Abstract
Dendritic cells (DCs) transfected with total amplified tumor cell RNA have the potential to induce broad antitumor immune responses. However, analytical methods required for quantitatively assessing the integrity, fidelity, and functionality of the amplified RNA are lacking. We have developed a series of assays including gel electrophoresis, northern blot, capping efficiency, and microarray analysis to determine integrity and fidelity and a model system to assess functionality after transfection into human DCs. We employed these tools to demonstrate that modifications to our previously reported total cellular RNA amplification process including the use of the Fast Start High Fidelity (FSHF) PCR enzyme, T7 Powerswitch primer, post-transcriptional capping and incorporation of a type 1 cap result in amplification of longer transcripts, greater translational competence, and a higher fidelity representation of the starting total RNA population. To study the properties of amplified RNA after transfection into human DCs, we measured protein expression levels of defined antigens coamplified with the starting total RNA populations and measured antigen-specific T cell expansion in autologous DC-T cell co-cultured in vitro. We conclude from these analyses that the improved RNA amplification process results in superior protein expression levels and a greater capacity of the transfected DCs to induce multifunctional antigen-specific memory T cells.
Keywords: dendritic cell, PCR, RNA, RNA amplification, RNA cap
Introduction
RNA is increasingly used in clinical applications.1,2 RNA-based therapeutics include inhibitors of messenger RNA (mRNA) translation (antisense RNA), the agents of RNA interference, catalytically active RNA molecules (ribozymes), and RNAs that bind proteins and other molecular ligands such as aptamers (reviewed in Burnett 2012). RNA can also be used to deliver the signals to attenuate or modify the phenotype or function of dendritic cells (DCs)3,4 or to express a protein to enhance a specific function directly in vivo.5,6 RNA is also used as a vehicle for antigen expression when injected by itself7,8 or in the context of DCs.9,10,11
The use of RNA as a source of antigen in autologous personalized immunotherapy has been demonstrated in a number of clinical trials.12,13 The use of amplified total cellular RNA as an antigen source precludes the need to identify or isolate specific tumor antigens, and enables targeting of unknown tumor antigens, including mutated antigens. In studies where the method of antigen delivery into DCs was by electroporation with RNA, more favorable antitumor immune responses could be demonstrated compared with other antigen delivery platforms, such as pulsing DCs with ultraviolet-irradiated tumor cells or tumor cell lysate.14,15 In addition, given the ability to amplify RNA, there is not a linear relationship between the amount of starting tumor material and the number of vaccine doses that can be manufactured, allowing applications in early stage disease in patients with minimal tumor burden.16
We have developed an RNA-loaded DC-based immunotherapy platform, which requires total tumor-amplified RNA.17 Previously, we reported improvements to the amplification process that eliminate amplification of antisense RNA species.18 The quality of RNA can be measured using standard biochemical and molecular biological techniques; however, to measure the impact of change in RNA quality on biological activity requires specialized approaches. For example, in one study, an increase in RNA purity was achieved by implementation of a novel purification method, high-performance liquid chromatography, which resulted in decreasing unwanted immune activation of the DCs and increased protein translation from that RNA.19 To demonstrate these improvements, defined model RNAs were used which allowed for detection of protein expression by conventional methods in conjunction with other assays that detect markers of immune activation.
Studies directed to optimize the quality of amplified RNA for maximal translation and antigen presentation in DCs are often conducted using model RNA, such as green fluorescent protein (GFP) RNA20,21,22,23 which upon transfection at high concentrations (microgram quantities per million of DCs) translates into GFP, easily detectable by standard flow cytometric techniques. However, the use of model RNAs does not address the translation of total amplified cellular RNA transfected into cells. The confounding issues of high molecular complexity make the detection of any single expressed protein from total cellular amplified RNA challenging.24,25 To circumvent some of these limitations, Grunebach et al. transfected GFP RNA into the renal cell carcinoma line, N43, extracted the RNA and subsequently used total RNA supplemented with the GFP RNA for transfection into DCs.26 Another group, Takahashi et al., used total RNA isolated from K562 cells which was stably transduced with a GFP-expressing vector and also was successful in detection of GFP in DCs several hours after transfection.27 However, GFP has a long half-life,28 and may not be a good surrogate for proteins encoded by total or amplified mammalian RNA. Therefore, new and better systems to assess the quality of amplified RNA in terms of translational competence are needed to explore the impact of amplification process modifications.
To achieve maximum protein expression from total amplified cellular RNA, the amplified RNA should maintain complexity, capture full-length transcripts, and preserve the relative abundance of various RNAs. We have developed a model system that not only allowed demonstration of protein expression from amplified RNA transfected into DCs but also provided a system to optimize the RNA amplification process to maximize protein expression thereby improving the immunopotency of RNA-transfected DCs.
Because of the high complexity of antigens present in the total RNA payload and difficulty with detection of individual proteins it was still necessary to utilize a model system, but instead of using GFP RNA our model system utilizes total cellular amplified RNA supplemented with a mixture of defined RNAs known to be overexpressed in tumor cells. The target antigens were tagged with a short hemagglutinin (HA) sequence and manipulated using various RNA amplification protocols. RNAs encoding HA-tagged proteins and total renal cell carcinoma (RCC) RNA was electroporated to DC. Following DC recovery post-electroporation, protein studies were conducted to measure the expression levels of selected target antigens as well as immunological assays directed toward one of the defined antigens, MART-1 (melanoma antigen recognized by T-cells 1). The impact of changes in the amplification protocol on biochemical quality of RNA structure, such as the capture of full-length transcripts and increases in capping efficiency of the RNA were demonstrated in a series of biochemical studies including northern blot, capping efficiency assay, and microarray studies. The protein studies together with immunological experiments, were in good agreement with the results of the biochemical analyses, demonstrating that the new amplification protocol results in an RNA payload which, after electroporation into the DCs, induces greater numbers of MART-1–specific T cells with an effector/memory cytotoxic T lymphocyte (CTL) phenotype (CD28+/CD45RA−) compared with the utilizing an RNA payload generated using the original protocol.
Results
Evaluation of improvements introduced in the amplification process in biochemical assays
Modifications made to the RNA amplification protocol are aimed at elevating protein expression levels in the RNA-electroporated DCs. A summary of all changes is shown as a schematic representation in Figure 1. In the new protocol, the PCR enzyme Advantage 2 is replaced by Fast Start High Fidelity (FSHF) enzyme as it offers similar properties as Advantage 2 and is available in an animal product-free formulation, which is preferred for manufacturing of clinical product intermediates. Also, the RNA-capping mechanism is changed from producing cotranscriptionally capped RNA to post-transcriptionally capped RNA. Previously, we have demonstrated that post-transcriptional capping of CD40L RNA results in a greater percentage of RNA that is capped and thus translation-competent.4 The post-transcriptional capping procedure, when applied to amplified RCC RNA, is expected to result in greater expression of the antigen RNAs. In addition, the post-transcriptionally added cap was treated with 2′-O-methyltransferase to generate a type 1 cap structure to increase translation efficiency of the RNA.
Figure 1.
Schematic of RNA amplification process. The total renal cell carcinoma (RCC) RNA (solid wave lines) is first converted into cDNA (dashed lines) in a reverse transcription reaction. In this reaction, the 5′ end is defined by the use of a Capswitch oligonucleotide and the fragment (solid straight line) is then amplified in a PCR reaction. The PCR step incorporates primers with modifications: 5′ primer contains T7 promoter sequences to allow for binding of RNA polymerase and transcription of cDNA template; 3′ primer contains (T)64 stretch which will convert to the polyA tail of RNA. Incorporation of a cap analogue occurs at or after the in vitro transcription stage. All variables which were considered as potential improvements in the process are indicated in italics. FSHF, Fast Start High Fidelity; IVT, in vitro transcribed.
