Abstract
A human single-chain variable fragment (scFv) antibody library was expressed on the surface of human T cells after transduction with lentiviral vectors (LVs). The repertoire was fused to a first-generation T cell receptor ζ (TCRζ)-based chimeric antigen receptor (CAR). We used this library to isolate antibodies termed CARbodies that recognize antigens expressed on the tumor cell surface in a proof-of-principle system. After three rounds of activation-selection there was a clear repertoire restriction, with the emergence dominant clones. The CARbodies were purified from bacterial cultures as soluble and active proteins. Furthermore, to validate its potential application for adoptive cell therapy, human T cells were transduced with a LV encoding a second-generation costimulatory CAR (CARv2) bearing the selected CARbodies. Transduced human primary T cells expressed significant levels of the CARbodies-based CARv2 fusion protein on the cell surface, and importantly could be specifically activated, after stimulation with tumor cells. This approach is a promising tool for the generation of antibodies fully adapted to the display format (CAR) and the selection context (cell synapse), which could extend the scope of current adoptive cell therapy strategies with CAR-redirected T cells.
Keywords: adoptive cell therapy, antibody, chimeric antigen receptor, repertoire selection, tumor-associated antigens
Introduction
Chimeric antigen receptors (CARs) are composed of an extracellular antigen recognition domain (usually a single-chain variable fragment (scFv) antibody) attached to transmembrane and cytoplasmic signaling domains.1 CAR-mediated recognition converts tumor-associated antigens (TAA) expressed on the cell surface, into recruitment points of effector functions, addressing the goal of major histocompatibility complex-independent activation of effector cells. First-generation CAR were constructed through the fusion of a scFv-based TAA-binding domain to a cytoplasmic signaling domain typically derived either from the ζ chain of the T cell receptor (TCR)/CD3 complex or from the γ chain associated with some Fc receptors.2 Second-generation CAR (CARv2) comprising the signaling region of the TCRζ in series with the signaling domain derived from the T cell costimulatory receptors CD28, 4-1BB (CD137) or OX40 (CD134).3
Upon encountering antigen, the interaction of the genetically transferred CAR triggers effector functions and can mediate cytolysis of tumor cells.1 The utility and effectiveness of the CAR approach have been demonstrated in a variety of animal models, and at the moment there are numerous ongoing clinical trials using CAR-based genetically engineered T lymphocytes for the treatment of cancer patients.4 CARs enable us to target effector cells toward any native extracellular antigen for which a suitable antibody exists. Engineered cells can be targeted not only to proteins but also to structures such as carbohydrate5 and glycolipid tumor antigens.6
Current methods for the generation of recombinant antibodies are mainly based on the use of purified proteins.7 However, for many cell surface molecules, such as G-protein coupled receptors and molecules with large extracellular domains, the use of recombinant proteins may not be feasible. Also direct immobilization of purified proteins onto plastic surface, may significantly alter protein conformation. For these reasons, antibodies selected on the basis of binding to a recombinant protein may not recognize the same protein in its native context.8 Furthermore, although the antibody in solution might recognize the antigen on the cell surface, it may not be effective when displayed in a CAR context.
Recently, we described a mammalian cell-based antibody display platform, which takes advantage of the functional capabilities of T lymphocytes. The display of antibodies on the surface of T lymphocytes, as a part of a CAR-mediating signaling, may ideally link the antigen–antibody interaction to a demonstrable change in cell phenotype, due to the surface expression of activation markers.9 By using a scFv-based CAR that recognizes a TAA, we have demonstrated that combining CAR-mediated activation with fluorescence-activated cell sorting (FACS) of CD69+ T cells, it is possible to isolate binders to surface TAA, with an enrichment factor of at least 103-fold after two rounds, resulting in a homogeneous population of T cells expressing TAA-specific CAR.9
Here, we generated a human scFv library displayed in a first-generation TCRζ CAR with a theoretical diversity of 1.5 × 105, and after three rounds of selection-activation in vitro against the human cervix carcinoma cell line HeLa, we selected several antibodies (termed CARbodies) that recognize antigens expressed on the tumor cell surface. The CARbodies were purified from the periplasmic fraction of Escherichia coli (E. Coli) as soluble and active proteins. Furthermore, to validate the potential application of selected CARbodies for adoptive cell transfer therapy with CAR-modified T cells, we designed a CARv2 containing the extracellular and transmembrane domains of the CD8α chain as a spacer/hinge region and the intracellular regions of CD28 and TCRζ. Transduced human T cells expressed significant levels of the CARbodies-based CARv2 fusion protein on the cell surface, and importantly could be specifically activated, after stimulation with HeLa cells.
Results
Construction of a lentiviral human scFv library in the context of a CAR (Griffin.CAR library)
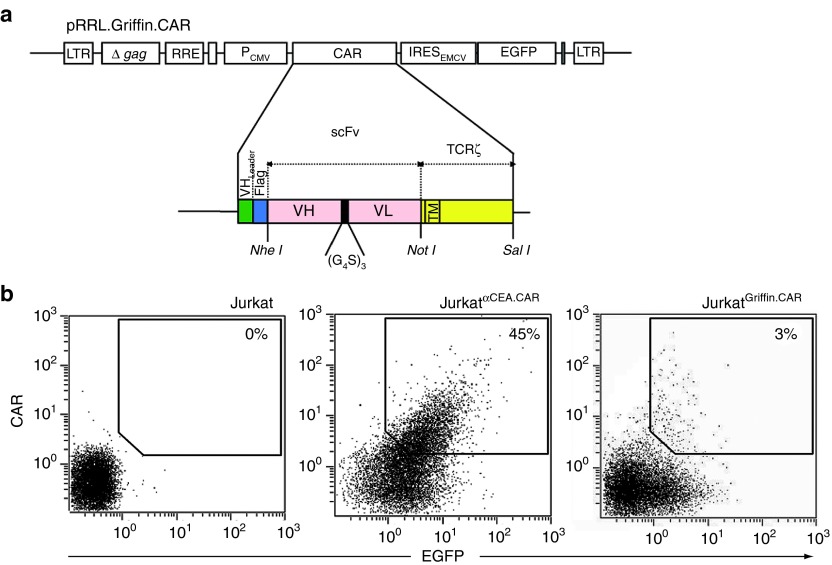
To construct a human scFv antibody library in the context of a CAR, the scFv-coding regions were amplified from the non-selected Griffin.1 library10 by PCR (Supplementary Table S1), and subcloned into the NheI/NotI digested backbone of plasmid pRRL.αNIP.TCRζ.IRES.EGFP,9 containing a bicistronic expression cassette encoding a first generation TCRζ-based CAR and the enhanced green fluorescent protein (EGFP), as reporter gene (Figure 1a). The CAR comprises a human immunoglobulin signal peptide, a FLAG epitope, and the scFv gene fused to the transmembrane and cytoplasmic regions of the human TCRζ chain.11 The diversity of the resulting plasmid library pRRL.Griffin.CAR was of 1.5 × 105 individual clones. The virus library (LVGriffin.CAR) was then produced on HEK-293T cells.
