon immunofluorescence microscope.
HRV plaque reduction neutralization assay. The plaque reduction neutralization assay was used to assess the neutralizing antibodies against HRV challenge. Briefly, 2 × 105 Hela cells were seeded in 12-well plates and cultured at 37 °C, 5% CO2 overnight. 100–200 PFU of HRV-14 wild-type virus (for each well) was mixed with serially diluted mouse sera for 1 hour at 37 °C, and then added onto the cell monolayers in 12-well plates. After 1 hour infection at 30 °C, the viral mixtures were removed and 0.42% agarose in complete Dulbecco's modified Eagle's medium was added to cover the cell monolayers. After 3 days of culture, the plates were fixed with 20% trichloroacetic acid and plaques were visualized by addition of 0.2% crystal violet in 20% ethanol. The titers were calculated as reciprocal of serum dilutions, and the IC50 titers were defined as serum dilutions showing 50% of plaque reduction compared with control (no sera added).
HIV-1 neutralization assay
Tat-regulated luciferase assay. HIVBaL (200 pg of p24) was incubated with serial dilutions of heat-inactivated sera for 1 hour and used to infect TZM-bl cells and 48 hours after infection, the cells were lysed and assayed for luciferase expression using Luciferase Assay System kit (Cat # 1500, Promega, Madison, WI) and the reporter luciferase activity was read on a Fluostar microplate reader (BMG Labtech, Chicago, IL) according to the instructions.
Neutralization after multi-round infection. Serum dilutions were incubated with HIVBaL and used to infect TZB-bl cells. After 1 hour incubation, the cells were washed five times with PBS and cultured with regular media. Culture supernatants collected 48 hours after infection were tested for p24 antigen using p24 ELISA kits (PerkinElmer, Melville, NY) according to the instructions.
Testing for potency and breadth of neutralization. Eleven HIV-1 international isolates were used to assess the potency and breadth of neutralization using a single-round Tat-regulated luciferase assay in TZM-bl cells. Briefly, the HIV-1 isolates (200 pg of p24) were preincubated with various dilutions of heat-inactivated mouse sera for 2 hours and then added to TZM-bl cells (5 × 104/well in 96-well plates) in the presence of 10 µg/ml of DEAE-Dextran. After 48 hours, the cells were lysed in lysis buffer and then Bright-Glo luciferase reagent was added to the wells, and the reporter luciferase activity was recorded in a luciferase reader. The neutralizing titers were calculated as the average of five mice sera (triplicates for each serum) and presented as the reciprocal of serum dilution yielding 50% inhibition of the reporter luciferase activity (compared with normal mouse serum values) after plotting the data with Sigmaplot software using a polynomial regression (quadratic) curve-fitting model.29
Statistical analysis. Statistical analyses were performed with GraphPad Prism version 6. Data between groups were compared using Unpaired Student T-tests. P < 0.05 was considered significant and P < 0.01 was considered extremely significant.
SUPPLEMENTARY MATERIAL Figure S1. Expression of 4E10 epitope in the lungs of wt HRV preimmunized mice following HRV-HIV infection. Figure S2. Plaque reduction neutralization assay of wt HRV-infected mice sera against chimeric 12B1 HRV-HIV. Figure S3. HIV-1 neutralization curves of five individual HRV-HIV 12B1 immunized mice sera. Table S1. Neutralization assay in TZM-bl cells using pseudoviruses was performed in Dr. Montefiori's reference lab at the Duke University.
Acknowledgments
We thank Eddy Arnold and Gail F. Arnold for critical reading of the manuscript and generous donation of the HRV-HIV chimeras, 4E10 epitope peptide, as well as 4E10 MAb. We also thank Victoria Polonis for providing the tier categorization information for the HIV primary isolates. We thank NIH AIDS Reagent Program for providing HIV-1 SF162 gp140 trimer and international panel of HIV-1 isolates. We thank David Montefiori and Celia Labranche for assistance with neutralization assay.
