Abstract
Progesterone, via the progesterone receptor (PGR), is essential for endometrial stromal cell decidualization, a cellular transformation event in which stromal fibroblasts differentiate into decidual cells. Uterine decidualization supports embryo implantation and placentation as well as subsequent events, which together ensure a successful pregnancy. Accordingly, impaired decidualization results not only in implantation failure or early fetal miscarriage, but also may lead to potential adverse outcomes in all three pregnancy trimesters. Transcriptional reprogramming on a genome-wide scale underlies progesterone dependent decidualization of the human endometrial stromal cell (hESC). However, identification of the functionally essential signals encoded by these global transcriptional changes remains incomplete. Importantly, this knowledge-gap undercuts future efforts to improve diagnosis and treatment of implantation failure based on a dysfunctional endometrium. By integrating genome-wide datasets derived from decidualization of hESCs in culture, we reveal that the promyelocytic leukemia zinc finger (PLZF) transcription factor is rapidly induced by progesterone and that this induction is indispensable for progesterone-dependent decidualization. Chromatin immunoprecipitation followed by next generation sequencing (ChIP-Seq) identified at least ten progesterone response elements within the PLZF gene, indicating that PLZF may act as a direct target of PGR signaling. The spatiotemporal expression profile for PLZF in both the human and mouse endometrium offers further support for stromal PLZF as a mediator of the progesterone decidual signal. To identify functional targets of PLZF, integration of PLZF ChIP-Seq and RNA Pol II RNA-Seq datasets revealed that the early growth response 1 (EGR1) transcription factor is a PLZF target for which its level of expression must be reduced to enable progesterone dependent hESC decidualization. Apart from furnishing essential insights into the molecular mechanisms by which progesterone drives hESC decidualization, our findings provide a new conceptual framework that could lead to new avenues for diagnosis and/or treatment of adverse reproductive outcomes associated with a dysfunctional uterus.
Author Summary
Following embryo attachment to the uterine epithelium, the underlying decidualized stroma is critical for further invasion by the conceptus into the maternal compartment. Because endometrial decidualization is required early in the continuum of events that lead to a successful pregnancy, abnormal decidualization can contribute not only to implantation failure or early miscarriage, but may initiate adverse reproductive outcomes that manifest in subsequent pregnancy trimesters. Genome-wide transcriptional changes by progesterone are known to underlie decidualization; however, the pivotal signals that functionally enable progesterone-driven decidualization are not fully known. Using an integrative analysis of genome-scale data along with studies on primary human endometrial stromal cells (hESCs), we reveal that the promyelocytic leukemia zinc finger (PLZF) transcription factor is rapidly induced by progesterone, and its induction is essential for progesterone-dependent hESC decidualization. Although PLZF in turn governs a remarkable array of target genes in the hESC, we demonstrate that PLZF tightly regulates the expression level of the early growth response 1 (EGR1) transcription factor, the perturbation of which compromises progesterone dependent decidualization. Together, our findings provide a new mechanistic perspective on progesterone action in the uterus which may furnish new opportunities for the formulation of more effective fertility solutions in the future.
Introduction
For healthy couples, the chance for conception per menstrual cycle is only thirty-to-forty percent [1], underscoring the remarkable inefficiency of human reproduction. Moreover, approximately a third of pregnancies detected early by human chorionic gonadotropin assay fail to result in live births [2, 3], implicating early implantation failure as a causal factor for preclinical pregnancy loss. Implantation failure and early embryo miscarriage also undercut the full potential of assisted reproductive technologies (ARTs) which rely on the transfer of healthy embryos into a uterus that must in turn execute the requisite cellular and molecular changes to ensure the establishment of the fetomaternal interface [4]. Early functional impairment of the uterus is also implicated in recurrent pregnancy loss when parental chromosomal abnormalities, maternal thrombophilic disorders and anatomical uterine defects are first eliminated as causal factors [5]. Apart from contributing to early implantation failure, insufficient execution of normal uterine changes around the time of implantation is also thought to underpin adverse outcomes—pre-eclampsia and placental insufficiency, intrauterine fetal growth restriction, and preterm birth—in later trimesters of pregnancy, termed the “adverse ripple effect”; reviewed in [6].
Successful advancement of embryo implantation into a receptive endometrium requires decidualization of the endometrial stromal cell layer, a critical cellular transformation process in which quiescent stromal fibroblasts proliferate and differentiate into transient epithelioid decidual cells [7]. In addition to promoting local resistance to oxidative insults and gestational immunotolerance for the developing fetal allograft, decidual cells are thought to modulate the invasion of the embryonic trophoblast to a sufficient depth to establish fetoplacental circulation between the chorionic villi and the maternal intervillus space. Therefore, due to the indispensability of the decidualized stroma for placentation, inadequate endometrial decidualization is considered a critical yet poorly understood etiologic factor not only for early implantation failure but also for a broad spectrum of pregnancy disorders that manifest later.
Along with exposure to preovulatory and nidatory estradiol-17β (E2), the endometrial stroma requires progesterone (P4) “the hormone of pregnancy” and its nuclear receptor (the progesterone receptor (PGR)) for decidualization [6]. While many of the cellular events that underpin endometrial stromal cell decidualization are known, the key molecular signals that mediate P4-dependent decidualization of the human endometrium remain unclear. Integrating datasets from chromatin immunoprecipitation followed by next generation sequencing (ChIP-Seq) and RNA sequencing (RNA-Seq), we reveal that the promyelocytic leukemia zinc finger (PLZF) transcription factor is rapidly induced by P4 in human endometrial stromal cells (hESCs) and that this induction is indispensable for P4-driven hESC decidualization. Additionally, we show that PLZF tightly regulates the expression levels of the early growth response 1 (EGR1) transcription factor, the perturbation of which impairs hESC decidualization.
Together, our studies have uncovered a new mediator pathway of the P4 signal that enables quiescent hESCs to differentiate into decidual cells, a cellular transformation process that is essential for early pregnancy establishment.
Results
Progestin induction of PLZF expression during decidualization of human endometrial stromal cells
Comparing undecidualized hESCs with decidualized hESCs in culture, we recently revealed by RNA-seq analysis that PLZF (also known as ZBTB 16, a Kruppel C2H2-type zinc-finger transcription factor) is listed in the top 20 genes which are significantly induced in hESCs during decidualization [8] (S1A Fig). Additionally, we previously showed by microarray analysis that Plzf is present in the top 100 genes induced in total uterine tissue of the mouse in response to acute P4 administration [9], suggesting PLZF is a P4-responsive gene and important for decidualization. To address this proposal, an established cell culture-based model for decidualization of primary hESCs was used to determine the transcriptional induction profile of PLZF (Fig 1A). In response to a hormone decidual stimulus (17β-estradiol (E2), medroxyprogesterone acetate (MPA), and cAMP (EPC)), rapid induction of high transcript levels for PLZF was observed by quantitative real time PCR (Fig 1A). The induction of transcripts encoding insulin-like growth factor binding protein 1 (IGFBP1) and prolactin (PRL) (both established decidual biomarkers [10]) confirmed decidualization of hESCs by EPC at the molecular level. Consistent with PLZF’s role as a transcription factor, most of the induced PLZF protein expression was present in the nucleus of decidual cells following EPC treatment (Fig 1B). Similar to findings in Fig 1A, PLZF protein expression was not detected in pre-decidual hESCs or with vehicle treatment (Fig 1B). Treatment of hESCs with E2, cAMP, or MPA revealed that only MPA significantly induced PLZF transcription in hESCs (Fig 1C), providing strong support for the progestin in the decidual hormone cocktail as the key driver of PLZF induction in decidualizing hESCs. Moreover, PLZF transcript levels are induced with acute progestin treatment (as early as 2 hours post MPA administration), which is not dependent on new protein synthesis (S1B and S1C Fig). These results support PLZF as an early progestin responsive gene in hESCs. As further support for this conclusion, immunohistochemical analysis of human endometrial biopsy samples show that PLZF protein is highly expressed in the nucleus of the endometrial stromal cell during the P4-dominant secretory phase of the menstrual cycle but is expressed at low levels in endometrial tissue biopsied during the E2-dominant proliferative phase of the cycle (Fig 1D–1F). In the uterus of the ovariectomized mouse, Plzf transcript and protein levels are also significantly and rapidly induced by P4 (S2 Fig), validating our previous microarray analysis of this tissue which showed Plzf transcription is induced by short-term P4 exposure [9]. Transcript and protein levels for Plzf were detected as early as six hours following P4 administration (S2A and S2B Fig). Furthermore, expression levels of Plzf protein are low in the endometrium of the adult virgin mouse but are significantly increased in the uterine stromal compartment of the early pregnant mouse (S2C Fig). It should be noted that global knockout of Plzf in the mouse results in a number of embryonic and developmental phenotypes [11] that preclude its use for uterine studies. However, the above results support the proposal that PLZF acts as a progestin/P4 early response transcription factor in human and mouse uterine stromal cells.
Fig 1. Progestin induction of PLZF expression in hESCs undergoing decidualization.