Figure 1.
Schematic representation of the lentiviral vector encoding a first generation chimeric antigen receptor (CAR). (a) Bicistronic vector (pRRL.Griffin.CAR) containing a human scFv repertoire in a first-generation TCRζ-based CAR and the EGFP gene. (b) FACS analysis of EGFP and cell surface CAR expression after transduction of Jurkat cells with either anti-CEA.CAR or Griffin.CAR encoding lentiviral vectors (LVαCEA.CAR and LVGriffin.CAR, respectively) at multiplicity of infectionI of ~1. Δ gag, ATG-deleted group specific antigen; CMV promoter, cytomegalovirus promoter; EGFP, enhanced green fluorescent protein; EMCV IRES, encephalomyocarditis virus internal ribosomal entry site; FACS, fluorescence-activated cell sorting; LTR, long terminal repeats; LV, lentiviral vector; RRE, Rev-responsive cis-acting element; scFv, single-chain variable fragment; TCR, T cell receptor.
Jurkat T cells were transduced with the polyclonal lentivirus library at multiplicity of infection (MOI) of ~1 to ensure the expression of no more than one antibody per cell. FACS analysis demonstrated that ~3% of the transduced cells (JurkatGriffin.CAR) were CAR+EGFP+ (Figure 1b). The percentage of CAR+EGFP+ cells increased to 50% when Jurkat T cells were transduced at the same MOI with a monoclonal lentivirus encoding an anti-CEA.CAR (Figure 1b). However, transduction of large numbers of Jurkat T cells (2 × 107) would ensure that the entire scFv repertoire of the LVGriffin.CAR library (1.5 × 105) is represented in the transduced population (JurkatGriffin.CAR).
Antigen-specific activation of human T cells transduced with the Griffin.CAR library
To study the reactivity of the transduced polyclonal population, JurkatGriffin.CAR cells were co-cultured at an effector to target ratio (E:T) of 1:1 in the presence of HeLa or HeLaCEA tumor cells, and the expression of CD69 evaluated by FACS (Supplementary Figure S1). Jurkat cells infected with monoclonal lentivirus (LVEGFP or LVαCEA.CAR) were used as controls. We found that about 1% of the JurkatGriffin.CAR cells expressed CD69 at the cell surface after co-culture with the HeLa cells. The percentage was lower (similar to the background) after co-culturing JurkatGriffin.CAR cells with HeLaCEA cells (Supplementary Figure S1). On average, CD69+ cells accounted for 40 ± 10% of monoclonal JurkatαCEA.CAR cells stimulated with CEA-positive cells. Importantly, T cell activation remained strictly antigen-specific with HeLa or HeLaCEA cells unable to induce CD69 expression in untransduced Jurkat cells (Supplementary Figure S1).
Selection of the Griffin.CAR library on human cancer cells
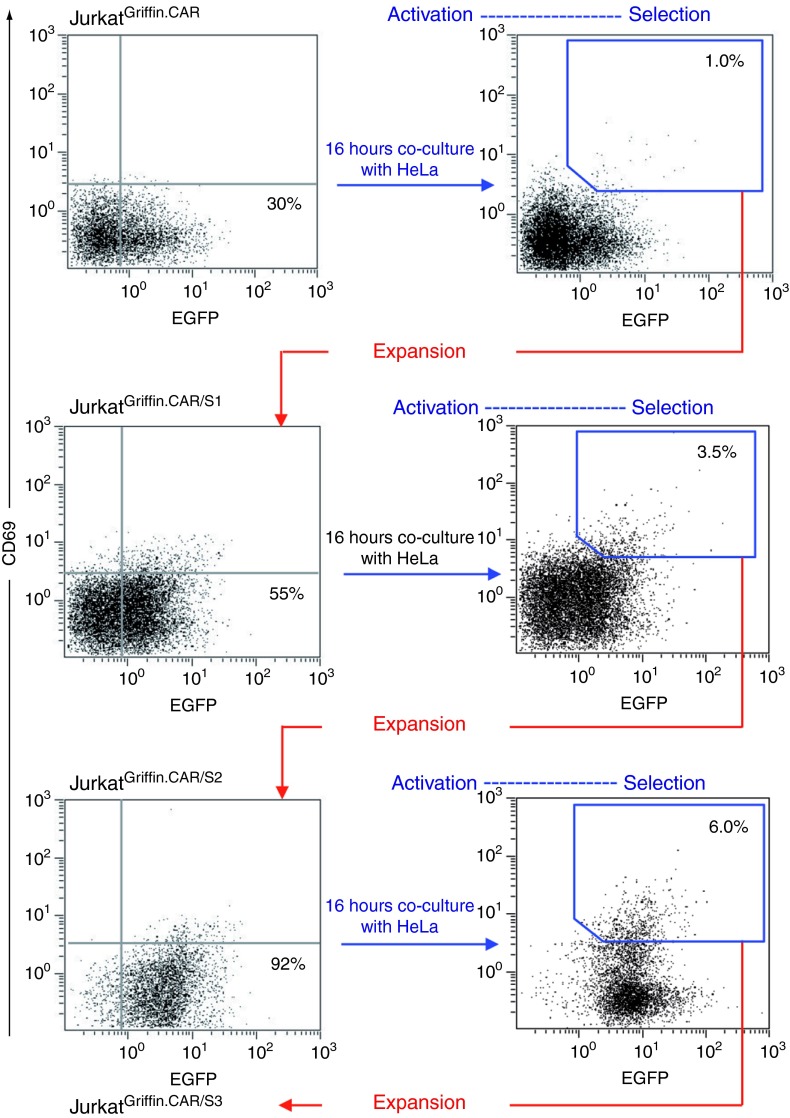
For the in vitro selection of antibodies against HeLa cell surface antigens, JurkatGriffin.CAR cells (3 × 107) were cultured with confluent monolayers of HeLa cells at an E:T ratio of 1:1. After 16 hours, T cells were recovered from the tumor cell monolayer by EDTA treatment, ficoll purified, washed twice with medium, and incubated with anti-CD69-PE monoclonal antibody (mAb). Double-positive CD69+EGFP+ cells were isolated by FACS sorting (Figure 2). The sorted population (JurkatGriffin.CAR/S1), which showed nearly twofold increase in the EGFP fluorescence intensity compared with the original JurkatGriffin.CAR population, was propagated and submitted for two additional rounds of activation/selection on HeLa cell monolayers (Figure 2). After three rounds, the resulting population (JurkatGriffin.CAR/S3) was almost 100% EGFP+ (Figure 2), and importantly 15% of JurkatGriffin.CAR/S3 cells expressed significant levels of CD69 in co-culture with HeLa cells (Supplementary Figure S2).
Figure 2.
Selection of chimeric antigen receptor (CAR)-activated Jurkat T cells. JurkatGriffin.CAR cells were stimulated with HeLa cells for 16 hours and further sorted on the basis of enhanced green fluorescent protein (EGFP) and CD69 expression. After a period of cell expansion, the activation/selection cycle was repeated two additional times.
