Abstract
Sunitinib is a multitargeting tyrosine kinase inhibitor used for metastatic renal cancer. There are no biomarkers that can predict sunitinib response. Such markers are needed to avoid administration of costly medication with side effects to patients who would not benefit from it. We compared global miRNA expression between patients with a short (≤12 months) versus prolonged (>12 months) progression-free survival (PFS) under sunitinib as first-line therapy for metastatic renal cell carcinoma. We identified a number of differentially expressed miRNAs and developed miRNA statistical models that can accurately distinguish between the two groups. We validated our models in the discovery set and an independent set of 57 patients. Target prediction and pathway analysis showed that these miRNAs are involved in vascular endothelial growth factor (VEGF), TGFβ, and mammalian target of rapamycin (mTOR)-mediated signaling and cell–cell communication. We tested the effect of these miRNAs on cellular proliferation and angiogenesis. We validated the negative correlation between miR-221 and its target, VEGFR2.miR-221 overexpression was associated with a poor PFS while its target, VEGFR2 was associated with longer survival. Gain of function experiments showed that miR-221 and miR-222 decreased angiogenesis and cellular proliferation in human umbilical vein endothelial cells (HUVEC) while increasing cellular proliferation in ACHN cells. miRNAs represent potential predictive markers for sunitinib response.
Introduction
Clear cell renal cell carcinoma (ccRCC) is one of the top 10 cancers in North America.1 It is an aggressive tumor with 20–30% of patients presenting with metastasis at diagnosis. A median overall survival (OS) of 28–29 months has been reported in recent targeted therapy trials.2,3
VEGFR-2 and PDGFR-β play a key role in the VHL-Hypoxia pathway. This is the main pathway involved in ccRCC pathogenesis, in which HIF1-α enhances the transcription of multiple proangiogenic and growth factors including the vascular endothelial growth factor (VEGF) and platelet-derived growth factor (PDGF). These factors subsequently activate a number of downstream pathways by binding mainly to VEGFR-2 and PDGFR-β.
Sunitinib, a first-line treatment for metastatic RCC (mRCC),4 is a multitarget receptor tyrosine kinase inhibitor, which inhibits the tyrosine kinase activity of several receptors including vascular endothelial growth factor receptors (VEGFR-1, VEGFR-2, and VEGFR-3), platelet-derived growth factor receptors (PDGFR-α and PDGFR-β), Fms-like tyrosine kinase 3 (FLT3), stem cell growth factor receptor KIT and rearranged during transfection (RET). The VEGFR2 encoding gene is kinase insert domain receptor “KDR” in humans. It is highly expressed in endothelial cells.5 Sunitinib is associated with a response rate of 30–45% and a progression-free survival (PFS) of 9.5–11 months.2,6 However, over 20% of patients are refractory to tyrosine kinase inhibitor (TKI) therapy.7 Causes of resistance are not fully understood but a number of possible mechanisms have been suggested.8,9,10 The discovery of biomarkers that could predict patients' response to sunitinib would improve cost-effectiveness by allowing selecting patients who are likely to respond while other patients could be directed to therapies with a different mechanism of action or for clinical trials of novel therapy. Furthermore, administration of expensive medications with significant side effects would be avoided in patients with low response probability.
miRNAs were shown to play crucial roles in different biological processes involved in tumor development, progression, and response to treatment including cellular differentiation, proliferation, apoptosis, DNA damage repair, and epithelial–mesenchymal transition. They are promising cancer biomarkers,11 since they are stable, and can be quantified from both formalin-fixed tissues and body fluids. Several groups have identified miRNA signatures that distinguish ccRCC from normal kidney,12,13,14 and documented the role of miRNAs in RCC pathogenesis,15,16,17,18 and their association with aggressive behavior.19,20 Also, the significance of miRNAs as a molecular classifier for RCC subtypes was recently reported.21,22
The use of miRNAs as predictive markers has been reported. In lung cancer, miR-34c is a promising predictive biomarker,23 and miR-21 was reported to correlate with sensitivity to chemotherapy.24 Takahashi et al.25 demonstrated that low miR-148a expression was associated with poor response to therapy in advanced colorectal cancer. In breast cancer, miR-342 expression was shown to positively correlate with response to tamoxifen.26
In this study, we identified miRNAs that can distinguish between mRCC patients with short versus long-term PFS under sunitinib as first-line treatment. We developed and validated miRNA predictive models. We show that miRNAs are involved in signaling pathways involved in angiogenesis. Overexpression of two of these miRNAs resulted in decreased angiogenesis and cellular proliferation in vascular endothelial cells while increased cellular proliferation in renal cancer cells.
Results
miRNAs are differentially expressed between metastatic clear cell RCC patients with short- versus long-term survival on sunitinib treatment
Seven hundred and fifty-three human miRNAs were screened in each of the 30 ccRCC cases. We identified a number of miRNAs that are differentially expressed in relation to the time to progression under sunitinib. As a dichotomous variable, patients were classified into two cohorts of short (≤12 months) versus long (>12 months) PFS under sunitinib. Median PFS for all 57 patients was 11 months. We identified 20 miRNAs to be differentially expressed between the two groups (Supplementary Table S1).
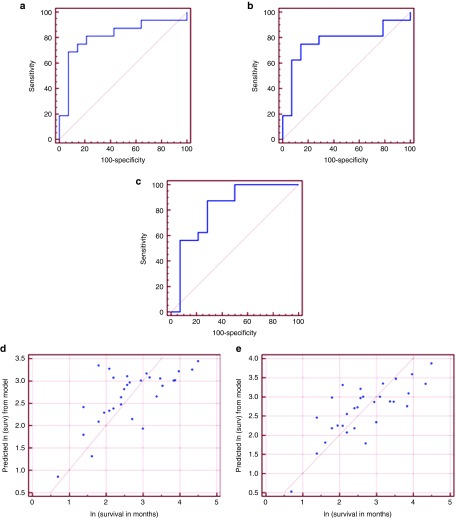
We developed three multivariate logistic regression models. The first model included miR-1225-3p and miR-208 (area under the curve (AUC) = 0.812, 95% CI: (0.629, 0.931), P = 0.0003). At a cut-off probability of 0.70, the model has a positive predictive value (PPV) of 92%. At a cut-off probability of 0.28, it has a negative predictive value (NPV) of 80% (Figure 1a).
Figure 1.
miRNA-based statistical models can predict renal cell carcinoma patients' survival under sunitinib treatment. (a–c) Multivariate logistic regression models to predict renal cell carcinoma patients' survival under sunitinib treatment as a dichotomous variable. Patients were divided into short-term (<12 months) versus long-term (≥ 12 months) survival under sunitinib. (a) Model 1: based on the expression of miR-1225-3p and miR-208 (AUC = 0.812, 95% CI: (0.629, 0.931), P = 0.0003). At a cut-off probability of 0.70, positive predictive value (PPV) = 92% and at a cutoff probability of 0.28, negative predictive value (NPV) = 80%. (b) Model 2: based on the expression ofmiR-1225-3P and miR-155-3p (AUC = 0.772, 95% CI: (0.583, 0.905), P = 0.0042). At a cut-off probability of 0.70, PPV = 91% and at a cut-off probability of 0.40, NPV = 77%. (c) Model 3: based on the expression of miR-597 and miR-1 (AUC = 0.812, 95% CI: (0.629, 0.931), P = 0.0003). At a cut-off probability of 0.80, PPV = 90% and at a cut-off probability of 0.21, NPV = 100%. (d,e) Multivariate regression models predict patients' survival under sunitinib treatment as a continuous variable. (d) miR-874 and miR-221 model (P = 0.0003). (e) miR-874, miR-221, and miR-424 model (P = 0.0001).
The second model included miR-1225-3p and miR-155-3p (AUC = 0.772, 95% CI: (0.583, 0.905), P = 0.0042). At a cut-off probability of 0.70, PPV = 91%. While at a cut-off probability of 0.40, NPV = 77% (Figure 1b). The third model included miR-597 and miR-1 (AUC = 0.812, 95% CI: (0.629, 0.931), P = 0.0003). At a cut-off probability of 0.80, PPV = 90%. At a cut-off probability of 0.21, NPV = 100% (Figure 1c).