Finally, the T7 Capswitch primer sequence was changed. The sequence of the T7 Capswitch primer described in Harris et al. is 5′-TAATACGACTCACTATAGGG+3AGGAAGCAGTGGTAACAACGCAGAGT-3′ and the sequence of the new T7 Powerswitch primer is 5′-GAATTT−17AATACGACTCACTATAG+1G+2T+3AGGAAGCAGTGGTAACAACGCAGAGT-3′ with the changed nucleotides indicated in bold. The numbers represent positions within a canonical T7 RNA polymerase (Pol) promoter where negative values indicate the non-transcribed nucleotides and positive values starting from G+1 represent the transcribed sequence. The extension of the canonical T7 RNA Pol promoter from −17 to −22 nucleotides using AT-rich sequences leads to an increase of T7 RNA Pol interaction with promoter sequences.29 In practice, this modification leads to an increased yield of transcribed RNA from the same mass of DNA templates (data not shown). To evaluate whether these changes to the amplification protocol represent an improvement, RCC RNA was amplified using either the original protocol described in Harris et al. (2005) or the newly developed protocol integrating FSHF, T7 Powerswitch primer, and a post-transcriptional method of RNA capping. Three biologically distinct RCC RNAs were used for this analysis and the biochemical characteristics of the RNA populations amplified using the two protocols were compared with each other.
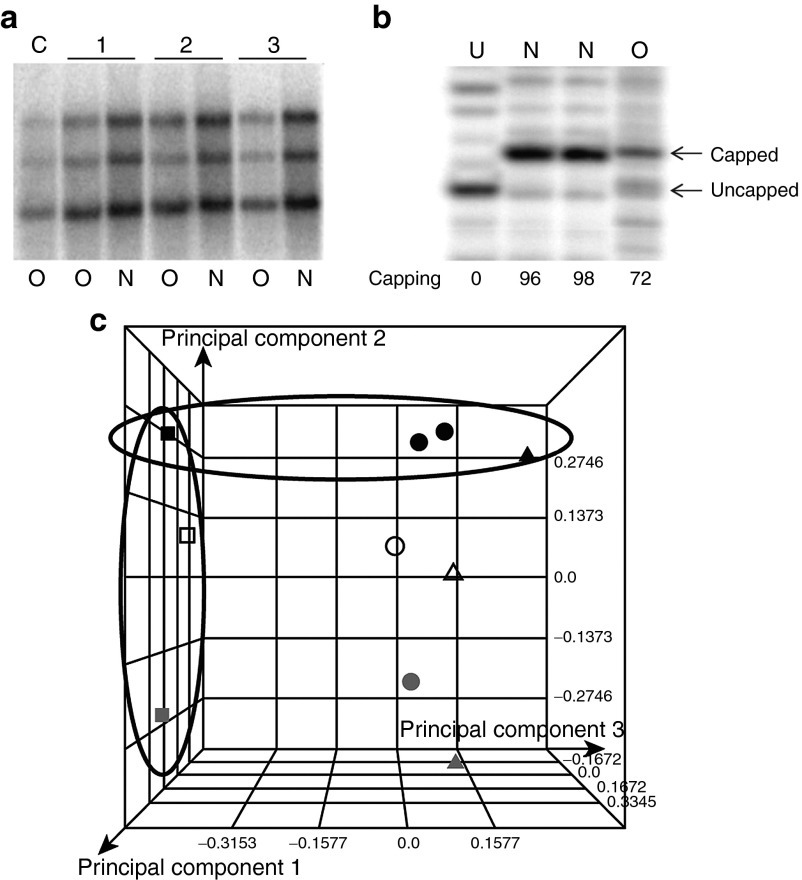
First, 10 µg of all amplified RNA samples were analyzed in a northern blot assay using a housekeeping gene probe complimentary to YWHAZ (Figure 2a). The northern blot analysis reveals the presence of distinct YWHAZ bands of expected sizes (1.0, 1.9, and 2.7 kb) in all RNA samples, indicating that both processes lead to the capture of mRNAs present in the starting total RNA population. However, the intensity of the YWHAZ transcripts is always greater in the samples generated using the new amplification protocol (Figure 2a, lanes N). Of particular note is the increased intensity of the highest molecular weight splice form. Next, the extent of RNA capping was tested using a capping efficiency assay and an example of the assay results is presented in Figure 2b. An uncapped counterpart RNA was generated which served as a control to position the cleaved product (Figure 2b, lane U). The percentage of capped RNA was determined for all three samples and is summarized in Table 1
Figure 2.
Biochemical characterization of RNAs amplified using the originally described protocol and newly developed amplification protocol. (a) Northern blot analysis of RNAs amplified using either original protocol (O) or new protocol (N) as indicated in the bottom of the panel. Lane C: interassay control for northern blot assay. Molecular weights of YWHAZ fragments recognized by the probe are indicated on the left. 1, 2, and 3 on top of the panel indicate three sets of RNA amplified from biologically distinct samples. (b) Capping efficiency assay on the RNAs amplified using original (old) and newly (new) developed protocol. The capping approaches for the RNAs are cotranscriptional and post-transcriptional, respectively. Lane U: sample obtained from RNA used to determine the migration of the cleaved product from uncapped RNA. Lanes O and N: samples obtained from final-capped RNA amplified using original and new protocols, respectively. Capping efficiency is expressed in percent of total RNA 5′ ends and indicated on the bottom of the panel. The position of cleaved products originating from capped RNA species and uncapped RNA species are indicated with arrows on the right. (c) Microarray data analyzed by principal component analysis of the variations found within total RNA and RNAs amplified using the two protocols. Squares circled as a group indicate the position of datasets obtained with three biologically distinct renal cell carcinoma RNA samples, not amplified. Triangles indicate the position of datasets obtained with RNA amplified using the original procedure from the three biological samples. Circles indicate the location datasets obtained with the three RNA samples amplified using the newly developed protocol. Black shapes circled indicate data sets originating from non-amplified RNA, amplified RNA using original and new protocol from the same biological sample. Duplicate black circles represent technical replicates in the microarray analyses conducted on one tumor RNA amplified using new protocol. The analysis is an average plot obtained over all three probes located along 5′–3′ axes of the transcript.
Table 1. Percent of capped RNA amplified from three biological samples.

Results of this analysis clearly demonstrate that all samples derived from the original method are capped with roughly 80% efficiency, consistent with expectations for the cotranscriptional capping method. However, all of the RNA samples generated using the new protocol have greater capping efficiency. The precision of the method developed to measure capping is 4–5%, so the capping efficiency obtained with RNA amplified using the new protocol is ~100%. This result is consistent within all three biological samples.
Microarray data (principal component analysis)
Initial experiments in microarray studies included overall quality control experiments. These control experiments included labeling total or amplified RNA and conducting duplicate hybridizations to the same or different slides. The data from all quality control experiments were within expectations for conventional gene expression microarrays. Replicate performance of experiments were within expectations: Technical replicate hybridizations feature identical starting sample (either total or amplified RNA), with independent target-labeling reactions and hybridizations and were reproducible (data not shown). When the technical variability was demonstrated to be highly consistent, the experimental design focused on biological replicates and on understanding the differences between the two amplification methods. The RNA probes were generated from three biologically distinct total tumor RNA or RNAs amplified using the original or newly developed RNA amplification protocol. Principal component analysis (Figure 2c) was used to plot the overall variability within the dataset. In this analysis, data points that are closest in space by dimension show the highest level of overall similarity to each other. The largest component of observed variation is along the axis that describes the amplification arms: PC1. The sample origin (biological differences) is expressed by the PC2 axis. This indicates that both amplification methods are highly reproducible and introduce a similar level of variation into each sample. This also indicates that both methods preserve the intrinsic biology of the samples. Comparison of the datasets obtained with RNA amplified using the original or modified protocol to the starting RNA population reveals that data points generated using the new amplification (Figure 2c, circles) protocol are positioned closer to the samples from which they originated (Figure 2c, squares).