Characterization of the selected antibodies
To confirm that the scFv consisted of VH and VL chains and to further analyze library diversity, DNA sequence analysis was performed on 200 randomly selected clones obtained after each round of selection. Sequence analysis showed that the majority of VH and VL chains had open-reading frames encoding full-length VH and VL chains (data not shown). Only 10% of the clones contained stop codons or frame shift mutations. In the original scFv library repertoire (JurkatGriffin.CAR) and in all the analyzed rounds, ~30% of the clones encoded an identical sequence, corresponding to the B1.8 scFv (anti-NIP) gene, present in the plasmid backbone used for library generation (pRRL.αNIP.TCRζ.IRES.EGFP). Sequence analysis confirmed that there were not repeated clones in the naive library, whereas the number of repeated clones increased after each successive round of selection (Figure 3a and Supplementary Tables S3–S6).
Figure 3.
Evolution of library diversity in the successive rounds of selection. (a) Sequence analysis confirmed that in the naive library there were not repeated sequences, except the B1.8 scFv gene that represents a 30% of the clones. Whereas the number of non-B1.8 repeated clones increased after each successive round of selection, the percentage of B1.8 clones remained constant along the selection process. (b) Enrichment observed in the different rounds of selection in the VL (upper) or the VH (lower). The lowest 100 P values in each of 500 simulations (psim) are sorted in ascending order (x-axis) and plotted in light gray. The observed lowest 100 P values (pobs) are overlaid in red for a better comparison (round 3, pobs are tangential to the x-axis). (c) The observed 500 lowest P values (pobs) of all the possible triplets of amino acids in the CDRs are plotted to compare the progressive enrichment in the different rounds (blue = S1, red = S2, green = S3) in the VL (upper) or the VH (lower). CDR, complementarity determining region; scFv, single-chain variable fragment.
We used a resampling approach to analyze the evolution of the diversity in the successive rounds of selection (Figure 3b). The enrichment during the first and second rounds progressively constrains the diversity of the repertoire (Figure 3c, blue and red lines). The enrichment occurs both in the VH and in the VL, although slightly uncoupled. The diversity of the third round (Figure 3c, green line) is considerably reduced by the presence of some predominant clones (Supplementary Tables S3–S6), rendering further rounds of panning unnecessary.
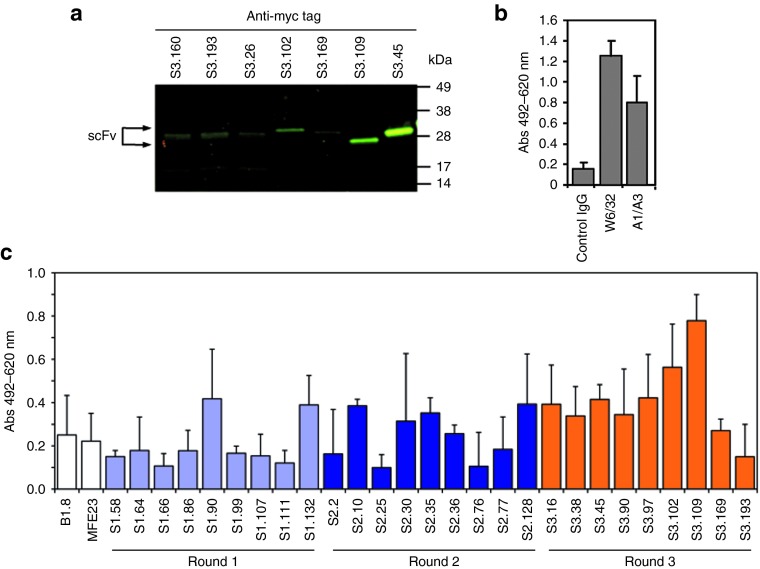
Based on the sequence analysis and on the number of repetitions, nine clones from each round of selection were selected for further characterization. The 27 selected scFvs (CARbodies) were cloned into bacterial expression vector pAB2 and transformed into E. coli TG1 cells. All periplasmic samples checked by western blotting were found to contain proteins of the correct size, ~28 kDa (Figure 4a). However, some scFvs showed lower molecular mass bands of ~15 kDa corresponding to VL domains produced by proteolytic cleavage of the scFvs during the isolation procedure. Cell-based ELISA demonstrated that 14 out of the 27 selected CARbodies specifically recognize antigens on HeLa cells (Figure 4c). The scFvs B1.8 (anti-NIP) and MFE23 (anti-CEA) were used as negative controls, whereas mouse mAb W6/32 (anti-major histocompatibility complex class I) and A1/A3 (anti-cytokeratins) were used as positive controls, to demonstrate the cell integrity after cell drying and fixing (Figure 4b). Interestingly, the number of HeLa-specific binders increased after each successive round of selection (Figure 4c).
Figure 4.
Evaluation of selected binders. (a) The presence of myc/6xHis-tagged soluble CARbodies in cell-free bacterial supernatant was demonstrated by western blot analysis, and ELISA against HeLa air-dried and fixed cells, using anti-c-myc mAb and HRP-conjugated goat anti-mouse IgG antibody. (b) The antigenic integrity was assessed with anti-MHC class I (W6/32) and anti-cytokeratin (A1/A3) mAbs. (c) The reactivity of a representative panel of soluble scFvs selected after one, two, and three rounds of selection was determined by ELISA. Myc/6xHis-tagged anti-NIP (B1.8) and anti-CEA (MFE23) scFvs were used as negative control to establish the background signal of the system (as shown in c). HRP, horseradish peroxidise; mAb, monoclonal antibody; MHC, major histocompatibility complex; scFv, single-chain variable fragment.
Antigen-specific activation of human T cells transduced with monoclonal scFv-CAR lentiviruses
To validate the selected HeLa-specific CARbodies on a second-generation CAR (CARv2) context, scFv genes were linked to the extracellular and transmembrane domains of the CD8α chain and the intracellular regions of CD28 and TCRζ chains (Supplementary Figure S3a). The functionality of the CARv2 was demonstrated using the anti-CEA scFv MFE23 as antigen-binding domain. The αCEA.CARv2 coding sequence was cloned into pRRL.FLAGαNIP.TCRζ.IRES.EGFP to generate the lentiviral vector (LV) pRRL.FLAGαCEA.CARv2 (Supplementary Figure S3a). The virus (LVαCEA.CARv2) was then produced on HEK-293T cells, and Jurkat T cells transduced at a MOI of ~30. Interestingly, CAR surface expression in JurkatαCEA.CARv2 cells was higher than observed in JurkatαCEA.CAR cells, transduced with the lentivirus LVαCEA.CAR at the same MOI (Supplementary Figure S3b). JurkatαCEA.CAR and JurkatαCEA.CARv2 cells were co-cultured with HeLa or HeLaCEA cells at an E:T ratio of 1:1, and the expression of CD69 was evaluated by FACS. Both populations expressed similar levels of CD69 at the cell surface after co-culture with HeLaCEA cells (Supplementary Figure S3c). T cell activation remained strictly antigen-specific with HeLa cells unable to induce CD69 expression in CAR-positive Jurkat cells (JurkatαCEA.CAR and JurkatαCEA.CARv2).