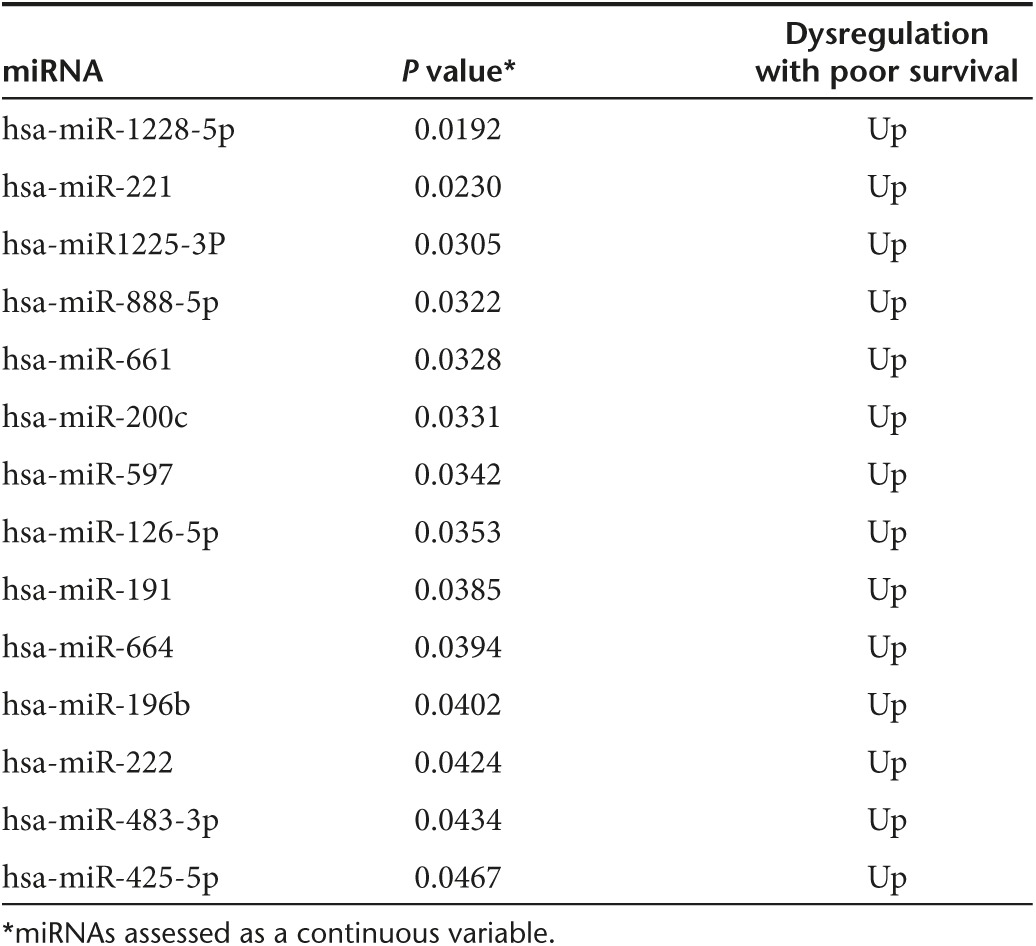
Analyzing miRNA expression as a continuous variable, we identified 14 miRNAs to be significantly differentially expressed between the two groups (Table 1). We constructed three multivariate regression models. The first model included miR-874 and miR-221 (P = 0.0003) (Figure 1d) and the second model included miR-874, miR-221, and miR-424 (P = 0.0001) (Figure 1e).
Table 1. Statistically significant differentially expressed miRNAs between patients with short- versus long-term progression-free survival on sunitinb therapy.
The correlation between miRNA models prognostic parameters
We evaluated the performance of our models before and after adjusting for Heng's criteria. Both PFS and OS were dichotomized, using a cutoff of 12 months for PFS (with 14 subjects ≤ 12 months PFS and 16 subjects > 12 months PFS) and a cutoff of 24 months for OS (with 13 subjects ≤ 24 months OS and 17 subjects > 24 months OS). Logistic regression models were employed, in order to determine the ability of each model to predict both PFS and OS, with Heng's score either included with or excluded from the model.
Correlation with OS is not a specific reflection of survival under sunitinib treatment because many patients received a second line treatment. The primary end point for this study was PFS under treatment. All our patients received sunitinib as first-line treatment and no other treatment was introduced during and up to developing progression under sunitinib.
As a univariate predictor, total Heng's score did not predict either PFS (P = 0.563) or OS (P = 0.707) using our dichotomous parameters. The multivariate model analyses showed that the first (miR-1225-3p and miR-208) and second (miR-1225-3p and miR-155-3p) models are significant predictors of OS after adjusting for Heng's score (P = 0.039 and 0.015 respectively), while the second (miR-1225-3p and miR-155-3p) and third (miR-597 and miR-1) models are significant predictors of PFS after adjusting for Heng's score (P = 0.032 and 0.037 respectively).
Furthermore, we tested the association between the expression levels of our miRNAs and OS. Our analysis showed that overexpression of miR-126-5p, miR-200c, miR-661, miR-664, miR-888-5p, and miR-1225-3p were associated with shorter OS (Supplementary Figure S1).
Also, we examined the association between miRNA expression and PFS in the subgroup of patients with stable disease and partial response (clinical benefit groups). When used as a dichotomous variable, overexpression of miR-191, miR-196b, miR-200c, miR-661, and miR-1228-5p were associated with decreased PFS (Supplementary Figure S2). In the subgroup of patients with stable disease, miR-22, miR-155-3p, miR-191, miR-196b, miR-200c, miR-222, miR-425-5p, miR-597, miR-623, miR-663b, miR-664, miR-888-5P, miR-1225-3p, and miR-1228-5p expression were associated with decreased PFS (data are not shown).
Predictive models validation
We validated our models using the gold-standard RT-qPCR with miRNA-specific primers on the discovery set and on an independent set of 27 patients. Validating on the discovery set, the third dichotomous model (miR-597 and miR-1) showed the best performance (AUC = 0.763, P = 0.004). At a cut-off probability of 0.83, PPV = 100% while at a cut-off probability of 0.366, NPV = 86%. The second model (miR-1225-3p and miR-155-3p), at a cut-off probability of 0.71, showed PPV = 100% and at a cutoff probability of 0.387, NPV = 83%. The first model (miR-1225-3p and miR-208) had an AUC = 0.679, at a cutoff probability of 0.72, PPV = 100% and at a cutoff probability of 0.42, NPV = 75%. Since achieving a very high combined sensitivity and specificity was not possible in our case, and since most mRCC patients are currently enrolled to sunitinib as first-line treatment, we focused more on the clinically relevant parameter for therapy decision making, which is either the inclusion or exclusion criteria (i.e., achieving high PPV or NPV).
When validated on the second set, the third dichotomous model (miR-597 and miR-1) showed the best performance (AUC = 0.763, P = 0.0035), followed by the first model (miR-1225-3p and miR-208) with AUC = 0.679 and finally the second model (miR-1225-3p and miR-155-3p) with AUC = 0.665, although the last two models did not reach significance (P = 0.076 and 0.123 respectively).
When performance was assessed on both sets combined, the (miR-597 and miR-1) model performed well (AUC = 0.713, P = 0.002). At a cut-off probability of 0.64, PPV = 88%. At a cut-off probability of 0.358, NPV = 87%. Although we lost significance for the other two models, miR-1225-3p and miR-208 model, showed AUC = 0.644, at a cut-off probability of 0.57, PPV = 100% and at a cutoff probability of 0.33, NPV = 80% and (miR-1225-3p and miR-155-3p) showed AUC = 0.634, at a cutoff probability of 0.606, PPV = 80% and at a cutoff probability of 0.317, NPV = 78%.
