Abstract
Hippocampal neurogenesis, the product of resident neural stem cell proliferation and differentiation, persists into adulthood but decreases with organismal aging, which may contribute to the age-related decline in cognitive function. The mechanisms that underlie this decrease in neurogenesis are not well understood, though evidence in general indicates that extrinsic changes in an aged stem cell niche can contribute to functional decline in old stem cells. Bone Morphogenetic Protein (BMP) family members are intercellular signaling proteins that regulate stem and progenitor cell quiescence, proliferation, and differentiation in various tissues and are likewise critical regulators of neurogenesis in young adults. Here, we establish that BMP signaling increases significantly in old murine hippocampi and inhibits neural progenitor cell proliferation. Furthermore, direct in vivo attenuation of BMP signaling via genetic and transgenic perturbations in aged mice led to elevated neural stem cell proliferation, and subsequent neurogenesis, in old hippocampi. Such advances in our understanding of mechanisms underlying decreased hippocampal neurogenesis with age may offer targets for the treatment of age-related cognitive decline.
Introduction
Neurogenesis occurs throughout our lifetime in the subgranular zone (SGZ) of the dentate gyrus (DG) of the hippocampus and the subventricular zone (SVZ) of the lateral ventricles in mammals, via differentiation of adult neural stem cells (NSCs) into excitatory granule neurons and inhibitory olfactory bulb interneurons, respectively [1]. Hippocampal neurogenesis is believed to aid new memory formation, while SVZ neurogenesis plays a role in sensory functions [2–4].
Neurogenesis, however, significantly declines with age, which is believed to result from both a reduction in the overall number of stem cells and in the ability of the remaining cells to function properly with age. In particular within the SGZ, type 1 and 2 NSCs and neural progenitor cells (NPCs) significantly decrease in number with increasing age [5–8], as demonstrated by the two-fold decline in Sox2+ cells and ninefold decline in proliferating BrdU+ and doublecortin (DCX)+ cells in neurogenic regions of aged mice. Additionally, an increase in quiescence of NSCs was also observed with aging in rats [9], and neurogenesis was also shown to decline in the hippocampi of primates [10, 11]. Moreover, SGZ neurogenesis, also active in humans [12], exhibits a steady decline with age [13]. The molecular mechanisms that underlie the loss of organ stem cell numbers with age, particularly in brain, are beginning to be elucidated. Both an elevation in the systemic levels of chemokines and a decrease in hippocampal Wnt signaling with age have been correlated with or demonstrated to hinder hippocampal neurogenesis [14–18]. Furthermore, TGF-β signaling has been implicated in the decline of neurogenesis with age in the subventricular zone (SVZ) [19], the other central nervous system (CNS) region that can exhibit adult neurogenesis.
Bone Morphogenic Protein (BMP) family members and most growth and differentiation factor (GDF) ligands, which can play roles in regulating stem cell function, activate cellular signaling by binding BMP type II receptors that in turn phosphorylate and activate type I receptor serine-threonine kinases. In the canonical pathway, the type I receptors then phosphorylate and activate specific R-Smads(1,5,8), which subsequently heterodimerize with Co-Smad4 and translocate to the nucleus, bind coactivators or corepressors, and thereby activate or inhibit gene expression [20, 21]. In particular, BMP induces expression of Id1 and Id3, bHLH transcriptional repressors that in some systems are necessary for inhibition of differentiation-inducing factors [22]. BMP signaling thereby regulates a variety of biological functions in various organ systems and during development, including within the CNS.
While BMP signaling has been extensively studied in embryonic pluripotent stem cells and NSCs [20, 23], its roles within the adult CNS are only beginning to be elucidated. BMP inhibits neurogenesis and promotes NSC glial differentiation in the adult SVZ [24], resulting in a depletion of the stem cell pool [25]. However, in the hippocampus BMP signaling through BMPRIA inhibits NSC proliferation and promotes their maintenance in an undifferentiated and quiescent state [26]. Additionally, overexpression of the BMP antagonist Noggin induces proliferation and enhanced the self-renewal of hippocampal stem cells in young adults, thereby expanding this pool [27]. Furthermore, BMP4 inhibition has been implicated in exercise-induced hippocampal neurogenesis and hippocampal dependent learning [28, 29]. In aged organisms, however, BMP involvement in the decline of stem cell function has in general only just begun to be elucidated. For example, in the hair follicle stem cell niche local elevation of BMP signaling was demonstrated to contribute to decline in hair regeneration [30]. However, the role of BMP signaling in NSC aging has not been addressed.
Here, we demonstrate that multiple BMP growth factors and downstream signaling effectors increase in expression with aging in the hippocampal NSC niche and as a result inhibit cell proliferation. Furthermore, we demonstrate a partial rescue of aged hippocampal neurogenesis via in vivo genetic and transgenic inhibition of BMP signaling.