A) Relative transcript levels of PLZF, IGFBP-1, and PRL in hESCs cultured with vehicle (black bar (control)) or the EPC cocktail (red bar) at the indicated time points of cell culture. Note: Day 0 is the first day that hESCs were treated with vehicle or the EPC cocktail. B) Immunofluorescent detection of PLZF protein expression in hESCs cultured with vehicle or the EPC cocktail at the indicated time points of culture. Left and right columns show images of the same cell field stained for DAPI and PLZF immunopositivity respectively. After 6 days of EPC treatment, note the prominent nuclear staining for PLZF expression in a decidualized hESC cell (arrowhead). C) Relative transcript levels of PLZF in hESCs cultured for 6 days in vehicle control, E2, cAMP, or MPA alone. Note: For each transcript type (panel A) or reagent (panel C), results are reported relative to day 0 or vehicle treatment group respectively as the mean ± standard error (SE) from triplicate samples from one subject (three subjects were tested in total). *P<0.05, **P<0.01, and ***P<0.001. D-F) Immunohistochemical analysis of PLZF expression in human endometrial tissue biopsied during proliferative phase (D) and secretory phase (E and F) of the menstrual cycle. Black and white arrowheads in panel D and E indicate glandular epithelial and stromal cells respectively; white arrowhead in panel F indicates the nuclear localization of PLZF protein in stromal cells. Scale bar in panel D denotes 100 μm and applies to panel E; scale bar in panel F represents 100 μm.
Progestin induction of PLZF in hESCs requires the progesterone receptor
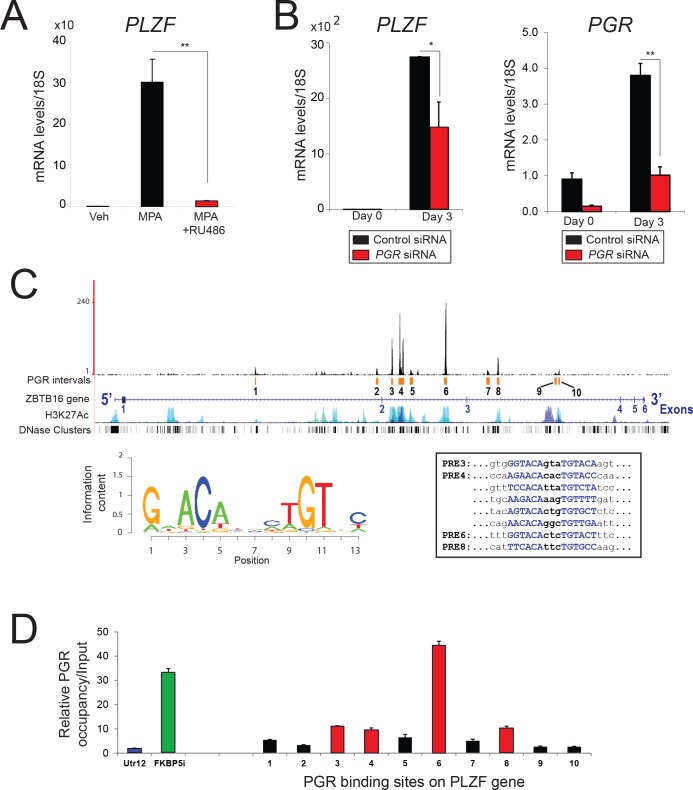
To confirm that PLZF is rapidly induced by progestin via the progesterone receptor (PGR) rather than by rapid non-genomic progestin effects [12], RU486 (a PGR antagonist) and small interfering (si) RNA-mediated knockdown of PGR were administered to hESCs (Fig 2). Following three days of culture with MPA or MPA plus RU486, PLZF transcript levels were examined in hESCs by quantitative real time PCR (Fig 2A). Compared to vehicle treatment, MPA significantly increased the transcript levels of PLZF; however, this level of induction was markedly attenuated with the co-administration of RU468 (Fig 2A). Further confirming the requirement of the PGR in this transcriptional induction, progestin induction of PLZF transcription was significantly attenuated with PGR knockdown (Fig 2B). In addition, acute hormone treatment of the ovariectomized Pgr knockout (PRKO) mouse revealed that rapid P4-induced Plzf transcription in the murine uterus requires endometrial Pgr (S2D Fig). Together, the above results strongly support an indispensable role for PGR in the rapid induction of PLZF transcription by progestin or P4 in human and murine uterine cells.
Fig 2. The progesterone receptor mediates progestin induction of PLZF expression in hESCs.
A) Relative transcript levels of PLZF in hESCs cultured with vehicle (control), MPA, or MPA in the presence of RU486 for three days in stripped media. Note the marked reduction in MPA-induced PLZF transcription when RU486 is included. B) PLZF and PR transcript levels in hESCs transfected with control siRNA or siRNA targeting PGR and cultured in EPC media at the indicated time points. Results are reported as the mean ± SE from triplicate samples from one subject (three subjects were tested in total); *P<0.05 and **P<0.01. C) Distribution of PGR binding intervals (designated 1 to 10) along the PLZF gene locus is shown along with H3K27Ac active sites and DNase hypersensitivity clusters. The sequence logo for progesterone-response element (PRE) is shown along with the sequence of PREs for sites 3, 4, 6, and 8. D) Validation of PGR binding to the PLZF gene using chromatin immunoprecipitation (ChIP). The histogram shows PCR results following PGR ChIP from hESCs on sites 1 to 10 as depicted in panel C. A non-binding untranslated region (Utr12 (blue bar)) and the FKBP5 intronic region (FKBP5i (green bar)) were used as negative and positive controls respectively for PGR binding.
Ten PGR binding sites (labeled 1 to 10 (Fig 2C)) in the PLZF gene were revealed from an analysis of a recently published chromatin immunoprecipitation-sequencing (ChIP-Seq) dataset derived from PGR cistrome studies on decidualized hESCs [8]. Four of these PGR binding sites (2, 4, 5, and 6) contain consensus progesterone-response elements (PREs) that are enriched in the hESC PGR-cistrome. Chromatin immunoprecipitation assay disclosed significant enrichment in PGR occupancy for sites 3, 4, 6, and 8 within the PLZF gene as compared to a negative control region (Fig 2D). Collectively, these results suggest that progestin transcriptional induction of PLZF involves binding of PGR to multiple sites across the PLZF gene.
Decidualization of hESCs requires PLZF
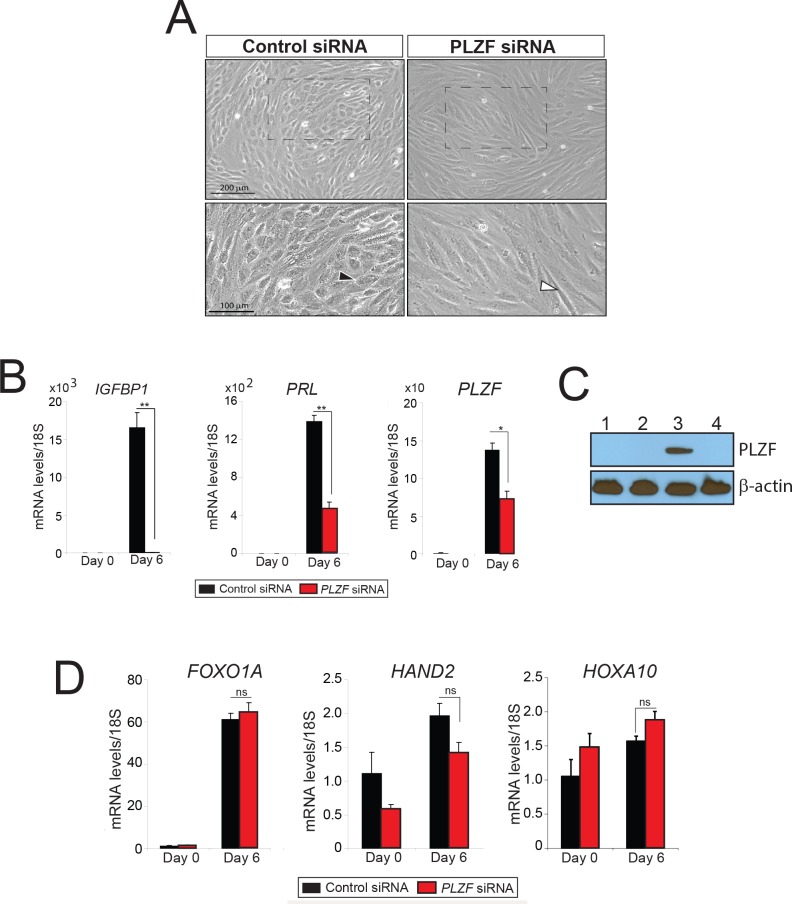
To determine whether PLZF is functionally required for in vitro hESC decidualization, siRNA mediated knockdown of PLZF transcription was performed in hESCs prior to EPC administration. Compared to control siRNA, hESCs treated with siRNA targeting PLZF failed to undergo the typical cellular transformation process in which elongated stromal fibroblasts transform into a polygonal epithelioid morphology, a morphological change which is indicative of stromal decidualization following EPC treatment (Fig 3A). Consistent with this block in stromal cell decidualization, a significant attenuation in the induction of decidual biomarkers (PRL and IGFBP-1) was observed in hESCs with PLZF knockdown (Fig 3B and 3C). Although these results underscore a critical functional role for PLZF in advancing progestin dependent hESC decidualization, surprisingly the expression level of a number of previously published PGR target genes in hESCs (FOXO1A, HAND2, and HOXA10 [13–15]) is not changed with PLZF knockdown (Fig 3D). These results indicate that the PLZF transcription factor is either positioned below these targets in the hierarchy of target genes for PGR action or PLZF operates in parallel to control its own gene target ensemble, which in turn underpins PGR dependent hESC decidualization.
Fig 3. Progestin-dependent decidualization of hESCs requires PLZF.