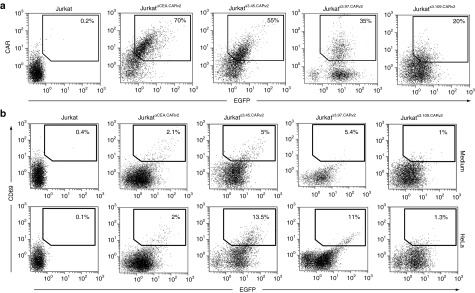
Once we demonstrated the functionality of the CARv2 construct, the selected HeLa-specific CARbodies S3.45, S3.97, and S3.109 were cloned into the LV to generate the pRRL.FLAGS3.45.CARv2, pRRL.FLAGS3.97.CARv2, and pRRL.FLAGS3.109.CARv2 plasmids. Jurkat T cells were transduced with the resulting lentiviral particles LVS3.45.CARv2, LVS3.97.CARv2 or LVS3.109.CARv2 at an MOI of ~30. Although EGFP was expressed at equivalent levels in all three populations, they expressed different levels of CARv2: JurkatS3.45.CARv2 (dim), JurkatS3.97.CARv2 (low/dim), and JurkatS3.109.CARv2 (low) (Figure 5a). To evaluate the functionality of the selected CARbody-based CARv2, JurkatS3.45.CARv2, JurkatS3.97.CARv2, and JurkatS3.109.CARv2 cells were co-cultured with HeLa cells at an E:T ratio of 1:1. Both JurkatS3.45.CARv2 and JurkatS3.97.CARv2 cells showed specific activation after co-culture with HeLa cells (Figure 5b). However, no activation was detected for JurkatS3.109.CARv1-EGFP as expected considering the limited expression of the S3.109 CARv2 in the cell surface (Figure 5b).
Figure 5.
Phenotypic and functional characterization of JurkatS3.45.CARv2, JurkatS3.97.CARv2, and JurkatS3.109.CARv2. (a) Comparative analysis of EGFP and cell surface expression of the selected CARbodies S3.45, S3.97, and S3.109 in the context of a second-generation CAR after transduction of Jurkat cells with LVS3.45.CARv2, LVS3.97.CARv2, and LVS3.109.CARv2 lentiviral vectors at multiplicity of infection of ~30. (b) FACS analysis of CD69 and EGFP expression by JurkatS3.45.CARv2, JurkatS3.97.CARv2, and JurkatS3.109.CARv2 stimulated either with HeLa (E:T = 1:1) for 16 hours. CAR, chimeric antigen receptor; EGFP, enhanced green fluorescent protein; E:T ratio, effector:target ratio; FACS, fluorescence-activated cell sorting; LV, lentiviral vector.
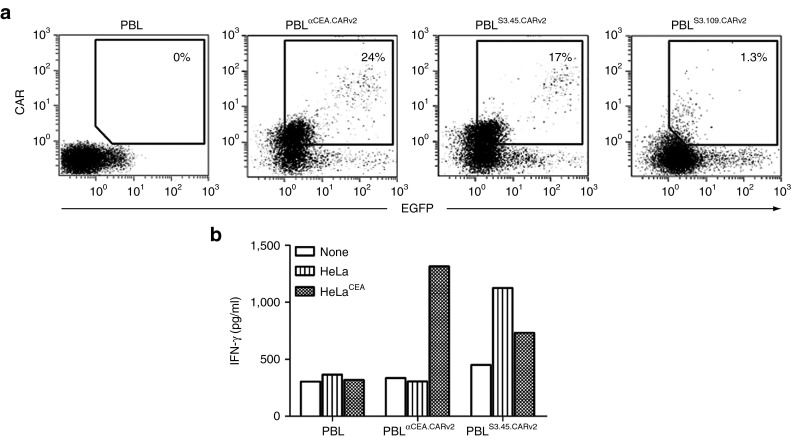
Next, primary human peripheral blood lymphocyte (PBL) were transduced with LVαCEA.CARv2, LVS3.45.CARv2 or LVS3.109.CARv2 at an MOI of ~20. Expression data were consistent with those observed in Jurkat cells: S3.109.CARv2 was almost undetectable on the cell surface, whereas αCEA.CARv2 and S3.45.CARv2 were expressed at similar levels (Figure 6a). Transduced human primary lymphocytes expressing αCEA.CARv2 (LTαCEA.CARv2) recognized CEA+ tumor cells and secreted high levels of interferon-γ (IFN-γ), but not when stimulated with CEA− tumor cells. PBLS3.45.CARv2 secreted IFN-γ when stimulated with both CEA+ and CEA− tumor cells, however, the levels were higher after co-culture with CEA cells (Figure 6b). In contrast, no secretion of IFN-γ was detected in the culture supernatants of untransduced PBL co-cultured with CEA+ or CEA− tumor cells (Figure 6b). These results combined with those obtained from cell-based ELISA demonstrate that the selected CARbodies bind antigens on the tumor cell surface and can induce the activation of human primary T cells in the context of a second-generation CAR.
Figure 6.
Phenotypic and functional characterization of human peripheral blood lymphocytes transduced with CARbodies-based CAR. (a) Comparative analysis of EGFP and cell surface expression of the selected CARbodies after transduction of human lymphocytes with LVαCEA.CARv2, LVS3.45.CARv2, and LVS3.109.CARv2 lentiviral vectors at multiplicity of infection of ~20. (b) IFN-γ secretion by untransduced PBL, or transduced PBL (PBLαCEA.CARv2 and PBLS3.45.CARv2) following incubation with HeLa or HeLaCEA cells. CAR, chimeric antigen receptor; EGFP, enhanced green fluorescent protein; IFN, interferon; LV, lentiviral vector; PBL, peripheral blood lymphocyte.
Discussion
Here, we demonstrated the feasibility of lymphocyte display to isolate functional human antibodies. We generated a human scFv library in the context of a first-generation CAR with a theoretical diversity of 1.5 × 105 and we demonstrated that after three rounds of selection-activation in vitro against a human epithelial carcinoma cell line, an enriched antibody population with presence of dominant clones was generated. Some of the selected scFv antibodies (CARbodies) were isolated in active form from periplasmic extracts of E. coli, and were easily purified. The purified CARbodies retained the ability to bind to antigen expressed on the tumor cell surface. Furthermore, some of the selected CARbodies were expressed on human Jurkat T cells and primary T lymphocytes in the context of a second-generation CAR composed of the CD28 and TCRζ signaling domain (CARv2), and we demonstrated that upon encountering specific antigen on tumor cell surface, human T cells harboring CARbodies-based CARv2 receptors are able to undergo specific activation and secreted high levels of IFN-γ. Because the selected CARbodies have unknown specificities, it will be mandatory to characterize and validate the target. An in-depth analysis of gene expression and protein expression in tissue arrays of the candidate antigen will be necessary to determine whether it may serve as potential target for antigen-directed immunotherapy approaches of cancer.