Results
BMP signaling increases in the aged murine hippocampus
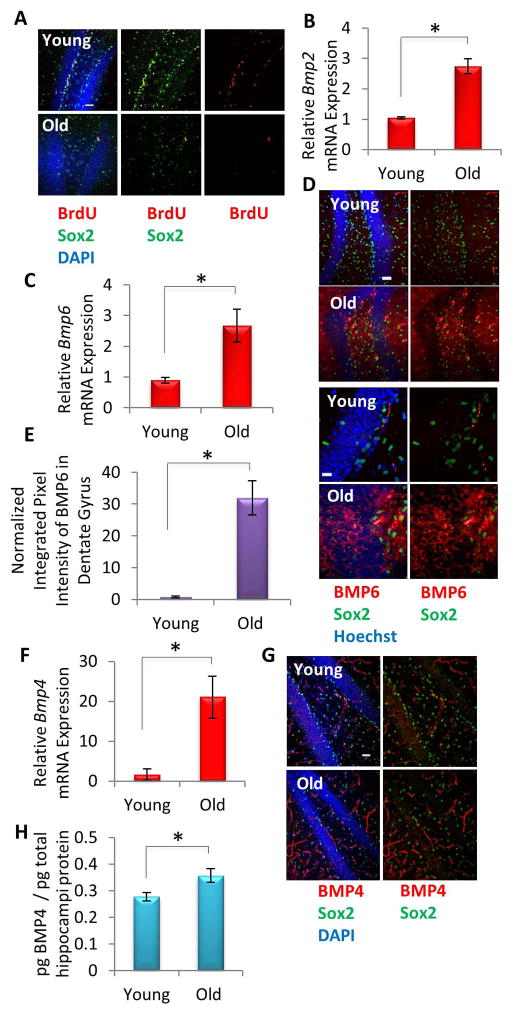
Consistent with prior reports [5–8], we found that Sox2+BrdU+ proliferating hippocampal cells, which include type 1 and 2a neural stem and progenitor cells, significantly decline with age (Figure 1A). To correlate this decrease in proliferating neural stem and progenitor cells with levels of BMPs, known negative regulators of cell proliferation and differentiation in the young dentate gyrus [26], we isolated hippocampi from young (2–4 mo) and old (22–24 mo) male C57BL6/J mice and analyzed mRNA and protein levels of various BMP family members using numerous available means. We observed an increase in Bmp2 and Bmp6 mRNA expression levels with age as assessed by qRT-PCR (Figure 1B,C), as well as an increase in BMP6 protein expression as assessed by immunofluorescence (Figure 1D,E). Bmp4 mRNA and protein expression levels – as assayed by qRT-PCR, immunofluorescence in whole tissue sections, and ELISA in tissue lysates – also increased in aged hippocampi (Figure 1F–H). BMP4 and BMP6 immunofluorescence staining was also more abundant in the cortex of old mice, indicating this brain region may also exhibit an increase in BMP signaling (Supplemental Figure 1A–C). Consistent with this observation, BMP6 pixel intensity quantification revealed significant increases in the cortex with age, but not the hypothalamus, suggesting BMP expression may increase spatially in particular brain regions with age (Supplemental Figure 1D,E). However, these results are consistent with a prior report that BMP6 expression increased in both the hippocampus and cortex of human AD patients and animal models of this disease [31]. Finally, a recent qPCR array study of dentate gyri from young (4 month) and aged (24 month) rats did not find changes in Bmp 2 and 4 mRNA levels [32], though differences in methodology and/or the longer lifespans of rats versus mice may account for this difference.
Figure 1. BMPs Increase with Age Locally in Mice Hippocampi.
(A) Young (2 month) and old (24 month) mice (n=3) were given daily IP injections of BrdU for 5 days, followed by perfusion and PFA fixation. Immunofluorescence (IF) was performed for Sox2 (green) and BrdU (red), with Dapi (blue) labeling all nuclei. Representative images are shown. Scale bar = 100 μM (B) Quantification of Bmp2 mRNA expression by qRT-PCR performed on RNA extracted from young and old hippocampi. The relative expression levels were normalized to GAPDH and presented relative to that of young hippocampi. Significant differences were identified by Student’s t-tests (*p<0.008). Error bars indicate standard error of the mean (n=4 mice per group). (C) qRT-PCR quantification of Bmp6 mRNA was performed on RNA extracted from young and old hippocampi. The expression levels were normalized to GAPDH and presented relative to that of young hippocampi. Significant differences were identified by Student’s t-tests (*p<0.05). Error bars indicate standard error of the mean (n=5 young, 4 old). (D) Immunofluorescence was performed on perfused young and old brain tissue sections (n=3 young, 3 old) for Sox2 (green) and BMP6 (red), with Hoechst (blue) labeling all nuclei. Representative low and high magnification images are shown. Scale bar = 50 μM (E) Integrated pixel intensity of BMP6 immunofluorescence in young and old dentate gyri tissue sections was calculated using ImageJ. Pixel intensities are presented relative to young dentate gyri. Significant differences were identified by Student’s t-tests (*p<0.003). Error bars indicate standard error of the mean (n=3 young, 3 old). (F) qRT-PCR quantification of Bmp4 mRNA was performed on RNA extracted from young and old hippocampi. The expression levels were normalized to GAPDH and presented relative to that of young hippocampi. Significant differences were identified by unpaired Student’s t-tests (*p<0.05). Error bars indicate standard error of the mean (n=3 young, 3 old mice per group). (G) Immunofluorescence was performed on perfused young and old brain tissue sections (n=3), for Sox2 (green) and BMP4 (red), with DAPI (blue) labeling all nuclei. Representative images are shown. Scale bar = 50 μM (H) An ELISA was performed on young and old hippocampal protein lysate to assess the level of BMP4. Significant differences were identified by Student’s t-tests (*p<0.05), and error bars indicate standard error of the mean (n=5 young, 5 old).
