Abstract
Cytotoxicity of transgenes carried by adeno-associated virus (AAV) vectors might be desired, for instance, in oncolytic virotherapy or occur unexpectedly in exploratory research when studying sparsely characterized genes. To date, most AAV-based studies use constitutively active promoters (e.g., the CMV promoter) to drive transgene expression, which often hampers efficient AAV production due to cytotoxic, antiproliferative, or unknown transgene effects interfering with producer cell performance. Therefore, we explored artificial riboswitches as novel tools to control transgene expression during AAV production in mammalian cells. Our results demonstrate that the guanine-responsive GuaM8HDV aptazyme efficiently attenuates transgene expression and associated detrimental effects, thereby boosting AAV vector yields up to 23-fold after a single addition of guanine. Importantly, riboswitch-harboring vectors preserved their ability to express functional transgene at high levels in the absence of ligand, as demonstrated in a mouse model of AAV-TGFβ1-induced pulmonary fibrosis. Thus, our study provides the first application-ready biotechnological system-based on aptazymes, which should enable high viral vector yields largely independent of the transgene used. Moreover, the RNA-intrinsic, small-molecule regulatable mode of action of riboswitches provides key advantages over conventional transcription factor–based regulatory systems. Therefore, such riboswitch vectors might be ultimately applied to temporally control therapeutic transgene expression in vivo.
Introduction
Adeno-associated virus (AAV) vectors represent potent tools for the genetic engineering of various organs and tissues and are currently evaluated as therapeutic agents in clinical trials for several indications.1,2,3 Besides therapeutic applications, viral vectors have also been extensively used for preclinical research, e.g., to study gene function in vivo or to setup disease models by modulating gene expression in different target organs such as liver,4 heart,5 brain,6 or lung.7 To date, the majority of AAV-based studies are carried out using constitutive promoters to drive ubiquitous (e.g., CMV promoter) or tissue-specific transgene expression. While constitutive transgene expression is sufficient for various applications and moreover is a desirable feature for the treatment of certain diseases like, e.g., monogenetic disorders, constitutive transgene expression can be unwanted in some instances, such as during viral vector production.
During AAV production by transient transfection in HEK-293 cells, AAV capsid- (cap), replication- (rep), and Adenoviral helper-proteins are expressed from one or two plasmids,8 leading to capsid formation and incorporation of a co-provided ITR-flanked single-stranded transgene DNA cassette.9,10 However, the application of constitutively active promoters ultimately results in transgene expression by the producer cells, representing an unnecessary byproduct of the production process. Transgene expression not only occupies the cells' translational machinery for the production of high levels of an unnecessary gene product, but might also induce proapoptotic, cell cycle-modulating or other unknown effects that interfere with producer cell performance and therefore reduce viral vector yield. While cytotoxic transgene effects can be expected and even desired in some instances, for example, oncolytic virotherapy, they also often occur unexpectedly when packaging sparsely characterized genes. Speculating that suppressing transgene expression during AAV production would increase viral vector yield, we were aiming to develop a vector system allowing transgene regulation in HEK-293 cells, the most commonly used cell culture system for AAV production. To enable broad applicability and minimal immunogenicity, we envisioned a system that works independently of additionally expressed transcription factors, allows rapid adaptability to different transgenes and promoters and is relatively small—an important property in the context of AAV vectors, which have a limited packaging capacity of ~4.7 kb.11 Therefore, we explored artificial riboswitches for the conditional regulation of transgene expression during AAV production (see scheme in Figure 1a).
Figure 1.
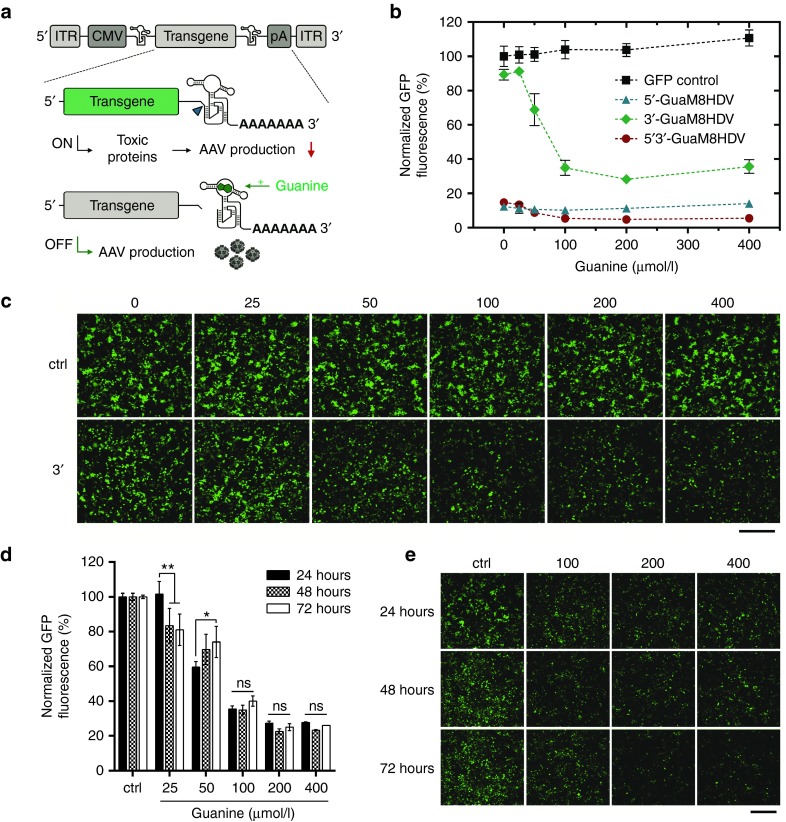
3′-GuaM8HDV enables efficient suppression of GFP expression in HEK-293 AAV producer cells. (a) Scheme of AAV-GuaM8HDV construct design and riboswitch-mediated transgene suppression. The riboswitch was inserted either upstream (5′) or downstream (3′) of the transgene, whose expression is driven by a CMV promoter. The transgene cassette is flanked by AAV2 ITR sequences, which define the DNA section that is packaged into the AAV particle. While transgene expression in absence of the ligand (guanine) might lead to toxic effects that can decrease AAV vector yield (upper panel), guanine addition triggers self-cleavage of the riboswitch, which attenuates transgene expression, thereby increasing AAV vector yield (lower panel). (b) Normalized GFP expression measured by flow cytometry and (c) fluorescence microscopic analysis of HEK-293 cells 24 h after transfection with either 5′-, 3′- or 5′3′-GuaM8HDV-harboring pAAV-GFP or a riboswitch-free pAAV-GFP control construct and addition of increasing concentrations of guanine. n = 4 biological replicates, mean ± SD. Bar = 400 µm. (d) Normalized GFP expression measured by flow cytometry and (e) fluorescence microscopic analysis of HEK-293 cells 24, 48, and 72 hours after transfection with the 3′-GuaM8HDV-harboring pAAV-GFP construct and addition of increasing concentrations of guanine. Ctrl = 0 µmol/l guanine. n = 3 biological replicates, mean ± SD. *P < 0.05; **P < 0.01. Bar = 400 µm.
Aptazyme riboswitches are composed of a ligand-binding aptamer fused to a self-cleaving ribozyme12 and represent novel tools to regulate transgene expression13,14 in a wide range of potential applications, including gene therapy vectors.15 Upon ligand binding, the aptamer domain undergoes a conformational change that, depending on the aptamer-ribozyme linker sequence, either induces or inhibits ribozyme self-cleavage. Accordingly, riboswitches can be engineered to act as an OFF- or ON-switch.16 When incorporated into the transgene's untranslated region (UTR), riboswitch activity induces mRNA cleavage resulting in loss of its 5′-cap or 3′-polyA structure, thus disrupting transcript stability and translation.17,18 Therefore, in contrast to classical repression systems such as inducible promoters (e.g., Tet-off19,20,21), riboswitch activity does neither require expression of additional transcription factors, nor rely on protein-nucleic acid interaction, but acts in a RNA-intrinsic manner. Moreover, only ~100 bp of plasmid space are required for riboswitch incorporation, representing a particular advantage for AAV vectors.
