Abstract
We have designed mitochondrially targeted transcription activator-like effector nucleases or mitoTALENs to cleave specific sequences in the mitochondrial DNA (mtDNA) with the goal of eliminating mtDNA carrying pathogenic point mutations. To test the generality of the approach, we designed mitoTALENs to target two relatively common pathogenic mtDNA point mutations associated with mitochondrial diseases: the m.8344A>G tRNALys gene mutation associated with myoclonic epilepsy with ragged red fibers (MERRF) and the m.13513G>A ND5 mutation associated with MELAS/Leigh syndrome. Transmitochondrial cybrid cells harbouring the respective heteroplasmic mtDNA mutations were transfected with the respective mitoTALEN and analyzed after different time periods. MitoTALENs efficiently reduced the levels of the targeted pathogenic mtDNAs in the respective cell lines. Functional assays showed that cells with heteroplasmic mutant mtDNA were able to recover respiratory capacity and oxidative phosphorylation enzymes activity after transfection with the mitoTALEN. To improve the design in the context of the low complexity of mtDNA, we designed shorter versions of the mitoTALEN specific for the MERRF m.8344A>G mutation. These shorter mitoTALENs also eliminated the mutant mtDNA. These reductions in size will improve our ability to package these large sequences into viral vectors, bringing the use of these genetic tools closer to clinical trials.
Introduction
Mitochondrial diseases impairing oxidative phosphorylation (OXPHOS) can affect multiple organs or single ones and can be caused by mutations in nuclear genes or in the mitochondrial DNA (mtDNA). Mutations in mtDNA are commonly in a heteroplasmic state, where mutant mtDNA co-exists with wild type. There are ~1,000 mtDNA molecules in a cell, and the wild-type mtDNA can compensate for the presence of mutant mtDNA, up to threshold levels, which are usually relatively high, 70–95%.1,2,3 This “recessive” feature of the mutant mtDNA means that by simply reducing the relative levels of mutant mitochondrial genomes, biochemical defects can be reversed.
TALENs are engineered nucleases based on the TALE DNA-binding domain from Xanthomonas fused to a FokI type-II nuclease domain. Because the FokI domains need to dimerize to cleave DNA, two TALEN monomers are required to bind adjacent recognition sequences on opposing sense/antisense strands of DNA.4,5 The DNA-binding specificity of TALE domains is given by tandem copies of 34 amino acid repeats that each preferentially bind one of the four DNA bases depending on the amino acids at positions 12 and 13 of the repeat. The nucleotide affinity of these repeat variable di-residues (RVDs) is well understood, providing the basis to engineer novel specificities.6,7 Because the last repeat contains only 20 amino acids (instead of 34), it is referred to as a 0.5 repeat.
Commonly, 15.5–22.5 TALE repeats are used per monomer to avoid off-target cleavage.5,8 Although TALEN can be very specific for genetic loci, this specificity is not perfect and off-site cleavages, in large and complex genomes, are a concern.9 Moreover, as mentioned above, the high specificity of TALENs is in great part due to the combination of both sequence specificity of TALE binding and the positional requirements of FokI cleavage. In this context, the ability of TALENs to differentiate targets with only single nucleotide differences could be difficult.
We have previously shown that for at least one point mutation (m.14459A>G), mitochondrial targeted TALEN (mitoTALEN) was able to reduce the mutation load.10 This is compatible with our previous observation that the mutant mtDNA undergoing a double-strand break by restriction endonucleases is followed by rapid degradation.11 The mtDNA copy number control ensures that the residual (wild type) intact mitochondrial genomes repopulate the cell.12
To test whether expression of mitoTALEN could be a general approach for the reduction of the levels of mtDNA point mutations, we developed mitoTALENs for two relatively common pathogenic mtDNA mutations. One is the m.8344A>G mutation in the tRNA lysine gene that accounts for 80 to 90% of cases presenting with myoclonus epilepsy with ragged red fibers (MERRF) syndrome13,14 and the other is the m.13513G>A mutation in the ND5 gene associated with MELAS/Leigh syndrome.15,16,17 We have also explored the reduction of the size of the mitoTALEN monomers to optimize their use for gene therapy.
Results
Designing mitoTALENs
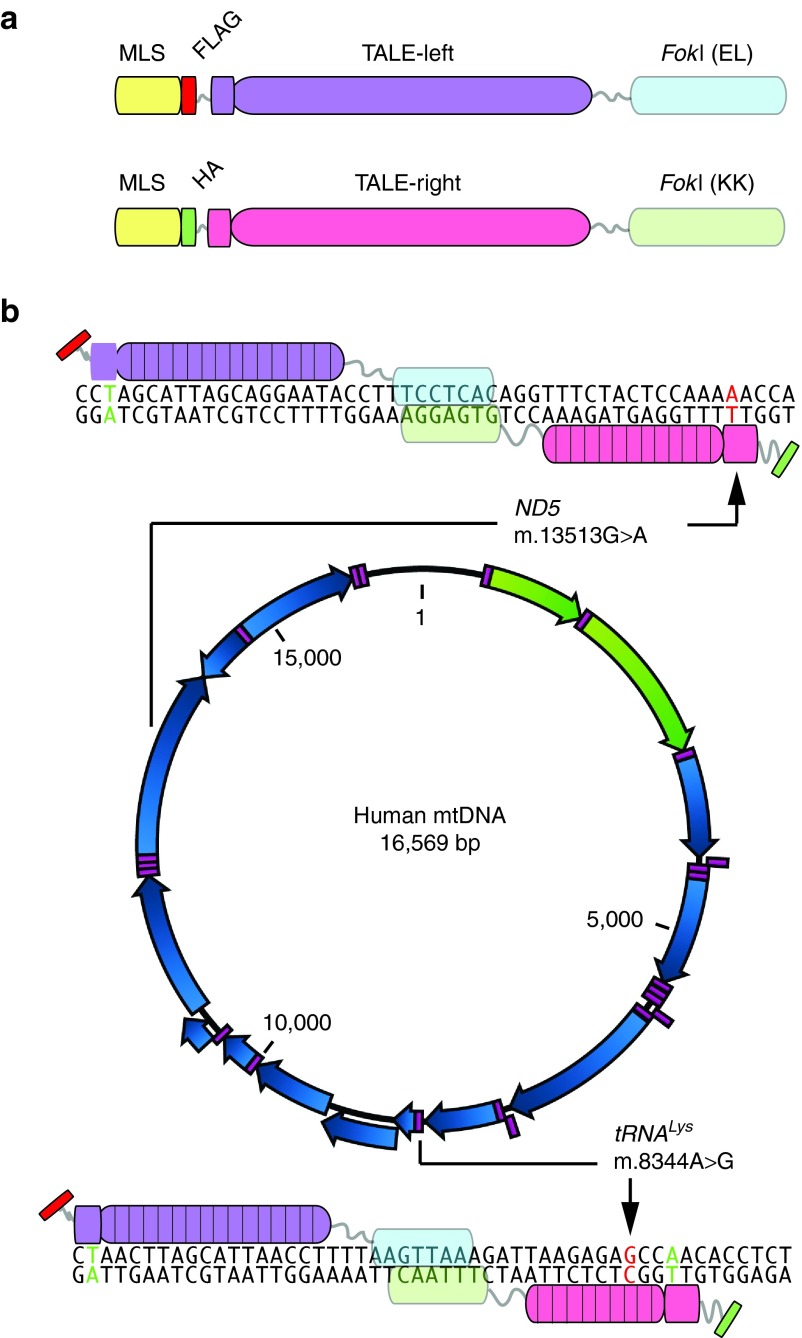
We developed TALENs against two distinct mtDNA point mutations at positions m.8344A>G and m.13513G>A (Figure 1). A required element for the binding of the N-terminus of most TALEN monomers is a T at position 0 in the DNA recognition sequence, immediately upstream (5′) to the region recognized by the RVDs.4,7 MtDNA has a well-recognized transition bias for naturally occurring point mutations (>90% transitions versus transversions18). For C>T and G>A “gain of T” transition mutations the requirement for T0 can be exploited to develop TALENs that can differentially recognize and cleave these mutations, as we have demonstrated in one previous case.10 For the m.13513G>A mutation in ND5, we further explored the utility of this approach, designing two different TALENs where the differentiating monomer binding site included a T0 of the antisense strand at the G>A mutation (Figure 1b and Supplementary Figure S1). The two TALENs differed from each other by the numbers of RVD repeats for the monomer binding the mutation, one having 9.5 RVDs and the other 12.5 RVDs.