A) Cultured for six days in EPC media, the comparative hESC morphology following transfection with control siRNA or PLZF siRNA is shown. A typical epithelioid hESC with a polygonal appearance is indicated with a black arrowhead in the presence of control siRNA. A white arrowhead shows a hESC that remains fibroblastic when cultured in the presence of PLZF siRNA. Bottom panels represent higher magnifications of regions demarcated by hatched boxes in the corresponding top panels. B) Transcript levels of IGFBP-1, PRL, and PLZF in hESCs transfected with control siRNA (black bar) or PLZF siRNA (red bar) and cultured in EPC media for the indicated time period. C) Western blot analysis of PLZF protein levels to determine the effectiveness of siRNA against PLZF from hESCs transfected with control siRNA or PLZF siRNA and cultured in EPC media. Lanes 1–3 and 2–4 represent hESCs transfected with control siRNA or PLZF siRNA respectively. Lanes 1–2 and 3–4 represent hESCs cultured in EPC media for 0 days and 6 days respectively; β-actin was used as loading control. D) Transcript levels of the progesterone-responsive genes: FOXOA1, HAND2, and HOXA10 in hESCs transfected with control siRNA or PLZF siRNA and cultured in EPC media. Results are reported as the mean ± SE from triplicate samples from one subject (three subjects were tested in total). *P<0.05, **P<0.01, and ns>0.05.
Identification of the PLZF cistrome in hESCs
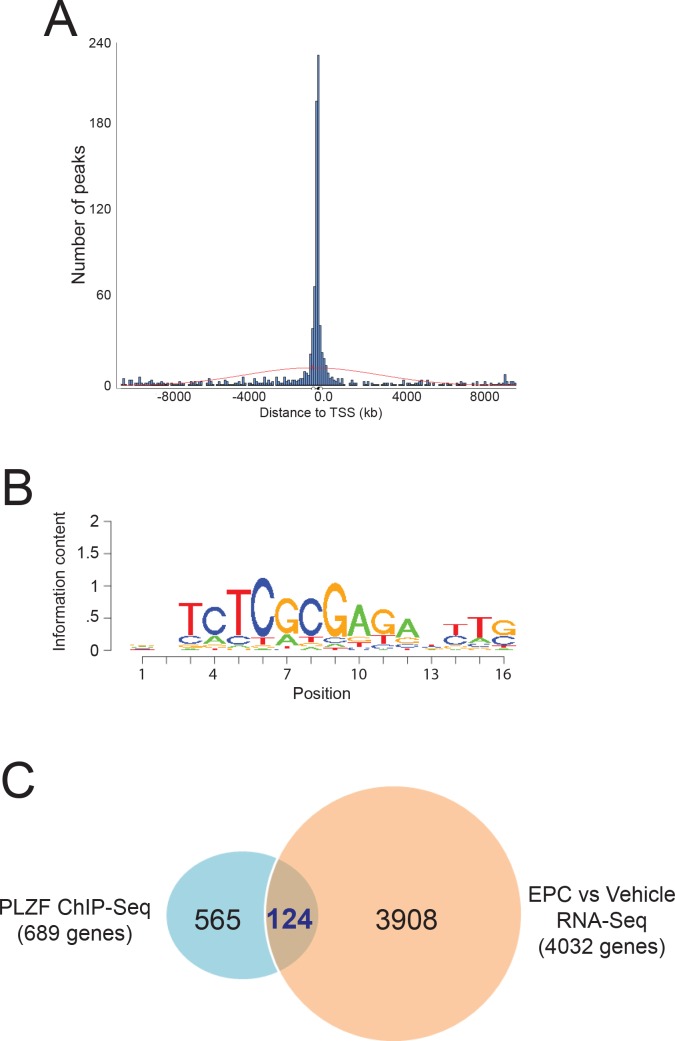
To identify possible PLZF target genes, ChIP-Seq was performed to identify the genome-wide binding events for PLZF in decidualizing hESCs (Fig 4). Having passed Illumina’s purity filter, the PLZF cistrome consists of 19,092,785 unique alignments (without duplicate reads) from more than 36 Million reads and aligns with no more than 2 mismatches to the genome. A total of 1,120 sequence intervals are enriched for PLZF binding across all chromosomes (supporting information). Peak distribution analyses revealed significant enrichment for PLZF binding near transcription start sites (TSS) with 48% of PLZF intervals located within 500 base pairs (b.p.) of TSS, indicating preferential binding of the PLZF transcription factor to gene promoter regions (Fig 4A and S1 Table). DNA sequence motif enrichment analysis revealed the zf-C2H2 DNA binding domain (MC00418) in the top enrichment cluster of motifs in the PLZF cistrome, a common DNA binding motif for many ZBTB transcription factors including PLZF [16] (Fig 4B). This cluster also contained DNA binding motifs for breast cancer 1 early onset (BRCA1) (MC00299) and v-ets avian erythroblastosis virus E26 oncogene homolog 1 (ETS1) (MC00018) (S2 Table). Following these enrichment groups, are motifs that are commonly found in cistromes of TSS-proximal binding transcription factors, such as motifs recognized by CCAAT-enhancer binding proteins (or C/EBPs), interferon-regulatory factors, SP1, and by leucine zipper family of transcription factors (i.e. JUN and FOS) (S2 Table).
Fig 4. Identification of the PLZF cistrome by ChIP-Seq.
A) Histogram representing the distance of PLZF binding peaks from the transcriptional start site (TSS) of all genes. B) Representative zf-C2H2 DNA binding domain motif (MC00418) identified by SeqPos motif enrichment analysis. C) Venn diagram comparing genes bound both by PLZF with genes for which their expression changes in hESCs following EPC induced decidualization [8].
To identify bona fide PLZF gene targets, we next compared 689 genes that have at least one PLZF ChIP binding region within 1kb of TSS with genes in hESCs for which expression changed with EPC treatment [8]. From this analysis, we identified 124 genes that are bound by PLZF and are differentially expressed during hESC decidualization (Fig 4C and S3 Table). By DAVID and GSEA analysis, the majority of these genes are associated with mitosis and cell cycle progression (S4 Table).
Transcriptional repression of early growth response 1 by PLZF is required for hESC decidualization
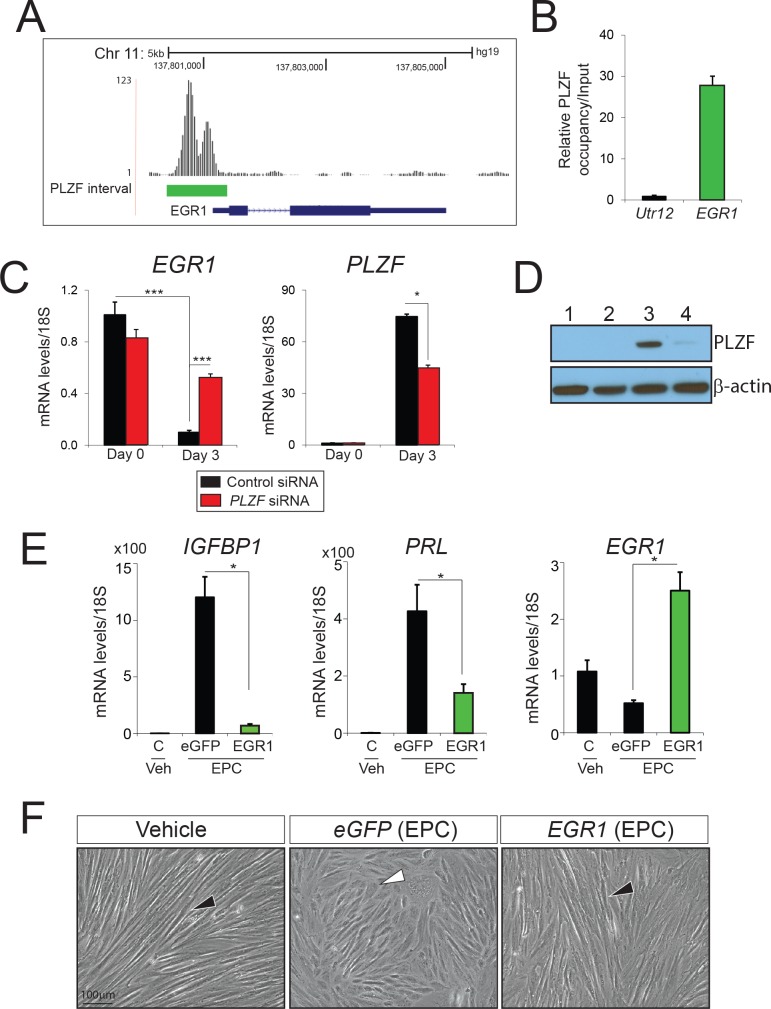
Because PLZF was previously shown to act as a transcriptional repressor of cellular proliferative programs in a number of physiological systems; reviewed in [16], we first focused on genes in the 124 gene list that promote early cellular growth responses and are downregulated during decidualization. The rationale for this prioritization is that a switch from a proliferative to a differentiative phenotype is an essential reprogramming step for hESCs to fully decidualize in response to EPC [7]. Based on the above criteria, we chose Early Growth Response 1 (EGR1) for further study because the gene: (a) ranks in the top five most downregulated gene in the 124 gene list (S3 Table); (b) belongs to a small family of transcription factors that enable early cellular growth responses [17]; and (c) exhibits strong PLZF ChIP binding at its promoter (Fig 5A and 5B).
Fig 5. PLZF-mediated transcriptional repression of EGR1 is critical for hESC decidualization.