Although the current repertoire is clearly below that of microbial display systems,7 it could be scaled up to generate libraries covering complexities of 106–107.12 This level of complexity is sufficient for the screening of enriched phage-displayed scFv repertoires or repertoires present in immunized donors. Other mammalian cell surface display platforms have been developed.13,14,15,16 However, such methods, similar to phage-display, bacterial-display, and retrovirus-display are affinity-based selection systems.12,17 One potential advantage of lymphocyte display is the selection of antibodies towards antigens present in their natural setup, in the synapse between the tumor cell and the T lymphocyte. Antibodies against transmembrane cell surface proteins that are difficult to purify in a native conformation and against membrane-proximal epitope that are not visible with the current display technologies might benefit from this platform. This may have important implications in CAR-mediated cancer therapy. In fact, it has been recently published that targeting membrane-distal CD22 epitope with CAR-expressing T lymphocytes revealed defects in both degranulation and lytic granule targeting.18 CD22-specific CAR-positive T cells exhibited lower levels of maximum lysis and lower antigen sensitivity than CAR-positive T cells targeting CD20, which has a shorter extracellular domain than CD22. This diminished sensitivity was not a result of reduced avidity of antigen engagement, but instead reflected weaker signaling triggered by CAR molecule when targeting membrane-distal epitopes of CD22.18 Therefore, targeting membrane-proximal epitopes on tumor cell surfaces might be convenient to improve the therapeutic efficacy of adoptively transferred T cell expressing CAR.
Previous studies by our group and others revealed that there is a dynamic equilibrium between the CAR and the target antigen cell surface densities that optimally triggers T cell activation.19,20,21 Studies in Jurkat T cells expressing the same TCRζ-based CAR revealed that, for a given target antigen concentration, an optimal CAR density existed to achieve maximal interleukin-2 (IL-2) secretion, with lower CAR densities inducing submaximal responses and higher densities promoting activation-induced cell death.19 These experiments suggest that CAR expression density may control both target recognition and cell survival decisions.19 According to these data, we could hypothesize that during the post-transduction culture period, some Jurkat T cells displaying scFvs specific for abundant and ubiquitous antigens expressed on the T cell surface could die via activation-induced cell death. The result of this potential “negative selection-like” process would be a mixed population of: (i) activated T cells expressing CAR (CAR+ T cells) expanding in response to antigens expressed on the T cell surface, (ii) CAR+ T cells that do not recognize antigens expressed on the T cell surface, and (iii) cells that do not express CAR (CAR T cells) or express a density of CAR insufficient to trigger T cell activation (CARlow T cells). The “negative selection-like” process is a potential advantage of the T lymphocyte display platform compared with conventional mammalian and non-mammalian repertoire selection platforms. A reduction in the number of antibodies recognizing antigens expressed on the T cell surface will reduce the background binding of nonspecific antibodies. Whether this mechanism was not operational, or if their impact on diversity of the repertoire was minimal, we could pre-incubate the original library with normal cells of various tissue types, to remove reactive CD69+ clones recognizing ubiquitous antigens.
In summary, this repertoire selection platform would allow: (i) the generation of antibodies adapted to the display format (CAR) and the cell context (synapse), (ii) identifying new TAA, and (iii) even the generation of antibodies against problematic cell surface receptors (e.g., G-protein coupled receptor). Furthermore, the selected CARbodies can be expressed in soluble form and purified to identify the target antigen. For these reasons, we believe that CARbodies can extend the scope of adoptive cell therapy with CAR-redirected T cells. The display on the repertoire on the surface of primary T cells, fused to a second- or third-generation multidomain CAR context, could simplify the selection process, since effective triggering would promote CAR-T cell survival and expansion. The approach could also be easily adapted to in vivo selection processes in tumor-bearing mice, and in other relevant disease-specific animal models.
Materials and Methods
Cells and culture conditions. 293T cells (human embryo kidney epithelia; CRL-11268), HeLa cells (human cervix carcinoma; CCL-2) were grown in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) heat-inactivated fetal calf serum, (Invitrogen Life Technologies, Carlsbad, CA), referred as to Dulbecco's modified Eagle's medium complete medium (DCM). Jurkat clone E6-1 (TIB-152) and all the cell lines derived from the transduction of Jurkat cells were maintained in RPMI-1640 (Invitrogen Life Technologies) supplemented with heat-inactivated 10% fetal calf serum, referred as to RPMI complete medium (RCM). All of these cell lines were obtained from the American Type Culture Collection (Rockville, MD). HeLaCEA cells22 were grown in RCM supplemented with 750 µg/ml G418 (Invitrogen Life Technologies). All cell lines were routinely screened for the absence of mycoplasm contamination by PCR using the Mycoplasm Plus TM Primer Set (Stratagene, Cedar Creek, TX).
Antibodies and reagents. The mAbs used included: M2 (anti-FLAG; Sigma-Aldrich, St Louis, MO); BD1690 (anti-HIV p24; Abcam,Cambridge, UK); SPVT3b (anti-human CD3ɛ Zymed, San Francisco, CA); W6/32 (anti-human major histocompatibility complex class I; Abcam); AE1/AE3 (anti-human cytokeratin (type I and II); Dako Denmark A/S, Glostrup Denmark); 9E10 (anti-c-myc; Abcam); and BG5 (biotinylated anti-human IL-2; Endogen, Woburn, MA). For direct staining, the following are used: PE-conjugated FN50 mAb (anti-human CD69; BD Biosciences, San José, CA); FITC-conjugated B3821F4A mAb (anti-human CD45-FITC); RD1-conjugated SFC112T4D11 mAb (anti-human CD4); ECD-conjugated SFCI21Thy2D3 mAb (anti-human CD8); and PC5-conjugated UCHT1 mAb (anti-human CD3ɛ), all from Beckman Coulter, Fullerton, CA. The polyclonal antibodies used included: rabbit anti-human IL2 (Endogen), horseradish peroxidase-conjugated goat anti-mouse IgG (Fc specific; Sigma-Aldrich); PE-conjugated goat F(ab́)2 fragment anti-mouse IgG (Fc specific, Jackson Immuno Research, Newmarket, UK); IRDye800-conjugated donkey anti-mouse IgG (Rockland Immunochemicals, Gilbertsville, PA); and biotinylated goat anti-HIV p24 (Abcam). Streptavidin-horseradish peroxidase polymer and human IL-2, were from Sigma-Aldrich and HIV-p24 core protein was from Jena Bioscience (Jena, Germany). Recombinant human fibronectin CH-296 fragment (Retronectin) was from Takara Bio (Otsu, Japan).