Figure 1.
Development of mitoTALEN for two pathogenic mtDNA mutations. (a) The structure of the mitoTALEN monomers is illustrated. They contain a mitochondrial localization signal (MLS), an immunological tag (FLAG or HA) and the TALE DNA-binding domain. The latter is fused to a FokI that works as a heterodimer (EL only dimerizes with KK). (b) Using the established TALE code, we designed TALE binding domains against mtDNA regions harboring two pathogenic mutations. The first region was at position m.8344A>G in the tRNALys gene, which is associated with the MERRF syndrome. The second region, surrounding position m.13513G>A in the ND5 gene is associated with MELAS/Leigh syndrome. We have designed two pairs for each region. The figure shows only one for each, but the second pair is described in Supplementary Figure S1. The base pair at position T0 is shown in green color and the mutated position in red (when they coincide they are labeled red). Arrows indicate the mutated base pair.
Unfortunately, the m.8344A>G mutation does not allow for the use of T0 design strategy as it is a T>C, A>G mutation, i.e. “loss of T.” In the sense strand, the transition A>G posed a further challenge for the design, as the conventional RVD for binding G is “NN” which cannot discriminate between A and G effectively,4,7 which constitute the wild type and mutant alleles for this mutation. However, in the antisense strand, the mutant gains a C, which can be recognized more specifically by an “HD” RVD that does not effectively bind T (the base present in the wild-type antisense). Therefore, for m.8344A>G, we chose to place the mutated G at position 3 of the antisense monomer, exploiting a “gain of C3” model (Figure 1b). We developed two m.8344A>G TALEN pairs with this general design, the main difference being that the monomer binding the mutated region had either 9.5 RVDs or 15.5 RVDs (Supplementary Figure S1). In both of them we designed the DNA-binding domain to include the mutant G at position C3 of the antisense strand (Figure 1 and Supplementary Figure S1). These TALENs were initially tested in yeast Single-Strand Annealing assays. Because one of the pairs (version A) showed similar efficiency in the binding and cleavage of both mutant and wild-type target sequences (Supplementary Figure S2), it was not used further and the subsequent experiments were performed with version B only (Figures 1 and Supplementary Figure S2).
For all TALENs the obligate heterodimeric FokI nuclease domains were used to reduce self-self off-site cleavage.19,20,21 Monomers were re-engineered to contain a mitochondrial localization signal (MLS, from SOD2 or COX8A/Su9), a specific immunological tag (HA or FLAG), a poly(A)/3′ UTR (SOD2 or ATP5B), and a fluorescent marker (mCherry or eGFP), the latter being separated from the TALEN coding region by a T2A picornavirus ribosome stuttering sequence.10,22 Mitochondrial localization was verified by immunocytochemistry (Figure 2). The presence of full-length proteins was determined by western blots (Figure 2). Some of the monomers showed two bands in western blots, and mitochondrial isolation and treatment with proteinase K indicates that the higher MW band corresponds to a small fraction of the monomer harboring the COX8A/Su9 that remains attached to the outside of the mitochondria (Supplementary Figure S3). Nonetheless, the mtDNA heteroplasmy changes (described below) show that the MLS are able to localize the mitoTALENs in the mitochondrial matrix with access to the mtDNA.
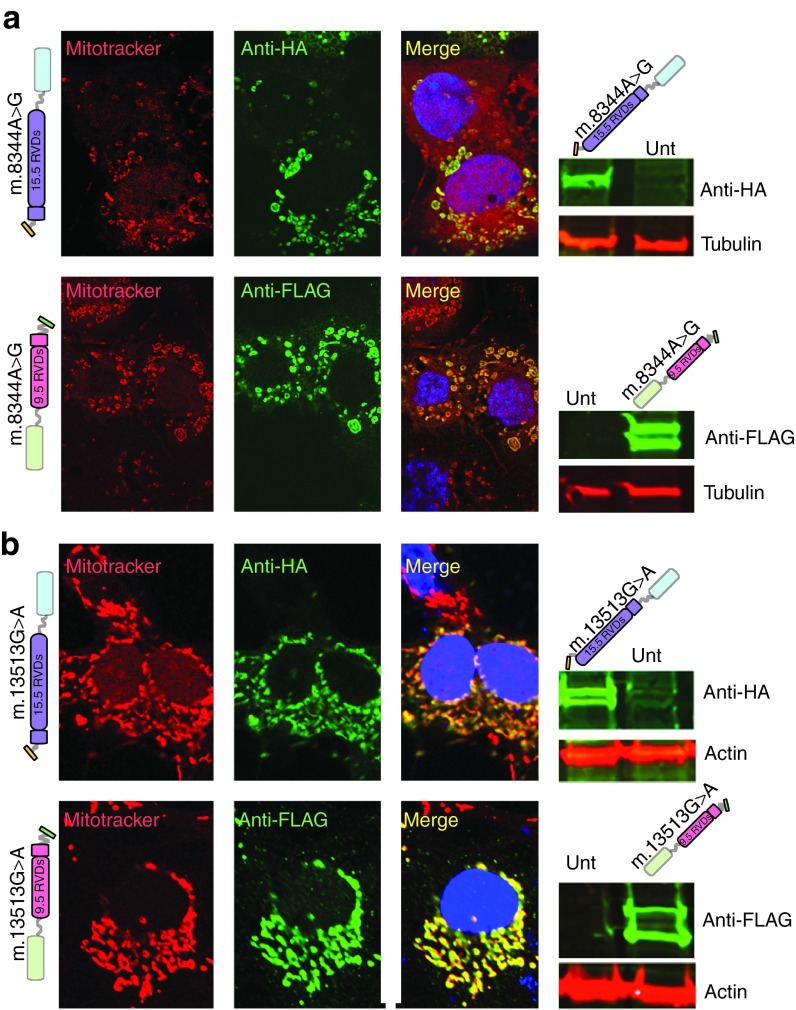
Figure 2.