When validating miRNA performance as a continuous variable, both (miR-874 and miR-221) and (miR-874, miR-221 and miR-424) models performed well in the same set of patients (P = 0.0003 and 0.0001 respectively) and on the second independent set of patients as well (P = 0.001 and 0.002 respectively).
miRNA involvement in sunitinib response
In order to explore the potential mechanisms by which miRNAs can be involved in sunitinib response, we performed target prediction and pathway analyses of the predicted targets of the miRNAs differentially expressed between short- and long-term PFS under sunitinib. We used a targeted approach for the identification of the significant miRNAs for functional analysis, based on a combined score of the degree of the miRNA dysregulation, the literature evidence of potential involvement of in angiogenic pathways, in-silico analysis showing the potential involvement of the targets of these miRNAs in pathways related to TKI response and resistance, literature evidence from other cancers showing the predictive ability of these miRNAs, and experimental evidence of the involvement of these miRNAs on treatment response.
miR-221 and miR-222 were among the most significant miRNAs identified. Target prediction identified kinase insert domain receptor (a type III receptor tyrosine kinase) (KDR), the VEGFR2 encoding gene, as potential target of these miRNAs, specially, miR-221 and miR-222 that were upregulated in patients with poor survival under sunitinib (Supplementary Table S2). VEGFR2 is one of the sunitinib-targeted receptors which are involved in the VEGF signaling pathway. Also, miR-221 and miR-222 were predicted to target genes involved in cell proliferation.
Therefore, we hypothesized that there are two potential mechanisms through which these miRNAs can affect survival under sunitinib treatment. The first is that the elevated expression levels of a number of miRNAs, including miR-221 and miR-222 can decrease KDR gene expression with subsequent decrease in VEGFR2 expression level which is required for sunitinib binding. The second is that miRNAs can exert an opposing effect to sunitinib by increasing the rate of kidney cancer cell proliferation, while sunitinib acts mainly on endothelial cells rather than RCC tumor cells at the clinically relevant concentrations.
In order to test our hypothesis, we validated the miRNA-target interaction by testing the effect of miRNAs on KDR expression. Also, we examined the correlation between miR-221 and VEGFR2 expression and their association with PFS under sunitinib in the same set of patients. Finally, we investigated the effect of these miRNAs on angiogenesis and cellular proliferation in both endothelial and cancer cells as detailed below.
miR-221/222 can target KDR
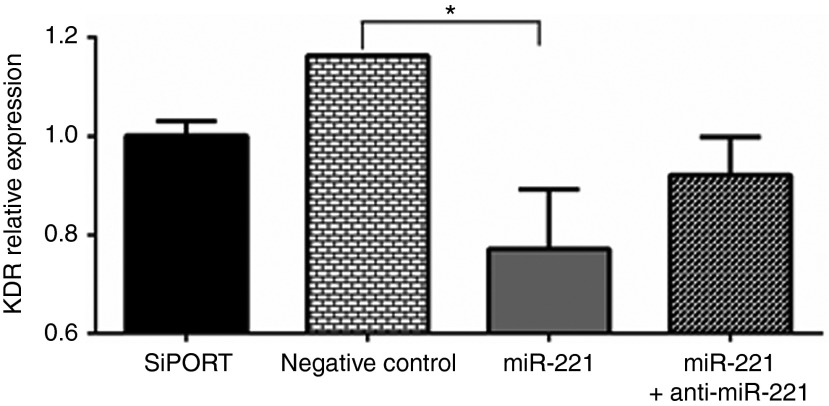
Our analyses showed that miR-221/222 target critical pathways and key molecules involved in tumor progression and cellular survival (Supplementary Table S2). KDR, the gene encoding for VEFGR2, one of the main receptors that mediate the antiangiogenic effects of sunitinib, was selected for experimental validation by measuring the effect of miRNA overexpression on the level of KDR. First, we screened a number of cell lines including human umbilical vascular endothelial cells (HUVEC), 786-O, ACHN, CAKI-1, and HEK-293 cells. KDR was highly expressed in HUVEC cells, in agreement with previous reports.27 We then compared the level of expression of KDR before and after transfection of each of these miRNAs. Overexpression of either of these miRNAs significantly decreased KDR expression in HUVEC cells (Figure 2).
Figure 2.
miR-221 can target kinase insert domain receptor (a type III receptor tyrosine kinase) (KDR). A representative bar graph showing that KDR, the gene encoding for VEGFR2, expression level was significantly decreased in human umbilical vein endothelial cells upon miR-221 transfection. This effect was partially restored with cotransfection of the miRNA and its inhibitor. Data are the means ± SEM of three experiments (*P < 0.05).
miR-221 and its predicted target, VEGFR2, significantly correlate to PFS under sunitinib
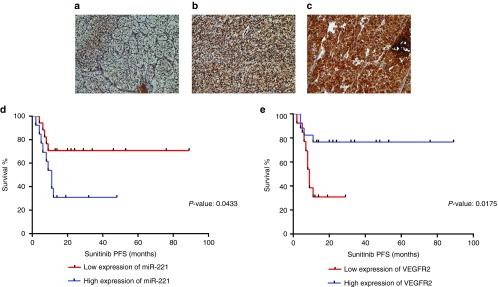
We assessed VEGFR2 protein expression using immunohistochemistry and quantified miR-221 expression in the same group of patients using qRT-PCR and correlated their expressions with PFS under sunitinib. miR-221 showed an inverse correlation with VEGFR2 expression (data not shown). Also, VEGFR2 was positively associated with better survival whereas miR-221 negatively correlated with survival (Figure 3).
Figure 3.
miR-221 and its predicted target, VEGFR2, significantly correlate to progression-free survival under sunitinib. (a–c) Representative photomicrographs showing the expression levels of VEGFR2 protein in renal cell carcinoma (RCC) by immunohistochemistry: (a) Weak, (b) moderate, (c) strong (all figures are original magnification ×400). (d,e) Kaplan–Meier curves of progression-free survival (PFS) under sunitinib for miR-221 (d) and VEGFR2 (e) expressions in mRCC patients. miR-221 and VEGFR2 expressions were dichotomized into high- and low-expression categories. Patients with higher miR-221 expression had significantly shorter PFS compared to those with low expression (P = 0.0433) while patients with higher VEGFR2 had significantly longer PFS compared to those with low expression (P = 0.0175).
miR-221 and miR-222 overexpression block angiogenesis
Based on target prediction analysis (Supplementary Table S2), we proposed that these two miRNAs function by blocking relevant signaling pathways involved in angiogenesis and cell proliferation (e.g., VEGF, TGF-β, mTOR, and MAPK signaling pathways. According to our proposed model, if these two miRNAs are overexpressed, they will suppress the VEGF pathway and as a result, tumors have to rely on “non-canonical” pathways that are not targeted by TKIs to maintain angiogenesis.28,29 This may explain the weak response and survival under sunitinib in these patients.