LVs construction and preparation of LV stocks. LVs encoding the αCEA.CAR and the (EGFP reporter gene (pRRL.αCEA.TCRζ.IRES.EGFP) or the EGFP alone (pRRL.IRES.EGFP) were constructed as previously described.9 For the construction of the LV encoding the Griffin.CAR library and (pRRL.Griffin.CAR), a DNA preparation of the pHEN2 Griffin.1 scFv phagemid library10 was digested with ClaI and SfiI and the scFv-coding sequence DNA band was gel purified (Wizard SV Gel and PCR Clean Up System; Promega, Madison, WI). The coding sequence of the scFv repertoire was PCR amplified by performing nine different reactions with the combination of the degenerated primers 1–9 as forward primers with the primer 10 as reverse primer (Supplementary Table S1). The PCR product was directly cloned into the pCR2.1 vector using the TOPO TA Cloning kit (Life Technologies, Gaithersburg, MD). Electro Ten Blue Cells (Stratagene, La Jolla, CA) were electroporated with the pCR2.1-TOPO ligation (pCR2.1-TOPOGriffin). Up to 10 electroporations were performed at 1.8 kV, 25 µF, 200 Ω. Different dilutions of the bacteria pool of all electroporations were plated on Amp-plates for titration, fingerprinting, and sequencing to determine the size and diversity of the library. From the residual bacteria suspension, an aliquot was stored as a glycerol stock at −80 °C, and the rest of the cells were grown in a total volume of 400 ml 2TY-Amp overnight at 37 °C. A Qiagen Midi-Prep kit (Qiagen, Hilden, Germany) was used for purification of plasmid DNA.
To construct the vector pRRL.Griffin.CAR, the NheI/NotI fragment from pCR2.1-TOPOGriffin plasmid was ligated into NheI/NotI digested backbone of plasmid pRRL.αNIP.TCRζ.IRES.EGFP.9 A total amount of 10 electroporations were performed with the Electro Ten Blue Cells at 1.8 kV, 25 µF, 200 Ω. The bacteria suspension was processed as described above. The diversity of the LV library was 1.5 × 105 different clones.
To generate the LVs pRRL.αNIP.CARv2 and pRRL.αCEA.CARv2, encoding a second-generation CAR (CARv2) with the anti-NIP scFv B1.8 and the anti-CEA scFv MFE23, respectively and the EGFP reporter gene, the CD8α.CD28.TCRζ coding sequence was digested with NotI/BstBI from pMA.CD8α.CD28.TCRζ plasmid (synthesized by GeneArt AG, Regensburg, Germany) and cloned into the pRRL.αNIP.TCRζ.IRES.EGFP or pRRL.αCEA.TCRζ.IRES.EGFP backbone digested with the same enzymes.
To generate the LVs encoding the selected antibodies in the context of a CARv2 (pRRL.S3.45.CARv2, pRRL.S3.97.CARv2, and pRRL.S3.109.CARv2), the selected scFvs (S3.45, S3.97, and S3.109) were digested from the intermediate vector pCR2.1-TOPO with NheI/NotI and inserted into the backbone pRRL.αNIP.CARv2 digested with the same enzymes. Lentiviral particles (Supplementary Table S2) were produced by cotransfection of 293T cells by calcium phosphate precipitation as previously described.23
Determination of vector titers. An ELISA directed against the p24 capsid protein was performed for titering the physical particles. The physical particles number was in the range of 1 × 107–1 × 108 physical particles/ml in all the LV preparations. Biological titration was performed as previously described.23 The infective particles (transducing units/ml) were in the range between 1 × 107–4 × 107 transducing units/ml in all the LV preparations.
Genetic modification of human Jurkat T cells. For transduction with lentiviral stocks (Supplementary Table S2) exponentially growing Jurkat T cells (1 × 105) were spinoculated. Briefly, the plates were continuously centrifuged at 1,000g for 2½ hours at 32 °C in a flat-bottom 96-well plate with the appropriated amount of virus particles to get MOI ranging from 1 to 30, in a final volume of 200 µl of appropriate medium. After the spinoculation, the cells were cultured for 48 hours with the virus. For transduction with the Griffin.CAR lentivirus (LVGriffin.CAR), exponentially growing Jurkat T cells (2 × 107) were plated in 4 wells of a 6-well plate and the lentiviral stock was added at an MOI of ~1 in a final volume of 2 ml/well of their appropriate medium. After the spinoculation, the cells were cultured for 48 hours with the virus. Cells were analyzed for EGFP and CAR expression by flow cytometry.
Genetic modification of human primary PBLs. Human PBLs were isolated from healthy donors peripheral blood by density gradient centrifugation. Cells were expanded with anti-CD3 and anti-CD28 coated beads (Invitrogen Life Technologies), according to the manufacturer's instructions, in the presence of 50 IU/ ml of human recombinant interleukin-2 (Sigma-Aldrich). Afterwards, cells were placed in wells coated with Retronectin (10 µg/cm2) and transduced with appropriate lentiviral stocks (Supplementary Table S2) at an MOI of ~20 for 4 hours at 37 °C and 5% CO2. Half the medium was then replaced with fresh medium and cultures were incubated overnight at 37 °C and 5% CO2. The same procedure was repeated once again. After 48 hours, the cells were expanded for three additional days under the same conditions, and then with 50 IU/ml IL-2 for additional 7 days. EGFP and CAR expression was determined by flow cytometry.
Flow cytometry. For phenotypic analysis, cells were incubated with appropriate dilutions of fluorochrome conjugated mAbs. Cell surface expression of amino-terminal FLAG-tagged CAR was evaluated after incubation of cells with anti-FLAG mAb followed by incubation with PE-conjugated goat F(ab́)2 anti-mouse IgG. All samples were analyzed using a Beckman Coulter FC500 flow cytometer (Beckman Coulter).
Jurkat T cell activation and selection. For small-scale co-culture assays, non-transduced or transduced Jurkat cells (JurkatEGFP, JurkatαCEA.CAR, JurkatGriffin.CAR, JurkatGriffin.CAR/S1, JurkatGriffin.CAR/S2, JurkatGriffin.CAR/S3, JurkatαCEA.CARv2, JurkatS3.45.CARv2, JurkatS3.97.CARv2, or JurkatS3.109.CARv2) were incubated with HeLa or HeLaCEA cells at various (E:T ratios in round-bottom 96-well plates (BD Biosciences). The plates were incubated at 37 °C for 16 hours. The cells were then collected and the surface expression of CD69 was examined by FACS analysis. For medium-scale co-cultures, non-transduced Jurkat, or transduced Jurkat cells (JurkatαCEA.CAR, JurkatGriffin.CAR, JurkatGriffin.CAR/S1, and JurkatGriffin.CAR/S2) were incubated on HeLa monolayers, harvested and stained with anti-CD69 mAb as previously described.9 Double-positive cells (EGFP+CD69+) were isolated by FACS using a FACSVantage cell sorter (BD Biosciences) as previously described.9
IFN-γ release assay. Rested engineered human T cells (5 × 104) were incubated with HeLa or HeLaCEA (5 × 104) in round-bottom 96-well plates in a final volume of 200 ml of RCM. After 24 hours, co-culture supernatants were assayed for presence of IFN-γ using an ELISA kit, according to the manufacturer's instructions (Gen-Probe Diaclone, Besançon, France).