mitoTALEN monomers are expressed and localized in mitochondria. (a) Taking advantage of the immune tags, we transfected COS7 cells with the respective pair of plasmids coding for the m.8344A>G mitoTALEN monomers. After 48 hours, we immunostained the cells with antibodies against FLAG or HA depicted in green. Cells were incubated with mitotracker red before fixation. DAPI was used for nuclear staining. Western blots (right panels) showed the presence of specific proteins. Double bands may indicate incomplete removal of the MLS. (b) The same experiment described above was performed for the mitoTALEN monomers against the m.13513G>A mtDNA mutation. Molecular weight of the different monomers were: m.8344A>G-15.5 RVDs=102.5 KDa; m.8344A>G-9.5 RVDs = 92.5 KDa; m.13513G>A-15.5 RVDs = 102.5KDa; m.13513G>A-9.5 RVDs = 92.5KDa. Normalization was done with monoclonal actin and tubulin antibodies.
mitoTALENs promote a robust change in mtDNA heteroplasmy
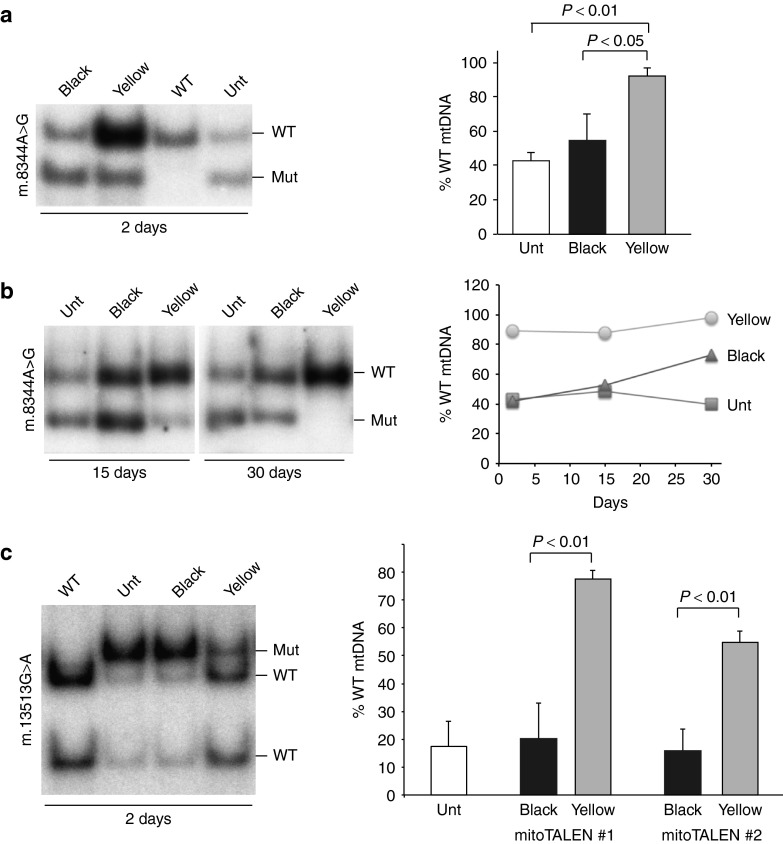
To test the efficacy of the mitoTALENs we produced transmitochondrial cybrid cell lines harboring different levels of the respective heteroplasmic mtDNA point mutations.23,24 Selected cybrid lines with a high level of mutant mtDNA were transfected with pairs of plasmids coding for each mitoTALEN monomer. Two days after transfection, cells were sorted for the fluorescent markers (mCherry and eGFP) and analyzed. Borrowing from the color effects seen in overlay images, cells expressing both green (eGFP) and red (mCherry) markers were termed “Yellow”, and cells sorted as negative for the markers were termed “Black” (Supplementary Figure S4). Most of subsequent experiments herein compare Yellow and Black cell populations. Figure 3a shows that the m.8344A>G mitoTALEN was able to markedly reduce the levels of mutant mtDNA in Yellow cells.
Figure 3.
mitoTALEN against single nucleotide changes can effectively reduce mutant mtDNA loads. (a) Transmitochondrial cybrids harboring heteroplasmic levels of the m.8344A>G mtDNA mutation were simultaneously transfected with two plasmids, coding for the two mitoTALEN monomers. After 48 hours, cells were sorted for the fluorescent markers and the sorted population expressing both monomers (mCherry and eGFP) were referred to as the “Yellow” population (both fluorescent markers). The mtDNA heteroplasmy in these cells was compared to the one in the “Black” population (sorted cells with no detectable fluorescence, Supplementary Figure S4). Sorted cell populations had their DNA extracted were analyzed by PCR/RFLP as described10 after digestion with BglI to differentiate the mutant from the WT load. (b) Comparison of change in heteroplasmy at 2, 15, or 30 days. (c) The same experiment was performed for transmitochondrial cybrids harboring the m.13513G>A mutation with two different pairs of mitoTALENs, using MboI digestion. The gel illustrates the results with one pair, whereas the quantification shows the results for both (mean ± SD; n = 3). The binding region for the different monomers is described in Figure 1 and Supplementary Figure S1. Unt, Untransfected.
Sorted cells were also cultured long term and we found that the robust change in mtDNA heteroplasmy levels in transmitochondrial cybrids (Figure 3a), which was maintained over time (Figure 3b). This finding is consistent with our previous study.10 We did notice a slight drift toward increasing wild-type mtDNA levels over time, including in the Black cell population (Figure 3b). We believe that the Black cell population may contain low levels of transfected cells and some cleavage of the mutant mtDNA may have occurred at low levels.