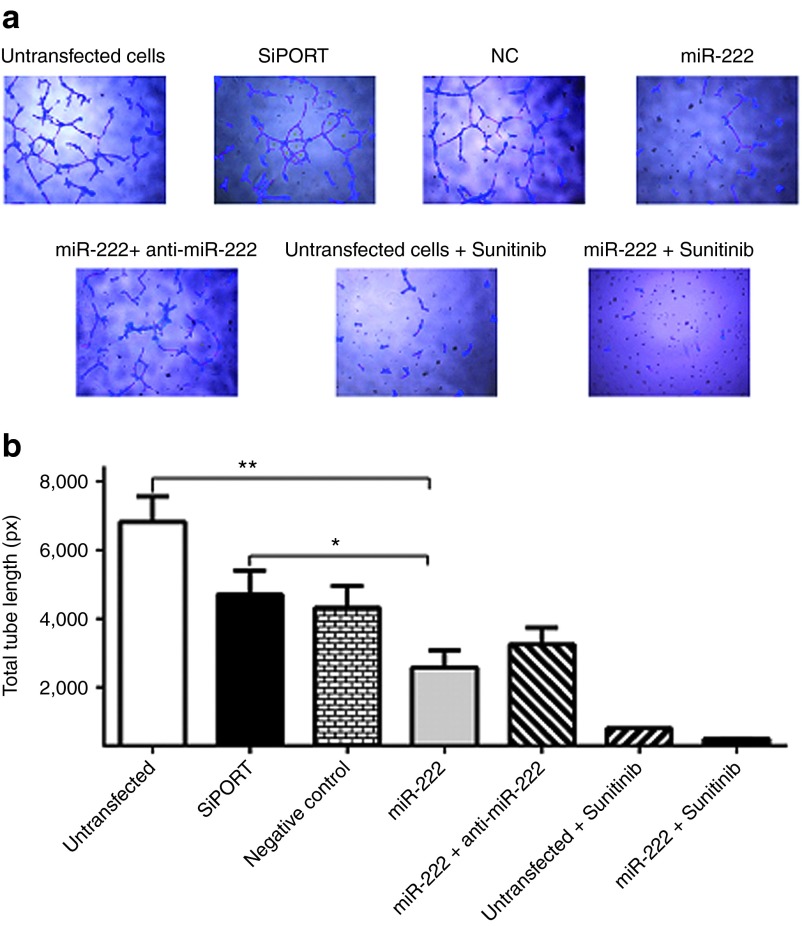
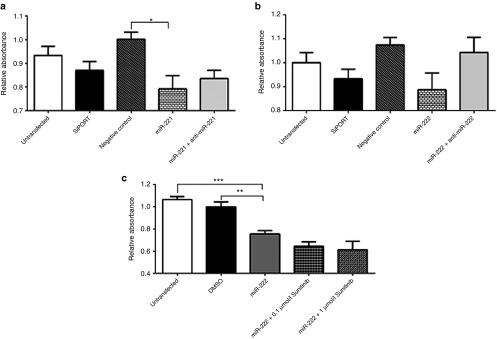
In order to experimentally validate this, we first checked the endogenous expression levels of miR-221 and miR-222 in HUVEC vascular endothelial cells, 786-O and ACHN kidney cancer cell lines and normal kidney tissue. Both miRNAs showed higher expression in HUVEC cells compared to normal kidney tissues and kidney cancer cell lines (Supplementary Figure S3). This is in agreement with previous reports showing their importance in regulating gene expression and function in endothelial cells.30 Gain of function experiments were then conducted by transfecting each of these miRNAs into HUVEC, 786-O, and ACHN cells. Controls included cells transfected with the miRNA inhibitors, cotransfected with each of these miRNAs and its inhibitor, untransfected cells, cells transfected with transfection agent only, and a random oligonucleotide negative control. Successful transfection was confirmed by qRT-PCR and fluorescent labeled oligonucleotide. We then investigated the effect of overexpression on angiogenesis using tube formation assay. Transfection with each of these two miRNAs resulted in significant reduction in the total tube length compared to controls. As shown in Figure 4a,b, overexpression of miR-222 significantly reduced total tube length compared to untransfected cells (P = 0.0012), cells transfected with transfection reagent only (P = 0.0359), negative control or cells cotransfected with miR-222 and its inhibitor. Similar results were obtained for miR-221, which decreased total tube length compared to negative controls (data not shown).
Figure 4.
Overexpression of miR-221 or miR-222 has negative effect on angiogenesis. (a) Representative photomicrographs showing the effect of miR-222 overexpression on tube formation using human umbilical vein endothelial cell line. Tube formation can be assessed using four parameters including cell covered area (blue), tubes (red), loops (yellow), and branching points (white). (b) Representative bar graph showing the effect of miR-222 on tube formation. Overexpression of miR-222 significantly reduced total tube length compared to untransfected cells (negative control (NC)), cells transfected with transfection reagent only or cells cotransfected with miR-222 and its inhibitor. Both untransfected and miR-222-transfected cells were then treated with sunitinib and showed decrease in total tube length compared to cells transfected with the miR-222 only. Data are the means ± SEM of three independent experiments (*P < 0.05 and **P < 0.01).
We then measured the combined effect of miR-222 or miR-221 and sunitinib on angiogenesis. Cells transfected with miR-221 or miR-222 and treated with sunitinib showed significant reduction in total tube length compared to cells transfected with the miRNA only or untransfected cells treated with sunitinib (P = 0.0125 and 0.0122 respectively). Similar results were obtained when the effect of miR-221 or miR-222 on other parameters such as total tube number and total branching points number were assessed (data not shown).
miR-221 and miR-222 overexpression decreased HUVEC cells proliferation
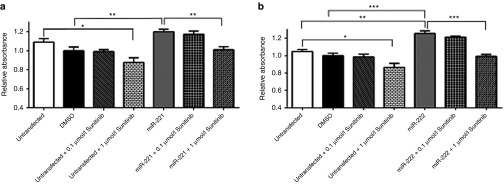
Next, we examined the effect of miR-221 and miR-222 on HUVEC cell proliferation. Appropriate controls were used as above. miR-221 or miR-222 transfection resulted in decreased endothelial cells proliferation compared to negative controls (Figure 5a,b). When the miR-221 or miR-222-transfected HUVEC cells were treated with sunitinib, cellular proliferation was further reduced at sunitinib concentrations of 0.1 and 1 µmol/l (Figure 5c). Together with prediction analysis, the results above indicate that both miR-221/222 and sunitinib share/compete for the same angiogenic and cell proliferation pathways of endothelial cells.
Figure 5.
The effect of miR-221 and miR-222 on HUVEC cells proliferation. (a,b) Overexpression of miR-221 or miR-222 has negative effect on endothelial cells proliferation. Representative bar graphs showing the effect of these two miRNAs on human umbilical vein endothelial cells proliferation. Cells transfected with either of miR-221 or miR-222 showed significantly reduced cellular proliferation compared to the control cells. This effect was partially restored by cotransfection of the miRNA and its inhibitor. (c) Sunitinib treatment resulted in further reduction of cell proliferation in miR-222-transfected endothelial cells. Cells were treated with sunitinib at concentrations of 0.1 and 1 µmol/l. Sunitinib resulted in reduced cellular proliferation at both concentrations. Data are the means ± SEM of three independent experiments (*P < 0.05 and ***P < 0.001). DMSO, dimethyl sulfoxide.
miR-221 and miR-222 overexpression increased ACHN cells proliferation
We then assessed the effect of miR-221 and miR-222 on cancer cell proliferation in vitro using the ACHN kidney cancer cell line. ACHN cells were transfected with miR-221 or miR-222 and/or treated with sunitinib. Unlike endothelial cells, sunitinib had no effect on cancer cell proliferation at a pharmacologically-relevant dose (0.1 µmol/l).27 At a high dose of 1 µmol/l, sunitinib treatment resulted in reduced cell proliferation. Interestingly, miR-221 and miR-222 overexpression enhanced cellular proliferation of ACHN cells. When miR-221 and miR-222 transfected ACHN cells were treated with sunitinib, sunitinib was not able to abolish the effect of miR-221/222 at pharmacologically-relevant concentrations (0.1 µmol/l). It was only able to partially counteract the effect of miRNAs at 1 µmol/l concentration. This, however, did not reach the baseline reduction of cellular proliferation caused by sunitinib on untransfected cells (Figure 6a,b).
Figure 6.
The effect of miR-221 and miR-222 on ACHN cells proliferation. (a,b) miR-221 or miR-222 overexpression increased kidney cancer cell proliferation while sunitinib decreased cellular proliferation at a higher concentration of 1 µmol/l. A representative bar graph showing the effect of miR-221 overexpression on renal cell carcinoma cell proliferation. Overexpression of miR-221 increased ACHN cells proliferation compared to the control cells. Comparable results were obtained for miR-222. There was no effect on cell proliferation when cells were treated with sunitinib at concentrations of 0.1 µmol/l. A reduction of cellular proliferation only was noticed at concentration of 1 µmol/l. Data are the means ± SEM of three independent experiments (*P < 0.05, **P < 0.01, and ***P < 0.001). DMSO, dimethyl sulfoxide.
Discussion
Sunitinib is currently a first-line treatment for mRCC.31 Despite the success of VEGF-targeted therapy in mRCC, over 20%of patients are refractory to first-line antivascular endothelial growth factor (VEGF) therapy and many patients progress within 1 year.7 A number of prognostic models have been developed to predict survival in patients treated with antiangiogenic therapy32,33,34 and a recent study suggested a value of pathological features to assess mRCC patients' survival and response to treatment.35 However, there is no accurate method that can predict outcome for each individual patient.