A) Location of PLZF binding sites in the promoter region of EGR1. B) The histogram shows the qPCR result following ChIP using the PLZF antibody on the EGR1 promoter region from panel A. C) Comparative transcript levels of EGR1 in hESCs transfected with control siRNA (black bar) or PLZF siRNA (red bar) and cultured in EPC media. D) Western blot analysis of PLZF protein levels from hESCs transfected with control siRNA (lanes 1–3) or PLZF siRNA (lanes 2–4) cultured in EPC media for 0 days (lanes 1–2) or 3 days (lanes 3–4).; β-actin was used as loading control. E) Relative transcript levels of IGFBP1, PRL, and EGR1 in non-transduced hESCs or hESCs virally transduced with control eGFP or EGR1 cDNAs (green bar) and cultured in EPC media. Results are reported as the mean ± SE from triplicate samples from one subject (a total of three subjects were tested). *P<0.05, **P<0.01, and ***P<0.001. F) Morphology of hESCs transduced with control eGFP or EGR1 cDNAs following six days of culture in the presence of the EPC cocktail; non-transduced hESCs cultured in vehicle were used as control.
In keeping with its role as a driver of cell growth and proliferation, EGR1 transcript levels are significantly reduced as hESCs differentiate into decidual cells (Fig 5C). Interestingly, analysis of our recent RNA-seq data from pre-decidual and decidualized hESCs shows that transcript levels of EGR1 along with EGR2 and EGR3 are significantly reduced with decidualization (S3A and S3B Fig); however, only EGR1 shows PLZF binding (S3 Table). Importantly, knockdown of PLZF expression results in a significant attenuation in the normal reduction of EGR1 transcript levels following three days of EPC treatment (Fig 5C and 5D), suggesting PLZF represses EGR1 transcription as hESCs decidualize. Moreover, lentiviral-mediated overexpression of EGR1 significantly attenuated hESC decidualization in response to EPC treatment as evidenced by a marked reduction in the levels of the decidual biomarkers: IGFBP1 and PRL at the molecular and cellular level (Fig 5E and 5F). Interestingly, immunohistochemistry disclosed an inverse correlation between stromal PLZF and EGR1 protein expression in the human endometrium during the E2 dominant proliferative and P4 dominant secretory phase of the menstrual cycle (S4 Fig). It should be noted that the human endometrial stroma can decidualize during the secretory phase of the cycle even in the absence of a conceptus [6]. In the late proliferative phase of the cycle, EGR1 expression is detected in both epithelial and stromal cells of the endometrium whereas PLZF expression is not detected (S4 Fig: compare A-C with G-I). During the secretory phase of the cycle, significant PLZF expression is found in endometrial stromal cells; however, EGR1 expression is negligible during this time period (S4 Fig: compare D-F with J-L). Together, these immunohistochemical findings further support the proposal that PLZF acts as a novel transcriptional repressor of EGR1 in endometrial stromal cells to enable decidualization.
In the uterus of the ovariectomized mouse, Egr1 is markedly induced with E2 treatment alone (E2) whereas this induction is significantly blunted with the inclusion of P4 (P4E2) (S5 Fig). Further confirming inhibition of E2 induction of Egr1 by P4, inclusion of RU486 in the P4E2 hormone treatment returns Egr1 levels to those observed with E2 alone (S5 Fig). Interestingly, P4 induction of PLZF expression during this period parallels P4 suppression of Egr1 expression (S5 Fig), providing correlative support for the proposal that P4 suppression of E2 induction of uterine Egr1 is mediated by PLZF. Together, our human and mouse studies support the proposal that E2-induced EGR-1 expression is suppressed by P4-induced PLZF in the endometrium.
In sum, recent genome-wide studies at the cistrome and transcriptome level underscore the regulatory complexity of P4-dependent hESC decidualization. Within this complexity, we have uncovered a new signaling pathway (PGR-PLZF-EGR1) that is crucial for P4-driven hESC decidualization.
Discussion
Extensive transcriptional reprogramming underpins hESC decidualization [8], we report here that induction of PLZF as part of these transcriptional changes is critical for P4-dependent hESC decidualization. Highly conserved in mammals, PLZF is a member of the poxvirus and zinc finger (POZ)/ broad-complex, tramtrack, and bric-`a-brac (BTB) and Kruppel zinc finger (or POK) family of transcription factors; reviewed in [16]. Previous investigations have demonstrated a pleiotropic regulatory role for PLZF in a myriad of developmental programs and physiological responses, from hind-limb skeletal patterning [11] to spermatogonial stem-cell maintenance [18, 19]. Due to its role in limiting cell-cycle progression and cellular proliferation [20, 21], PLZF is known to act as a tumor suppressor in a broad spectrum of malignancies [22–26]. At the transcriptional level, PLZF is known primarily as a repressor; reviewed in [16]; however, an increasing number of studies describe PLZF as an activator of transcription [27–31]. Recruitment of corepressors and subsequent chromatin remodeling has been shown to underlie the repressor function of PLZF [32–34].
In different physiological contexts, PLZF is induced by the steroid hormones: cortisone [35], testosterone [36], and aldosterone [37]. These steroids along with P4 mediate their actions through closely related nuclear receptors belonging to subfamily 3 (Group C) of the nuclear receptor superfamily [38]; note: members of this receptor subfamily bind to a similar DNA response element containing the consensus half-site: AGAACA. Therefore, PLZF most likely evolved as one of a small number of specific target genes for this subclass of steroid hormones, which are known to exert wide-ranging physiological and pathophysiological responses. Although, a previous study reported that glucocorticoids and P4 can induce PLZF transcription in both hESCs and myometrial smooth muscle cells in vitro [39], a functional role for PLZF in these uterine cell types was not addressed.
Observed both in-vitro and in-vivo, endometrial stromal fibroblasts undergo proliferation and differentiation before their transformation into polygonal epithelioid decidual cells [7]. Using an integrative genome-wide approach, we reveal that PLZF is indispensable for P4-dependent hESC decidualization. Indeed, the requirement for PLZF in the morphological and functional transformation of an endometrial stromal fibroblast to an epithelioid decidual cell is reminiscent of its previously reported role in the morphological transformation of embryonic fibroblasts to a more flattened polygonal morphology [40, 41], a change in cell shape which correlates with the acquisition of cellular resistance to oncogenic transformation. Furthermore, the terminal differentiation which results from decidualization is in keeping with PLZF’s role in other physiological systems in which this transcription factor suppresses cell growth and proliferation in favor of differentiation; reviewed in [16].
The rapid P4-induction of PLZF transcription in uterine stromal cells suggests that this transcription factor acts as a direct functional mediator of the P4 signal. This proposal is supported by our ChIP-Seq studies which identified at least ten potential PGR binding sites that span introns 1–3 of the human PLZF gene. This DNA binding pattern concurs with previous studies which show that nuclear receptor binding sites are commonly found in introns located far beyond the confines of the promoter-proximal region of target genes [42, 43]. Looping of the intervening DNA has been posited as one possible mechanism to enable long-range interaction by transcription factors with the promoter-proximal region [44, 45]. Early PLZF induction following progestin/P4 administration also suggests that this transcription factor regulates target gene expression changes that occur early in the P4 signaling cascade. Through the integration of ChIP-Seq and RNA-Seq datasets, we revealed that EGR-1 represents one of these target genes for which its expression level is tightly controlled by PLZF to enable normal hESC decidualization by P4 exposure.
Recognizing the GC-rich consensus sequence GCG(T/G)GGGCG in promoter regions of target genes, EGR1 (also known as NGFI-A, Zif 268, or Krox 24) is a Cys2-His2 zinc finger transcription factor, which belongs to an immediate early response gene family that includes EGR 2–4 [46]. A wide spectrum of extracellular signaling cues activates EGR1, which in turn modulates cellular proliferation, differentiation, and apoptosis in diverse target tissues [17]. For example, EGR1 is induced by E2 in MCF-7 cells [47], suggesting that EGR1 is a mediator of the established E2 mitogenic effects in this human breast cancer cell line. Studies on the Egr1 knockout mouse reveal that Egr1 is required for normal follicular development, ovulation, and luteinization [48]; however, uterine functionality was not assessed. Despite the aforementioned, earlier studies demonstrated that Egr1 is rapidly induced in the rat uterus by E2 [49, 50], suggesting that Egr1 may mediate the known mitogenic effects of E2 in this tissue. Interestingly, results from these studies are similar to our findings in the mouse in which Egr1 is rapidly induced to high levels by E2 in the uterus; however, we further showed that this induction is also markedly reduced by P4 with simultaneous induction of PLZF. Intriguingly, recent investigations show that unscheduled elevation of EGR1 levels compromises hESC viability in vitro [51], indicating that strict controls on the levels of this transcription factor are critical for normal hESC function. These data may explain the need to reduce the expression levels of EGR1 as the pre-decidual fibroblast transforms into a decidual cell and underscores the importance of the P4:PGR: PLZF signaling axis in this regulatory process. Recent mouse studies further support the above by showing that while Egr1 is strongly expressed in the subluminal stroma surrounding the implanting blastocyst during early pregnancy [52], Egr1 expression is significantly reduced following in vivo and in vitro artificial decidualization [52]. Moreover, Egr1 overexpression downregulates the expression of the decidual marker decidual/trophoblast prolactin-related protein (Dtprp) in murine uterine stromal cells, while inhibition of Egr1 upregulated the expression of Dtprp under in vitro decidualization conditions [52]. Collectively, these results agree with our findings in hESCs in which overexpression of Egr1 blocks decidualization and significantly attenuates the induction of the decidual biomarkers, PRL and IGFBP-1.