Both the 9.5 and 12.5 RVD mitoTALENs against the m.13513G>A mutation triggered a robust shift in heteroplasmy. Figure 3c illustrates the heteroplasmy shift obtained with one of them (version B) and shows the quantitative determinations for both mitoTALEN versions.
Heteroplasmic cells treated with mitoTALEN improve their OXPHOS function
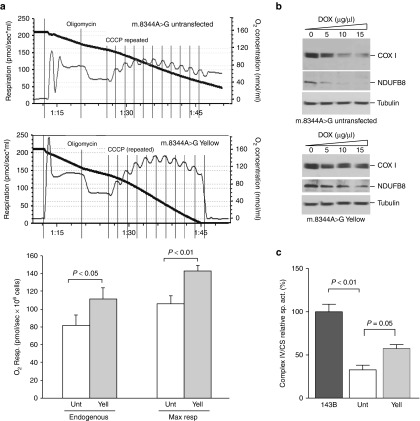
To determine whether the change in heteroplasmy improved the biochemical OXPHOS defect, respiration of m.8344A>G Yellow cells was compared to the one in untransfected cells. Respiration was inhibited by Oligomycin and the stimulated by multiple additions of the uncoupler CCCP. Maximum respiration was increased in Yellow cells (Figure 4a). The m.8344A>G mutation in tRNALys is known to impair mitochondrial translation due to its defective aminoacylation.25 We measured the steady-state levels of two OXPHOS proteins: (i) NDUFB8, a nuclear encoded subunit of Complex I and (ii) COXI, an mtDNA-coded subunit of Complex IV. Because the protein synthesis defect is relatively mild in heteroplasmic cells, we included low concentrations of the mitochondrial protein synthesis inhibitor doxycycline to stress the defect. As shown in Figure 4b, the levels of these proteins were decreased at lower concentrations of doxycycline in untransfected cells. Although NDUFB8 is nuclear encoded, the steady-state levels of this protein are reduced because it is rapidly degraded if not incorporated into the final complex I assembly.26 Finally, we measured the activity of Complex IV and found that Yellow cells had a significant increase in Complex IV activity (Figure 4c).
Figure 4.
Functional recovery of mitoTALEN-treated MERRF m.8344A>G tRNA lysine transmitochondrial cybrids. (a) Transmitochondrial cybrids harboring heteroplasmic levels of the m.8344A>G mtDNA mutation were treated with mitoTALEN and the “Yellow” population expanded for 14 days. Cells were assayed for oxygen consumption and compared with the parental line. Oligomycin was used to inhibit respiration. To determine the maximum respiration, the uncoupler CCCP was added in a stepwise manner. Quantification of the respiratory rates (endogenous and maximal) is depicted in the lower part of panel a. (b) The same cells were analyzed for the steady-state levels of OXPHOS proteins by western blot. To exacerbate the defect, cells were treated with different concentrations of doxycycline for 72 hours, a mitochondrial protein synthesis inhibitor. (c) Enzymatic activity of complex IV (CIV) and citrate synthase (CS) were measured spectrophotometrically in mitochondria from cultured cells treated (Yell) and untreated (Unt) with mitoTALENs and in 143B osteosarcoma control line. Values of specific activity of CIV/CS in the ND5 mutant clone were compared with the values in mutant clone after TALEN treatment. Panel a: mean±SD; n = 3; t-test. Panel c: mean ± SEM; n = 3; ANOVA, Tukey post hoc.
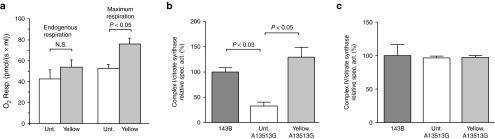
Similar results were obtained for when transfecting the m.13531G>A ND5 heteroplasmic cybrid cells with our mitoTALEN. Yellow cells respired more robustly (Figure 5a) and had restored Complex I activity (Figure 5b) when compared to the parental cell line harboring the m.13531G>A mutation in ND5, which impairs complex I activity and respiration.16,17 As expected, there was no change in complex IV activity in Yellow cells (Figure 5c) because ND5 is a subunit of Complex I.
Figure 5.
Functional recovery of mitoTALEN-treated MELAS/Leigh Syndrome m.13513G>A ND5 transmitochondrial cybrids. (a) Transmitochondrial cybrids harboring heteroplasmic levels of the m.13513G>A mtDNA mutation were treated with mitoTALENs and the “Yellow” (Yell) population expanded for 14 days. Cells were assayed for oxygen consumption and compared with the untreated parental line (Unt.). (b) The activity of Complex I and cytochrome oxidase (panel c) was measured in these cells, plus a 143B osteosarcoma control line, using spectrophotometric assays. Values were normalized to citrate synthase activity. Error bars correspond to mean ± SEM; n = 3; ANOVA, with Tukey post hoc.
Optimizing mitoTALENs for gene therapy
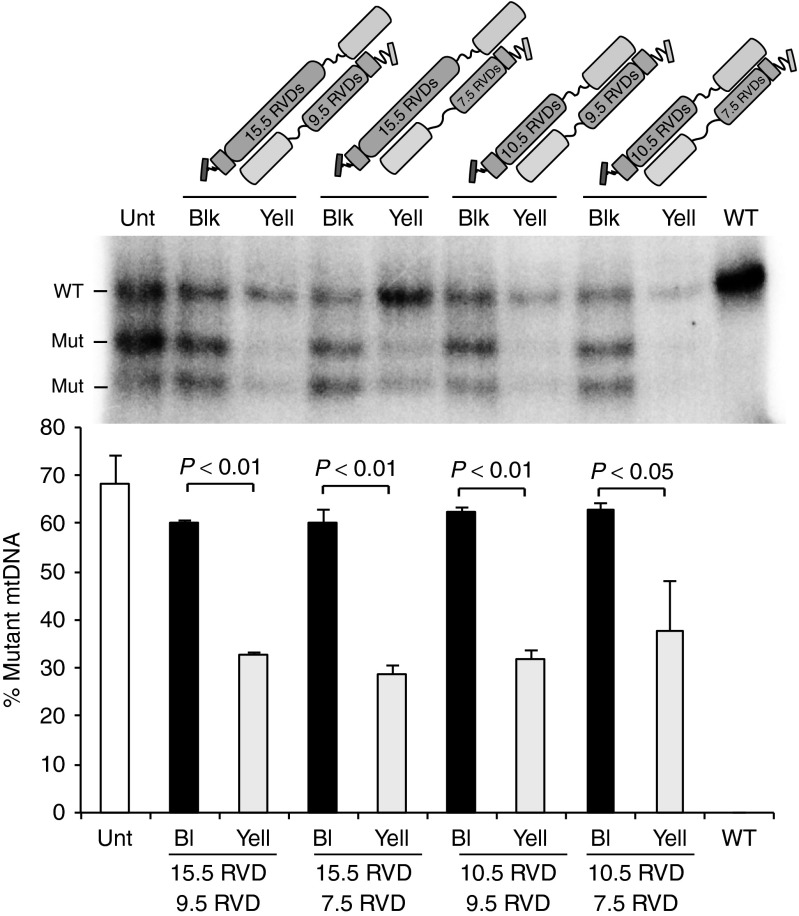
Although mitoTALENs are promising tools for the genetic therapy of patients with heteroplasmic mtDNA mutations, some challenges remain. A technical one is related to the large size of mitoTALENs and the requirement for two monomers that make it difficult to package them in many vector systems. Because mtDNA is a low complexity genome compared to the nuclear genome, we reasoned that smaller TALE DNA-binding domains would still be effective in shifting heteroplasmy, without increasing off-site cleavage to the point at which differential cleavage would be lost. The original m.8344A>G mitoTALEN sense and antisense monomers, which had 15.5 and 9.5 RVDs, were re-designed to have 10.5 and 7.5 RVDs, respectively. The binding regions are depicted in Supplementary Figure S5. These shorter monomers were tested for synthesis and for mitochondrial localization after transfection into COS7 cells (Supplementary Figure S6). Heteroplasmic transmitochondrial cybrid cell lines carrying the m.8344A>G mtDNA mutation were transfected with different combinations of sense and antisense mitoTALEN monomers and Yellow cells compared to Black cells to examine potential changes mtDNA heteroplasmy. As shown in Figure 6, all combinations resulted in significant reductions in the mutant load. The combination of the shortest monomers (10.5 and 7.5 RVDs) was perhaps the least robust, with more variability among independent experiments, but was still able to significantly eliminate mutant mitochondrial genomes (Figure 6).