Development of the model system
The mass of single-coding mRNA in a complex mixture of total RNA is vanishingly small and the use of model systems is unavoidable. To overcome limitations of model systems previously described in the literature, that employ large quantities of a defined RNA22 or foreign GFP RNA,27 we utilized four transcripts known to be frequently overexpressed in tumor tissue and known to be capable of eliciting immune responses in vitro. The four candidate RNAs were also selected for their diverse molecular weights which enable the investigation of the fidelity of the amplification process to amplify RNAs of different lengths. The RNAs used for this analysis were MAGE-3 (melanoma antigen family A, 3), PSA (prostate-specific antigen), survivin, and MART-1. The length of these RNAs, cDNAs, and molecular weights of the respective proteins are listed in Table 2. All four antigen cDNAs were modified with an HA-tag which allows the detection and enrichment of the protein using anti-HA tag antibody in downstream studies. The in vitro-transcribed RNAs were prepared and mixed to obtain a HA-tagged RNA mix. To study the ability of these RNAs to be amplified from the same matrix as total RCC RNA, the mixture of HA-tagged transcripts synthesized in vitro was added to the total RCC RNA (1:10 ratio). RCC total RNA, as well as HA-tagged RNAs added to RCC total RNA were amplified using the two protocols described in the Materials and Methods. The quality of the cDNA intermediate product was analyzed using non-denaturing agarose gel electrophoresis (Figure 3a). cDNA amplified from unmanipulated RCC RNA migrates as a smear on an agarose gel with some weak identifiable bands present (Figure 3a, lanes RCC), a common signature of a cDNA libraries. When comparing the quality of amplified RNA without HA antigens, more identifiable bands are present when the new amplification protocol was used. Amplified RCC RNA containing the mixture of HA-tagged RNAs displays a pattern with clearly identifiable bands of cDNAs on the RCC RNA background (Figure 3a, lanes RCC+HA). When comparing the two lanes containing amplified RCC RNA with the HA antigen mixture, the highest molecular weight band of 1.1 kb, corresponding to MAGE3 transcript, is present only when the new protocol is applied and not in the RCC RNA amplified using the old protocol. The cDNAs obtained in this experiment were in vitro-transcribed and the RNAs were analyzed using denaturing gel electrophoresis. Amplified RCC RNA migrates as a smear with some identifiable bands present (Figure 3b, lane RCC), which is consistent with a pattern present in a typical polyA+ RNA population. The pattern obtained on the denaturing gel when four HA-tagged RNAs were added to the RCC RNA before the amplification step reveals three clearly identifiable bands in the lane containing the RNA amplified using old process and four bands in the lane amplified using the newly developed protocol (Figure 3b, lane RCC+HA). Overall, the data obtained using this model system is in agreement with the data generated during the biochemical characterization of the RNAs by northern blot and microarray analyses.
Table 2. Sizes of RNA and proteins for HA-tagged proteins used in the study, as well as percentage of GC content of each cDNA.

Figure 3.
Model system to monitor amplification of model RNAs and their expression in the dendritic cells. (a) Non-denaturing agarose gel analysis of amplified cDNA. L: molecular weight ladder, lanes RCC: cDNA amplified from total RCC RNA. Lane RCC+HA: cDNA amplified from the HA-tagged RNAs mixed with RCC RNA before amplification. The cDNA were amplified using either the old (O) or new (N) protocol as indicated at the bottom. (b) Denaturing agarose gel analysis of in vitro-transcribed RNA. L: molecular weight ladder, lanes RCC: RNA amplified from total RCC RNA. Lane RCC+HA: in vitro-transcribed RNA amplified from the HA-tagged RNAs mixed with RCC RNA before amplification. The RNAs were amplified using either the old (O) or new (N) protocol as indicated at the bottom. HA, hemagglutinin; RCC, renal cell carcinoma.
The new amplification process for RNA results in greater antigen translation after electroporation to DCs
To study expression of proteins encoded by the amplified RNAs in DCs, the prepared RNAs were electroporated into DCs matured via the PME-CD40L DC maturation protocol30 with and without CD40L RNA added. Four hours post-electroporation recovery, the cells were washed and used to prepare total DC lysates. The DC lysates underwent immunoprecipitation using agarose-conjugated anti-HA tag antibody followed by detection by western blot using antigen– or HA-tag–specific antibodies. The negative control consisted of DCs electroporated with RCC RNA amplified using the newly developed protocol but without the HA RNAs added (Figure 4, lane NC). The positive control consisted of DCs electroporated with a mixture of the four in vitro-transcribed, capped, and polyadenylated HA-RNA transcripts (Figure 4, lane PC).
Figure 4.
Expression analysis of hemagglutinin (HA)-tagged model RNAs amplified using the two protocols in the electroporated dendritic cells (DCs). (a) Western blot analysis of immunoprecipitated (IP) complexes obtained from DC lysates electroporated with RNAs, using anti-HA tag antibodies. Protein molecular weight marker positions are indicated on the left. (b) Western blot analysis of IP complexes with specific antibodies as indicated on the right. Lanes contain IP complexes obtained from DCs electroporated as follows: Lane NC: using RCC RNA alone amplified via new protocol. Lane PC: using a mixture of the four model antigen RNAs transcribed in vitro and not amplified. Lanes Old: using RNA amplified via old amplification protocol from HA-tagged RNA-RCC mix. Lanes New: using RNA amplified via new amplification protocol from HA-tagged RNA-RCC mix. Plus and minus symbols at the top of the panel indicate the presence or absence of CD40L RNA in the RNA mix, respectively. RCC, renal cell carcinoma.
First, all of the immunoprecipitated complexes were analyzed with anti-HA antibody in a western blot assay (Figure 4a) and the four bands detected with anti-HA antibody are clearly identifiable in the positive control lane (Figure 4a, lane PC, indicated with arrows). In experimental conditions, only 14 and 17 kDa bands are detected in the four DC preparations electroporated with amplified RNA. The higher molecular weight bands of 33 and 36 kDa proteins are absent in the lysates generated using RNA amplified via old protocol and are present in the lysates generated using RNA amplified via the new protocol. To confirm the identity of the protein bands detected with anti-HA antibody, additional western blot analyses were conducted (Figure 4b). These experiments revealed that expression of survivin, migrating at 17 kDa can be detected in all DC preparations. Expression of MART-1 protein of 14 kDa is likewise detectable in all DC lysates electroporated with the RCC RNA amplified with the mixture of HA RNAs. However, the level of MART-1 expression is higher in the lysates prepared with the DCs electroporated with the RNAs amplified with the new protocol. An even more dramatic difference in the level of protein detection is evident with the use of anti-MAGE-3–specific antibodies, where only a weak trace is present in the lysates of DC electroporated with the RCC RNA amplified using the old method, and a stronger signal is detected in the lysates prepared with the RNA amplified using the new amplification protocol. The weak signal for MAGE-3 protein in the DC lysates generated with the RNA amplified using the old protocol is an expected result because of the lack of MAGE-3 cDNA and RNA in the amplified cDNA and RNA population, respectively. PSA antigen is detectable only when the new amplification protocol is utilized. These data confirm the expression of all four specific antigens in the model system as evidenced by the positive control. This validates our model system for the detection of protein expression in the electroporated DCs. Also, the expression of specific antigens is detected as well as in the lysates prepared with the amplified RNAs. This confirms that the process of RNA amplification results in the population of RNAs which are translation-competent after electroporation into DCs. In addition, these data demonstrate that the RNAs amplified using the new protocol result in greater expression of the antigens used. It is apparent that the use of the FSHF PCR enzyme in conjunction with post-transcriptional capping of the RNA results in both improved biochemical properties and increased translation of antigens encoded by the RNA. Lastly, these data demonstrate that the CD40L RNA present in the RCC RNA (3 µg of CD40L for every 2 µg of RCC RNA) does not inhibit the translation of the antigens encoded in the RCC RNA (Figure 4, compare positive and negative lanes). The amount of any model antigen detected in the sample without CD40L RNA is higher than the amount of same protein detected in the lane with CD40L RNA (Figure 4; compare positive and negative lane in the new process to each other). However, we believe that this is a consequence of the absolute mass of RCC RNA delivered per cell number, since cells prepared without CD40L RNA were electroporated using 5 µg of RCC RNA per million of DC whereas the cells prepared with CD40L RNA used only 2 µg of RCC RNA per million of DC with the difference of 3 µg corresponding to the CD40L RNA. Therefore, the difference in the signal is most likely a consequence of the RCC RNA mass added per million of cells and not due to inhibition by the expression of other proteins.