Interestingly, we demonstrate that PLZF knockdown does not change the expression levels of FOXO1A, HOXA10 or HAND2 during hESC decidualization; these three genes have been shown to be important for the hESC decidual response [14, 53–57]. We also reveal that knockdown of FOXO1A, HAND2 or HOXA10 expression levels does not affect PLZF induction during hESC decidualization, suggesting that PLZF operates in a separate parallel pathway to FOXO1A, HAND2 and HOXA10 signaling. As further support for this conclusion, previous global transcript profiling of hESCs following FOXO1A or HOXA10 knockdown did not show changes in PLZF expression levels [58, 59]. In related studies, our ChIP-Seq analysis did not detect PLZF binding sites within the IGFBP1 or PRL genes, indicating that the downregulation of IGFBP1 and PRL expression following PLZF knockdown is through an indirect signaling axis and is not through FOXOA1 regulation of these decidual biomarkers [60].
In conclusion, pregnancy success relies on the execution of a programmed sequence of events, with uterine decidualization following embryo implantation representing a crucial early event which ensures not only the establishment but also the long-term maintenance of the fetomaternal interface. Advancement in our mechanistic understanding of P4’s involvement in decidualization is predicated upon identifying the key effectors that mediate the P4 signal into a decidual response. An integrative analysis of genome-scale data, together with primary hESC studies, we disclose PLZF as a potent mediator of P4 responsiveness which is essential for hESC decidualization. We also reveal that PLZF controls EGR-1 expression levels, the perturbation of which can compromise decidual progression. Given that further understanding of the molecular mechanisms that underlie P4-driven decidualization is essential if we are to formulate new clinical modalities in the diagnosis, prognosis, and treatment of infertility based on a defective endometrium, we believe our findings offer a new mechanistic perspective on P4 responsiveness in the uterus which may contribute to novel fertility solutions in the future.
Materials and Methods
Human endometrial stromal cell isolation
For in vitro decidualization studies, endometrial biopsies were obtained using a pipelle catheter or curette under sterile conditions from the uterine fundus of healthy women of reproductive age during the proliferative phase of their menstrual cycle. Subjects ranged in age between 27–38 years and had a normal uterus as evaluated by transvaginal ultrasound. Volunteers provided written informed consent prior to endometrial tissue biopsy, which was conducted in accordance with a protocol approved by the Institutional Review Board (IRB) at Baylor College of Medicine and the guidelines of the Declaration of Helsinki [61]. A portion of the endometrial biopsy was used for immunohistochemistry (see below). The remainder of the biopsy was used to prepare human endometrial stromal cells (hESCs) as previously described [9, 62]. Isolated hESCs were cultured in DMEM/F-12 media containing 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 0.1 mg/ml streptomycin (termed hESC media); studies were conducted with early passage primary hESCs (≤ four passages). For each experiment, at least three individual subjects provided the endometrial biopsy samples for cell preparation. For immunohistochemical analysis, endometrial biopsies also were taken from volunteers during the mid-secretory phase of their cycle (day 19–23) and fixed in formalin overnight (along with proliferative phase endometrial tissue) before paraffin embedding the following day.
Decidualization of hESCs and siRNA transfection
For RNA interference based studies, hESCs were transfected in triplicate with non-targeting (or control) small interfering (si)RNAs (D-001810-10-05), siRNAs targeting PLZF (L-005196-00-0005), or PGR (L-006763-00-0005) or FOXO1A (L-003006-00-0005) or HAND2 (L-008698-02-0005) or HOXA10 (L-006336-00-0005) (Thermo Scientific, Dharmacon RNAi Technologies, Chicago, IL) in six-well culture plates. In each well, siRNAs (60 pmol) were transfected into hESCs using Lipofectamine 2000 reagent in 1× Opti-MEM I reduced-serum media (Invitrogen Corporation). Forty-eight hours after transfection, in vitro decidualization of hESCs was initiated by treating cells with 1× Opti-MEM I reduced-serum media containing 2% FBS, 10nM E2, 1μM MPA (Sigma-Aldrich), and 50 μM cAMP (Sigma-Aldrich) (termed EPC media). The EPC medium was changed every two days, and hESCs harvested at defined time points as indicated [62]. Quantitation of PRL and IGFBP-1 transcript levels served as a positive molecular readout for hESC decidualization.
ChIP-seq data analyses and validation
Human ESCs were cultured in EPC media for 72 hours before PLZF and Input ChIP-Seq were performed by Active Motif, Inc. (Carlsbad, CA). Human ESCs derived from six subjects were pooled before being fixed in 4% formaldehyde, snap-frozen, and shipped on dry ice to Active Motif. A goat polyclonal anti-human PLZF antibody (sc-11146; Santa Cruz Biotechnology, Inc.) was used for the PLZF ChIP assay. Enriched DNA from the ChIP assay was amplified and DNA libraries were sequenced on an Illumina platform, followed by alignment of sequenced reads (24 million reads) to the human genome (GRCh37/hg19, February 2009). Peak calling was performed by applying a threshold of 18 (5 consecutive bins containing 0.18 aligns) before the resultant-called peaks were stored as Browser Extendable Data (BED) files. Due to the pronounced enrichment for TSS-proximal binding of PLZF by Cis-regulatory Element Annotation System (CEAS) analysis, genes associated with PLZF binding were called if the ChIP binding intervals were located within 950 bp from the TSS. Galaxy/Cistrome’s implementation of the Cis-regulatory Element Annotation System (CEAS) and Genomic Regions Enrichment of Annotations Tool (GREAT) tools were used for enrichment analyses on chromosomes and genomic annotations. Database for Annotation, Visualization, and Integrated Discovery Bioinformatics Resources (DAVID) v6.7 (http://david.abcc.ncifcrf.gov/)) and Gene Set Enrichment Analysis (GSEA) on Molecular Signatures Databases (MSigDB) collection (http://software.broadinstitute.org/gsea/index.jsp) analyses were performed on protein coding genes to identify enriched biological themes. Integration of PLZF ChIP-Seq data with RNA-Seq data sets was performed using a relational database system [63]. ChIP validation assays were performed on hESCs cultured in EPC media for seventy-two hours (Active Motif). A goat polyclonal anti-human PLZF antibody and a rabbit anti-human PGR (H-190) antibody (sc-7208, Santa Cruz Biotechnology, Inc.) were used to immunoprecipitate PLZF and PR proteins with associated bound DNA from fragmented chromatin respectively. Enriched DNA fragments from the PLZF and PGR ChIP assays were subjected to standard PCR analyses using specific primers (S5 Table) for specific binding regions. Reverse cross-linked chromatin fragments that were not immunoprecipitated (input DNA) were used as an internal control.
Molecular analyses
For quantitative real time PCR studies, total RNA was isolated from cells or tissue using the RNeasy total RNA isolation kit (Qiagen Inc., Valencia, CA). Complementary DNA (cDNA) was synthesized using the Bio-Rad Reverse Transcription kit (Bio-Rad Laboratories, Inc., Hercules, CA). Quantitative real time PCR analyses were performed using validated primers (Applied Biosystems/Life Technologies, Grand Island, NY) and TaqMan 2× master mix; ribosomal RNA (18S) was used as an internal control for gene specific primers; the primer list is shown in S6 Table.
For western analyses, total protein was extracted from uterine tissue using 1× protein lysis buffer (0.5% sodium deoxycholate, 1% Nonidet P-40, 0.1% SDS, phosphate-buffered saline (PBS), pH 7.4). An equal amount of protein extract (20 μg) was run on 4–15% gradient SDS-PAGE gels (Bio-Rad Laboratories, Inc.) and transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were blocked with 1× TBS containing 0.5% Tween-20 and 5% non-fat dry milk powder. Subsequently, membranes were incubated with rabbit polyclonal anti-PLZF (Santa Cruz Biotechnology, Inc.) or rabbit anti-human PGR (H-190) antibody (Santa Cruz Biotechnology, Inc.) or monoclonal anti-β-actin (Sigma-Aldrich) antibodies to detect PLZF, PGR and β-actin proteins respectively. Horseradish peroxidase-conjugated antibodies (Santa Cruz Biotechnology, Inc.) were used as secondary antibodies and the SuperSignal West Pico Chemiluminescent Substrate kit (Pierce Biotechnology, Rockford, IL) was used to detect the chemiluminescent signal.
Immunofluorescence cytology
When hESCs reached 60–70% confluence on eight-well chamber slides in triplicate wells, media was changed to EPC media. The EPC media was changed every two days for a total of six days before hESCs were fixed in 2% paraformaldehyde and permeabilized in 0.2% Triton-X detergent. Cells were washed and blocked in 1× PBS with 0.5% normal goat serum before incubation overnight with the rabbit polyclonal anti-human PLZF primary antibody ((H-300) Santa Cruz Biotechnology, Inc.). To detect immunoreactivity, hESCs were incubated with the Alexa Fluor 488-conjugated goat anti-rabbit secondary antibody (A-11008; Life Technologies) to visualize immunopositive cells (green fluorescence). Cell nuclei were visualized using the 4, 6-diamidino-2-phenylindole stain (DAPI; Sigma-Aldrich). Finally, slides were separated from the chambers and mounted using Slowfade mounting media before imaging.
Lentivirus transduction
A lentiviral vector expressing EGR1 (EX-OL00487-LX304) and a control vector expressing EGFP (EX-EGFP-LX304-B) were purchased from GeneCopoeia (Rockville, MD). Lentivirus was produced in the Gene Vector Core at Baylor College of Medicine. Twenty four hours following transduction of hESCs with control virus harboring empty vector or lentivirus expressing EGR1, media was replaced with EPC media to induce decidualization; lentiviral-mediated EGR1 overexpression was confirmed by real-time PCR.