In our study, we explored a recently engineered, guanine-responsive riboswitch called GuaM8HDV22 as a tool to regulate transgene expression in the AAV vector genome. This switch is based on a guanine aptamer derived from the 5′-UTR of the Bacillus subtilis xpt-pbuX operon23 which was fused to the hepatitis delta virus ribozyme.22,24 We first generated GuaM8HDV-harboring pAAV-GFP plasmids to characterize GuaM8HDV's switching behavior under the conditions relevant for AAV production. Next, we investigated whether riboswitch-mediated transgene regulation can be used to attenuate the functional effects of genes known to interfere with producer cell performance in an either proapoptotic (BAX, TNFα), cell-cycle-regulating (TGFβ1) or unknown (LOXL2) manner. In a proof-of-concept experiment, we then explored whether transient suppression of these genes during AAV production would increase the yield of respective AAV preparations. To validate that riboswitch-containing vectors preserve the ability to express the transgene in the absence of ligand, we finally tested the 3′-GuaM8HDV-containing AAV-TGFβ1 vector in a mouse model of pulmonary fibrosis.
Results
3′-GuaM8HDV enables efficient suppression of reporter gene expression in HEK-293 AAV producer cells
To identify a riboswitch construct that enables efficient regulation of transgene expression under the conditions used for AAV production, we generated pAAV plasmids harboring an eGFP gene under the control of a CMV-promoter and the guanine-responsive GuaM8HDV riboswitch in either the 3′-UTR, 5′-UTR, or at both positions. Using the calcium phosphate procedure, HEK-293 cells were transiently transfected with these constructs and after 5 hours the medium was replaced with fresh medium containing increasing concentrations of guanine of up to 400 µmol/l. Twenty-four hours after transfection, GFP fluorescence was analyzed by microscopy and flow cytometry. While transfection with the riboswitch-free control construct led to stable GFP expression, which was not affected by increasing guanine concentrations, both the 5′- and 5′3′-GuaM8HDV constructs showed strongly decreased GFP expression even without addition of guanine, which further decreased with increasing guanine concentration (Figure 1b). In contrast, the 3′-GuaM8HDV construct retained ~90 % of GFP expression when no guanine was present, which could be dose dependently suppressed by increasing guanine concentrations, approaching a minimum of ~25% residual GFP expression at 200 µmol/l guanine (Figure 1b). Microscopic analysis further underscored these results (Figure 1c). Moreover, microscopy revealed a slight decrease in cell numbers at 400 µmol/l guanine that, however, was not associated with increased LDH release (Supplementary Figure S1), suggesting that guanine concentrations >400 µmol/l might exert slight antiproliferative but no prominent cytotoxic effects. For AAV production, transfected HEK-293 cells are usually kept in culture for 72 hours. Importantly, using the 3′-GuaM8HDV construct and a single addition of guanine, GFP expression remained stably suppressed over the required time period of 72 hours (Figure 1d) which again was also validated by microscopy (Figure 1e). When the cells were further incubated for an additional 4 days, a slight continuous increase in residual GFP expression was observed; however, this increase could be fully blocked by a second addition of guanine 72 hours after transfection (Supplementary Figure S2). Taken together, the pAAV construct harboring GuaM8HDV in its 3′-UTR effectively attenuates transgene expression in a guanine dose-dependent manner.
3′-GuaM8HDV attenuates transgene-mediated impairment of producer cell integrity
After having identified the 3′-GuaM8HDV construct as a suitable vector backbone, we next asked whether riboswitch-mediated transgene regulation would allow suppressing the functional effects of genes known to impair host cell integrity upon (over)expression and thus might negatively influence AAV vector production. To this end, we replaced the GFP gene by the genes encoding BAX, TNFα, TGFβ1, and LOXL2, respectively. While BAX and TNFα are well-known inducers of apoptosis,25,26 TGFβ1 is involved in many processes such as cell growth, cell-cycle regulation and cell differentiation.27 In contrast, LOXL2 has not been reported in the context of cytotoxicity so far, however, our attempts to produce LOXL2-expressing AAV vectors for protein characterization studies failed due to very low viral vector yield, indicating producer cell impairment of yet unknown cause.
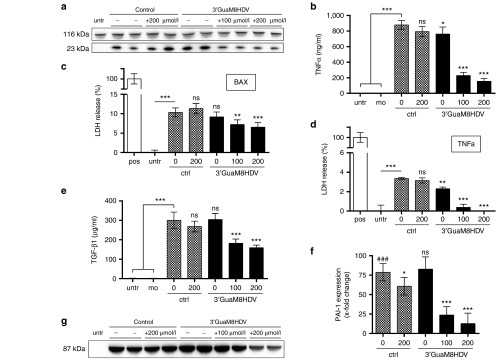
As expected, guanine addition to transfected cells dose dependently downregulated transgene expression in all cases tested (Figure 2a,b,e,g). Importantly, this decrease translated into an attenuation of cytotoxic and cell performance-impairing effects: specifically, while TNFα and BAX expression induced cytotoxicity, as measured by an increased release of LDH, GuaM8HDV-mediated suppression (Figure 2a,b) decreased cytotoxic effects by about 40% in the case of BAX (Figure 2c) and to baseline levels in the case of TNFα (Figure 2d). Moreover, riboswitch activation successfully reduced TGFβ1 protein levels (Figure 2e) and associated downstream signaling as assessed by gene expression measurement of the surrogate marker28 gene PAI-1 (Figure 2f). Finally, GuaM8HDV activation successfully reduced LOXL2 protein expression in a dose-dependent manner (Figure 2g). Thus, GuaM8HDV-mediated regulation of transgene expression effectively suppressed unwanted effects exerted by the selected genes.
Figure 2.
3′-GuaM8HDV attenuates transgene-mediated impairment of producer cell integrity. (a) Western blot analysis of BAX (23 kDa) expression in HEK-293 cell lysates 16 hours after transfection with either the 3′-GuaM8HDV-harboring pAAV-BAX construct or a riboswitch-free pAAV-BAX control construct and addition of indicated amounts of guanine. anti-Vinculin (116 kDa) staining was used as a loading control. (b) ELISA measurement of TNFα protein levels in the supernatant of HEK-293 cells, 24 hours after transfection with either the 3′-GuaM8HDV-harboring pAAV-TNFα construct or a riboswitch-free pAAV-TNFα control construct and addition of indicated amounts of guanine. n = 6 biological replicates, mean ± SD. (c,d) Cytotoxicity analysis by lactate dehydrogenase (LDH) measurement in the supernatant of HEK-293 cells 24 hours after transfection with either (c) the 3′-GuaM8HDV-harboring pAAV-BAX construct or a riboswitch-free pAAV-BAX control construct or (d) the 3′-GuaM8HDV-harboring pAAV-TNFα construct or a riboswitch-free pAAV-TNFα control construct and addition of indicated amounts of guanine. LDH levels measured in triton-lysed cells served as a positive control and were set 100%. n = 5 biological replicates, mean ± SD. (e) ELISA measurement of TGFβ1 protein levels in the supernatant and (f) qPCR-based measurement of PAI-1 gene expression in total RNA samples of HEK-293 cells, 24 hours after transfection with either the 3′-GuaM8HDV-harboring pAAV-TGFβ1 construct or a riboswitch-free pAAV-TGFβ1 control construct and addition of indicated amounts of guanine. n = 4 biological replicates, mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001 relative to the ctrl 0 µmol/l sample or as indicated. ###P < 0.001 relative to untreated cells. (g) Western blot analysis of LOXL2 (87 kDa) expression in the supernatant of HEK-293 cells 48 hours after transfection with either the 3′-GuaM8HDV-harboring pAAV-LOXL2 construct or a riboswitch-free pAAV-LOXL2 control construct and addition of indicated amounts of guanine. Ponceau S staining was used as a loading control (see Supplementary Figure S3).