Sequence analysis. Total RNA was isolated from the original JurkatGriffin.CAR population and after each round of selection (JurkatGriffin.CAR/S1, JurkatGriffin.CAR/S2, and JurkatGriffin.CAR/S3) with the RNeasy Plus Mini Kit (Qiagen) and used for reverse transcription with cDNA Synthesis Kit for RT-PCR (AMV) (Roche Diagnostics, Mannheim, Germany). The scFv gene fragments were PCR amplified with primers 11 and 12 (Supplementary Table S1) and cloned into pCR2.1 using the TOPO TA Cloning kit. One Shot Stbl3 chemically competent E. coli cells (Invitrogen Life Technologies) were transformed with the pCR2.1 plasmid containing the repertoires (pre- and post-selections). From each population, a representative sample of 200 different clones was selected to perform a sequence analysis of the antibody variable domains with primers 13 and 14 (Supplementary Table S1). Sequences were analyzed using the SIB bioinformatics resource portal for proteomics; UTOPIA's CINEMA software to approximate framework and complementarity determining regions (CDR) and for protein alignments; and V-BASE2 database24 to determine the VH and VL family and genes of each scFv.
Statistical analysis of the composition of the CDRs. The frequencies of all possible combinations of three residues within the CDRs of either the VH or the VL were calculated using a Python (version 3.2) module that yields a multidimensional matrix with the composition of the three amino acids motif, the relative position in the CDR of each residue in the motif, and the number of cases. Because the interaction between the antibody and its ligand may involve non-contiguous amino acids from the different CDRs, all the possible combinations of three residues among the CDRs were considered. We analyzed the enrichment by comparing the relative abundance in the preselected repertoire and in the output of each round of selection by a Fisher's exact test. This P value of an amino acid triplet represents the probability that we would observe this or an even more imbalanced ratio (amino acid triplet in the preselected repertoire or in the output of the selection) by chance. Statistical significance was considered for P values <0.05. Due to the sheer number of triplets analyzed, it is expected that, by chance, some differences would be statistically significant between the preselected repertoire and the output. Therefore these differences were further assessed by a resampling simulation.25 Sequences from the output and the preselected repertoire were pooled and resampled into 500 permutated groups with the same size of the observed repertoires, which were later re-evaluated for the multidimensional matrix. Fisher's exact test was performed in the 500 randomized datasets, and distributions of the 100 lowest P values generated in each simulated test were graphically compared with the distribution of the 100 lowest P values from the experimental data.
Expression and purification of soluble scFv antibody fragments. For the expression of the selected scFv antibodies in bacteria, the plasmid pAB1 was digested with NcoI/NotI to introduce an oligonucleotide pair (Supplementary Table S1, oligonucleotides 15 and 16) carrying a polylinker between the promoter and the myc/6xHis tag; the resulting plasmid was named pAB2. The coding sequence of each of the selected scFvs, were extracted from plasmids pCR2.1 with NheI/NotI and cloned into the pAB2 backbone, digested with the same enzymes. E. coli TG1 cells were transformed with the appropriate vectors and soluble scFv fragments were obtained as described.26 Purification was performed using the AKTAprime plus system (affinity step: HisTrap or HiTrap rProtein A FF columns (GE Healthcare, Uppsala, Sweden)) according to the manufacturer's protocol and checked by ELISA and sodium dodecyl sulfate polyacrylamide gel electrophoresis. The purified scFvs were dialyzed against phosphate-buffered saline, quantified by Coomassie Plus Protein assay (Thermo Fisher Scientific Pierce, Rockford, IL) and analyzed by ELISA and sodium dodecyl sulfate polyacrylamide gel electrophoresis under reducing conditions using 4–12% gradient polyacrylamide gels (Invitrogen Life Technologies). For western blotting, protein samples were blotted onto nitrocellulose membranes (Invitrogen Life Technologies) and reacted with anti-c-myc mAb, followed by incubation with an IRDye800-conjugated donkey anti-mouse IgG. Visualization and quantitative analysis of protein bands were carried out with the Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE).
Cell ELISA for screening of soluble scFv antibody fragments. HeLa cells (2.6 × 104 cells/well) were plated in flat-bottom 96-well plates (BD Biosciences) and cultured overnight in 200 µl of appropriate medium to establish cell monolayers. Plates were washed with phosphate-buffered saline, fixed by air-drying and blocked for 1 hour at room temperature with 1% skimmed milk/phosphate-buffered saline. Then cell-free bacterial supernatant or purified scFvs, diluted at 50% with 4% skimmed milk, were added and incubated for 1 hour at room temperature. After washing six times with phosphate-buffered saline, binding of soluble scFv to the cell monolayers was detected with anti-c-myc mAb, followed by incubation with horseradish peroxidase-conjugated goat anti-mouse IgG antibody.
SUPPLEMENTARY MATERIAL Figure S1. Functional characterization of naive JurkatGriffin.CAR cells. Figure S2. Functional characterization of selected JurkatGriffin.CAR cells. Figure S3. Schematic representation of lentiviral vector encoding a second-generation CAR (CARv2). Table S1. Oligonucleotides used for library construction and sequencing. Table S2. Lentiviral vectors. Table S3. V gene usage and deduced amino acid sequences of CDR3 of unselected Griffin.CAR library. Table S4. V gene usage and deduced amino acid sequences of CDR3 after one round of activation/selection on HeLa cells. Table S5. V gene usage and deduced amino acid sequences of CDR3 after two rounds of activation/selection on HeLa cells. Table S6. V gene usage and deduced amino acid sequences of CDR3 after three rounds of activation/selection on HeLa cells.
Acknowledgments
This study was supported by grants from Ministerio de Ciencia e Innovación (BIO2008-03233), Ministerio de Economía y Competitividad (BIO2011-22738), and Comunidad de Madrid (S-BIO-0236-2006 and S2010/BMD-2312) to L.A.-V. V.A.-C. was supported by a predoctoral fellowship (BFI07.132) from the Gobierno Vasco. D.S.-M. was supported by a Comunidad de Madrid/Fondo Social Europeo Training grant (FPI-000531). M.C. was supported by Instituto de Salud Carlos III (Contrato Rio Hortega, CM06/00055). The authors declared no conflict of interest.
Supplementary material
Functional characterization of naive JurkatGriffin.CAR cells.
Functional characterization of selected JurkatGriffin.CAR cells.
Schematic representation of lentiviral vector encoding a second-generation CAR (CARv2).
Oligonucleotides used for library construction and sequencing.
Lentiviral vectors.
V gene usage and deduced amino acid sequences of CDR3 of unselected Griffin.CAR library.
V gene usage and deduced amino acid sequences of CDR3 after one round of activation/selection on HeLa cells.
V gene usage and deduced amino acid sequences of CDR3 after two rounds of activation/selection on HeLa cells.
V gene usage and deduced amino acid sequences of CDR3 after three rounds of activation/selection on HeLa cells.