We provide evidence suggesting the clinical utility of miRNAs as predictive markers for mRCC patients' PFS receiving sunitinib first line, and show that this predicts PFS and OS independent of Heng score.
It has to be noted, however, that the logestic models did not necessarily utilize the highly differentially expressed miRNAs but rather combination of miRNAs, in certain situations, some of them have low discriminatory power but in combination with other miRNAs perform better with much higher probability of discrimination between the two groups. In other words, the models are not a reflection of the highly differentially expressed but rather of combinations of miRNAs that can accurately distinguish between the two outcomes.
Our data are in keeping with recent literature. miR-942 was recently shown to predict sunitinib efficacy in renal cell carcinoma36 and Berkers et al.37 demonstrated that miR-141 downregulation was associated with poor response to sunitinib in patients with mRCC. Also, Gámez-Pozo et al.38 constructed models that can predict sunitinib resistance based on miRNAs expression. Differences between the studies can be attributed to experimental design and clinical end points that were used in the analysis. Berkers et al. analyzed 20 fresh frozen ccRCC tissues from patients with synchronous metastasis and patients with PFS more than 1 year were considered good responders while patients with PFS less than 6 months were considered poor responders. Gámez-Pozo et al. analyzed patients' blood samples that were taken before the initiation of treatment and 2 weeks after therapy. Also, they assigned their patients into three different groups; poor response group <6 months, prolonged response group > 18 months, and moderate response group 6–18 months. In the present study, we analyzed pretreatment formalin-fixed paraffin-embedded tumors. It has to be noted, however, that resistance to TKI's including sunitinib is likely multifactorial and is a continuum rather than an endpoint process. This is difficult to assess in a single specimen.
Our data showed that miR-221 and miR-222 were significantly upregulated in mRCC patients with poor PFS on sunitinib treatment. These two miRNAs share the same seed sequence and their genes are located closely on Chromosome Xp11.3. The finding that miR-222 and miR-221 promote kidney cancer cell proliferation is not surprising. They were reported to be overexpressed in many cancers including breast, thyroid, colon, pancreatic, prostate, and bladder cancers.12,39 In our previous work, miR-221 and miR-222 were found to be upregulated in metastatic compared to primary ccRCC14 and Teixeira et al.40 showed that the increased miR-221 was associated with poor survival in RCC. In hepatocellular carcinoma, miR-221 overexpression was associated with decreased OS, metastasis, and advanced stage.41
We identified a number of miR-221 and miR-222 predicted targets and pathways that are involved in cancer promoting pathways. In agreement with our results, Lupini et al.42 identified several oncogenic pathways through which miR-221 promotes cancer. Overexpression of miR-221 was shown to increase ZEB2 and vimentin expression and decrease E-cadherin in pancreatic cancer.43 In addition, miR-221/222 inhibition enhanced apoptosis and decreased cellular proliferation and invasion by upregulating PTEN expression in gastric cancer.44 miR-221/222 was also shown to enhance epithelial–mesenchymal transition by targeting adiponectin receptor 1 which negatively control NF-κB, IL6, and JAK2/STAT3 signaling axis45 and their overexpression was shown to be associated with tumor metastasis in breast cancer.46
Our results show that these two miRNAs have opposing effects on cancer and endothelial cells. Whereas their overexpression inhibits the angiogenesis in endothelial cells, it enhances proliferation of cancer cells, in agreement with a recent study which showed their ability to target the cell cycle inhibitor p27.47 It is also possible that one of these miRNAs is been secreted by one cell type and been up taken by the other cell type.
The connection between miR-221 and miR-222 and treatment response is clear. We showed that miR-221 and miR-222 target VEGFR2, one of the sunitinib targeting receptors. Our results are in agreement with previous reports that demonstrated that resistance to treatment may develop when the targeted protein is not accessible for the drug binding.48 In addition, these two miRNAs were predicted to target c-KIT, which is another sunitinib-targeting receptor.49,50,51
The effect of sunitinib on cancer and endothelial cells is dose-dependent. Huang et al.27 provided evidence that, at the pharmacologically relevant concentrations, sunitinib acts mainly on endothelial rather than tumor cells in RCC, and that it inhibits tumor cell growth only at higher concentrations. These findings can explain the association of miR-221/222 overexpression with poor survival in mRCC patients receiving sunitinib. Their overexpression downregulates the traditional angiogenesis pathways that are utilized by sunitinib, thus minimizing its effect. Moreover, these two miRNAs increase tumor cell proliferation, which is not significantly affected by sunitinib at clinically relevant doses.
In agreement with previous reports,52,53 our analysis show that VEGFR2 expression is positively associated with PFS under sunitinib treatment. Our findings may also have therapeutic implications as inhibiting miR-221 and miR-222 might improve mRCC patients' survival if given as an adjuvant therapy in combination with sunitinib. Recently, it was shown that circular miR-221 sponge has anticancer effects in malignant melanoma. Another group developed an apatamer-based delivery system with selectivity to cancer cells and showed an antitumor therapeutic effects by inhibiting miR-221.54
In conclusion, we provide evidence that miRNAs are promising predictive markers for sunitinib treatment in metastatic RCC. Specifically, we show that miR-221 and miR-222 target the VEGFR with subsequent effect on angiogenesis. They also exert an opposite, tumor promoting effect on RCC cells and as such, they have potential as combination with TKI's therapy to prevent drug resistance.
Materials and Methods
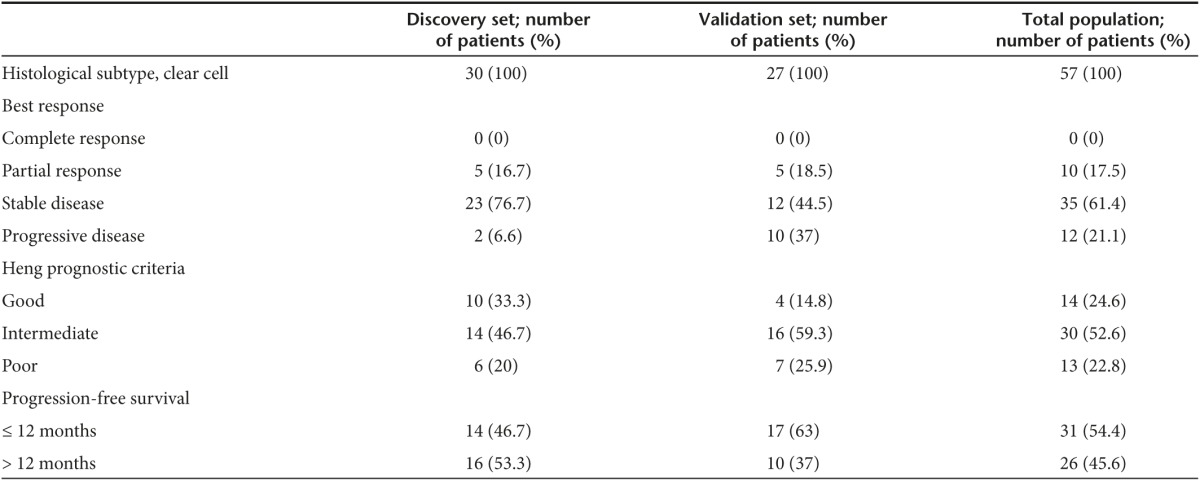
Patient population and specimen collection. Fifty-seven primary pretreatment ccRCC formalin-fixed paraffin-embedded tissue specimens were collected from Sunnybrook Health Sciences Center and St. Michael's Hospital, Toronto, Canada. Thirty cases were used as a discovery set and 27 cases were included as an independent validation set (Table 2). Areas of pure tumor tissues with no hemorrhage or necrosis were selected by a pathologist. Multiple sections were mixed from the same tumor to compensate for tumor heterogeneity. Pure tumor areas were excised using Laser capture microdissection. Tumor classification and staging were done according to the 2002 TNM System and the 2004 World Health Organization Classification. Study was approved by the Research Ethics Board at St. Michael's Hospital and The Sunnybrook Health Science Center.