Figure 6.
Shorter mitoTALENs can effectively reduce mutant mtDNA loads. We compared the original functional m.8344A>G mitoTALEN monomers, having 15.5 or 9.5 RVDs, with shorter monomers of 10.5 and 7.5 RVDs, respectively. The specific binding sites are described in Supplementary Figure S5 and Supplementary Text. All different combinations between sense and antisense monomers promoted reduction in mutant loads, as shown by comparing heteroplasmy in “Black” and “Yellow” sorted cell populations. The bar graph represents three independent experiments (mean ± SEM; n = 3).
Discussion
Through these experiments we were able to establish mitoTALEN as a general approach to reduce point mutant mtDNA loads in a heteroplasmic environment. We were able to reduce the mutation load in cultured cells harboring two clinically important mtDNA mutations that cause severe encephalopathies. The elimination of the target mutant mtDNA was not complete, and we do not have an explanation for that. Mitochondrial targeted restriction endonucleases gave us a more robust elimination of the targeted mitochondrial genome,27 suggesting that mitoTALENs are less efficient than restriction nucleases to cleave the mtDNA. Although the TALEN architecture involves specific binding to 30–40 bases (15–20 per monomer) in most cases,4 the low mtDNA complexity allowed us to reduce the binding site down to a total of 17 bases. It appears particularly important to make the binding site to the mutated region small, thereby increasing the impact of a single base change. This was illustrated by the fact that the m.8344A>G mitoTALEN version A, which had the monomer binding to the mutation harboring 15.5 RVDs, was unable to differentiate mutant and wild-type targets. On the other hand, monomers with ≤12.5 RVDs appear to be able to discriminate single base differences. However, it is important to emphasize that the location of the discriminating base has a major impact on the binding. The importance of binding at different positions of TALEN monomers has been the subject of many studies,4,28 and it is generally accepted that the proximal bases have a higher impact on binding.29 The presence of a T at position 0 is a required feature for most TALEN architectures, although in some cases a C at this position was reported as permissible to binding,4 and, different specificities for N-terminus binding are being developed.30 Therefore, whenever possible, we took advantage of position T0 as the discriminating one. However, as exemplified by the m.8344A>G mutation, this is not always possible. In this particular mutation, we were able to place a C at position 3 and found that it was able to effectively discriminate between mutant and wild-type mtDNA. However, as mentioned above, this was true only when the mitoTALEN monomer binding to the m.8344A>G mutant was relatively short (9.5 instead of 15.5 RVDs). This can be explained by the relative importance of a position 3 binding among 15 specific others. It is also important to remember that the other (sense in this case) monomer has a perfect binding, making cleavage of the mutated region likely. The use of non-conventional RVDs may also help in the future design of mitoTALENs.31
The functional assays confirmed the prediction that mitoTALEN-induced heteroplasmy shift leads to improved OXPHOS capacity by reverting the original biochemical defect, which can be a defective protein synthesis in the case of tRNA gene mutations or enzyme activity of single OXPHOS complexes because of a mutation in a protein coding gene. These results open the path for using this approach to treat patients with mtDNA mutations.
To bring this technology into clinical trials, it is important to improve technical hurdles, such as the large size of mitoTALEN monomers. A mitoTALEN monomer with 15.5 RVDs, a CMV promoter and a short poly(A) site is approximately 3,200 bp in length. If using recombinant viral delivery, ideally all the required information for both monomers should be expressed from a single virion. This creates a challenge for smaller vectors such as AAV. We started addressing this problem by reducing the size of the monomers, particularly the discriminating monomer, to very short binding domains. We found that even a monomer with 7.5 RVDs could efficiently reduce m.8344A>G mtDNA heteroplasmy ex vivo. Although this short monomer worked well with the complementary short monomer of 10.5 RVDs, it worked more consistently with the second (common) monomer containing 15.5 RVDs.
Although the reduction in size will facilitate technical issues related to vector design, alternative architectures, such as zinc finger nucleases that are intrinsically shorter, could also be used.32 In our hands zinc finger nucleases are more complex to design and modify although it is an architecture also shown to work in reducing mtDNA heteroplasmy.33,34 The other alternative, CRISPR poses a challenge for mtDNA use, as it requires the import of an RNA component. Our attempts to use this latter platform to change mtDNA heteroplasmy have failed (unpublished observations).
In summary, mitoTALENs employing gain of T or gain of C design principles have the capacity to target many clinically relevant mtDNA point mutations. Moreover the use of discriminating monomers in the range of 7.5–15.5 RVDs can robustly reduce the levels of mtDNA genomes with pathogenic point mutations in heteroplasmic cells and revert the defective OXPHOS phenotypes. This approach has the potential of providing an effective treatment to a subgroup of patients with mitochondrial diseases.