The new amplification process results in greater expansion of antigen-specific CTL
To evaluate the immunopotency of the RNA-transfected DC, cells were electroporated with the RCC RNA spiked with MART-1 RNA alone and amplified using either the original or improved method of RNA amplification. Following T cell co-culture, CTLs were analyzed using MART-1–specific pentamer and surface markers. Representative fluorescence-activated cell sorting plot of MART-1–specific pentamer staining is shown in Figure 5. The data demonstrate that the DC electroporated with the RNA amplified using improved protocol leads to increased absolute numbers of MART-1+ CTL. Data from one of two representative experiments is given in Table 3.
Figure 5.
Frequency of MART-1–specific cytotoxic T lymphocyte (CTL) after dendritic cell (DC) stimulation. The percentage of MART-1–specific CTL present after stimulation with DC containing RNA amplified using the (a) original process or the (b) novel process (B) was determined 3 days after the third round of DC stimulation. MART-1–specific CTLs were identified by labeling co-cultures with MART-1 PE-Pentamer and anti-CD8-PerCP-Cy5.5 antibody. One of two representative experiments is shown. MART-1, melanoma antigen recognized by T-cells 1.
Table 3. Evaluation of MART-1–specific cells and phenotype.

The numbers of MART-1–specific CTL were increased 3.35-fold from 13,153 to 31,353 cells when the new process to generate RNA was used. Furthermore, the RNA amplified using the new RNA amplification protocol lead to an increase in the overall percentage of CD28+/CD45RA− MART-1+ CTL from 74.2 to 91.5%. Next, the effector function of the MART-1–specific CTLs was assessed by intracellular cytokine staining (interferon-γ (IFN-γ), tumor necrosis factor-α (TNF-α), and interleukin-2 (IL-2)) and surface expression of CD107, a marker of lytic function. A 2.7-fold increase in the number of MART-1–specific CTL that express the degranulation marker CD107a as well as all three cytokines IFN-γ, TNF-α, and IL-2 was observed. There is a 2.1-fold increase in the number of MART-1+ CTL that express the degranulation marker CD107a as well as two cytokines IFN-γ and TNF-α. These two subsets of MART-1–specific CTL represent a population of CTL that have the capacity to mount a strong response upon antigen challenge.31,32
These analyses demonstrate that the RNA generated using the new process was also capable of enhancing the overall quality of the antigen-specific immune response by increasing the absolute numbers of multifunctional MART-1–specific CTL with lytic activity. Data from one of two representative experiments is shown (Table 4).
Table 4. Evaluation of MART-1–specific multifunctionality.

Discussion
The expanding role of RNA use in therapeutics necessitates the need for new and better analyses for both integrity and function. Ideally, methods of analysis will produce information on suitability of the material for the intended purpose. Traditionally, RNA integrity and size is evaluated by denaturing agarose slab gel electrophoresis to resolve and visualize RNA, and RNA integrity is evaluated either by eye or by gel imaging software. When total RNA is analyzed, some standardization of integrity assessment can be achieved by using the ratio of 28S and 18S ribosomal RNA. However, visual assessment of the 28S:18S ribosomal RNA ratio on agarose gels is subjective as the appearance of ribosomal RNA bands have been shown to be inconsistent as a consequence of electrophoresis conditions, amount of RNA loaded, and saturation of ethidium bromide fluorescence. We and others have found that measurements based on ribosomal RNA do not reflect the quality of mRNA (data not shown).33 More recently, a microfluidics-based platform, introduced by Agilent and followed by others, has provided a more standardized assay method for total nucleic acids for integrity and size determination.34 Still relying on gel electrophoresis principles, the sample is separated on microfabricated chips and detected via laser-induced fluorescence by the Bioanalyzer 2100 (Agilent, Santa Clara, CA). To standardize quality control for total RNA, Agilent has developed the RNA Integrity Number (RIN) software algorithm.35,36 However, these methods are not applicable to the mRNA populations that do not have distinct RNA bands nor do RNA integrity-based tests betray biological function of the RNA. To demonstrate antigen expression from RNA, analysis of translated protein detection should be employed. However, the use of amplified total tumor RNA results in the dilution of any one RNA transfected into DCs, resulting in limited level of protein expression. The use of immunological assays have demonstrated that transfection of DCs with amplified total tumor RNA is sufficient to generate antigenic peptides to tumor antigens recognized by T-lymphocytes,17,24,25,26,37 but direct detection of translated proteins in transfected cells remains elusive.
Therefore, we employed a variety of methods to evaluate the quality of amplified RNA and the level of translational competence. In the course of this work, we also implemented improvements which would lead to better RNA amplification, translation of antigens, and subsequent improvement in immunopotency of transfected DCs. To demonstrate improved RNA amplification due to change of 5′ T7 Capswitch primer and PCR enzyme, we employed agarose gel electrophoresis, northern blot, and microarray analyses. To analyze improvements in capping efficiency resulting from the change to the post-transcriptional capping method, we developed a capping efficiency assay. Increased protein translation which was anticipated as a result of the capping mechanism for amplified RNA was assessed by western blot detection of HA tag-mediated immunoprecipitated target proteins. Finally, the improvements in the RNA amplification protocol were tested in DC-T cell co-cell culture experiments to evaluate whether the improvements made to the biochemical properties of the RNA molecules such as abundance and increased capping efficiency combined with the new type of cap (type 1 cap) leads to a better antigen presentation and immunopotency in vitro. We developed a model system which involved the amplification and detection of four HA antigen RNAs allowing for assessment of whether specific RNAs of different lengths/sequences during the amplification procedure were captured but form the matrix of total cellular RNA. To overcome the issue of diluted mass of any single RNA in total population, this model system relies on enrichment of translated antigens via immunoprecipitation using anti-HA-tag antibody
Biochemical characterization of total amplified RNA was evaluated by the northern blot signal strength of the representative housekeeping gene, YWHAZ. The higher intensity signal specific for this probe was obtained in the RNA amplified using the new protocol compared with the original protocol. The increase in signal indicates an increase in the abundance of transcripts captured using the new amplification protocol and its superiority in comparison to the original protocol.
Microarray analyses conducted to study the composition and complexity of amplified RNA populations using three probes per gene analyzed and the data represented in the principal component analysis analysis is an average over all three probe types. This analysis demonstrated the superiority of the new amplification protocol which results in an amplified RNA population more closely resembling the starting total RNA population. The data for each individual probe type were interrogated as well (data not shown). Results demonstrate that the largest bias is introduced by the probe located at the 5′ end of the transcript. This is not unexpected since the process of RNA amplification is directional from 3′ to 5′ and the failure to produce the full-length transcripts due to the processivity of the PCR enzyme would result in this difference. It is noteworthy to mention that the target-labeling protocol utilized oligo dT for priming. Therefore, some of the differences in the detection of 5′ probes can be attributed to differences of the amplification protocols under investigation but also compounded by the nature of the target labeling for the analysis. Taken together, all of the results from the biochemical RNA characterization demonstrate that the new RNA amplification process results in higher quality RNA.