Table 2. Descriptive statistics of the study population.
miRNA expression screening. Total RNA isolation was done using the miRNeasy Kit (Qiagen, Mississauga, Canada) according to the manufacture's protocol. RNA quality and concentration were determined spectrophotometrically (NanoDrop 1000 Spectrophotometer; NanoDrop Technologies, Wilmington, DE). Samples optimal for analysis were stored at −80 °C.
Five hundred nanograms of total RNA from each sample were reverse transcribed using a Megaplex Primer Pool Human Set A+B (Life Technologies, Burlington, Canada) with a TaqMan miRNA reverse-transcription kit as suggested by the manufacturer. cDNA samples of individual patients were analyzed by a TaqMan low-density array human microRNA card set A+B.
Quantitative reverse transcription polymerase chain reaction (RT-qPCR). For validation, miRNA-specific reverse transcription was performed with 500 ng total RNA using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) as recommended by the manufacture. RT-qPCR was performed using the TaqMan microRNA Assay Kit on the Step One Plus Real-Time PCR System (Applied Biosystems). Thermal cycling conditions were according to the manufacture's fast protocol and all reactions were performed in triplicate. Relative expression was determined using the ▵▵Ct method and expression values were normalized to small nuclear RNA, U6 snRNA, RNU48, and RNU44 (Applied Biosystems).
For the expression analyses of KDR, primer sequence was as follows: forward-5′-CCCAGGAAAAGACGAACTTG-3′ and reverse-5′-TCCAATGGGAGTTCATCTGG-3′. Reverse transcription was performed with high-capacity RNA-to-cDNA kit (Applied Biosystems) as per the manufacturer's instructions. qRT-PCR was performed using the Fast Syber Green Master Mix (Applied Biosystems). Peptidylprolylisomerase A (cyclophilin A) (PPIA) was used as endogenous controls. The PPIA primer sequences were as follows: forward-5′-ATGCTGGCCCCAACACAA-3′ and reverse-5′-TCTCCACCAATTACTTTTATGTCC-3′.
Statistical analysis. PFS under treatment was calculated from starting date of sunitinib therapy to the date of tumor progression or death while on treatment. OS was calculated from starting date of any systemic therapy to the date of death from any cause. RT-PCR measurements (Ct values) for 753 miRNAs were obtained from each specimen. miRNA Ct data were normalized to three endogenous controls (U6, RNU44, RNU48). Data were analyzed as both continuous and dichotomous variables, with Ct values greater than 32.0 considered to be nonexpressed and truncated to 32.0. A cut-off of 12 months was used to distinguish short survival (≤ 12 months) from longer-term PFS (> 12 months). Receiver operating characteristic curves were generated to identify individual miRNAs that were significant predictors of survival. Stepwise logistic regression analyses were conducted, in order to construct models relating survival category to Ct values from two or more miRNAs. As a continuous variable, univariate regression analyses were conducted to identify individual miRNAs that were significant predictors of PFS under treatment; multivariate stepwise regression analyses were then conducted in order to construct models relating expression levels of miRNAs to survival.
The predictive abilities of the models obtained from the above analyses were assessed on a separate validation cohort of 27 metastatic kidney cancer patients. The data from the original and validation cohorts were then combined, and the predictive abilities of the models obtained from the above analyses were assessed using the larger patient cohort (total n = 57).
Cell culture and miRNA transfection. ACHN kidney cancer cell lines and human HUVEC cells were obtained from American Type Culture Collection (Manassas, VA) and were grown according to manufacturer's protocol. Pre-miR precursors and anti-miR miR inhibitors for miR-221 and miR-222 were purchased from Applied Biosystems. Cells were transfected using siPORT NeoFX transfection agent (Ambion, Austin, TX) as described in our previous publication.55 siPORT control resulted in some degree of reduction of proliferation and tube formation. This can be attributed to little toxic effect and subtracted as a background change that has to be subtracted from the intervention arm. miRNA precursors and inhibitors were diluted in the same media to a final concentration of 30 nmol/l. Three separate transfections were performed and each was analyzed in triplicate. Transfection efficiency was confirmed using BLOCK-IT Fluorescent Oligo (Invitrogen).
Tube formation assay. The effects of miR-221 and miR-222 on angiogenesis were examined using in vitro matrigel tube formation assay. HUVEC cells were transfected either with SiPORT NeoFX transfection agent, scrambled miRNA, miR-221, miR-222 or their inhibitors, or cotransfected with the miRNA and its inhibitor. Twenty-four hours later, cells were trypsinized and resuspended. One hundred and fifty microliters of cell suspension (3.0 × 104 cells) per well were added to the solidified matrigel. Sunitinib was purchased from SelleckChemand and was added to the untransfected cells; cells were transfected with either miR-221 or miR-222 in a concentration of 1 µmol/l. Cells were incubated for 18 hours at 37 °C and 5% CO2 in a humidified tissue culture incubator. The experiment was performed in triplicate. Photomicrographs in three fields were taken. Data were analyzed by using WimTube image analysis tool (http://ibidi.com/xtproducts/en/Software-and-Image-Analysis/Automated-Image-Analysis/Tube-Formation-Image-Analysis-WimTube).
Cell proliferation assay. Cellular proliferation was measured by using cell proliferation reagent WST-1 (Roche Applied Science) colorimetric assay. Cells were plated at 6.0 × 103 cells per well in a 96-well plate and transfected either with SiPORT NeoFX transfection agent, miR-221 and miR-222 or cotransfected with the miRNA and its inhibitor. Cells were also treated with sunitinib in two concentrations (0.1 and 1 µmol/l). Proliferation reagent WST-1 was added to each well and incubated for 2 hours at 37 °C. The absorbance of each well was measured at a wavelength of 440 nm. Each test was repeated in six replicates.
Tissue microarray construction and immunohistochemistry. Tissue microarrays were built using the 30 tumors of the discovery set. Two 1 mm cores were obtained from two different blocks to account for tumor heterogeneity. Paraffin sections of the tissue microarrays were cut for immunohistochemistry in 4-μm thickness for VEGFR2 immunostaining. A combination of a proportion and intensity scores was used to assess VEGFR2 expression: the proportion score (proportion of positive tumor cells) was: 0: none, 1: 1–24%, 2: 25–49%, 3: 50–74, 4: ≥75%. The intensity score (intensity of staining by tumor cells) was: 0: none, 1: weak, 2: moderate, 3: strong. A total score was obtained by the combining both scores. Moderate or strong staining intensity in >24% was considered strong expression. Two cores were evaluated for each patient, and the arithmetic average score was reported.
For the immunohistochemical demonstration of VEGFR2, C-terminal (Sigma-Aldrich GmbH, Seelze, Germany; Cat# SAB4500965; dil'n 1:30), the streptavidin-biotin-peroxidase complex protocol using the LSAB+ Kit (DAKO, Carpenteria, CA) was employed. After routine deparaffinization, rehydration and blocking of endogenous peroxidase activity, sections were pretreated for antigen retrieval by microwaving in 0.1 mM sodium citrate buffer (pH 6.0). Sections were incubated overnight at room temperature. Diaminobenzidine served as chromogen. Cases were interpreted by a pathology fellow unaware of clinical information of the tissue samples.
Target prediction and pathway analyses. Target prediction was done using TargetScanHuman 6.2 (http://www.targetscan.org/) and miRecords (http://mirecords.biolead.org/) softwares. Only predictions by at least three programs were included in the analysis. We filtered the predicted gene targets list through extensive literature search and pathway analysis using DIANA-mirPath (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=mirpath/index).
SUPPLEMENTARY MATERIAL Figure S1. Higher expression of miR-126-5p, miR-200c, miR-661, miR-664, miR-888-5p, and miR-1225-3p are associated with shorter overall survival in mRCC patients on sunitinib treatment. Figure S2. Kaplan-Meier curves for progression free survival in the combined clinical benefit subgroup of patients (stable disease and partial response). Figure S3. miR-221 and miR-222 endogenous expression levels are higher in HUVEC endothelial cells compared to 786-O and ACHN kidney cancer cell lines. Table S1. Top differentially expressed miRNAs between patients with short versus long term progression free survival on sunitinb therapy. Table S2. miR-221/222 pathway analysis of predicted targets.