3′-GuaM8HDV-mediated suppression of toxic transgene expression increases AAV yields
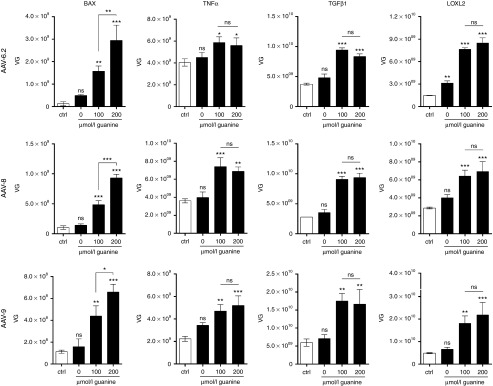
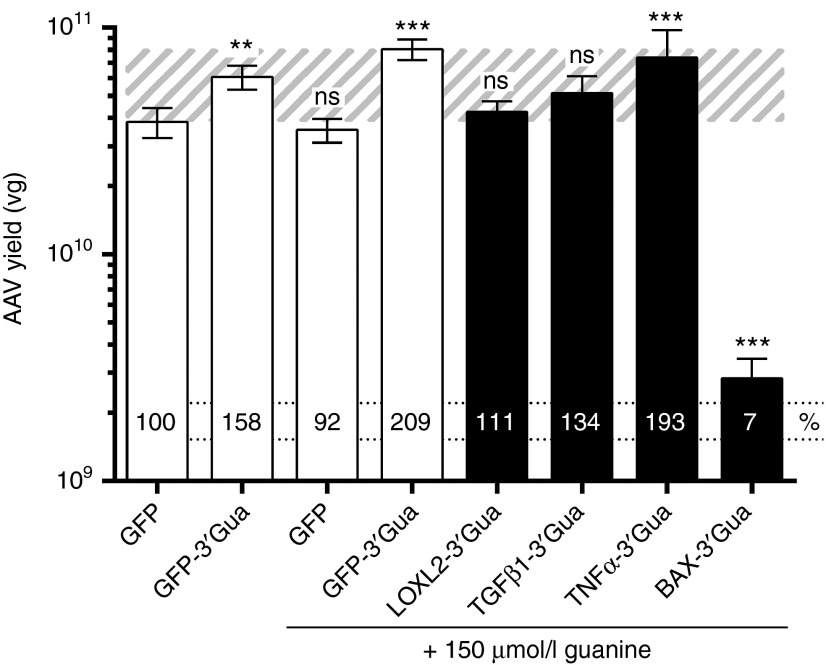
During the production of AAV vectors in HEK-293 cells, transgenes are expressed as an unnecessary byproduct, which might interfere with producer cell performance. We thus asked, whether riboswitch-mediated attenuation of transgene expression during viral vector production increases the yield of respective AAV vectors. In a proof-of-concept approach, we produced AAV vectors harboring the genes encoding TNFα, BAX, TGFβ1, and LOXL2 under the control of a CMV promoter with and without the GuaM8HDV riboswitch placed in the 3′-UTR. To examine potential differences between AAV variants, we packaged each of these genes in AAV6.2, AAV8, and AAV9—three of the most efficient and commonly used vectors for gene transfer to the lung, liver, and brain,29,30 respectively. For AAV production, HEK-293 cells were transfected with plasmids encoding the AAV cap and adenoviral helper genes along with a construct carrying the gene of interest flanked by AAV2 ITRs. Five hours after transfection, the medium was replaced with fresh medium, optionally supplemented with 100 or 200 µmol/l guanine. Our results show that in all cases tested, transgene suppression led to an increase in AAV yield (Figure 3). Specifically, using the riboswitch-free control construct, AAV-BAX yields were very low (1 × 108 vg) due to strong cytotoxicity (Figure 2c), whereas upon guanine addition a dose-dependent, up to 23-fold increase (AAV6.2) could be achieved. Interestingly, TNFα seemed to be less potent regarding apoptosis induction (Figure 2d), which translated into initially higher AAV yields using the control construct (2–4 × 109 vg). Nevertheless, by GuaM8HDV-mediated suppression, yields could be further increased by up to 2.3-fold (AAV8). Moreover, while both AAV-TGFβ1 and -LOXL2 yields were in the range of 1–5 × 109 vg using conventional constructs, a threefold to fivefold increase of AAV yields could be obtained by riboswitch activation. Notably, while the absolute yields of AAV6.2, -8, and -9 were partly different, riboswitch-mediated effects appeared to be independent of the AAV capsid variant used. We finally directly compared the yields of AAV-BAX-, -TNFα-, -TGFβ1-, and -LOXL2-riboswitch vectors to those obtained using conventional or riboswitch-carrying AAV-GFP constructs. In the first place, our data show that neither guanine nor the presence of GuaM8HDV per se negatively influence AAV vector yield. In fact, it rather seems that also in the case of GFP, riboswitch-activation might be beneficial, as a twofold increase in AAV-GFP yields was observed (Figure 4). Furthermore, by using the riboswitch system, the yields of AAV-TNFα, -TGFβ1, and -LOXL2 vectors could be increased to or beyond the levels of conventional AAV-GFP vectors (Figure 4). Despite a strong increase, AAV-BAX levels only reached 7% of the titer of the AAV-GFP vector. Taken together, riboswitch-mediated suppression of toxic transgene expression increased AAV yields, while the effect size seemed to be dependent on the severity of initial toxicity.
Figure 3.
3′-GuaM8HDV-mediated suppression of toxic transgene expression increases AAV yields. Benzonase-resistant AAV vector genomes (VG) were quantified by qPCR in HEK-293 cell lysate, 72 hours after transfection with either the 3′-GuaM8HDV pAAV-BAX, -TNFα, -TGFβ1, or -LOXL2 construct or respective riboswitch-free control constructs (and further plasmids necessary for the production of AAV6.2, AAV8, or AAV9—see Materials and Methods section for details) in presence of indicated concentrations of guanine, which were added during the medium exchange step about 5 hours after transfection. n = 3 biological replicates, mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Figure 4.
The 3′-GuaM8HDV riboswitch system enables high-titer vector production. AAV6.2 vectors carrying various transgene constructs were produced as described in Figure 3 in the presence or absence of 150 µmol/l guanine (as indicated). Seventy-two hours after transfection, benzonase-resistant AAV vector genomes (VG) were quantified by qPCR in HEK-293 cell lysate. The percentage of vector titers relative to the riboswitch-free GFP control construct is depicted on each bar. The AAV-GFP yield range is indicated by the striped background. 3′-Gua = 3′GuaM8HDV. n = 6 biological replicates, mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, relative to GFP without guanine.