The change in RNA-capping approach from the cotranscriptional to the post-transcriptional method to derive a type 1 cap resulted in a pronounced increase in antigen translation as demonstrated by western blot analyses of the immunoprecipitated proteins from the DCs electroporated with RNAs amplified using two different protocols. The cap structure plays a role in several aspects of RNA metabolism including protection of the transcript from decay and initiation of cap-dependent translation. During cap-dependent translation, the 5′ cap functions as a recognition element for the eukaryotic translation initiation factor eIF4E which in turn helps recruit the small ribosomal subunit to the mRNA for the initiation of protein synthesis.38 In the originally described RNA amplification protocol, the cap is inserted via a cotranscriptional capping mechanism where a cap predecessor such as m7G(5′)ppp(5′)G (m7G) is substituted for a portion of the guanosine triphosphate in the reaction. Due to its close resemblance to the guanine base (G), a portion of the resulting fragments in such reaction is uncapped. Our data using the cotranscriptional capping method results in only 80% of all RNA transcripts being capped. The effective concentration of cotranscriptionally capped RNAs produced using the originally described RNA amplification protocol is further reduced because of the ability of the standard m7G cap analog to incorporate at the 5′ end of the RNA molecules in the reverse orientation 30–50% of the time.39 Based on the structural characterization of the interaction between the 5′ mRNA cap and the protein synthesis machinery, it is expected that RNAs with a reverse cap orientation are not competent for translation.40 This further reduces the pool of translation-competent RNA in the final RNA population to half of 80%, bringing it to ~40%. The use of anti-reverse cap analogue or other cap analogues instead of m7G cap can overcome this deficiency,41,42 but because they are incorporated as part of transcription step it will still not reach 100% capping efficiency. However, the new approach for capping tested here in a context of the amplified tumor RNA results in effectively 100% capping efficiency (Table 1 and Figure 2b). The new approach takes advantage of commercially available vaccinia virus capping enzyme which can modify RNA molecules synthesized in vitro in the absence of any cap and attach the cap in a post-transcriptional fashion, a process that more closely resembles the capping process in living cells.43 Additional enzyme, 2′-O-methyltransferase, which can also be added to the capping reaction catalyzes 2′-O-methylation of the first transcribed nucleotide in a capped RNA resulting in formation of a type 1 cap structure indistinguishable from the cap formed on RNA transcripts in the nuclei of eukaryotic cells. This enzyme can be used to convert capped RNAs from type 0 to type 1 structures. Kuge et al. have demonstrated that type 1 cap ribose methylation can enhance the efficiency of translation of an mRNA injected into Xenopus oocytes.44 The method of post-transcriptional capping and formation of type 1 cap structure was previously tested in our laboratory using CD40L mRNA and similar to results reported here lead to increased capping efficiency of CD40L RNA in the DCs. The 100% capping efficiency of CD40L RNA and type 1 cap in turn lead to increase of translation.4 The experiments conducted here demonstrate the antigen transcripts mixed with RCC RNA and then amplified using the new protocol and capping method lead to marked increase of antigen protein levels in electroporated DCs. Interestingly, the enhanced translation of RNA amplified using the new protocol correlated with an increase in the quality of this RNA when it was analyzed by the traditional method of denaturing agarose gel electrophoresis (Figure 3a). This highlights the need to supplement the established techniques for RNA quality assessment with the new methodologies. The difference in the amplified RNAs are detectable in the HA RNA-RCC RNA mix by the absence of MAGE-3 transcript in the HA RNA-RCC RNA mix amplified using the original protocol versus the newly developed protocol. This assessment was possible only due to a relatively large abundance of the MAGE-3 transcript in our model system. The MAGE-3 transcript was readily detectable only when the new amplification protocol was applied at the level of cDNA and in vitro-transcribed RNA (Figure 3). MAGE-3 GC content is 56% (Table 2) and is slightly elevated compared with the MART and survivin sequences 48 and 52%, respectively. The highest percent of GC content among four antigens studied was in the PSA sequence, 58%. It is possible that a combination of the length and a relatively high GC content prevented MAGE-3 from efficient amplification using the original protocol. It is important, however, that this deficiency in RNA amplification was overcome with the implementation of the new amplification protocol. Most likely the combination of new PCR enzyme and cycling conditions during the cDNA amplification step was responsible for correcting this deficiency.
The changes to the RNA amplification protocol leading to the increase of protein expression levels were also demonstrated to induce a more robust antigen-specific T cell response when transfected into DC. Previously reported results31 have shown that mature DCs electroporated with CD40L RNA can prime a unique population of antigen-reactive CTL that maintain CD28 receptor expression as well as exhibit both cytolytic function and the ability to secrete multiple types of cytokines. Therefore, improvements to the RNA antigen payload would be advantageous for enhancing antigen-specific immune responses. Results presented here show that the improved RNA payload generated using the new RNA amplification protocol can enhance the expansion of MART-1–specific CTL as well as increase the numbers of multifunction MART-1–specific CTL that exhibit a memory T cell phenotype. Improving the effector memory function of an antigen-specific response can impact the ability to generate strong immunity in vivo.45 Therefore, the new improved CD40L RNA has the potential to greatly impact the overall immune-generating potential of electroprated DCs.
The improvements evaluated here were conducted in a context of RCC RNA, but the conclusions are applicable to RNA of any biological origin where the starting RNA material is limiting and in need of high fidelity amplification. These situations include tumor samples obtained via needle biopsy or laser capture techniques when used for cell-free or RNA-transfected cellular immunization. The improved amplification protocol may also be useful for techniques such as whole transcriptome shotgun sequencing (RNAseq) or for the cloning of rare transcripts from isolated RNA.
Materials and Methods
Antigen templates. The HA-tag–coding sequence (YPYDVPDYA) was cloned into a pGEM4Z64A plasmid backbone at the 5′ end of coding regions of four antigens including MAGE-3, PSA, survivin, and MART-1.
Cloning manipulations left several cryptic ATG codons present between the T7 promoter and initiator methionine codon of each coding sequence. Therefore, a PCR fragment encoding each antigen lacking cryptic ATG codons was amplified using the plasmid template with coding regions and the following primers: 5′ middle UTR-HA tag (5′-GAATTTAATACGACTCACTATAGGA AAACTTCTCCCCGATCTGCGGCCACTGGACTGCCCATCAGCATGGCATACCCATACGACGTCCCAGACTACGCT-3′, T7 promoter in bold, CD40L 5′-UTR in plain text, HA-tag underlined) and 3′-UTR-64T (5′-T64AATTAAGAATTCGAGCTCGGTACC-3′). The resulting PCR fragments carried the T7 promoter, the CD40L 5′-UTR sequence, the HA-tag followed by the specific antigen-coding sequence and a T64 tail that will form a polyA tail of 64 adenosine residues in the RNA after in vitro transcription. These PCR fragments served as in vitro transcription templates to generate specific antigen RNAs that have CD40L 5′-UTRs, HA tags, and polyA tails of 64 adenosine residues.
In vitro transcription and capping. For the generation of cotranscriptionally capped RNA, in vitro transcription was carried out from the T7 promoter in each PCR antigen template using AmpliCap T7 High Yield Message Maker Kit from Cellscript (Madison, WI) as per the manufacturer's instructions. For the generation of uncapped RNA, in vitro transcription was carried out using AmpliScribe T7-Flash Transcription Kit from Cellscript as per the manufacturer's instructions. Uncapped RNAs were then post-transcriptionally capped using Cellscript ScriptCap m7G Capping System in conjunction with ScriptCap 2′-O-Methyltransferase to generate a type 1 cap structure.