Histological analyses
Briefly, paraformaldehyde-fixed tissues were processed and embedded in paraffin as previously described [62]. Tissue blocks were serially sectioned into 5-μm thick sections that were placed on Superfrost Plus glass slides (Fisher Scientific, Inc., Pittsburgh, PA). For immunohistochemical analyses, sections were deparaffinized before being rehydrated and boiled in a citric acid-based antigen unmasking solution (Vector Laboratories, Inc., Burlingame, CA). After blocking, sections were incubated overnight at 4°C with a rabbit polyclonal anti-human PLZF (H-300) antibody (1:150 dilution; sc-22839, Santa Cruz Biotechnology Inc., Dallas, TX) or anti-human EGR1 (#15F7) antibody (1:150 dilution; Cell Signaling, Danvers, MA). After incubation with the primary antibodies, sections were incubated with the goat anti-rabbit IgG secondary antibody (Vector Laboratories, Inc.), followed by incubation with ZyMax streptavidin-horseradish peroxidase conjugate (Invitrogen Corporation, Carlsbad, CA). The 3, 3’-diaminobenzidine (DAB) peroxidase substrate kit (Vector Laboratories, Inc.) was used to visualize immunoreactivity before sections were counterstained with hematoxylin. Cover slips were mounted onto stained sections using Slowfade mounting media (Fisher Scientific, Inc.).
Mice and hormone treatments
In a recurrent photocycle of 12h lights on-off, mice were housed in temperature controlled (22°C ± 2°C) rooms in an AAALAC accredited vivarium maintained by the Center for Comparative Medicine at the Baylor College of Medicine. A diet of irradiated Tekland global soy protein-free extruded rodent food pellets (Harlan Laboratories Inc., Indianapolis, IN) and fresh water were provided ad libitum to mice. Animal studies were undertaken following the guidelines described in the Guide for the Care and Use of Laboratory Animals (published by the National Research Council (Eighth Edition 2011)). Animal protocols (AN-1513; AN-544; and AN-4203) used in this research received prior approval from the Institutional Animal Care and Use Committee (IACUC) at Baylor College of Medicine. The PRKO mouse has been previously described [64]. The following anesthesia was used for surgery: ketamine 37.5mg, xylazine 1.9mg, and acepromazine 0.37 mg sq to 5ml with 2.45ml sterile water given at 0.75–1.5ml/kg BW, IP. Mice were euthanized by cervical disarticulation while under the plane of anesthesia; CO2 euthanasia was achieved with automated CO2 euthanasia chambers (EUTHANEX).
To evaluate uterine responsiveness to steroid hormone, mice were ovariectomized at six weeks-of-age and then rested for two weeks to ensure removal of serum ovarian steroid hormones before administration of exogenous hormone. Subcutaneous (s.c.) injection within the interscapular region of 100 μl sesame oil (vehicle control), 1 mg P4 (Sigma-Aldrich, St. Louis, MO), 100 ng E2 (Sigma), or 1mg RU486 (Sigma) in 100 μl vehicle was used to administer hormones and the antiprogestin. Following hormone treatment, whole uterine tissue was processed from euthanized mice for RNA, protein, and/or histological analysis.
Statistical analyses
For statistical analyses, a two-tailed student’s t test and one-way ANOVA with Tukey’s post-hoc test were performed using the Instat tool package version 3.0 (GraphPad software Inc., La Jolla, CA). Data with p values less than 0.05 were considered statistically significant.
Supporting Information
A) Table shows the change in ZBTB16 (PLZF), IGFBP1, and PRL expression in hESCs during decidualization from a recently published RNA-seq dataset [8]. The relative gene expression change in hESCs following treatment with the decidual stimulus (EPC) compared to vehicle treatment is represented as a Log2 transformed fold change value. B) Relative transcript levels of PLZF in hESCs treated with MPA alone at indicated time points. C) Relative transcript levels of PLZF in hESCs pre-treated with cyclohexamide (10ug/ML) for an hour and treated with MPA for four hours as indicated. D) Comparative protein levels of PGR in hESCs cultured for zero days (lanes 1 and 2) or four days (lanes 3 and 4) or six days (lanes 5 and 6) with the decidual stimulus (EPC; lanes 3 and 5) or MPA and cAMP (PC; lanes 4 and 6) compared to vehicle treatment cells (lanes 1 and 2). Note: The PGR isoforms (A and B) are elevated in EPC and PC treated cells; β-actin was used as loading control.
(TIF)
A) Relative Plzf transcript levels in uteri from ovariectomized wild-type mice treated with sesame oil (vehicle control) or progesterone for the indicated times. B) Western blot analyses for Plzf and β-actin protein levels in uteri from ovariectomized wild-type mice treated with sesame oil (vehicle control) or progesterone for the indicated times. Note: uteri from ovariectomized PRKO mice treated with progesterone for 6 hours were used as Pgr negative controls. C) Immunohistochemical detection of Plzf in uteri from adult virgin and early pregnant (5 days post coitum (d.p.c)) mice. Note the low levels of Plzf expression in the uterus of the virgin mouse compared to striking expression levels of Plzf in the stromal compartment of uterus of the early pregnant mouse (white arrow). At this stage of pregnancy, however, expression of Plzf was not detected in the luminal or glandular epithelium (LE and GE respectively); scale bar denotes 100μm and applies to all panels. D) Relative Plzf transcript levels in uteri from ovariectomized wild type control and PRKO mice injected with sesame oil (vehicle control) or progesterone and euthanized 6 hours post-injection. Results represent the mean ± SE; n = 3 mice/group. *P<0.05, **P<0.01, ns = not significant.
(TIF)
A) Table depicting EGR1, EGR2, and EGR3 transcript expression changes during EPC-driven decidualization [8]; gene expression changes are represented as Log2 transformed fold change. B) Relative transcript levels of EGR1, EGR2, EGR3, and EGR4 in hESCs cultured for 3 days in vehicle control (red bar) or EPC cocktail (black bar).
(TIF)
Immunohistochemical detection of PLZF (A to F) and EGR1 (G to L) expression in human endometrial tissue biopsied during proliferative phase (A-C and G-I) and secretory phase (D-F and J-I) of the menstrual cycle. Black and white arrowheads indicate glandular epithelium and stroma respectively; scale bar in panel A indicates 100 μm and applies to all panels.
(TIF)
Relative Plzf and Egr1 transcript levels in uteri from ovariectomized wild-type mice treated with sesame oil (vehicle control) or E2 (3 h) or P4E2 in presence or absence of RU486. Note: The RU486 antagonist was added 30 min prior to P4 treatment (P4 was administered 3 hours prior to E2 treatment). Results represent the mean ± SE; n = 3 mice/group. *P<0.05, **P<0.01, ns = not significant.
(TIF)
A) Transcript levels of PLZF in hESCs transfected with control siRNA or FOXO1A siRNA, HAND2 siRNA, or HOXA10 siRNA and cultured in EPC media for the indicated time period. B) Efficient knockdown of FOXOA1, HAND2, and HOXA10 was confirmed with the measurement of transcript levels of FOXOA1, HAND2, and HOXA10 in hESCs transfected with siRNA against respective protein as indicated. Results are reported as the mean ± SE from triplicates. *P<0.05, **P<0.01and ns>0.05.
(TIF)
(XLS)
(XLS)
(XLS)
(XLS)
(DOC)
(DOC)
(ZIP)
Acknowledgments
The technical expertise of Jie Li, Yan Ying, and Rong Zhao is gratefully acknowledged.
Data Availability
All relevant data are within the paper and its Supporting Information files. All metadata files are available from the Gene Expression Omnibus database (accession number GSE75115).