3′-GuaM8HDV-harboring AAV vectors preserve bioactivity in vivo
Since the observed increase in viral vector yield is only valuable, if riboswitch-containing AAV vectors preserve functionality, i.e., their ability to express the transgene in vivo, we finally explored transgene expression and functional downstream effects by a GuaM8HDV-harboring AAV vector in mice. For this purpose, we made use of a well-established preclinical disease model, where AAV-mediated TGFβ1 expression induces pulmonary fibrosis characterized by increased immune cell influx and structural remodeling that ultimately results in decreased lung function.7 To this end, we first conducted a larger lab-scale production of the 3′GuaM8HDV-harboring AAV6.2-TGFβ1 vector, which was subsequently purified by an iodixanol density gradient. Notably, our results obtained in six-well microplates (Figure 3) could be successfully scaled up to larger lab scale (i.e., 40 × 15 cm plates), yielding 3.9-fold higher AAV titers (2.2 × 1013 vg) with the riboswitch in presence of 150 µmol/l guanine as compared to previous productions (5.6 × 1012 vg) with the control construct (Supplementary Figure S4). For the in vivo assessment of TGFβ1 expression, AAV6.2-TGFβ1 vectors with or without the GuaM8HDV riboswitch in their 3′-UTR were then applied to the lung of mice by intratracheal administration and fibrosis manifestation was assessed 3 weeks after application. Anticipating that the riboswitch-harboring construct (due to the minimally constitutive riboswitch activity in vitro (Figure 1b) might show slightly lower expression than the control construct, we applied both an equal and a 1.5-fold higher dose of the 3′GuaM8HDV vector as compared to the TGFβ1 control vector.
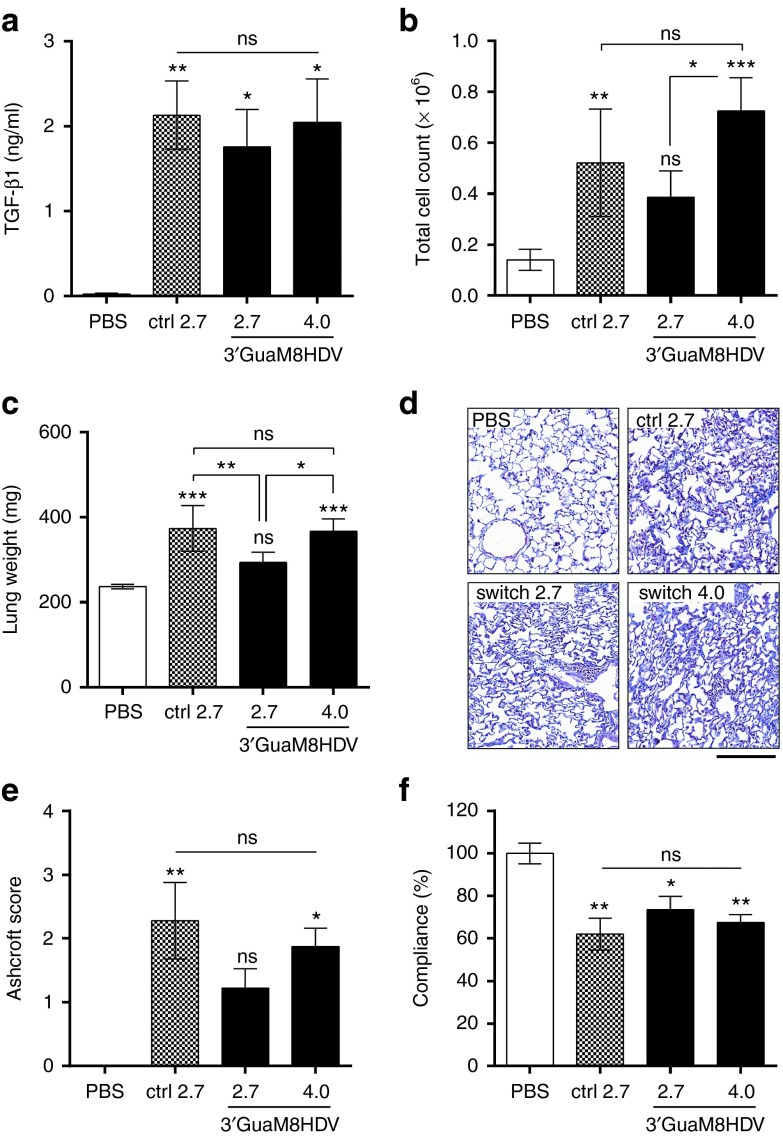
Our results demonstrate that TGFβ1 protein levels in lung lavage samples were slightly higher in mice that received the conventional AAV-TGFβ1 vector (ctrl) as compared to the same dose of the riboswitch-harboring vector (Figure 5a). However, this difference could be fully compensated by applying a 1.5-fold higher dose of the 3′-GuaM8HDV vector. The differences in TGFβ1 levels also translated into differences in immune cell influx, where both the control and high-dose riboswitch group showed higher levels than the low dose switch vector (Figure 5b). As expected, the cellular composition of the immune cell populations was similar in all cases, with monocytes being the major cell type followed by neutrophils and lymphocytes (Supplementary Figure S5). Directly evident of increased extracellular matrix (ECM) deposition, which is a hallmark of tissue remodeling in pulmonary fibrosis, an increase in lung weight was observed in all TGFβ1-overexpressing animals (Figure 5c). Again, the increase triggered by the TGFβ1 control vector (1.55-fold) was significantly higher than that induced by the switch-harboring construct (1.24-fold). However, by using a 1.5-fold higher dose of the switch vector, these differences could be completely abolished (Figure 5c). To assess the structural changes associated with fibrotic tissue remodeling, lung tissue slices were analyzed by Masson-trichrome staining (Figure 5d) and subsequent pathological scoring (“Ashcroft score”) of disease severity (Figure 5e), which further validated the observed differences and similarities among the treatment groups. Finally, as the ultimate consequence of fibrotic lung tissue scarring, impairment of lung function was observed in all TGFβ1 treated animals (Figure 5f). While the decrease in lung compliance was higher in TGFβ1 control vector treated animals (−38.0%) than in animals receiving an equal dose of riboswitch-TGFβ1 vector (−26.4%), no statistically significant differences were observed when a 1.5-fold higher dose of riboswitch-vector (−32%) was applied. Taken together, the 3′-GuaM8HDV-harboring AAV vector successfully induced TGFβ1 expression in the lung of mice, which triggered expected pathological changes at similar efficiency as the riboswitch-free control vector.
Figure 5.
3′-GuaM8HDV-harboring AAV vectors are functional in vivo. Mice received a single application of either 2.7 × 1011 vg of a riboswitch-free AAV6.2-CMV-TGFβ1 control vector, the same dose or a 1.5-fold higher dose (4.0 × 1011 vg) of the 3′-GuaM8HDV-containing AAV6.2-CMV-TGFβ1 vector or PBS via intratracheal application. Analyses were conducted 21 days after application. (a) ELISA measurement of TGFβ1 protein levels in bronchoalveolar lavage (BAL) samples. (b) Total immune cell counts measured in BAL samples. (c) Wet lung weight. (d) Masson-trichrome staining of formalin-fixed paraffin-embedded lung tissue sections and (e) corresponding Ashcroft score. (f) Lung function (lung compliance) analysis. n = 5 animals per group, mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, relative to PBS-treated animals or as indicated. Bar = 200 µm.
Discussion
Our study demonstrates that the GuaM8HDV riboswitch efficiently attenuates expression of producer cell performance-impairing transgenes during AAV vector production in HEK-293 cells, which translates into significantly increased AAV yields. Importantly, in the absence of ligand, GuaM8HDV-harboring AAV vectors preserved the ability to express the transgene at high levels, which was demonstrated by AAV-mediated TGFβ1 expression that induced expected fibrotic changes in the lung of mice. Thus, this study is the first to provide an application-ready biotechnological system based on riboswitch-mediated expression control.