Amplification of HA antigen RNA and RCC RNA. A detailed explanation of the original amplification procedure including primer sequences can be found in Harris et al.18
Briefly, either total RCC RNA, or HA antigen RNAs mixed with total RCC RNA at 1:10 ratio were amplified according to the following procedure. A total of 2 µg of RNA, 64T + oligo primer, and Capswitch RNA block primer (2 µmol/l final concentration each) were mixed in 40 µl total volume reaction. Primers were allowed to anneal to the RNA templates at 72 °C for 2 minutes before addition of 16 µl of 5X First Strand Buffer, 8 µl of 100 mmol/l DTT, 8 µl of 10 mmol/l dNTPs, and 8 µl Superscript II Reverse Transcriptase (200 U/µl; Invitrogen, Carlsbad, CA). Reverse transcription was carried out for 1 hour at 42 °C. A total of 20 µl of the reverse transcription reaction was added to a mixture containing 819 µl water, 105 µl of 10X Advantage 2 PCR Buffer, 21 µl of 10 mmol/l dNTPs, 42 µl of 10 µmol/l 64T + oligo primer, 21 µl of 20 µmol/l T7 Capswitch primer, and 21 µl of Advantage 2 Polymerase (Clontech, Mountain View, CA). A total of 100 µl of PCR reaction mix were aliquoted into 10 PCR tubes and PCR was carried out using the following program: 95 °C, 1 minute then 20 cycles of 95 °C, 5 seconds; 65 °C, 5 seconds; 68 °C, 6 minutes; 4 °C hold.
The modified amplification protocol utilized a different PCR enzyme, FSHF (Roche, Indianapolis, IN) as well as T7 Powerswitch primer instead of the T7 Capswitch primer. T7 Powerswitch primer (5′-GAATTTAATACG ACTCACTATAGG TAGGAAGCAGTGGTAACAACGCAGAG T-3′) contains an additional GAATT sequence at the 5′ end of the primer to stabilize the T7 RNA polymerase–DNA interaction29 and increases the yield of the RNA transcript synthesized by T7 RNA polymerase (I.Y. Tcherepanova, unpublished data). The amplification procedure with FSHF enzyme was as follows: 2 µg of RNA was annealed to Capswitch RNA block primer and 64T + oligo primer in a 10 µl reaction. After annealing, 4 µl of 5X First Strand Buffer, 2 µl of 100 mmol/l DTT, 2 µl of 10 mmol/l dNTPs, and 2 µl of Superscript II Reverse Transcriptase (200 U/µl; Invitrogen) were added to the annealing reaction. The first strand synthesis was carried out at 45 °C for 1 hour. The complete reverse transcription reaction volume was taken further into a 1,000 µl PCR reaction (aliquotted over 10 PCR tubes) containing 100 µl of FSHF reaction buffer with MgCl2, 40 µl of 10 µmol/l 64T+ oligo, 20 µl of 20 µmol/l Powerswitch T7 primer, 20 µl of PCR nucleotide mix (10 mmol/l each), and 20 µl of FSHF enzyme. PCR was carried out using the following program: 95 °C for 5 minutes followed by 10 cycles of 95 °C, 30 seconds; 65 °C, 30 seconds; and 67 °C, 6 minutes, then 10 cycles of 95 °C, 30 seconds; 65 °C, 30 seconds; 67 °C, 6 minutes with 5-second increase in each cycle, followed by 67 °C hold for 7 minutes and 4 °C hold. All amplified cDNAs were purified by Dynabeads SILANE genomic DNA (Invitrogen) kit and eluted in water.
Analysis of HA antigen RNA expression in DCs. A total of 75 µg amplified RNA was electroporated into 15 million matured DCs as described previously.30,31 After 5 hours incubation in AIM-V media (Invitrogen) containing the proteosome inhibitor lactacystin, cells were lysed and recovered protein was quantitated by the bicinchoninic acid assay. For each sample, 1.5 mg protein was immunoprecipitated with 50 µl of goat anti-HA antibodies conjugated to agarose (ab1233; Abcam, Cambridge, MA) in a total volume of 1 ml lysis buffer while rotating overnight at 4 °C. Immunoprecipitated proteins were analyzed by western blot analysis using antigen-specific rabbit polyclonal primary antibodies (PSA, NBP1-67675; Mart-1, NB 100-91859 (both from Novus Biologicals, Littleton, CO); Mage3, GTX 118244 (GeneTex, Irvine, CA); Survivin, NB 500-201 (Novus Biologicals); HA tag, mouse monoclonal MMS-101P (Covance, Dedham, MA); 1:500 of PSA, Mart1, and Mage3 antibodies, 1:1,000 of Survivin and HA-tag antibodies; overnight incubation) and mouse anti-rabbit IgG conjugated HRP secondary antibodies (sc-2357; Santa Cruz Biotechnologies, Santa Cruz, CA) or goat anti-mouse HRP secondary antibodies (sc-2055; Santa Cruz Biotechnologies). Membrane blocking and incubation with the primary antibody was carried out using 3% BSA (Sigma, St Louis, MO) in phosphate-buffered saline (PBS; Lonza, Allendale, NJ) supplemented with 0.1% Tween-20 (PBS-T). Incubation with secondary antibody was carried out using PBS-T. Detection was achieved using ECL Plus (Pierce, Rockford, IL) and the Storm 840 Imager with the fluorescence settings.
Northern blot analysis of amplified RNA. Ten micrograms of amplified RCC RNA was resolved on a denaturing agarose gel and transferred to a membrane according to the standard procedures. YWHAZ transcripts were detected using specific probe generated by DECAprime II (Ambion, Austin, TX) labeling of the respective human phospholipase A2 cDNA fragment.
Capping efficiency measurement of amplified RCC RNA. The capping efficiency of amplified RNA was measured essentially as described in Tcherepanova et al.4 with the following modifications: amplified RCC RNA and HA antigen RCC RNA 1:10 were used instead of CD40L RNA as a substrate and the DNA oligonucleotide 5′-GCGTACTCTGCGTTG-3′ was used in lieu of 5′-CGCTCGAGCATGCAT/3ddC/-3′. Briefly, in this assay, RNAse H directed cleavage is carried out with an oligo that hybridizes near the 5′ end of the transcript. The common sequence in all transcripts was defined by a T7 Powerswitch primer used in the amplification protocol. Subsequently, the cleavage products are radioactively end-labeled with T4 RNA ligase and visualized by electrophoresing on an 8 mol/l urea, 15% polyacrylamide gel followed by scanning of the image by Storm 840 scanner (Amersham Biosciences, Piscataway, NJ). 5′ RNA cleavage products resulting from capped RNA migrate one nucleotide slower on the denaturing 15% polyacrylamide gel compared with those derived from the uncapped RNA. The position of the fragments from uncapped RNAs are obtained from treatment using uncapped RNA population. The capped product migrates one nucleotide slower. The sum of the signal intensity (measured by ImageQuant Software, version 5.2; GE Healthcare Bio-Sciences, Piscataway, NJ) derived from capped and uncapped bands within capped RNA lane is taken as 100% and the percent of capped RNA (higher molecular weight band) is described in relation to the total density from the two cleavage products.
Microarray analysis of the relative abundance of amplified transcripts and principal component analysis. To evaluate the quality of RNA captured by amplification protocol on population level, microarray analysis was conducted. Custom microarray was generated as follows:
Randomly chosen transcripts with RefSeq IDs collection was nominated which also contained the list of genes deposited into the Center for Biotechnology Information Gene Expression Omnibus database under the accession number GSE6344.46 That data set contained a list of genes overexpressed in RCC clear type, generated in a genomic profiling of RCC clear type and matched normal tissue samples in gene expression between tumor tissue and normal kidney. Total number of genes nominated was limited to 15,000. Each reference sequence was divided into three segments of roughly equal length and hybridization probe was designed for each sequence segment resulting in the array design with three probes per transcript spaced along the 3′/5′ axis. Microarrays were printed using ~45,000 probes (Agilent). Labeling of RNA, hybridization, and data collection were done according to the manufacturer's protocol (Agilent). For more information, refer to Supplementary Materials and Methods and GEO study accession number GSE37866.