Funding Statement
This research was supported by the National Institute of Child Health & Human Development/National Institutes of Health R01 HD042311 to JPL and K99 HD080742 to RK. These studies also were supported in part by the Baylor College of Medicine Gene Vector Core (NIDDK P30DK079638). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Macklon NS, Geraedts JP, Fauser BC. Conception to ongoing pregnancy: the 'black box' of early pregnancy loss. Human reproduction update. 2002;8(4):333–43. . [DOI] [PubMed] [Google Scholar]
- 2.Wilcox AJ, Weinberg CR, O'Connor JF, Baird DD, Schlatterer JP, Canfield RE, et al. Incidence of early loss of pregnancy. N Engl J Med. 1988;319(4):189–94. 10.1056/NEJM198807283190401 . [DOI] [PubMed] [Google Scholar]
- 3.Zinaman MJ, Clegg ED, Brown CC, O'Connor J, Selevan SG. Estimates of human fertility and pregnancy loss. Fertility and sterility. 1996;65(3):503–9. . [PubMed] [Google Scholar]
- 4.Blesa D, Ruiz-Alonso M, Simon C. Clinical management of endometrial receptivity. Seminars in reproductive medicine. 2014;32(5):410–3. 10.1055/s-0034-1376360 . [DOI] [PubMed] [Google Scholar]
- 5.Rai R, Regan L. Recurrent miscarriage. Lancet. 2006;368(9535):601–11. 10.1016/S0140-6736(06)69204-0 . [DOI] [PubMed] [Google Scholar]
- 6.Cha J, Sun X, Dey SK. Mechanisms of implantation: strategies for successful pregnancy. Nature medicine. 2012;18(12):1754–67. Epub 2012/12/12. 10.1038/nm.3012 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gellersen B, Brosens JJ. Cyclic decidualization of the human endometrium in reproductive health and failure. Endocr Rev. 2014;35(6):851–905. 10.1210/er.2014-1045 . [DOI] [PubMed] [Google Scholar]
- 8.Mazur EC, Vasquez YM, Li X, Kommagani R, Jiang L, Chen R, et al. Progesterone receptor transcriptome and cistrome in decidualized human endometrial stromal cells. Endocrinology. 2015;156(6):2239–53. Epub 2015/03/18. 10.1210/en.2014-1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kommagani R, Szwarc MM, Kovanci E, Creighton CJ, O'Malley BW, Demayo FJ, et al. A murine uterine transcriptome, responsive to steroid receptor coactivator-2, reveals transcription factor 23 as essential for decidualization of human endometrial stromal cells. Biology of reproduction. 2014;90(4):75 10.1095/biolreprod.114.117531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brosens JJ, Hayashi N, White JO. Progesterone receptor regulates decidual prolactin expression in differentiating human endometrial stromal cells. Endocrinology. 1999;140(10):4809–20. Epub 1999/09/28. 10.1210/endo.140.10.7070 . [DOI] [PubMed] [Google Scholar]
- 11.Barna M, Hawe N, Niswander L, Pandolfi PP. Plzf regulates limb and axial skeletal patterning. Nat Genet. 2000;25(2):166–72. 10.1038/76014 . [DOI] [PubMed] [Google Scholar]
- 12.Peluso JJ, Pru JK. Non-canonical progesterone signaling in granulosa cell function. Reproduction. 2014;147(5):R169–78. 10.1530/REP-13-0582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daftary GS, Taylor HS. Implantation in the human: the role of HOX genes. Seminars in reproductive medicine. 2000;18(3):311–20. 10.1055/s-2000-12568 . [DOI] [PubMed] [Google Scholar]
- 14.Shindoh H, Okada H, Tsuzuki T, Nishigaki A, Kanzaki H. Requirement of heart and neural crest derivatives-expressed transcript 2 during decidualization of human endometrial stromal cells in vitro. Fertility and sterility. 2014;101(6):1781–90 e1-5. 10.1016/j.fertnstert.2014.03.013 . [DOI] [PubMed] [Google Scholar]
- 15.Vasquez YM, Mazur EC, Li X, Kommagani R, Jiang L, Chen R, et al. FOXO1 is required for binding of PR on IRF4, novel transcriptional regulator of endometrial stromal decidualization. Molecular endocrinology. 2015:me20141292 10.1210/me.2014-1292 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suliman BA, Xu D, Williams BR. The promyelocytic leukemia zinc finger protein: two decades of molecular oncology. Frontiers in oncology. 2012;2:74 10.3389/fonc.2012.00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sukhatme VP. Early transcriptional events in cell growth: the Egr family. J Am Soc Nephrol. 1990;1(6):859–66. . [DOI] [PubMed] [Google Scholar]
- 18.Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, et al. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36(6):647–52. 10.1038/ng1366 . [DOI] [PubMed] [Google Scholar]
- 19.Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, et al. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36(6):653–9. 10.1038/ng1367 . [DOI] [PubMed] [Google Scholar]
- 20.Choi WI, Kim MY, Jeon BN, Koh DI, Yun CO, Li Y, et al. Role of promyelocytic leukemia zinc finger (PLZF) in cell proliferation and cyclin-dependent kinase inhibitor 1A (p21WAF/CDKN1A) gene repression. The Journal of biological chemistry. 2014;289(27):18625–40. 10.1074/jbc.M113.538751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costoya JA, Hobbs RM, Pandolfi PP. Cyclin-dependent kinase antagonizes promyelocytic leukemia zinc-finger through phosphorylation. Oncogene. 2008;27(27):3789–96. 10.1038/onc.2008.7 . [DOI] [PubMed] [Google Scholar]
- 22.Cheung M, Pei J, Pei Y, Jhanwar SC, Pass HI, Testa JR. The promyelocytic leukemia zinc-finger gene, PLZF, is frequently downregulated in malignant mesothelioma cells and contributes to cell survival. Oncogene. 2010;29(11):1633–40. 10.1038/onc.2009.455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsieh CL, Botta G, Gao S, Li T, Van Allen EM, Treacy DJ, et al. PLZF, a tumor suppressor genetically lost in metastatic castration-resistant prostate cancer, is a mediator of resistance to androgen deprivation therapy. Cancer Res. 2015;75(10):1944–8. 10.1158/0008-5472.CAN-14-3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hui AW, Lau HW, Cao CY, Zhou JW, Lai PB, Tsui SK. Downregulation of PLZF in human hepatocellular carcinoma and its clinical significance. Oncology reports. 2015;33(1):397–402. 10.3892/or.2014.3578 . [DOI] [PubMed] [Google Scholar]
- 25.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–28. 10.1016/j.cell.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao GQ, Unger P, Yang Q, Kinoshita Y, Singh K, McMahon L, et al. Loss of PLZF expression in prostate cancer by immunohistochemistry correlates with tumor aggressiveness and metastasis. PloS one. 2015;10(3):e0121318 10.1371/journal.pone.0121318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaber ZB, Butler SJ, Novitch BG. PLZF regulates fibroblast growth factor responsiveness and maintenance of neural progenitors. PLoS Biol. 2013;11(10):e1001676 10.1371/journal.pbio.1001676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikeda R, Yoshida K, Tsukahara S, Sakamoto Y, Tanaka H, Furukawa K, et al. The promyelotic leukemia zinc finger promotes osteoblastic differentiation of human mesenchymal stem cells as an upstream regulator of CBFA1. The Journal of biological chemistry. 2005;280(9):8523–30. 10.1074/jbc.M409442200 . [DOI] [PubMed] [Google Scholar]
- 29.Labbaye C, Quaranta MT, Pagliuca A, Militi S, Licht JD, Testa U, et al. PLZF induces megakaryocytic development, activates Tpo receptor expression and interacts with GATA1 protein. Oncogene. 2002;21(43):6669–79. 10.1038/sj.onc.1205884 . [DOI] [PubMed] [Google Scholar]
- 30.Singh SP, Zhang HH, Tsang H, Gardina PJ, Myers TG, Nagarajan V, et al. PLZF regulates CCR6 and is critical for the acquisition and maintenance of the Th17 phenotype in human cells. J Immunol. 2015;194(9):4350–61. 10.4049/jimmunol.1401093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang N, Frank GD, Ding R, Tan Z, Rachakonda A, Pandolfi PP, et al. Promyelocytic leukemia zinc finger protein activates GATA4 transcription and mediates cardiac hypertrophic signaling from angiotensin II receptor 2. PloS one. 2012;7(4):e35632 10.1371/journal.pone.0035632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barna M, Merghoub T, Costoya JA, Ruggero D, Branford M, Bergia A, et al. Plzf mediates transcriptional repression of HoxD gene expression through chromatin remodeling. Dev Cell. 2002;3(4):499–510. . [DOI] [PubMed] [Google Scholar]
- 33.Chauchereau A, Mathieu M, de Saintignon J, Ferreira R, Pritchard LL, Mishal Z, et al. HDAC4 mediates transcriptional repression by the acute promyelocytic leukaemia-associated protein PLZF. Oncogene. 2004;23(54):8777–84. 10.1038/sj.onc.1208128 . [DOI] [PubMed] [Google Scholar]
- 34.Hong SH, David G, Wong CW, Dejean A, Privalsky ML. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor alpha (RARalpha) and PLZF-RARalpha oncoproteins associated with acute promyelocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(17):9028–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peppi M, Kujawa SG, Sewell WF. A corticosteroid-responsive transcription factor, promyelocytic leukemia zinc finger protein, mediates protection of the cochlea from acoustic trauma. J Neurosci. 2011;31(2):735–41. 10.1523/JNEUROSCI.3955-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang F, Wang Z. Identification and characterization of PLZF as a prostatic androgen-responsive gene. Prostate. 2004;59(4):426–35. 10.1002/pros.20000 . [DOI] [PubMed] [Google Scholar]
- 37.Naray-Fejes-Toth A, Boyd C, Fejes-Toth G. Regulation of epithelial sodium transport by promyelocytic leukemia zinc finger protein. Am J Physiol Renal Physiol. 2008;295(1):F18–26. 10.1152/ajprenal.00573.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83(6):835–9. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fahnenstich J, Nandy A, Milde-Langosch K, Schneider-Merck T, Walther N, Gellersen B. Promyelocytic leukaemia zinc finger protein (PLZF) is a glucocorticoid- and progesterone-induced transcription factor in human endometrial stromal cells and myometrial smooth muscle cells. Molecular human reproduction. 2003;9(10):611–23. . [DOI] [PubMed] [Google Scholar]