References
- Alvarez-Vallina, L (2001). Genetic approaches for antigen-selective cell therapy. Curr Gene Ther 1: 385–397. [DOI] [PubMed] [Google Scholar]
- Gross, G, Waks, T and Eshhar, Z (1989). Expression of immunoglobulin-T-cell receptor chimeric molecules as functional receptors with antibody-type specificity. Proc Natl Acad Sci USA 86: 10024–10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz, L, Blanco, B and Alvarez-Vallina, L (2004). Antibodies and gene therapy: teaching old ‘magic bullets' new tricks. Trends Immunol 25: 85–91. [DOI] [PubMed] [Google Scholar]
- Lipowska-Bhalla, G, Gilham, DE, Hawkins, RE and Rothwell, DG (2012). Targeted immunotherapy of cancer with CAR T cells: achievements and challenges. Cancer Immunol Immunother 61: 953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzanzanica, D, Canevari, S, Mazzoni, A, Figini, M, Colnaghi, MI, Waks, T et al. (1998). Transfer of chimeric receptor gene made of variable regions of tumor-specific antibody confers anticarbohydrate specificity on T cells. Cancer Gene Ther 5: 401–407. [PubMed] [Google Scholar]
- Kershaw, MH, Teng, MW, Smyth, MJ and Darcy, PK (2005). Supernatural T cells: genetic modification of T cells for cancer therapy. Nat Rev Immunol 5: 928–940. [DOI] [PubMed] [Google Scholar]
- Hoogenboom, HR (2005). Selecting and screening recombinant antibody libraries. Nat Biotechnol 23: 1105–1116. [DOI] [PubMed] [Google Scholar]
- Lipes, BD, Chen, YH, Ma, H, Staats, HF, Kenan, DJ and Gunn, MD (2008). An entirely cell-based system to generate single-chain antibodies against cell surface receptors. J Mol Biol 379: 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Camino, V, Sánchez-Martín, D, Compte, M, Sanz, L and Alvarez-Vallina, L (2009). Lymphocyte display: a novel antibody selection platform based on T cell activation. PLoS ONE 4: e7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths, AD, Williams, SC, Hartley, O, Tomlinson, IM, Waterhouse, P, Crosby, WL et al. (1994). Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J 13: 3245–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Vallina, L and Hawkins, RE (1996). Antigen-specific targeting of CD28-mediated T cell co-stimulation using chimeric single-chain antibody variable fragment-CD28 receptors. Eur J Immunol 26: 2304–2309. [DOI] [PubMed] [Google Scholar]
- Urban, JH, Schneider, RM, Compte, M, Finger, C, Cichutek, K, Alvarez-Vallina, L et al. (2005). Selection of functional human antibodies from retroviral display libraries. Nucleic Acids Res 33: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli, RR, Bauer, M, Buser, RB, Gwerder, M, Muntwiler, S, Maurer, P et al. (2008). Isolation of human monoclonal antibodies by mammalian cell display. Proc Natl Acad Sci USA 105: 14336–14341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, M, Nagata, S and Pastan, I (2006). Isolation of anti-CD22 Fv with high affinity by Fv display on human cells. Proc Natl Acad Sci USA 103: 9637–9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taube, R, Zhu, Q, Xu, C, Diaz-Griffero, F, Sui, J, Kamau, E et al. (2008). Lentivirus display: stable expression of human antibodies on the surface of human cells and virus particles. PLoS ONE 3: e3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkowicz, R, Jager, GC and Nolan, GP (2005). A random peptide library fused to CCR5 for selection of mimetopes expressed on the mammalian cell surface via retroviral vectors. J Biol Chem 280: 15195–15201. [DOI] [PubMed] [Google Scholar]
- Winter, G, Griffiths, AD, Hawkins, RE and Hoogenboom, HR (1994). Making antibodies by phage display technology. Annu Rev Immunol 12: 433–455. [DOI] [PubMed] [Google Scholar]
- James, SE, Greenberg, PD, Jensen, MC, Lin, Y, Wang, J, Till, BG et al. (2008). Antigen sensitivity of CD22-specific chimeric TCR is modulated by target epitope distance from the cell membrane. J Immunol 180: 7028–7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Vallina, L and Russell, SJ (1999). Efficient discrimination between different densities of target antigen by tetracycline-regulatable T bodies. Hum Gene Ther 10: 559–563. [DOI] [PubMed] [Google Scholar]
- Turatti, F, Figini, M, Balladore, E, Alberti, P, Casalini, P, Marks, JD et al. (2007). Redirected activity of human antitumor chimeric immune receptors is governed by antigen and receptor expression levels and affinity of interaction. J Immunother 30: 684–693. [DOI] [PubMed] [Google Scholar]
- Alvarez-Vallina, L, Yañez, R, Blanco, B, Gil, M and Russell, SJ (2000). Pharmacologic suppression of target cell recognition by engineered T cells expressing chimeric T-cell receptors. Cancer Gene Ther 7: 526–529. [DOI] [PubMed] [Google Scholar]
- Hefta, LJ, Chen, FS, Ronk, M, Sauter, SL, Sarin, V, Oikawa, S et al. (1992). Expression of carcinoembryonic antigen and its predicted immunoglobulin-like domains in HeLa cells for epitope analysis. Cancer Res 52: 5647–5655. [PubMed] [Google Scholar]
- Compte, M, Blanco, B, Serrano, F, Cuesta, AM, Sanz, L, Bernad, A et al. (2007). Inhibition of tumor growth in vivo by in situ secretion of bispecific anti-CEA x anti-CD3 diabodies from lentivirally transduced human lymphocytes. Cancer Gene Ther 14: 380–388. [DOI] [PubMed] [Google Scholar]
- Retter, I, Althaus, HH, Münch, R and Müller, W (2005). VBASE2, an integrative V gene database. Nucleic Acids Res 33(Database issue): D671–D674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolonin, MG, Sun, J, Do, KA, Vidal, CI, Ji, Y, Baggerly, KA et al. (2006). Synchronous selection of homing peptides for multiple tissues by in vivo phage display. FASEB J 20: 979–981. [DOI] [PubMed] [Google Scholar]
- Sanz, L, Kristensen, P, Russell, SJ, Ramirez García, JR and Alvarez-Vallina, L (2001). Generation and characterization of recombinant human antibodies specific for native laminin epitopes: potential application in cancer therapy. Cancer Immunol Immunother 50: 557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Functional characterization of naive JurkatGriffin.CAR cells.
Functional characterization of selected JurkatGriffin.CAR cells.
Schematic representation of lentiviral vector encoding a second-generation CAR (CARv2).
Oligonucleotides used for library construction and sequencing.
Lentiviral vectors.
V gene usage and deduced amino acid sequences of CDR3 of unselected Griffin.CAR library.
V gene usage and deduced amino acid sequences of CDR3 after one round of activation/selection on HeLa cells.
V gene usage and deduced amino acid sequences of CDR3 after two rounds of activation/selection on HeLa cells.
V gene usage and deduced amino acid sequences of CDR3 after three rounds of activation/selection on HeLa cells.