Acknowledgments
This work was supported by grants from the Canadian Institute of Health Research (MOP 119606), Kidney Foundation of Canada (KFOC130030), the Kidney Cancer Research Network of Canada, and Prostate Cancer Canada Movember Discovery Grants (D2013-39). The funding was obtained from The Anna-Liisa Farquharson Chair in Renal Cell Cancer Research to Dr. Bjarnason. The authors declared no conflict of interest.
Supplementary Material
Higher expression of miR-126-5p, miR-200c, miR-661, miR-664, miR-888-5p, and miR-1225-3p are associated with shorter overall survival in mRCC patients on sunitinib treatment.
Kaplan-Meier curves for progression free survival in the combined clinical benefit subgroup of patients (stable disease and partial response).
miR-221 and miR-222 endogenous expression levels are higher in HUVEC endothelial cells compared to 786-O and ACHN kidney cancer cell lines.
References
- Siegel, R, Ma, J, Zou, Z and Jemal, A (2014). Cancer statistics, 2014. CA Cancer J Clin 64: 9–29. [DOI] [PubMed] [Google Scholar]
- Motzer, RJ, Hutson, TE, McCann, L, Deen, K and Choueiri, TK (2014). Overall survival in renal-cell carcinoma with pazopanib versus sunitinib. N Engl J Med 370: 1769–1770. [DOI] [PubMed] [Google Scholar]
- Motzer, RJ, Nosov, D, Eisen, T, Bondarenko, I, Lesovoy, V, Lipatov, O et al. (2013). Tivozanib versus sorafenib as initial targeted therapy for patients with metastatic renal cell carcinoma: results from a phase III trial. J Clin Oncol 31: 3791–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motzer, RJ, Hutson, TE, Tomczak, P, Michaelson, MD, Bukowski, RM, Oudard, S et al. (2009). Overall survival and updated results for sunitinib compared with interferon alfa in patients with metastatic renal cell carcinoma. J Clin Oncol 27: 3584–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, S (2011). Vascular endothelial growth factor (VEGF), VEGF receptors and their inhibitors for antiangiogenic tumor therapy. Biol Pharm Bull 34: 1785–1788. [DOI] [PubMed] [Google Scholar]
- Motzer, RJ, Hutson, TE, Tomczak, P, Michaelson, MD, Bukowski, RM, Rixe, O et al. (2007). Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med 356: 115–124. [DOI] [PubMed] [Google Scholar]
- Heng, DY, Mackenzie, MJ, Vaishampayan, UN, Bjarnason, GA, Knox, JJ, Tan, MH et al. (2012). Primary anti-vascular endothelial growth factor (VEGF)-refractory metastatic renal cell carcinoma: clinical characteristics, risk factors, and subsequent therapy. Ann Oncol 23: 1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergers, G and Hanahan, D (2008). Modes of resistance to anti-angiogenic therapy. Nat Rev Cancer 8: 592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammers, HJ, Verheul, HM, Salumbides, B, Sharma, R, Rudek, M, Jaspers, J et al. (2010). Reversible epithelial to mesenchymal transition and acquired resistance to sunitinib in patients with renal cell carcinoma: evidence from a xenograft study. Mol Cancer Ther 9: 1525–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa, T, Tsuchida, R, Muramatsu, M, Shimamura, T, Wang, F, Suehiro, J et al. (2013). Inhibition of histone demethylase JMJD1A improves anti-angiogenic therapy and reduces tumor-associated macrophages. Cancer Res 73: 3019–3028. [DOI] [PubMed] [Google Scholar]
- Khella, HW, Bakhet, M, Lichner, Z, Romaschin, AD, Jewett, MA and Yousef, GM (2013). MicroRNAs in kidney disease: an emerging understanding. Am J Kidney Dis 61: 798–808. [DOI] [PubMed] [Google Scholar]
- Gottardo, F, Liu, CG, Ferracin, M, Calin, GA, Fassan, M, Bassi, P et al. (2007). Micro-RNA profiling in kidney and bladder cancers. Urol Oncol 25: 387–392. [DOI] [PubMed] [Google Scholar]
- Jung, M, Mollenkopf, HJ, Grimm, C, Wagner, I, Albrecht, M, Waller, T et al. (2009). MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J Cell Mol Med 13(9B): 3918–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, NM, Bao, TT, Grigull, J, Youssef, YM, Girgis, A, Diamandis, M et al. (2011). miRNA profiling for clear cell renal cell carcinoma: biomarker discovery and identification of potential controls and consequences of miRNA dysregulation. J Urol 186: 1077–1083. [DOI] [PubMed] [Google Scholar]
- Chen, X, Wang, X, Ruan, A, Han, W, Zhao, Y, Lu, X et al. (2014). miR-141 is a key regulator of renal cell carcinoma proliferation and metastasis by controlling EphA2 expression. Clin Cancer Res 20: 2617–2630. [DOI] [PubMed] [Google Scholar]
- Chow, TF, Mankaruos, M, Scorilas, A, Youssef, Y, Girgis, A, Mossad, S et al. (2010). The miR-17-92 cluster is over expressed in and has an oncogenic effect on renal cell carcinoma. J Urol 183: 743–751. [DOI] [PubMed] [Google Scholar]
- Khella, HW, Bakhet, M, Allo, G, Jewett, MA, Girgis, AH, Latif, A et al. (2013). miR-192, miR-194 and miR-215: a convergent microRNA network suppressing tumor progression in renal cell carcinoma. Carcinogenesis 34: 2231–2239. [DOI] [PubMed] [Google Scholar]
- Lichner, Z, Mejia-Guerrero, S, Ignacak, M, Krizova, A, Bao, TT, Girgis, AH et al. (2012). Pleiotropic action of renal cell carcinoma-dysregulated miRNAs on hypoxia-related signaling pathways. Am J Pathol 180: 1675–1687. [DOI] [PubMed] [Google Scholar]
- Heinzelmann, J, Unrein, A, Wickmann, U, Baumgart, S, Stapf, M, Szendroi, A et al. (2014). MicroRNAs with prognostic potential for metastasis in clear cell renal cell carcinoma: a comparison of primary tumors and distant metastases. Ann Surg Oncol 21: 1046–1054. [DOI] [PubMed] [Google Scholar]
- White, NM, Khella, HW, Grigull, J, Adzovic, S, Youssef, YM, Honey, RJ et al. (2011). miRNA profiling in metastatic renal cell carcinoma reveals a tumour-suppressor effect for miR-215. Br J Cancer 105: 1741–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman, E, Dotan, Z, Barshack, I, David, MB, Dov, A, Tabak, S et al. (2010). Accurate molecular classification of renal tumors using microRNA expression. J Mol Diagn 12: 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef, YM, White, NM, Grigull, J, Krizova, A, Samy, C, Mejia-Guerrero, S et al. (2011). Accurate molecular classification of kidney cancer subtypes using microRNA signature. Eur Urol 59: 721–730. [DOI] [PubMed] [Google Scholar]
- Mascaux, C, Feser, WJ, Lewis, MT, Barón, AE, Coldren, CD, Merrick, DT et al. (2013). Endobronchial miRNAs as biomarkers in lung cancer chemoprevention. Cancer Prev Res (Phila) 6: 100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, J, Gao, W, Zhu, CJ, Liu, YQ, Mei, Z, Cheng, T et al. (2011). Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non-small cell lung cancer. Chin J Cancer 30: 407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, M, Cuatrecasas, M, Balaguer, F, Hur, K, Toiyama, Y, Castells, A et al. (2012). The clinical significance of MiR-148a as a predictive biomarker in patients with advanced colorectal cancer. PLoS One 7: e46684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, YJ, Wu, JZ, Ji, MH, Ma, T, Qiao, EQ, Ma, R et al. (2013). miR-342 is associated with estrogen receptor-α expression and response to tamoxifen in breast cancer. Exp Ther Med 5: 813–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D, Ding, Y, Li, Y, Luo, WM, Zhang, ZF, Snider, J et al. (2010). Sunitinib acts primarily on tumor endothelium rather than tumor cells to inhibit the growth of renal cell carcinoma. Cancer Res 70: 1053–1062. [DOI] [PubMed] [Google Scholar]