Expansion of MART-1–specific cytotoxic T cells. RNA-electroporated mature DCs were used to expand MART-1–specific CTL as previously described.30 Briefly, mature DC electroporated with MART-1 RNA were co-cultured with CD8+ T cells at a 10:1 CD8+ T cell to DC ratio in 6-well tissue culture dishes (Corning, Corning, NY) in R-10 medium (10% FBS (Atlanta Biologicals, Norcross, GA), RPMI-1640 (supplemented with 10 mmol/l HEPES, pH 7.4; 1 mmol/l sodium pyruvate; 0.1 mmol/l Non-essential amino acids; 2 mmol/l sodium glutamate, 55 µmol/l 2-mercaptoethanol (Invitrogen))). Co-cultures were maintained in medium supplemented with 0.2 U/ml IL-2 and 10 ng/ml IL-7 (R&D Systems, Minneapolis, MN) for the first 3 days. On day 3, the medium was supplemented with 20 U/ml IL-2. CD8+ T cells were stimulated on days 7 and 14 with fresh DC stimulators at a 10:1 ratio in R10 medium supplemented with 20 U/ml IL-2 and 10 ng/ml IL-7. Detection of MART-1–specific CTL present in the co-cultures as well as all functional assays were performed 3 days after the last cycle of stimulation on day 17.
Flow cytometry and intracellular cytokine labeling. To determine CTL intracellular cytokine labeling, in combination with cell surface labeling, responder CTL were analyzed as previously described.32 Briefly on day 17, CTL were harvested and stimulated with MART-1 peptide-pulsed T2 target cells or T2 target cells pulsed with an irrelevant peptide at a ratio of 10:1 for 4 hours. During this incubation time, 5 µl of anti-CD107a-PEcy5 antibody (eBioscience, San Diego, CA) with Brefeldin A and Monensin (BD Biosciences, San Jose, CA) were added to determine degranulation. After the 4-hour timepoint, MART-1–specific CTL were identified by labeling with PE-labeled HLA-A*201/ELAGIGILTV Pentamer (MART-1 PE-Pentamer) (Proimmune, Sarasota, FL) for 10 minutes at room temperature. MART-1 PE-Pentamer–labeled cells were washed with FACS buffer (PBS (Cambrex, Walkersville, MD), 2% FBS, 0.01% sodium azide (Sigma), and 2 mmol/l EDTA (Sigma)), then labeled with the following human-specific antibodies, anti-CD8-ECD (Beckman Coulter, Brea, CA), anti-CD28-APC, anti-CD45RA-FITC (BD Biosciences). For intracellular cytokine determination, CTL were fixed then permeabilized using the eBioscience buffer sets. Cells were then labeled with anti-IL-2-PerCP-cy5.5 (BioLegend, San Diego, CA), anti-IFN-γ-PE-cy7 (eBioscience), and anti-TNF-α–AF700 (BD Biosciences) antibodies in the presence of Permeabilization buffer (eBioscience) to detect intracellular cytokine expression. Cells were washed in FACS buffer and fluorescence absolute cells' counts were determined using TruCOUNT counting beads (BD Biosciences). A total of 300,000–400,000 lymphocyte gated events for each sample were collected using a LSRII (BD Biosciences). Data was analyzed using Boolean gating with FlowJo software (Tree Star, Ashland, OR) to define the MART-1+/CD28+/CD45RA− CTL subset positive for CD107, IFN-γ, TNF-α or IL-2 background gating was determined by setting gates on the MART-1+ CD28+ CTL population that was restimulated in the intracellular staining assay with T2 target cells pulsed with an irrelevant peptide.
Acknowledgments
We thank Bob Walker for help in preparation of the manuscript. Argos Therapeutics is a privately held company. All authors are present- or past-salaried employees of Argos Therapeutics, Inc.
Supplementary Material
References
- Castanotto, D and Rossi, JJ (2009). The promises and pitfalls of RNA-interference-based therapeutics. Nature 457: 426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett, JC and Rossi, JJ (2012). RNA-based therapeutics: current progress and future prospects. Chem Biol 19: 60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creusot, RJ, Chang, P, Healey, DG, Tcherepanova, IY, Nicolette, CA and Fathman, CG (2010). A short pulse of IL-4 delivered by DCs electroporated with modified mRNA can both prevent and treat autoimmune diabetes in NOD mice. Mol Ther 18: 2112–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherepanova, IY, Adams, MD, Feng, X, Hinohara, A, Horvatinovich, J, Calderhead, D et al. (2008). Ectopic expression of a truncated CD40L protein from synthetic post-transcriptionally capped RNA in dendritic cells induces high levels of IL-12 secretion. BMC Mol Biol 9: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó, K, Muramatsu, H, Keller, JM and Weissman, D (2012). Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol Ther 20: 948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kormann, MS, Hasenpusch, G, Aneja, MK, Nica, G, Flemmer, AW, Herber-Jonat, S et al. (2011). Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat Biotechnol 29: 154–157. [DOI] [PubMed] [Google Scholar]
- Rittig, SM, Haentschel, M, Weimer, KJ, Heine, A, Muller, MR, Brugger, W et al. (2011). Intradermal vaccinations with RNA coding for TAA generate CD8+ and CD4+ immune responses and induce clinical benefit in vaccinated patients. Mol Ther 19: 990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weide, B, Carralot, JP, Reese, A, Scheel, B, Eigentler, TK, Hoerr, I et al. (2008). Results of the first phase I/II clinical vaccination trial with direct injection of mRNA. J Immunother 31: 180–188. [DOI] [PubMed] [Google Scholar]
- Bringmann, A, Held, SA, Heine, A and Brossart, P (2010). RNA vaccines in cancer treatment. J Biomed Biotechnol 2010: 623687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gulck, E, Vlieghe, E, Vekemans, M, Van Tendeloo, VF, Van De Velde, A, Smits, E et al. (2012). mRNA-based dendritic cell vaccination induces potent antiviral T-cell responses in HIV-1-infected patients. AIDS 26: F1–12. [DOI] [PubMed] [Google Scholar]
- Routy, JP, Boulassel, MR, Yassine-Diab, B, Nicolette, C, Healey, D, Jain, R et al. (2010). Immunologic activity and safety of autologous HIV RNA-electroporated dendritic cells in HIV-1 infected patients receiving antiretroviral therapy. Clin Immunol 134: 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Z, Dannull, J, Yang, BK, Dahm, P, Coleman, D, Yancey, D et al. (2005). Telomerase mRNA-transfected dendritic cells stimulate antigen-specific CD8+ and CD4+ T cell responses in patients with metastatic prostate cancer. J Immunol 174: 3798–3807. [DOI] [PubMed] [Google Scholar]
- Heiser, A, Coleman, D, Dannull, J, Yancey, D, Maurice, MA, Lallas, CD et al. (2002). Autologous dendritic cells transfected with prostate-specific antigen RNA stimulate CTL responses against metastatic prostate tumors. J Clin Invest 109: 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benencia, F, Courrèges, MC and Coukos, G (2008). Whole tumor antigen vaccination using dendritic cells: comparison of RNA electroporation and pulsing with UV-irradiated tumor cells. J Transl Med 6: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, K, Zhao, JJ, Wang, H, Li, JJ, Liang, XT, Sun, JC et al. (2010). Comparative analysis of cytotoxic T lymphocyte response induced by dendritic cells loaded with hepatocellular carcinoma -derived RNA or cell lysate. Int J Biol Sci 6: 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiser, A, Maurice, MA, Yancey, DR, Wu, NZ, Dahm, P, Pruitt, SK et al. (2001). Induction of polyclonal prostate cancer-specific CTL using dendritic cells transfected with amplified tumor RNA. J Immunol 166: 2953–2960. [DOI] [PubMed] [Google Scholar]