- 40.Hobbs RM, Pandolfi PP. Shape-shifting and tumor suppression by PLZF. Oncotarget. 2010;1(1):3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi J, Sun M, Vogt PK. Smooth muscle alpha-actin is a direct target of PLZF: effects on the cytoskeleton and on susceptibility to oncogenic transformation. Oncotarget. 2010;1(1):9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122(1):33–43. 10.1016/j.cell.2005.05.008 . [DOI] [PubMed] [Google Scholar]
- 43.Hewitt SC, Li L, Grimm SA, Chen Y, Liu L, Li Y, et al. Research resource: whole-genome estrogen receptor alpha binding in mouse uterine tissue revealed by ChIP-seq. Molecular endocrinology. 2012;26(5):887–98. 10.1210/me.2011-1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biddie SC, John S, Hager GL. Genome-wide mechanisms of nuclear receptor action. Trends Endocrinol Metab. 2010;21(1):3–9. 10.1016/j.tem.2009.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144(3):327–39. 10.1016/j.cell.2011.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Donovan KJ, Tourtellotte WG, Millbrandt J, Baraban JM. The EGR family of transcription-regulatory factors: progress at the interface of molecular and systems neuroscience. Trends Neurosci. 1999;22(4):167–73. . [DOI] [PubMed] [Google Scholar]
- 47.Chen CC, Lee WR, Safe S. Egr-1 is activated by 17beta-estradiol in MCF-7 cells by mitogen-activated protein kinase-dependent phosphorylation of ELK-1. J Cell Biochem. 2004;93(5):1063–74. 10.1002/jcb.20257 . [DOI] [PubMed] [Google Scholar]
- 48.Lee SL, Sadovsky Y, Swirnoff AH, Polish JA, Goda P, Gavrilina G, et al. Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1). Science. 1996;273(5279):1219–21. . [DOI] [PubMed] [Google Scholar]
- 49.Cicatiello L, Sica V, Bresciani F, Weisz A. Identification of a specific pattern of "immediate-early" gene activation induced by estrogen during mitogenic stimulation of rat uterine cells. Receptor. 1993;3(1):17–30. . [PubMed] [Google Scholar]
- 50.Suva LJ, Harm SC, Gardner RM, Thiede MA. In vivo regulation of Zif268 messenger RNA expression by 17 beta-estradiol in the rat uterus. Molecular endocrinology. 1991;5(6):829–35. 10.1210/mend-5-6-829 . [DOI] [PubMed] [Google Scholar]
- 51.Chang SJ, Tzeng CR, Lee YH, Tai CJ. Extracellular ATP activates the PLC/PKC/ERK signaling pathway through the P2Y2 purinergic receptor leading to the induction of early growth response 1 expression and the inhibition of viability in human endometrial stromal cells. Cell Signal. 2008;20(7):1248–55. 10.1016/j.cellsig.2008.02.011 . [DOI] [PubMed] [Google Scholar]
- 52.Guo B, Tian XC, Li DD, Yang ZQ, Cao H, Zhang QL, et al. Expression, regulation and function of Egr1 during implantation and decidualization in mice. Cell cycle. 2014;13(16):2626–40. 10.4161/15384101.2014.943581 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benson GV, Lim H, Paria BC, Satokata I, Dey SK, Maas RL. Mechanisms of reduced fertility in Hoxa-10 mutant mice: uterine homeosis and loss of maternal Hoxa-10 expression. Development. 1996;122(9):2687–96. . [DOI] [PubMed] [Google Scholar]
- 54.Grinius L, Kessler C, Schroeder J, Handwerger S. Forkhead transcription factor FOXO1A is critical for induction of human decidualization. The Journal of endocrinology. 2006;189(1):179–87. 10.1677/joe.1.06451 . [DOI] [PubMed] [Google Scholar]
- 55.Labied S, Kajihara T, Madureira PA, Fusi L, Jones MC, Higham JM, et al. Progestins regulate the expression and activity of the forkhead transcription factor FOXO1 in differentiating human endometrium. Molecular endocrinology. 2006;20(1):35–44. 10.1210/me.2005-0275 . [DOI] [PubMed] [Google Scholar]
- 56.Lim H, Ma L, Ma WG, Maas RL, Dey SK. Hoxa-10 regulates uterine stromal cell responsiveness to progesterone during implantation and decidualization in the mouse. Molecular endocrinology. 1999;13(6):1005–17. 10.1210/mend.13.6.0284 . [DOI] [PubMed] [Google Scholar]
- 57.Takano M, Lu Z, Goto T, Fusi L, Higham J, Francis J, et al. Transcriptional cross talk between the forkhead transcription factor forkhead box O1A and the progesterone receptor coordinates cell cycle regulation and differentiation in human endometrial stromal cells. Molecular endocrinology. 2007;21(10):2334–49. 10.1210/me.2007-0058 . [DOI] [PubMed] [Google Scholar]
- 58.Buzzio OL, Lu Z, Miller CD, Unterman TG, Kim JJ. FOXO1A differentially regulates genes of decidualization. Endocrinology. 2006;147(8):3870–6. 10.1210/en.2006-0167 . [DOI] [PubMed] [Google Scholar]
- 59.Lu Z, Hardt J, Kim JJ. Global analysis of genes regulated by HOXA10 in decidualization reveals a role in cell proliferation. Molecular human reproduction. 2008;14(6):357–66. 10.1093/molehr/gan023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim JJ, Buzzio OL, Li S, Lu Z. Role of FOXO1A in the regulation of insulin-like growth factor-binding protein-1 in human endometrial cells: interaction with progesterone receptor. Biology of reproduction. 2005;73(4):833–9. 10.1095/biolreprod.105.043182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.WMA Declaration of Helsinki Serves as Guide to Physicians. Calif Med. 1966;105(2):149–50. Epub 1966/08/01. [PMC free article] [PubMed] [Google Scholar]
- 62.Kommagani R, Szwarc MM, Kovanci E, Gibbons WE, Putluri N, Maity S, et al. Acceleration of the glycolytic flux by steroid receptor coactivator-2 is essential for endometrial decidualization. PLoS Genet. 2013;9(10):e1003900 Epub 2013/11/10. 10.1371/journal.pgen.1003900 PGENETICS-D-13-01567 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stashi E, Lanz RB, Mao J, Michailidis G, Zhu B, Kettner NM, et al. SRC-2 is an essential coactivator for orchestrating metabolism and circadian rhythm. Cell reports. 2014;6(4):633–45. Epub 2014/02/18. 10.1016/j.celrep.2014.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lydon JP, DeMayo FJ, Funk CR, Mani SK, Hughes AR, Montgomery CA Jr., et al. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev. 1995;9(18):2266–78. Epub 1995/09/15. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Table shows the change in ZBTB16 (PLZF), IGFBP1, and PRL expression in hESCs during decidualization from a recently published RNA-seq dataset [8]. The relative gene expression change in hESCs following treatment with the decidual stimulus (EPC) compared to vehicle treatment is represented as a Log2 transformed fold change value. B) Relative transcript levels of PLZF in hESCs treated with MPA alone at indicated time points. C) Relative transcript levels of PLZF in hESCs pre-treated with cyclohexamide (10ug/ML) for an hour and treated with MPA for four hours as indicated. D) Comparative protein levels of PGR in hESCs cultured for zero days (lanes 1 and 2) or four days (lanes 3 and 4) or six days (lanes 5 and 6) with the decidual stimulus (EPC; lanes 3 and 5) or MPA and cAMP (PC; lanes 4 and 6) compared to vehicle treatment cells (lanes 1 and 2). Note: The PGR isoforms (A and B) are elevated in EPC and PC treated cells; β-actin was used as loading control.
(TIF)
A) Relative Plzf transcript levels in uteri from ovariectomized wild-type mice treated with sesame oil (vehicle control) or progesterone for the indicated times. B) Western blot analyses for Plzf and β-actin protein levels in uteri from ovariectomized wild-type mice treated with sesame oil (vehicle control) or progesterone for the indicated times. Note: uteri from ovariectomized PRKO mice treated with progesterone for 6 hours were used as Pgr negative controls. C) Immunohistochemical detection of Plzf in uteri from adult virgin and early pregnant (5 days post coitum (d.p.c)) mice. Note the low levels of Plzf expression in the uterus of the virgin mouse compared to striking expression levels of Plzf in the stromal compartment of uterus of the early pregnant mouse (white arrow). At this stage of pregnancy, however, expression of Plzf was not detected in the luminal or glandular epithelium (LE and GE respectively); scale bar denotes 100μm and applies to all panels. D) Relative Plzf transcript levels in uteri from ovariectomized wild type control and PRKO mice injected with sesame oil (vehicle control) or progesterone and euthanized 6 hours post-injection. Results represent the mean ± SE; n = 3 mice/group. *P<0.05, **P<0.01, ns = not significant.
(TIF)
A) Table depicting EGR1, EGR2, and EGR3 transcript expression changes during EPC-driven decidualization [8]; gene expression changes are represented as Log2 transformed fold change. B) Relative transcript levels of EGR1, EGR2, EGR3, and EGR4 in hESCs cultured for 3 days in vehicle control (red bar) or EPC cocktail (black bar).
(TIF)
Immunohistochemical detection of PLZF (A to F) and EGR1 (G to L) expression in human endometrial tissue biopsied during proliferative phase (A-C and G-I) and secretory phase (D-F and J-I) of the menstrual cycle. Black and white arrowheads indicate glandular epithelium and stroma respectively; scale bar in panel A indicates 100 μm and applies to all panels.
(TIF)
Relative Plzf and Egr1 transcript levels in uteri from ovariectomized wild-type mice treated with sesame oil (vehicle control) or E2 (3 h) or P4E2 in presence or absence of RU486. Note: The RU486 antagonist was added 30 min prior to P4 treatment (P4 was administered 3 hours prior to E2 treatment). Results represent the mean ± SE; n = 3 mice/group. *P<0.05, **P<0.01, ns = not significant.
(TIF)
A) Transcript levels of PLZF in hESCs transfected with control siRNA or FOXO1A siRNA, HAND2 siRNA, or HOXA10 siRNA and cultured in EPC media for the indicated time period. B) Efficient knockdown of FOXOA1, HAND2, and HOXA10 was confirmed with the measurement of transcript levels of FOXOA1, HAND2, and HOXA10 in hESCs transfected with siRNA against respective protein as indicated. Results are reported as the mean ± SE from triplicates. *P<0.05, **P<0.01and ns>0.05.
(TIF)
(XLS)
(XLS)
(XLS)
(XLS)
(DOC)
(DOC)
(ZIP)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. All metadata files are available from the Gene Expression Omnibus database (accession number GSE75115).