While incorporation of the GuaM8HDV riboswitch into the 3′-UTR of an AAV vector allowed for efficient guanine dose-dependent regulation of transgene expression, which is in line with the original publication,22 5′-positioning of the same riboswitch resulted in a strong decrease of transgene expression independent of guanine addition. This is most likely due to the fact that the GuaM8HDV switch contains four artificial start codons22 that reduce translation from the proper GFP start codon following further downstream. Previous work suggests that removal of these start codons improves switching performance at the 5′ position.13 Notably, in our study the exact position of the switch has not been optimized, which might explain why the reported ON/OFF-ratio of up to 29.522 could not be reproduced in our study. In fact, the maximum ON/OFF-ratio in our GFP experiments only was 4.4. Moreover, although well-established in literature,13,31 the theophylline-responsive P1-F5 hammerhead ribozyme did not show any switching behavior when integrated in our AAV vector system (data not shown). This further supports the notion that position-based effects might be crucial for structural integrity and optimal switch performance, which was previously demonstrated by Yen et al.18 In this regard, the exceptional structural stability of HDV-derived aptazymes22 might offer a particular advantage for their use in technological applications such as the one described herein. Besides optimization of the position, we are currently exploring whether multiple integration of GuaM8HDV further increases the off-rate, as previously reported for other aptazymes.31,32 Moreover, results obtained with constitutively active ribozymes show that expression can be changed by >100-fold,31 demonstrating the principle power of riboswitches, which might be achieved with novel screening strategies and further structural engineering in the future. Nevertheless, our current results suggest that the degree of transgene suppression achieved with our approach can be sufficient to fully block functional outcomes such as toxicity and other cell integrity-impairing effects. This holds particularly true for only moderately toxic genes or when toxicity is specific for overexpression, whereas highly potent toxic transgenes might require the use of alternative gene regulation systems. In fact, while BAX seemed to exert highly potent proapoptotic effects which could not be fully blocked by the riboswitch, apoptosis induced by the less potent TNFα could be fully reversed by switch activation. Notably, this phenomenon also translated into AAV yields, where AAV-BAX (despite a drastic increase of up to 23-fold) did not fully reach normal levels (7% of the titer obtained with an AAV-GFP control vector), but AAV-TNFα titers could be boosted to the normal yield range. Similarly, TGFβ1 signaling was effectively blocked by the riboswitch, resulting in an excellent AAV yield (2.1 × 1013 vg from 40 transfected 15 cm cell culture dishes) that resembles the titers obtained for AAV-GFP. Moreover, also AAV-LOXL2 titers could be increased to the AAV-GFP yield range by suppressing LOXL2 expression during AAV production. However, in contrast to the previously mentioned genes, the reasons for the low initial AAV-LOXL2 yields remain unclear, since LOXL2 has neither been associated with cytotoxicity nor antiproliferative effects so far. These results nicely illustrate the value of the riboswitch approach, especially for sparsely characterized genes—a very commonly observed situation, particularly in basic research. Moreover, it also cannot be excluded that well-characterized, therapeutically relevant genes might unexpectedly turn out to compromise viral vector production for clinical studies.
Improving viral vector yields during production is obviously only valuable, if riboswitch-harboring constructs preserve functionality, i.e., the capacity to express the transgene at high levels as long as no ligand is present. As shown by our in vitro experiments, GuaM8HDV-harboring constructs retained about 90% protein expression as compared to the riboswitch-free controls, representing an excellent value which is in accordance with previous findings for this aptazyme.22 More importantly, our preclinical mouse study demonstrates that the 3′-GuaM8HDV-harboring AAV-TGFβ1 vector successfully induced expected pathological changes upon pulmonary expression of TGFβ1, a cytokine that has been previously shown to induce pulmonary fibrosis in a dose-dependent manner.7,33,34 However, a 1.5-fold higher AAV dose had to be used in order to fully reach the functional outcome observed with the riboswitch-free construct. Reasons for the slightly lower initial expression might include ligand-independent basal activity of the riboswitch, which might be more pronounced in vivo as compared to the only slight effects observed in vitro, and minor riboswitch activation by endogenously present guanine or cellular disturbing factors such as RNA-binding proteins.35 Nevertheless, even a 1.5-fold higher AAV dose can be easily tolerated given that the average riboswitch-mediated increase in viral vector yield of 12 test-productions (Figure 3) was 5.4-fold. Moreover, for transgenes where particularly low AAV yields are obtained, the riboswitch approach might make it possible to conduct in vivo studies in the first place. Finally, we did not observe any differences between the normal dose of AAV-TGFβ1 and the 1.5-fold higher dose of the riboswitch-containing TGFβ1 vector with regard to the magnitude or cellular composition of immune cell influx following vector application. Still, it cannot be ruled out that, in some instances, a 1.5-fold vector dose could lead to enhanced cellular or humoral immune responses, higher transgene expression in off-target cells or other detrimental effects.
Besides the use of an inducible expression system, transgene expression during viral vector production could be avoided by using tissue-specific promoters that are inactive in HEK-293 producer cells. In fact, respective promoters have been identified and successfully applied to transcriptionally target viral vector-mediated transgene expression to specific tissues and cell types, such as the liver/hepatocytes (LP1 promoter36), heart/cardiomyocytes (cTnT promoter37), CNS/neurons (synapsin-1 promoter38), lung/clara cells (CC10 promoter39), and adipose tissue/adipocytes (Adiponectin promoter40), to name a few. However, strong ubiquitous expression as mediated by the CMV promoter has been used in the majority of viral vector-based studies and is desired in many cases, for example, for the broad transduction of whole organs consisting of many different cell types like the lung and the brain, for studying gene function in multiple tissues of interest, or in therapeutic applications, where transcriptional targeting is difficult or impossible, for instance, oncolytic therapy. Another option to circumvent transgene toxicity-associated issues in viral vector production would be to use a nonmammalian production platform, such as the well-described AAV production system in SF9 insect cells,41 where most mammalian promoters are inactive. However, given that this system requires the relatively time-consuming preparation of bacmids and baculoviruses prior to actual AAV production, the HEK-293 system remains preferred for most AAV applications in basic research, because of its ease of use.
The riboswitch approach described herein also offers several advantages over alternative methods to regulate transgene expression during viral vector production such as transcription factor- and dimerizer-inducible promoters (e.g., tetracyclin- and rapamycin-dependent systems19,20,21,42,43) or siRNA-mediated gene knockdown. First, aptazymes do not rely on the additional expression of transcription factors or RNA, but can be controlled by simple addition of a small molecule to the culture medium during the routine medium exchange step. Second, due to its RNA-intrinsic mode of action (i.e., mRNA destabilization and inhibition of translation), transgene regulation is independent of protein-protein or protein-DNA interactions. This is a particular advantage given that stoichiometric imbalance between protein and DNA might be responsible for the loss of regulation with inducible promoters at high copy numbers.44 Third, small molecule-mediated regulation allows for immediate regulation that can be simply reversed by replacing the medium. Fourth, after initial cloning of a 3′-GuaM8HDV- and multiple cloning site-harboring AAV standard vector, the system can be rapidly adapted for the regulated expression of virtually any target gene. Fifth, the riboswitch sequence only occupies about 100 nucleotides of plasmid space, which is particularly advantageous for AAV vectors, which have a packaging capacity of only ~4.7 kb of DNA.11
In addition to their application in viral vector production, riboswitches represent highly attractive tools for the control of gene expression in vivo, particularly as a safety switch for viral vector gene therapy. However, to date there are only two studies demonstrating successful transgene regulation by an aptazyme in vivo,18,32 which is likely due to the fact that most aptazymes present today were identified by screening approaches in vitro, in bacteria or Saccharomyces cerevisiae,45 which seems to complicate the transfer to higher-order eukaryotes. However, advanced strategies combining rational design in silico and optimized screening platforms13,46,47,48 should in the future lead to the discovery of novel switches with improved functionality in vivo. Although clinical proof-of-concept is still lacking, the principle of aptazyme-mediated control of viral replication and infectivity has been recently demonstrated in vitro.15 With respect to gene therapy, riboswitches possess two additional advantageous properties: First, due to their RNA-intrinsic mode of action, they are likely to behave nonimmunogenic. Second, riboswitch activity can in principle be engineered to respond to small molecule drugs with preferred pharmacological properties. By altering the aptamer binding domain, several studies have proven this principle, for instance to obtain antibiotic-responsive switches.47,49 Furthermore, a long term goal could be the development of aptazymes that specifically sense disease biomarkers such as proteins, small RNAs50 or chemical metabolites and respond by inducing/blocking therapeutic gene expression according to current disease status.