- Boczkowski, D, Nair, SK, Nam, JH, Lyerly, HK and Gilboa, E (2000). Induction of tumor immunity and cytotoxic T lymphocyte responses using dendritic cells transfected with messenger RNA amplified from tumor cells. Cancer Res 60: 1028–1034. [PubMed] [Google Scholar]
- Harris, J, Monesmith, T, Ubben, A, Norris, M, Freedman, JH and Tcherepanova, I (2005). An improved RNA amplification procedure results in increased yield of autologous RNA transfected dendritic cell-based vaccine. Biochim Biophys Acta 1724: 127–136. [DOI] [PubMed] [Google Scholar]
- Karikó, K, Muramatsu, H, Ludwig, J and Weissman, D (2011). Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res 39: e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami, K, Yamaguchi, Y, Ohshita, A, Kawabuchi, Y, Ohta, K, Hihara, J et al. (2005). Generation of antigen-presenting cells using cultured dendritic cells and amplified autologous tumor mRNA. Oncology 69: 399–407. [DOI] [PubMed] [Google Scholar]
- Markovic, SN, Dietz, AB, Greiner, CW, Maas, ML, Butler, GW, Padley, DJ et al. (2006). Preparing clinical-grade myeloid dendritic cells by electroporation-mediated transfection of in vitro amplified tumor-derived mRNA and safety testing in stage IV malignant melanoma. J Transl Med 4: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalady, MF, Onaitis, MW, Padilla, KM, Emani, S, Tyler, DS and Pruitt, SK (2002). Enhanced dendritic cell antigen presentation in RNA-based immunotherapy. J Surg Res 105: 17–24. [DOI] [PubMed] [Google Scholar]
- Van Tendeloo, VF, Ponsaerts, P, Lardon, F, Nijs, G, Lenjou, M, Van Broeckhoven, C et al. (2001). Highly efficient gene delivery by mRNA electroporation in human hematopoietic cells: superiority to lipofection and passive pulsing of mRNA and to electroporation of plasmid cDNA for tumor antigen loading of dendritic cells. Blood 98: 49–56. [DOI] [PubMed] [Google Scholar]
- Van Tendeloo, VF, Ponsaerts, P and Berneman, ZN (2007). mRNA-based gene transfer as a tool for gene and cell therapy. Curr Opin Mol Ther 9: 423–431. [PubMed] [Google Scholar]
- Mitchell, DA and Nair, SK (2000). RNA-transfected dendritic cells in cancer immunotherapy. J Clin Invest 106: 1065–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünebach, F, Müller, MR, Nencioni, A and Brossart, P (2003). Delivery of tumor-derived RNA for the induction of cytotoxic T-lymphocytes. Gene Ther 10: 367–374. [DOI] [PubMed] [Google Scholar]
- Takahashi, M, Narita, M, Ayres, F, Satoh, N, Abe, T, Yanao, T et al. (2003). Cytoplasmic expression of EGFP in dendritic cells transfected with in vitro transcribed mRNA or cellular total RNA extracted from EGFP expressing leukemia cells. Med Oncol 20: 335–348. [DOI] [PubMed] [Google Scholar]
- Corish, P and Tyler-Smith, C (1999). Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng 12: 1035–1040. [DOI] [PubMed] [Google Scholar]
- Tang, GQ, Bandwar, RP and Patel, SS (2005). Extended upstream A-T sequence increases T7 promoter strength. J Biol Chem 280: 40707–40713. [DOI] [PubMed] [Google Scholar]
- Calderhead, DM, DeBenedette, MA, Ketteringham, H, Gamble, AH, Horvatinovich, JM, Tcherepanova, IY et al. (2008). Cytokine maturation followed by CD40L mRNA electroporation results in a clinically relevant dendritic cell product capable of inducing a potent proinflammatory CTL response. J Immunother 31: 731–741. [DOI] [PubMed] [Google Scholar]
- DeBenedette, MA, Calderhead, DM, Ketteringham, H, Gamble, AH, Horvatinovich, JM, Tcherepanova, IY et al. (2008). Priming of a novel subset of CD28+ rapidly expanding high-avidity effector memory CTL by post maturation electroporation-CD40L dendritic cells is IL-12 dependent. J Immunol 181: 5296–5305. [DOI] [PubMed] [Google Scholar]
- DeBenedette, MA, Calderhead, DM, Tcherepanova, IY, Nicolette, CA and Healey, DG (2011). Potency of mature CD40L RNA electroporated dendritic cells correlates with IL-12 secretion by tracking multifunctional CD8(+)/CD28(+) cytotoxic T-cell responses in vitro. J Immunother 34: 45–57. [DOI] [PubMed] [Google Scholar]
- Godler, DE, Loesch, DZ, Huggins, R, Gordon, L, Slater, HR, Gehling, F et al. (2009). Improved methodology for assessment of mRNA levels in blood of patients with FMR1 related disorders. BMC Clin Pathol 9: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleige, S and Pfaffl, MW (2006). RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med 27: 126–139. [DOI] [PubMed] [Google Scholar]
- Imbeaud, S, Graudens, E, Boulanger, V, Barlet, X, Zaborski, P, Eveno, E et al. (2005). Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res 33: e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, A, Mueller, O, Stocker, S, Salowsky, R, Leiber, M, Gassmann, M et al. (2006). The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa, E and Vieweg, J (2004). Cancer immunotherapy with mRNA-transfected dendritic cells. Immunol Rev 199: 251–263. [DOI] [PubMed] [Google Scholar]
- Gingras, AC, Raught, B and Sonenberg, N (1999). eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68: 913–963. [DOI] [PubMed] [Google Scholar]
- Pasquinelli, AE, Dahlberg, JE and Lund, E (1995). Reverse 5' caps in RNAs made in vitro by phage RNA polymerases. RNA 1: 957–967. [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano, J, Gingras, AC, Sonenberg, N and Burley, SK (1997). Cocrystal structure of the messenger RNA 5' cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89: 951–961. [DOI] [PubMed] [Google Scholar]
- Stepinski, J, Waddell, C, Stolarski, R, Darzynkiewicz, E and Rhoads, RE (2001). Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogs 7-methyl(3'-O-methyl)GpppG and 7-methyl (3'-deoxy)GpppG. RNA 7: 1486–1495. [PMC free article] [PubMed] [Google Scholar]
- Grudzien-Nogalska, E, Kowalska, J, Su, W, Kuhn, AN, Slepenkov, SV, Darzynkiewicz, E et al. (2013). Synthetic mRNAs with superior translation and stability properties. In: Rabinovich, PM (ed.). Synthetic Messenger RNA and Cell Metabolism Modulation: Methods and Protocols, Methods in Molecular Biology, vol. 969. Springer Science+Business Media: New York. pp. 55–73. [DOI] [PubMed] [Google Scholar]
- Venkatesan, S, Gershowitz, A and Moss, B (1980). Modification of the 5' end of mRNA. Association of RNA triphosphatase with the RNA guanylyltransferase-RNA (guanine-7-)methyltransferase complex from vaccinia virus. J Biol Chem 255: 903–908. [PubMed] [Google Scholar]
- Kuge, H, Brownlee, GG, Gershon, PD and Richter, JD (1998). Cap ribose methylation of c-mos mRNA stimulates translation and oocyte maturation in Xenopus laevis. Nucleic Acids Res 26: 3208–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seder, RA, Darrah, PA and Roederer, M (2008). T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol 8: 247–258. [DOI] [PubMed] [Google Scholar]
- Gumz, ML, Zou, H, Kreinest, PA, Childs, AC, Belmonte, LS, LeGrand, SN et al. (2007). Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res 13: 4740–4749. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.