Materials and Methods
MitoTALEN constructs. Custom-made TALE domains were assembled by Cellectis Bioresearch (Paris, France). We designed and constructed plasmids coding for mitoTALEN monomers using the In-Fusion HD cloning kit from Clontech Laboratories according to Bacman et al.10 Modifications included removal of the nuclear localization signal, inclusion of a mitochondrial localization signal derived from SOD2 or modified COX8A/Su9 tandem MLS, inclusion of a unique immuno-tag in the N-terminus of the mature protein (Hemagglutinin (HA) or Flag), inclusion of a 3′ untranslated region from a mitochondrial gene known to localize mRNA to ribosomes on mitochondria, inclusion of an independent fluorescent marker to select for expression (eGFP in one monomer and mCherry in the other) and inclusion of a recoded picornaviral 2A-like sequence (T2A) between the mitoTALEN and the fluorescent marker.10,22
Single-strand annealing. Single-strand annealing in Saccharomyces cerevisiae was performed as previously described.35
Cells: transfection and sorting. Heteroplasmic cybrids harboring the m.8344A>G/tRNALys/MERRF point mutation were derived from fusions of an osteosarcoma cell line devoid of mtDNA (143B/206 line20) and dermal fibroblasts from a patient harboring the point mutation.36 Likewise, cybrids cells harboring the ND5 m.13513G>A mutation were produced by fusing dermal fibroblasts with 143B/206 line20. The clinical features of the patient and manipulations to obtain clones with high levels of heteroplasmy are described in Supplementary Text.
Cells were transfected with GenJet DNA In Vitro Transfection Reagent version II (SL100489; SignaGen Laboratories) following the protocol suggested by the manufacturers. After 48 hours, cells were sorted using a FACSAria IIU by gating on single-cell fluorescence using a 561-nm laser and 600LP, 610/20 filter set for mCherry and a 488-nm laser and 505LP, 530/30 filter set for eGFP.
Mitochondrial expression of the mitoTALENs. For western blot studies, we subjected 40 μg of total proteins from cellular homogenates of COS7 cells collected 48 hours after transfection to electrophoresis in 4% Tris–HCl polyacrylamide gels and transferred the separated proteins to nitrocellulose membranes (162-0115, Bio-Rad, Hercules, CA), as described.27 Antibodies used for blotting were rat monoclonal antibody to HA (1867423; Roche Biochemical; Indianapolis, IN 1:1,000), mouse monoclonal antibody to Flag (F3165; Sigma-Aldrich, St. Louis, MO; 1:1,000) and mouse monoclonal antibody to tubulin (T9026; Sigma-Aldrich, Indianapolis, IN; 1:1,000) and polyclonal antibody against actin (A2066; Sigma-Aldrich 1:1,000). Secondary antibodies were IRDye 800–conjugated antibody to rat IgG (612-732-120; 1:5,000), IRDye 800–conjugated antibody to mouse IgG (610-732-124; 1:5,000), and IRDye 700–conjugated antibody to mouse IgG (610-430-002; 1:5,000) from Rockland. An Odyssey Infrared Imaging System (LI-COR, Lincoln, NE) was used to scan the blots.
To test for mitochondrial internalization of mitoTALEN monomers, HEK293T cells (6 × T75 flasks) were tranfected with a plasmid coding for a FLAG-tagged MERRF mitoTALEN monomer (version C, 7.5 RVDs). After 24 hours, cells were disrupted by nitrogen cavitation and mitochondria were isolated by differential centrifugation as described in Adachi et al.37 Isolated mitochondria was treated with 5 or 50 µg/ml proteinase K on ice for 30 minutes. Samples were analyzed by western blot as described above. Antibodies used for blotting were mouse monoclonal antibody to Flag (F3165; Sigma-Aldrich; 1:1,000) and rabbit monoclonal antibody to VDAC1 (#4661; Cell Signaling, Danvers, MA 1:2,000).
For immunocytochemical studies, we plated COS7 cells onto coverslips and then transfected them with mitoTALEN plasmids as described above. At 24 hours posttransfection, cells were incubated for 30 minutes at 37 °C with 200 nmol/l MitoTracker Red CMXRos (Invitrogen, Waltham, MA) and fixed with 2% paraformaldehyde in phosphate-buffered saline for 20 minutes. After a brief treatment with methanol (5 minutes), primary antibodies to HA (Roche; 1:200) or Flag (Sigma-Aldrich; 1:200) in 2% BSA in phosphate-buffered saline were incubated overnight at 4 °C in a humid chamber. The next day, coverslips were incubated for 2 hours at room temperature with Alexa Fluor 488–conjugated goat antibody to rat IgG (A-11006; Invitrogen; 1:200) or Alexa Fluor 488–conjugated goat antibody to mouse IgG (A-11001; Invitrogen; 1:200), as previously described.27 Images were recorded using a Zeiss LSM510 confocal microscope.
Quantification of m.8344A>G and m.13513G>A mutation load by “last-cycle hot” PCR and RFLP. We determined the levels of the m.8344A>G and m.13513G>A mutations by “last-cycle hot” PCR38 which visualizes only nascent amplicons and removes interference from point mutation heteroduplexes formed during melting and annealing cycles. Total genomic DNA was extracted from FACS sorted cells with the NucleoSpin Tissue XS kit (740901.50; Macherey-Nagel, Clontech, Mountain View, CA) according to the manufacturer's instructions. DNA was used as template, and PCR was performed with the following mtDNA primers:
The mutant allele for the m.8344A>G mutation completes a mismatched primer with a BglI half-site after PCR amplification, creating a restriction-fragment length polymorphism. The following primers were used: B-5′-GTAGTATTTAGTTTGGGGCATTTCACTGTAAAGCCGTGTTGG-3′; F- 5′-CTACCCCCTCTAGAGCCCAC-3′ and PCR products were digested with BglI.
For the m.13513G>A, the mutant allele m.13513G>A completes a mismatched primer with a MboI half-site after PCR amplification, creating a mismatch sequence allowing a restriction-fragment length polymorphism. B-5′-ATAGATAGGGCTCAGGCGTTTGTGTATGATATGTTTGCGGTTTCGATGATGTGAT-3′; F-5′-TCCATCATCCACAACCTTAACAATGA-3′. The resulted amplicons were digested with MboI. The digested products were resolved in a 12% polyacrylamide gel. Radioactive signal was quantified using a Cyclone phosphorimaging system (PerkinElmer) and OptiQuant software (PerkinElmer, Waltham, MA).
Sequencing. Samples were sequenced using ABI BigDye chemistry according to manufacturer's procedure. Sequences were aligned using CLCBio Main Workbench (Qiagen, Hilden, Germany).