In summary, our study demonstrates applicability of the artificial guanine-responsive GuaM8HDV riboswitch for the transient suppression of toxic or cellular integrity-impairing transgenes during viral vector production, which led to up to 23-fold higher AAV yields after a single addition of guanine to the culture medium. Importantly, GuaM8HDV-harboring AAV vectors preserved their ability to express the transgene in the absence of ligand in vivo, as demonstrated in a mouse model of AAV-TGFβ1-induced pulmonary fibrosis. We thus propose riboswitches as innovative tools enabling high-titer viral vector production in mammalian cell culture systems. Moreover, such riboswitch vectors might be ultimately used to temporally control the expression of therapeutic genes in vivo.
Materials and Methods
Cell culture. HEK-293h cells were cultured in DMEM medium (#31966, Life Technologies, Darmstadt, Germany) supplemented with 10% FCS (#10500, Life Technologies, Darmstadt, Germany) at 37 °C and 5% CO2.
Plasmid constructs. An eGFP containing plasmid harboring the GuaM8HDV sequence (flanked by Bsp119I and EcoRI restriction sites) 49 nt upstream of the eGFP start codon and 12 nt downstream of the stop codon (flanked by HindIII and BglII sites) was synthesized (Life technologies, Karlsruhe, Germany). For construction of the 5′-GuaM8HDV construct, the eGFP cassette was cloned into a pAAV-MCS vector using Bsp119I and HindIII. The pAAV-MCS vector contains AAV2 ITRs flanking a CMV-promoter-multiple cloning site-SV40 polyA cassette. For the 3′-GuaM8HDV construct, the eGFP cassette was cloned using EcoRI and BglII. For the 5′3′-GuaM8HDV construct, Bsp119I and BglII were used. As a control, eGFP was cloned into pAAV-MCS using EcoRI and HindIII. For the functional pAAV-eGFP-3′GuaM8HDV plasmid DNA sequence, see Supplementary Materials and Methods. The murine TGFβ1 and LOXL2-V5, and the human BAX and TNF cDNAs were synthesized (Life Technologies, Karlsruhe, Germany) and used to replace the eGFP sequence in either the control or 3′-GuaM8HDV construct via BamHI/Bsp119I + HindIII.
Transient transfection of HEK-293 cells. The day before transfection, HEK-293h cells were seeded in multiwell plates to reach ~70% confluence on the day of transfection. For transfection, 0.5 µg DNA per cm2 of culture area were mixed with 1/10 culture volume of 300 mmol/l CaCl2 and added drop by drop to an equal volume of 2× HBS buffer (50 mmol/l HEPES, 280 mmol/l NaCl, 1.5 mmol/l Na2HPO4). After incubation for 2 minutes at room temperature, the mix was added to the cells. After 5 hours of incubation, the culture medium was replaced by fresh medium, optionally supplemented with guanine. Guanine (#G11950; Sigma Aldrich, St Louis, MO) was previously dissolved in 0.1 mol/l NaOH at 20 mmol/l and diluted to the desired concentration in cell culture medium.
GFP assay. HEK-293 cells were transfected as described above and incubated for 24 to 72 hours, as specified in the text. At indicated time points, fluorescence microscopic pictures were taken using the IN Cell Analyzer 2000 HCA imaging device (GE Healthcare Life Sciences, Little Chalfont, Buckinghamshire, UK) at ×10 magnification. Cells were then washed with PBS, detached using trypsin-EDTA, washed and resuspended in PBS containing 10% FCS. Cells were analyzed for eGFP fluorescence using a FACSCanto II flow cytometer (BD Biosciences, San Jose, CA), and data were analyzed using FACSDiva software (BD Biosciences). Mean fluorescence intensity values were recorded, the fluorescence of untreated cells was subtracted, and the resulting values were divided by the ratio of GFP-positive cells to compensate for potential differences in transfection efficiency. The corrected fluorescence values of the GFP-control construct without guanine addition was set 100%, and mean fluorescence of the riboswitch construct-transfected samples were calculated accordingly.
Measurement of direct fluorescence. Cells were GFP-transfected as described above, using black multiwell plates. At specified time points, cells were washed with PBS, and fluorescence was measured using a PowerWave HT Microplate Spectrophotometer (BioTek, Winooski, VT) at 488 nm (excitation) and 535 nm (emission). Fluorescence values were normalized as described above.
Immunodetection. Western blotting was conducted using standard methods and following antibodies: polyclonal rabbit anti-human BAS (#a3533; Dako, Carpinteria, CA; 1:1,000), anti-V5 tag antibody (#ab95038; Abcam, Cambridge, UK; 1:1,000), and monoclonal anti-vinculin antibody (#V9131; Sigma-Aldrich, St. Louis, MO; 1:2,000). ELISA measurements were conducted using the human TNF-alpha Quantikine ELISA Kit (#DTA00C; R&D Systems, Minneapolis, MN) and the Mouse/Rat/Porcine/Canine TGF-beta 1 Quantikine ELISA Kit (MB100B; R&D Systems, Minneapolis, MN).
qPCR analysis. Total RNA was isolated using the RNeasy Mini Kit (#74104; Qiagen, Hilden, Germany) and reversely transcribed to cDNA using the High-Capacity cDNA Reverse Transcription Kit (#4368814, Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. qPCR reactions were set up using the QuantiFast Probe RT-PCR Kit (#204456, Qiagen, Hilden, Germany) and gene-specific primer/probe sets for the human SERPINE1 and POLR2A gene, respectively. Gene expression analysis was carried out using the 7900HT Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA) and associated SDS 2.4 software. SERPINE1 (PAI-1) gene expression was normalized (ΔΔCt method) to the expression of POLR2A (RNA polymerase II) which served as a housekeeping gene.
Cytotoxicity assay. Cytotoxicity was assessed by measuring lactate dehydrogenase (LDH) release in the cell supernatant using the CytoTox-ONE Homogeneous Membrane Integrity Assay (#G7890; Promega, Madison, WI) according to the manufacturer's instructions. LDH release measured using untreated cells served as negative control and was set to 0%. Triton-lysed untransfected HEK-293 cells served as a positive control and were set to 100%.
AAV production. HEK-293 cells were transfected as described above using 0.5 µg total plasmid DNA per cm2 of culture area. For the cotransfection, equimolar amounts of plasmids were used. For production of AAV6.2 and AAV9 vectors, the AAV2 cap sequence in pAAV-RC (Agilent Technologies, Waldbronn, Germany) was replaced by that of AAV6.2 (EU368910.1) or AAV9 (AX753250) which had been synthesized at Life Technologies (Karlsruhe, Germany). The resulting pAAV-AAV6.2 or pAAV-AAV9 plasmids were co-transfected with pHELPER (Agilent Technologies, Waldbronn, Germany) and a plasmid harboring the transgene cassette flanked by AAV2 ITRs. For AAV8 production, pDP8rs (Plasmid Factory, Heidelberg, Germany) was used instead of the pAAV plasmid and pHELPER. Five hours after transfection, the medium was replaced with fresh medium (optionally supplemented with guanine), and the cells were incubated for 72 hours. For cell detachment, EDTA was added to a final concentration of 6.25 mmol/l. Cells were collected by centrifugation and resuspended in 50 mmol/l Tris, 150 mmol/l NaCl, 2 mmol/l MgCl2, pH8.5. Cells were then lysed by three freeze/thaw cycles using liquid nitrogen and a 37 °C water bath. For small scale (6-well microplates) productions, the lysate was incubated with 250 U/ml benzonase (#70746, Merck Millipore, Darmstadt, Germany) for 24 hours at 37 °C to deplete residual plasmid DNA and centrifuged at 2,500xg for 10 minutes to remove cell debris. NaOH was then added to 100 mol/l final concentration, and the mix was incubated at 70 °C for 30 minutes which both inactivates benzonase and lyses the AAV particles to release viral DNA. HCl was then added to neutralize the solution for qPCR analysis (see below). For purification of AAVs produced at lab scale, the cell lysate was benzonase digested (100 U/ml, 1 hour at 37 °C) and centrifuged at 2500xg for 10 minutes to remove cell debris. The supernatant was purified using a discontinuous iodixanol-gradient (#D1556, Sigma-Aldrich, St. Louis, MO) as described previously,51 rebuffered, and concentrated using Amicon-15 MWCO 100,000 (Merck Millipore) devices, sterile filtered, and stored in PBS + 10% glycerol at −80 °C.