- Korn, C, Scholz, B, Hu, J, Srivastava, K, Wojtarowicz, J, Arnsperger, T et al. (2014). Endothelial cell-derived non-canonical Wnt ligands control vascular pruning in angiogenesis. Development 141: 1757–1766. [DOI] [PubMed] [Google Scholar]
- Stefater, JA 3rd, Lewkowich, I, Rao, S, Mariggi, G, Carpenter, AC, Burr, AR et al. (2011). Regulation of angiogenesis by a non-canonical Wnt-Flt1 pathway in myeloid cells. Nature 474: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez, Y, Fernández-Hernando, C, Pober, JS and Sessa, WC (2007). Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res 100: 1164–1173. [DOI] [PubMed] [Google Scholar]
- Lee-Ying, R, Lester, R and Heng, D (2014). Current management and future perspectives of metastatic renal cell carcinoma. Int J Urol 21: 847–855. [DOI] [PubMed] [Google Scholar]
- Choueiri, TK, Garcia, JA, Elson, P, Khasawneh, M, Usman, S, Golshayan, AR et al. (2007). Clinical factors associated with outcome in patients with metastatic clear-cell renal cell carcinoma treated with vascular endothelial growth factor-targeted therapy. Cancer 110: 543–550. [DOI] [PubMed] [Google Scholar]
- Heng, DY, Xie, W, Regan, MM, Warren, MA, Golshayan, AR, Sahi, C et al. (2009). Prognostic factors for overall survival in patients with metastatic renal cell carcinoma treated with vascular endothelial growth factor-targeted agents: results from a large, multicenter study. J Clin Oncol 27: 5794–5799. [DOI] [PubMed] [Google Scholar]
- Motzer, RJ, Bukowski, RM, Figlin, RA, Hutson, TE, Michaelson, MD, Kim, ST et al. (2008). Prognostic nomogram for sunitinib in patients with metastatic renal cell carcinoma. Cancer 113: 1552–1558. [DOI] [PubMed] [Google Scholar]
- Park, JY, Lee, JL, Baek, S, Eo, SH, Go, H, Ro, JY et al. (2014). Sarcomatoid features, necrosis, and grade are prognostic factors in metastatic clear cell renal cell carcinoma with vascular endothelial growth factor-targeted therapy. Hum Pathol 45: 1437–1444. [DOI] [PubMed] [Google Scholar]
- Prior, C, Perez-Gracia, JL, Garcia-Donas, J, Rodriguez-Antona, C, Guruceaga, E, Esteban, E et al. (2014). Identification of tissue microRNAs predictive of sunitinib activity in patients with metastatic renal cell carcinoma. PLoS One 9: e86263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkers, J, Govaere, O, Wolter, P, Beuselinck, B, Schöffski, P, van Kempen, LC et al. (2013). A possible role for microRNA-141 down-regulation in sunitinib resistant metastatic clear cell renal cell carcinoma through induction of epithelial-to-mesenchymal transition and hypoxia resistance. J Urol 189: 1930–1938. [DOI] [PubMed] [Google Scholar]
- Gámez-Pozo, A, Antón-Aparicio, LM, Bayona, C, Borrega, P, Gallegos Sancho, MI, García-Domínguez, R et al. (2012). MicroRNA expression profiling of peripheral blood samples predicts resistance to first-line sunitinib in advanced renal cell carcinoma patients. Neoplasia 14: 1144–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramantieri, L, Fornari, F, Callegari, E, Sabbioni, S, Lanza, G, Croce, CM et al. (2008). MicroRNA involvement in hepatocellular carcinoma. J Cell Mol Med 12(6A): 2189–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira, AL, Ferreira, M, Silva, J, Gomes, M, Dias, F, Santos, JI et al. (2014). Higher circulating expression levels of miR-221 associated with poor overall survival in renal cell carcinoma patients. Tumour Biol 35: 4057–4066. [DOI] [PubMed] [Google Scholar]
- Rong, M, Chen, G and Dang, Y (2013). Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer 13: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupini, L, Bassi, C, Ferracin, M, Bartonicek, N, D'Abundo, L, Zagatti, B et al. (2013). miR-221 affects multiple cancer pathways by modulating the level of hundreds messenger RNAs. Front Genet 4: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, A, He, S, Tian, B, Hu, W and Zhang, Z (2013). MicroRNA-221 mediates the effects of PDGF-BB on migration, proliferation, and the epithelial-mesenchymal transition in pancreatic cancer cells. PLoS One 8: e71309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun-Zhi, Z, Lei, H, An-Ling, Z, Yan-Chao, F, Xiao, Y, Guang-Xiu, W et al. (2010). MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer 10: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, MS, Yu, N, Stinson, SY, Yue, P, Newman, RJ, Allan, BB et al. (2013). miR-221/222 targets adiponectin receptor 1 to promote the epithelial-to-mesenchymal transition in breast cancer. PLoS One 8: e66502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenberg, N, Anastasov, N, Rappl, K, Braselmann, H, Auer, G, Walch, A et al. (2013). MiR-221/-222 differentiate prognostic groups in advanced breast cancers and influence cell invasion. Br J Cancer 109: 2714–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehbacher, A, Urbich, C and Dimmeler, S (2008). Targeting microRNA expression to regulate angiogenesis. Trends Pharmacol Sci 29: 12–15. [DOI] [PubMed] [Google Scholar]
- Buczek, M, Escudier, B, Bartnik, E, Szczylik, C and Czarnecka, A (2014). Resistance to tyrosine kinase inhibitors in clear cell renal cell carcinoma: from the patient's bed to molecular mechanisms. Biochim Biophys Acta 1845: 31–41. [DOI] [PubMed] [Google Scholar]
- Poliseno, L, Tuccoli, A, Mariani, L, Evangelista, M, Citti, L, Woods, K et al. (2006). MicroRNAs modulate the angiogenic properties of HUVECs. Blood 108: 3068–3071. [DOI] [PubMed] [Google Scholar]
- Staszel, T, Zapała, B, Polus, A, Sadakierska-Chudy, A, Kieć-Wilk, B, Stępień, E et al. (2011). Role of microRNAs in endothelial cell pathophysiology. Pol Arch Med Wewn 121: 361–366. [PubMed] [Google Scholar]
- Urbich, C, Kuehbacher, A and Dimmeler, S (2008). Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res 79: 581–588. [DOI] [PubMed] [Google Scholar]
- Dornbusch, J, Zacharis, A, Meinhardt, M, Erdmann, K, Wolff, I, Froehner, M et al. (2013). Analyses of potential predictive markers and survival data for a response to sunitinib in patients with metastatic renal cell carcinoma. PLoS One 8: e76386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakawa, T, Miyake, H, Kusuda, Y and Fujisawa, M (2013). Expression level of vascular endothelial growth factor receptor-2 in radical nephrectomy specimens as a prognostic predictor in patients with metastatic renal cell carcinoma treated with sunitinib. Urol Oncol 31: 493–498. [DOI] [PubMed] [Google Scholar]
- Kim, JK, Choi, KJ, Lee, M, Jo, MH and Kim, S (2012). Molecular imaging of a cancer-targeting theragnostics probe using a nucleolin aptamer- and microRNA-221 molecular beacon-conjugated nanoparticle. Biomaterials 33: 207–217. [DOI] [PubMed] [Google Scholar]
- White, NM, Bui, A, Mejia-Guerrero, S, Chao, J, Soosaipillai, A, Youssef, Y et al. (2010). Dysregulation of kallikrein-related peptidases in renal cell carcinoma: potential targets of miRNAs. Biol Chem 391: 411–423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Higher expression of miR-126-5p, miR-200c, miR-661, miR-664, miR-888-5p, and miR-1225-3p are associated with shorter overall survival in mRCC patients on sunitinib treatment.
Kaplan-Meier curves for progression free survival in the combined clinical benefit subgroup of patients (stable disease and partial response).
miR-221 and miR-222 endogenous expression levels are higher in HUVEC endothelial cells compared to 786-O and ACHN kidney cancer cell lines.