OXPHOS proteins expression under doxycycline treatment. Sorted cells after the m.8344A>G/MERRF mitoTALEN transfection were allowed to grow for 14 days. Both m.8344A>G untransfected and “Yellow” sorted cells were treated with increasing concentrations of Doxycycline (D-9891; Sigma-Aldrich; 0–15 μg/ml) for 72 hours. Cell homogenates (40 μg of total proteins) were subject to electrophoresis in 4–20% Tris–HCl polyacrylamide gels (4561094; Bio-Rad Laboratories) and transferred to nitrocellulose membranes (162-0115; Bio-Rad), then blotted with MTCO1 (ab-14705; Abcam, Cambridge, UK), NDUFB8 (ab-110242; Abcam) and α-Tubulin (T9026; Sigma-Aldrich) primary antibodies followed by anti-mouse IgG-HRP linked secondary antibody (7076; Cell Signaling).
Measurement of cell respiration. Cells in exponential growing phase were collected and respiration rate of intact cells was measured at 37 °C with Oroboros high-resolution respirometer Oxygraph-2k (Oroboros Instruments GmbH, Austria).39 Besides the endogenous respiration, oligomycin-resistant respiration in the presence of 1 μg/ml Oligomycin (O-4876; Sigma-Aldrich) and uncoupled respiration with 1 μl of 0.1 mmol/l of CCCP were also recorded (0.05 μmol/l per step).40 Data was analyzed with DatLab 4 software (Oroboros Instruments, Innsbruck, Austria).
Enzyme activities. For spectrophotometric analysis, we subjected 9 × 106 exponentially growing cells to centrifugation and washed them once with phosphate-buffered saline. The pellets were resuspended in 500 μl of 20 mmol/l MOPS (pH 7.4), 100 mmol/l Sucrose, 1 mmol/l EGTA, 10 mmol/l Triethanolamine, 5% Percoll, proteases inhibitors and Bovine Serum albumin 1 g/l. Cells were disrupted by cavitation (600 psi for 30 minutes) and mitochondria were obtained as described in Adachi et al.37 Complex I (NADH CoQ oxidoreductase) reaction was started with coenzyme Q1 (100 µmol/l) (C7956; Sigma-Aldrich) and inhibited with rotenone (10 µmol/l) (R8875; Sigma-Aldrich).41 Complex IV reaction was assayed using reduced Cytochrome C (2 mmol/l) (C2506; Sigma-Aldrich) and Citrate synthase reaction was started with Oxalacetate (0.5 mmol/l) (O-4126; Sigma-Aldrich)).42 Assay results were normalized to protein concentration obtained using the BioRad Bradford Assay Kit and BSA as standard, and subsequently Complex I and IV activities were normalized to Citrate synthase activity. Assays were performed in a BioTek Synergy H1 hybrid plate reader.
Statistical analyses. Pairwise comparisons were performed using two-tailed Student's t-test using GraphPad Prism (GraphPad Software) and SPSS package (v.22, IBM). ANOVA was performed with SPSS.
SUPPLEMENTARY MATERIAL Figure S1. Design and binding of mitoTALENs against two different pathogenic mtDNA mutations. Figure S2. Single Strand Annealing assay of m.8344A>GTALENs. Figure S3. A small fraction of mitoTALEN monomer remains attached to the outside of the mitochondria. Figure S4. Sorting of cells transfected with mitoTALEN. Figure S5. Design and binding of short mitoTALENs against the m.8344A>G mtDNA mutation. Figure S6. Shortened mitoTALEN monomers are expressed and localized in mitochondria. Supplementary Text
Acknowledgments
We are grateful to the skilled assistance of the Flow Cytometry Core Facility for the cell sorting services and the Oncogenomics Core Facility for the Sanger sequencing services at the Sylvester Comprehensive Cancer Center, University of Miami.
This work was supported by donations from Mr. Ron Biscardi, the JDM Fund, and grants from the NIH (5R01EY010804 and 5R01NS079965), the Muscular Dystrophy Association, and the United Mitochondrial Disease Foundation.
Supplementary Material
References
- Thorburn, DR and Dahl, HH (2001). Mitochondrial disorders: genetics, counseling, prenatal diagnosis and reproductive options. Am J Med Genet 106: 102–114. [DOI] [PubMed] [Google Scholar]
- DiMauro, S and Schon, EA (2003). Mitochondrial respiratory-chain diseases. N Engl J Med 348: 2656–2668. [DOI] [PubMed] [Google Scholar]
- Gardner, JL, Craven, L, Turnbull, DM and Taylor, RW (2007). Experimental strategies towards treating mitochondrial DNA disorders. Biosci Rep 27: 139–150. [DOI] [PubMed] [Google Scholar]
- Cermak, T, Doyle, EL, Christian, M, Wang, L, Zhang, Y, Schmidt, C et al. (2011). Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer, D, Wang, H, Kiani, S, Lai, CS, Gao, Q, Cassady, JP et al. (2011). Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 29: 731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou, MJ and Bogdanove, AJ (2009). A simple cipher governs DNA recognition by TAL effectors. Science 326: 1501. [DOI] [PubMed] [Google Scholar]
- Boch, J, Scholze, H, Schornack, S, Landgraf, A, Hahn, S, Kay, S et al. (2009). Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326: 1509–1512. [DOI] [PubMed] [Google Scholar]
- Valton, J, Cabaniols, JP, Galetto, R, Delacote, F, Duhamel, M, Paris, S et al. (2014). Efficient strategies for TALEN-mediated genome editing in mammalian cell lines. Methods 69: 151–170. [DOI] [PubMed] [Google Scholar]
- Watanabe, T, Ochiai, H, Sakuma, T, Horch, HW, Hamaguchi, N, Nakamura, T et al. (2012). Non-transgenic genome modifications in a hemimetabolous insect using zinc-finger and TAL effector nucleases. Nat Commun 3: 1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacman, SR, Williams, SL, Pinto, M, Peralta, S and Moraes, CT (2013). Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nat Med 19: 1111–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayona-Bafaluy, MP, Blits, B, Battersby, BJ, Shoubridge, EA and Moraes, CT (2005). Rapid directional shift of mitochondrial DNA heteroplasmy in animal tissues by a mitochondrially targeted restriction endonuclease. Proc Natl Acad Sci USA 102: 14392–14397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz, F, Bayona-Bafaluy, MP, Rana, M, Mora, M, Hao, H and Moraes, CT (2002). Human mitochondrial DNA with large deletions repopulates organelles faster than full-length genomes under relaxed copy number control. Nucleic Acids Res 30: 4626–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoffner, JM, Lott, MT, Lezza, AM, Seibel, P, Ballinger, SW and Wallace, DC (1990). Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell 61: 931–937. [DOI] [PubMed] [Google Scholar]
- Berkovic, SF, Shoubridge, EA, Andermann, F, Andermann, E, Carpenter, S and Karpati, G (1991). Clinical spectrum of mitochondrial DNA mutation at base pair 8344. Lancet 338: 457. [DOI] [PubMed] [Google Scholar]