AAV genomic titer determination. For the determination of genomic titers of purified AAV stocks, viral DNA was isolated using the Viral Xpress DNA/RNA Extraction Reagent (#3095, Merck Millipore, Darmstadt, Germany) and quantified by subsequent qPCR analysis using the QuantiFast Probe RT-PCR Kit (#204456, Qiagen) and primers specific for the CMV-promoter sequence (forward: 5′-CGTCAATGGGTGGAGTATTTACG-3′, reverse: 5′- AGGTCATGTACTGGGCATAATGC-3′, probe: 5′- AGTACATCAAGTGTATCATATGCCAAGTACGCCC-3′). A CMV-promoter containing plasmid was routinely used to prepare a standard curve for quantification. For the determination of AAV titers in cell lysate, AAV cell lysate was processed as described above, and samples were diluted 1:100 in water prior to qPCR analysis.
Animals. Male C57Bl/6J mice (9–12 weeks of age) were purchased from Charles River Laboratories (Sulzfeld, Germany). The experiment was conducted in accordance with the German law on animal welfare (TierSchG) and has been approved by the Regierungspräsidium Tübingen (approval no. 12–030).
AAV in vivo experiment. Under light anesthesia (3–4% isoflurane; Abbott, Wiesbaden, Germany), mice were administered 50 µl of AAV suspension or PBS into the trachea. AAV concentrations were 2.7 × 1011 or 4.0 × 1011 vg, as specified in the respective text sections and figures. Twenty-one days after application, mice were anesthetized and subjected to lung function measurement (see Supplementary Materials and Methods) before being killed by a pentobarbital overdose (800 mg/kg, i.p.). After determining lung weight, lungs were flushed ex vivo two times with 0.7 ml PBS to obtain bronchoalveolar lavage (BAL) fluid. BAL immune cell counts were measured using the Sysmex XT1800 iVet cell analyzer (Sysmex, Norderstedt, Germany). Finally, lungs were formalin fixed and paraffin embedded for histological analysis (see Supplementary Materials and Methods for detailed procedure).
Statistical analysis. Statistical analysis was performed using GraphPad Prism 6.01 (GraphPad, La Jolla, CA). Differences between test groups were assessed by one-way ANOVA and Tukey's posttest for multiple comparisons, except from Figure 1d, where a two-way ANOVA was used and Supplementary Figure S4, where a t-test was applied.
SUPPLEMENTARY MATERIAL Figure S1. LDH-release following treatment with increasing guanine concentrations. Figure S2. GuaM8HDV-mediated attenuation of GFP expression over time. Figure S3. Ponceau S staining of the western blot membrane as a total protein loading control for the LOXL2 western blot shown in Figure 2g. Figure S4. AAV6.2-CMV-TGFβ1 vector yields obtained using a conventional or 3′-GuaM8HDV-harboring transgene cassette. Figure S5. Differential immune cell counts in bronchoalveolar lavage (BAL) samples. Materials and Methods
Acknowledgments
This work was supported by Boehringer Ingelheim Pharma GmbH & Co. KG, Biberach, Germany. B.S., T.L., F.G., and S.K. are employees of Boehringer Ingelheim Pharma GmbH & Co. KG, Biberach, Germany. The authors thank Eva Martin, Felix Miller, Janine Beier and Helene Lichius (Boehringer Ingelheim) for excellent technical assistance. Further, the authors are indebted to Birgit Stierstorfer (Boehringer Ingelheim) who carried out histological analysis.
Supplementary Material
References
- Kotterman, MA and Schaffer, DV (2014). Engineering adeno-associated viruses for clinical gene therapy. Nat Rev Genet 15: 445–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieger, JC and Samulski, RJ (2012). Adeno-associated virus vectorology, manufacturing, and clinical applications. Methods Enzymol 507: 229–254. [DOI] [PubMed] [Google Scholar]
- Mingozzi, F and High, KA (2011). Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat Rev Genet 12: 341–355. [DOI] [PubMed] [Google Scholar]
- Roche-Molina, M, Sanz-Rosa, D, Cruz, FM, García-Prieto, J, López, S, Abia, R et al. (2015). Induction of sustained hypercholesterolemia by single adeno-associated virus-mediated gene transfer of mutant hPCSK9. Arterioscler Thromb Vasc Biol 35: 50–59. [DOI] [PubMed] [Google Scholar]
- Werfel, S, Jungmann, A, Lehmann, L, Ksienzyk, J, Bekeredjian, R, Kaya, Z et al. (2014). Rapid and highly efficient inducible cardiac gene knockout in adult mice using AAV-mediated expression of Cre recombinase. Cardiovasc Res 104: 15–23. [DOI] [PubMed] [Google Scholar]
- Chung, CY, Koprich, JB, Siddiqi, H and Isacson, O (2009). Dynamic changes in presynaptic and axonal transport proteins combined with striatal neuroinflammation precede dopaminergic neuronal loss in a rat model of AAV alpha-synucleinopathy. J Neurosci 29: 3365–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel, B, Duechs, MJ, Schmid, R, Stierstorfer, BE, Bucher, H, Quast, K et al. (2015). Modeling pulmonary disease pathways using recombinant adeno-associated virus 6.2. Am J Respir Cell Mol Biol (epub ahead of print). [DOI] [PubMed]
- Grimm, D (2002). Production methods for gene transfer vectors based on adeno-associated virus serotypes. Methods 28: 146–157. [DOI] [PubMed] [Google Scholar]
- Myers, MW and Carter, BJ (1980). Assembly of adeno-associated virus. Virology 102: 71–82. [DOI] [PubMed] [Google Scholar]
- Naumer, M, Sonntag, F, Schmidt, K, Nieto, K, Panke, C, Davey, NE et al. (2012). Properties of the adeno-associated virus assembly-activating protein. J Virol 86: 13038–13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, JY, Fan, PD and Frizzell, RA (1996). Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum Gene Ther 7: 2101–2112. [DOI] [PubMed] [Google Scholar]
- Tang, J and Breaker, RR (1997). Rational design of allosteric ribozymes. Chem Biol 4: 453–459. [DOI] [PubMed] [Google Scholar]
- Ausländer, S, Ketzer, P and Hartig, JS (2010). A ligand-dependent hammerhead ribozyme switch for controlling mammalian gene expression. Mol Biosyst 6: 807–814. [DOI] [PubMed] [Google Scholar]
- Frommer, J, Appel, B and Müller, S (2015). Ribozymes that can be regulated by external stimuli. Curr Opin Biotechnol 31: 35–41. [DOI] [PubMed] [Google Scholar]
- Ketzer, P, Kaufmann, JK, Engelhardt, S, Bossow, S, von Kalle, C, Hartig, JS et al. (2014). Artificial riboswitches for gene expression and replication control of DNA and RNA viruses. Proc Natl Acad Sci USA 111: E554–E562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link, KH and Breaker, RR (2009). Engineering ligand-responsive gene-control elements: lessons learned from natural riboswitches. Gene Ther 16: 1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaux, S and Van Hoof, A (2006). Yeast transcripts cleaved by an internal ribozyme provide new insight into the role of the cap and poly(A) tail in translation and mRNA decay. RNA 12: 1323–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen, L, Svendsen, J, Lee, JS, Gray, JT, Magnier, M, Baba, T et al. (2004). Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature 431: 471–476. [DOI] [PubMed] [Google Scholar]
- Baron, U and Bujard, H (2000). Tet repressor-based system for regulated gene expression in eukaryotic cells: principles and advances. Methods Enzymol 327: 401–421. [DOI] [PubMed] [Google Scholar]