Supplementary Material
References
- Koff, WC, Burton, DR, Johnson, PR, Walker, BD, King, CR, Nabel, GJ et al. (2013). Accelerating next-generation vaccine development for global disease prevention. Science 340: 1232910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes, BF, Gilbert, PB, McElrath, MJ, Zolla-Pazner, S, Tomaras, GD, Alam, SM et al. (2012). Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 366: 1275–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koff, WC (2012). HIV vaccine development: challenges and opportunities towards solving the HIV vaccine-neutralizing antibody problem. Vaccine 30: 4310–4315. [DOI] [PubMed] [Google Scholar]
- Correia, BE, Bates, JT, Loomis, RJ, Baneyx, G, Carrico, C, Jardine, JG et al. (2014). Proof of principle for epitope-focused vaccine design. Nature 507: 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan, JS, Chen, M, Joyce, MG, Sastry, M, Stewart-Jones, GB, Yang, Y et al. (2013). Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342: 592–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek, G, Guenaga, FJ, Schief, WR, Skinner, J, Baker, D, Wyatt, R et al. (2010). Elicitation of structure-specific antibodies by epitope scaffolds. Proc Natl Acad Sci USA 107: 17880–17887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia, BE, Ban, YE, Holmes, MA, Xu, H, Ellingson, K, Kraft, Z et al. (2010). Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure 18: 1116–1126. [DOI] [PubMed] [Google Scholar]
- Ofek, G, Tang, M, Sambor, A, Katinger, H, Mascola, JR, Wyatt, R et al. (2004). Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol 78: 10724–10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick, MB, Labrijn, AF, Wang, M, Spenlehauer, C, Saphire, EO, Binley, JM et al. (2001). Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol 75: 10892–10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley, JM, Wrin, T, Korber, B, Zwick, MB, Wang, M, Chappey, C et al. (2004). Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol 78: 13232–13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, GF, Velasco, PK, Holmes, AK, Wrin, T, Geisler, SC, Phung, P et al. (2009). Broad neutralization of human immunodeficiency virus type 1 (HIV-1) elicited from human rhinoviruses that display the HIV-1 gp41 ELDKWA epitope. J Virol 83: 5087–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, G, Lapelosa, M, Bradley, R, Mariano, TM, Dietz, DE, Hughes, S et al. (2013). Chimeric rhinoviruses displaying MPER epitopes elicit anti-HIV neutralizing responses. PLoS One 8: e72205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings, GZ, Francis, MJ, Rowlands, DJ and Chain, BM (1993). Antigen processing and presentation of human rhinovirus to CD4 T cells is facilitated by binding to cellular receptors for virus. Eur J Immunol 23: 1340–1345. [DOI] [PubMed] [Google Scholar]
- Smith, AD, Geisler, SC, Chen, AA, Resnick, DA, Roy, BM, Lewi, PJ et al. (1998). Human rhinovirus type 14:human immunodeficiency virus type 1 (HIV-1) V3 loop chimeras from a combinatorial library induce potent neutralizing antibody responses against HIV-1. J Virol 72: 651–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett, NW, Walton, RP, Edwards, MR, Aniscenko, J, Caramori, G, Zhu, J et al. (2008). Mouse models of rhinovirus-induced disease and exacerbation of allergic airway inflammation. Nat Med 14: 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croyle, MA, Patel, A, Tran, KN, Gray, M, Zhang, Y, Strong, JE et al. (2008). Nasal delivery of an adenovirus-based vaccine bypasses pre-existing immunity to the vaccine carrier and improves the immune response in mice. PLoS One 3: e3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, JS, Pillet, S, Bello, AJ and Kobinger, GP (2013). Airway delivery of an adenovirus-based Ebola virus vaccine bypasses existing immunity to homologous adenovirus in nonhuman primates. J Virol 87: 3668–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellhorn, G, Caldwell, Z, Mineart, C and Stamatatos, L (2009). Improving the expression of recombinant soluble HIV Envelope glycoproteins using pseudo-stable transient transfection. Vaccine 28: 430–436. [DOI] [PubMed] [Google Scholar]
- Cardoso, RM, Zwick, MB, Stanfield, RL, Kunert, R, Binley, JM, Katinger, H et al. (2005). Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22: 163–173. [DOI] [PubMed] [Google Scholar]
- Yang, G, Holl, TM, Liu, Y, Li, Y, Lu, X, Nicely, NI et al. (2013). Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies. J Exp Med 210: 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finton, KA, Larimore, K, Larman, HB, Friend, D, Correnti, C, Rupert, PB et al. (2013). Autoreactivity and exceptional CDR plasticity (but not unusual polyspecificity) hinder elicitation of the anti-HIV antibody 4E10. PLoS Pathog 9: e1003639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle-Cooper, C, Hudson, KE, Cooper, AB, Ota, T, Skog, P, Dawson, PE et al. (2013). Immune tolerance negatively regulates B cells in knock-in mice expressing broadly neutralizing HIV antibody 4E10. J Immunol 191: 3186–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, H, Henry, KA, Wu, SS, Chruscinski, A, Utz, PJ and Scott, JK (2011). Reactivity profiles of broadly neutralizing anti-HIV-1 antibodies are distinct from those of pathogenic autoantibodies. AIDS 25: 1247–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y, Tong, P, Li, Y, Lu, Z and Chen, Y (2014). 10E8-like neutralizing antibodies against HIV-1 induced using a precisely designed conformational peptide as a vaccine prime. Sci China Life Sci 57: 117–127. [DOI] [PubMed] [Google Scholar]
- Richardson, JS, Abou, MC, Tran, KN, Kumar, A, Sahai, BM and Kobinger, GP (2011). Impact of systemic or mucosal immunity to adenovirus on Ad-based Ebola virus vaccine efficacy in guinea pigs. J Infect Dis 204 Suppl 3: S1032–S1042. [DOI] [PubMed] [Google Scholar]
- Sather, DN, Carbonetti, S, Kehayia, J, Kraft, Z, Mikell, I, Scheid, JF et al. (2012). Broadly neutralizing antibodies developed by an HIV-positive elite neutralizer exact a replication fitness cost on the contemporaneous virus. J Virol 86: 12676–12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch, DH, Whitney, JB, Moldt, B, Klein, F, Oliveira, TY, Liu, J et al. (2013). Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature 503: 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, F, Halper-Stromberg, A, Horwitz, JA, Gruell, H, Scheid, JF, Bournazos, S et al. (2012). HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 492: 118–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hioe, CE, Wrin, T, Seaman, MS, Yu, X, Wood, B, Self, S et al. (2010). Anti-V3 monoclonal antibodies display broad neutralizing activities against multiple HIV-1 subtypes. PLoS One 5: e10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.