- Chol, M, Lebon, S, Bénit, P, Chretien, D, de Lonlay, P, Goldenberg, A et al. (2003). The mitochondrial DNA G13513A MELAS mutation in the NADH dehydrogenase 5 gene is a frequent cause of Leigh-like syndrome with isolated complex I deficiency. J Med Genet 40: 188–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santorelli, FM, Tanji, K, Kulikova, R, Shanske, S, Vilarinho, L, Hays, AP et al. (1997). Identification of a novel mutation in the mtDNA ND5 gene associated with MELAS. Biochem Biophys Res Commun 238: 326–328. [DOI] [PubMed] [Google Scholar]
- Shanske, S, Coku, J, Lu, J, Ganesh, J, Krishna, S, Tanji, K et al. (2008). The G13513A mutation in the ND5 gene of mitochondrial DNA as a common cause of MELAS or Leigh syndrome: evidence from 12 cases. Arch Neurol 65: 368–372. [DOI] [PubMed] [Google Scholar]
- Brown, WM, Prager, EM, Wang, A and Wilson, AC (1982). Mitochondrial DNA sequences of primates: tempo and mode of evolution. J Mol Evol 18: 225–239. [DOI] [PubMed] [Google Scholar]
- Miller, JC, Holmes, MC, Wang, J, Guschin, DY, Lee, YL, Rupniewski, I et al. (2007). An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol 25: 778–785. [DOI] [PubMed] [Google Scholar]
- Doyon, Y, Vo, TD, Mendel, MC, Greenberg, SG, Wang, J, Xia, DF et al. (2011). Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods 8: 74–79. [DOI] [PubMed] [Google Scholar]
- Li, T, Huang, S, Jiang, WZ, Wright, D, Spalding, MH, Weeks, DP et al. (2011). TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res 39: 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczak, AL, Workman, CJ, Wang, Y, Vignali, KM, Dilioglou, S, Vanin, EF et al. (2004). Correction of multi-gene deficiency in vivo using a single ‘self-cleaving' 2A peptide-based retroviral vector. Nat Biotechnol 22: 589–594. [DOI] [PubMed] [Google Scholar]
- King, MP and Attardi, G (1989). Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science 246: 500–503. [DOI] [PubMed] [Google Scholar]
- Bacman, SR and Moraes, CT (2007). Transmitochondrial technology in animal cells. Methods Cell Biol 80: 503–524. [DOI] [PubMed] [Google Scholar]
- Enriquez, JA, Chomyn, A and Attardi, G (1995). MtDNA mutation in MERRF syndrome causes defective aminoacylation of tRNA(Lys) and premature translation termination. Nat Genet 10: 47–55. [DOI] [PubMed] [Google Scholar]
- Diaz, F, Garcia, S, Padgett, KR and Moraes, CT (2012). A defect in the mitochondrial complex III, but not complex IV, triggers early ROS-dependent damage in defined brain regions. Hum Mol Genet 21: 5066–5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacman, SR, Williams, SL, Garcia, S and Moraes, CT (2010). Organ-specific shifts in mtDNA heteroplasmy following systemic delivery of a mitochondria-targeted restriction endonuclease. Gene Ther 17: 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, D, Yan, C, Wu, J, Pan, X and Yan, N (2014). Revisiting the TALE repeat. Protein Cell 5: 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger, JP, Pattanayak, V, Reyon, D, Tsai, SQ, Sander, JD, Joung, JK et al. (2014). Broad specificity profiling of TALENs results in engineered nucleases with improved DNA-cleavage specificity. Nat Methods 11: 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, BM, Mercer, AC and Barbas, CF 3rd (2013). Directed evolution of the TALE N-terminal domain for recognition of all 5' bases. Nucleic Acids Res 41: 9779–9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juillerat, A, Pessereau, C, Dubois, G, Guyot, V, Maréchal, A, Valton, J et al. (2015). Optimized tuning of TALEN specificity using non-conventional RVDs. Sci Rep 5: 8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacman, SR, Williams, SL, Pinto, M and Moraes, CT (2014). The use of mitochondria-targeted endonucleases to manipulate mtDNA. Methods Enzymol 547: 373–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minczuk, M, Papworth, MA, Miller, JC, Murphy, MP and Klug, A (2008). Development of a single-chain, quasi-dimeric zinc-finger nuclease for the selective degradation of mutated human mitochondrial DNA. Nucleic Acids Res 36: 3926–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammage, PA, Rorbach, J, Vincent, AI, Rebar, EJ and Minczuk, M (2014). Mitochondrially targeted ZFNs for selective degradation of pathogenic mitochondrial genomes bearing large-scale deletions or point mutations. EMBO Mol Med 6: 458–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddell, L, Manthey, G, Pannunzio, N and Bailis, A (2011). Quantitation and analysis of the formation of HO-endonuclease stimulated chromosomal translocations by single-strand annealing in Saccharomyces cerevisiae. J Vis Exp 55: 3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci, JP, Schon, EA and King, MP (1997). Point mutations in the mitochondrial tRNA(Lys) gene: implications for pathogenesis and mechanism. Mol Cell Biochem 174: 215–219. [PubMed] [Google Scholar]
- Adachi, S, Gottlieb, RA and Babior, BM (1998). Lack of release of cytochrome C from mitochondria into cytosol early in the course of Fas-mediated apoptosis of Jurkat cells. J Biol Chem 273: 19892–19894. [DOI] [PubMed] [Google Scholar]
- Moraes, CT, Ricci, E, Bonilla, E, DiMauro, S and Schon, EA (1992). The mitochondrial tRNA(Leu(UUR)) mutation in mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes (MELAS): genetic, biochemical, and morphological correlations in skeletal muscle. Am J Hum Genet 50: 934–949. [PMC free article] [PubMed] [Google Scholar]
- Wang, X and Moraes, CT (2011). Increases in mitochondrial biogenesis impair carcinogenesis at multiple levels. Mol Oncol 5: 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesta, D and Gnaiger, E (2012). High-resolution respirometry: OXPHOS protocols for human cells and permeabilized fibers from small biopsies of human muscle. Methods Mol Biol 810: 25–58. [DOI] [PubMed] [Google Scholar]
- Martinez, B, del Hoyo, P, Martin, MA, Arenas, J, Perez-Castillo, A and Santos, A (2001). Thyroid hormone regulates oxidative phosphorylation in the cerebral cortex and striatum of neonatal rats. J Neurochem 78: 1054–1063. [DOI] [PubMed] [Google Scholar]
- Barrientos, A, Fontanesi, F and Díaz, F (2009). Evaluation of the mitochondrial respiratory chain and oxidative phosphorylation system using polarography and spectrophotometric enzyme assays. Curr Protoc Hum Genet Chapter 19: Unit19.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.