- Toniatti, C, Bujard, H, Cortese, R and Ciliberto, G (2004). Gene therapy progress and prospects: transcription regulatory systems. Gene Ther 11: 649–657. [DOI] [PubMed] [Google Scholar]
- Clackson, T (1997). Controlling mammalian gene expression with small molecules. Curr Opin Chem Biol 1: 210–218. [DOI] [PubMed] [Google Scholar]
- Nomura, Y, Zhou, L, Miu, A and Yokobayashi, Y (2013). Controlling mammalian gene expression by allosteric hepatitis delta virus ribozymes. ACS Synth Biol 2: 684–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal, M, Boese, B, Barrick, JE, Winkler, WC and Breaker, RR (2003). Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113: 577–586. [DOI] [PubMed] [Google Scholar]
- Wu, HN, Lin, YJ, Lin, FP, Makino, S, Chang, MF and Lai, MM (1989). Human hepatitis delta virus RNA subfragments contain an autocleavage activity. Proc Natl Acad Sci USA 86: 1831–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault, TT and Manon, S (2011). Bax: addressed to kill. Biochimie 93: 1379–1391. [DOI] [PubMed] [Google Scholar]
- Chen, G and Goeddel, DV (2002). TNF-R1 signaling: a beautiful pathway. Science 296: 1634–1635. [DOI] [PubMed] [Google Scholar]
- Shi, Y and Massagué, J (2003). Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113: 685–700. [DOI] [PubMed] [Google Scholar]
- Keski-Oja, J, Raghow, R, Sawdey, M, Loskutoff, DJ, Postlethwaite, AE, Kang, AH et al. (1988). Regulation of mRNAs for type-1 plasminogen activator inhibitor, fibronectin, and type I procollagen by transforming growth factor-beta. Divergent responses in lung fibroblasts and carcinoma cells. J Biol Chem 263: 3111–3115. [PubMed] [Google Scholar]
- Limberis, MP, Vandenberghe, LH, Zhang, L, Pickles, RJ and Wilson, JM (2009). Transduction efficiencies of novel AAV vectors in mouse airway epithelium in vivo and human ciliated airway epithelium in vitro. Mol Ther 17: 294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zincarelli, C, Soltys, S, Rengo, G and Rabinowitz, JE (2008). Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther 16: 1073–1080. [DOI] [PubMed] [Google Scholar]
- Ketzer, P, Haas, SF, Engelhardt, S, Hartig, JS and Nettelbeck, DM (2012). Synthetic riboswitches for external regulation of genes transferred by replication-deficient and oncolytic adenoviruses. Nucleic Acids Res 40: e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, YY, Jensen, MC and Smolke, CD (2010). Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc Natl Acad Sci USA 107: 8531–8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sime, PJ, Xing, Z, Graham, FL, Csaky, KG and Gauldie, J (1997). Adenovector-mediated gene transfer of active transforming growth factor-beta1 induces prolonged severe fibrosis in rat lung. J Clin Invest 100: 768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ask, K, Labiris, R, Farkas, L, Moeller, A, Froese, A, Farncombe, T et al. (2008). Comparison between conventional and “clinical” assessment of experimental lung fibrosis. J Transl Med 6: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groher, F and Suess, B (2014). Synthetic riboswitches - a tool comes of age. Biochim Biophys Acta 1839: 964–973. [DOI] [PubMed] [Google Scholar]
- Graham, T, McIntosh, J, Work, LM, Nathwani, A and Baker, AH (2008). Performance of AAV8 vectors expressing human factor IX from a hepatic-selective promoter following intravenous injection into rats. Genet Vaccines Ther 6: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad, KM, Xu, Y, Yang, Z, Acton, ST and French, BA (2011). Robust cardiomyocyte-specific gene expression following systemic injection of AAV: in vivo gene delivery follows a Poisson distribution. Gene Ther 18: 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kügler, S, Lingor, P, Schöll, U, Zolotukhin, S and Bähr, M (2003). Differential transgene expression in brain cells in vivo and in vitro from AAV-2 vectors with small transcriptional control units. Virology 311: 89–95. [DOI] [PubMed] [Google Scholar]
- Auricchio, A, O'Connor, E, Weiner, D, Gao, GP, Hildinger, M, Wang, L et al. (2002). Noninvasive gene transfer to the lung for systemic delivery of therapeutic proteins. J Clin Invest 110: 499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig-Schmidt, S, Geiger, M, Luippold, G, Birk, G, Mennerich, D, Neubauer, H et al. (2014). Gene delivery to adipose tissue using transcriptionally targeted rAAV8 vectors. PLoS One 9: e116288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urabe, M, Ding, C and Kotin, RM (2002). Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum Gene Ther 13: 1935–1943. [DOI] [PubMed] [Google Scholar]
- Sanftner, LM, Rivera, VM, Suzuki, BM, Feng, L, Berk, L, Zhou, S et al. (2006). Dimerizer regulation of AADC expression and behavioral response in AAV-transduced 6-OHDA lesioned rats. Mol Ther 13: 167–174. [DOI] [PubMed] [Google Scholar]
- Chen, SJ, Johnston, J, Sandhu, A, Bish, LT, Hovhannisyan, R, Jno-Charles, O et al. (2013). Enhancing the utility of adeno-associated virus gene transfer through inducible tissue-specific expression. Hum Gene Ther Methods 24: 270–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fechner, H, Wang, X, Srour, M, Siemetzki, U, Seltmann, H, Sutter, AP et al. (2003). A novel tetracycline-controlled transactivator-transrepressor system enables external control of oncolytic adenovirus replication. Gene Ther 10: 1680–1690. [DOI] [PubMed] [Google Scholar]
- Berens, C and Suess, B (2015). Riboswitch engineering - making the all-important second and third steps. Curr Opin Biotechnol 31: 10–15. [DOI] [PubMed] [Google Scholar]
- Wachsmuth, M, Findeiß, S, Weissheimer, N, Stadler, PF and Mörl, M (2013). De novo design of a synthetic riboswitch that regulates transcription termination. Nucleic Acids Res 41: 2541–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauser, B, Atanasov, J, Siewert, LK and Hartig, JS (2015). Ribozyme-based aminoglycoside switches of gene expression engineered by genetic selection in S. cerevisiae. ACS Synth Biol 4: 516–525. [DOI] [PubMed] [Google Scholar]
- Wittmann, A and Suess, B (2011). Selection of tetracycline inducible self-cleaving ribozymes as synthetic devices for gene regulation in yeast. Mol Biosyst 7: 2419–2427. [DOI] [PubMed] [Google Scholar]
- Beilstein, K, Wittmann, A, Grez, M and Suess, B (2015). Conditional control of mammalian gene expression by tetracycline-dependent hammerhead ribozymes. ACS Synth Biol 4: 526–534. [DOI] [PubMed] [Google Scholar]
- Klauser, B and Hartig, JS (2013). An engineered small RNA-mediated genetic switch based on a ribozyme expression platform. Nucleic Acids Res 41: 5542–5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock, M, Alvira, M, Vandenberghe, LH, Samanta, A, Toelen, J, Debyser, Z et al. (2010). Rapid, simple, and versatile manufacturing of recombinant adeno-associated viral vectors at scale. Hum Gene Ther 21: 1259–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.