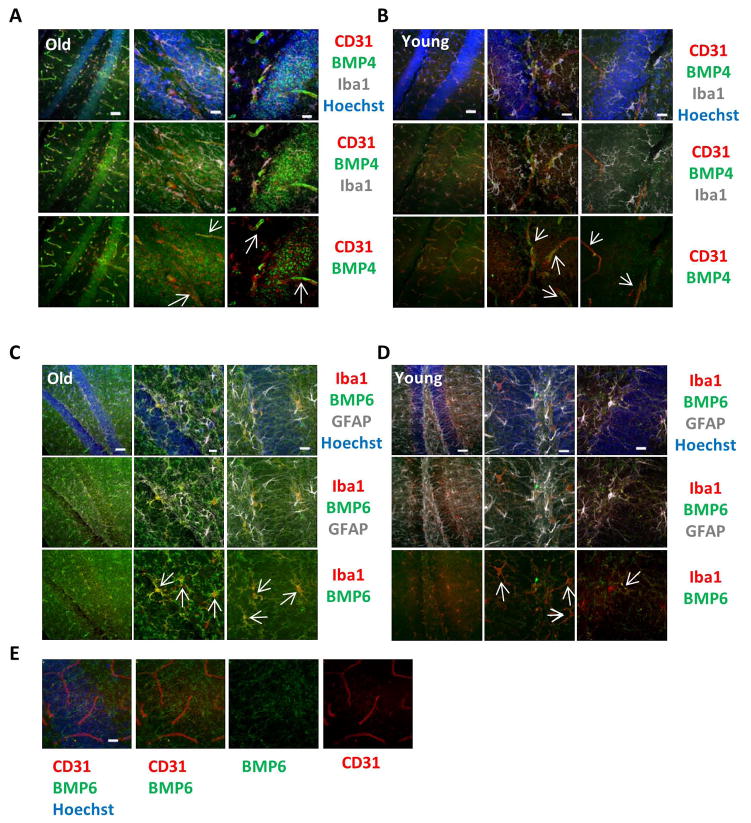
To assess the source of BMP production, costaining of BMP 4 and 6 with specific markers of cells known to interact with hippocampal NSCs – including endothelial cells, astrocytes, and microglia – was performed. BMP4 was expressed in a vascular-like pattern and colocalized with CD31+ endothelium in both old and young dentate gyri (Figure 2A, B), consistent with findings that BMP4 is expressed in endothelial cells in response to stress-inducing cues [33]. BMP4 also colocalized with CD31+ endothelium in the cortex of young and old mice (Supplemental Figure 1A), and increased in expression with age. BMP6, interestingly, was primarily colocalized in Iba1+ microglia rather than endothelial cells (Figure 2C–E) in the dentate gyrus. Likewise, in the cortex of aged, but not young, mice BMP6 localized in Iba1+ microglia, but was also found to localize with CD31+ endothelial cells (Supplemental Figure 1B,C).
Figure 2. BMP4 and BMP6 colocalization with endothelial cells or microglia.
(A) Immunofluorescence was performed on perfused old and young brain tissue sections for BMP4 (green), CD31 (red), and Iba1 (gray), with Hoechst (blue) labeling all nuclei. Representative low and high magnification images are shown. Scale bars = 50 μm. Arrows indicate areas of BMP4 and CD31 colocalization. (B) Immunofluorescence was performed on perfused old and young brain tissue sections for BMP6 (green), Iba1 (red), and GFAP (gray), with Hoechst (blue) labeling all nuclei. Representative low and high magnification images are shown. Scale bars = 50 μm. Arrows indicate areas of BMP6 and Iba1 colocalization. (C) Immunofluorescence was performed on perfused young brain tissue sections for BMP6 (green) and CD31 (red), with Hoechst (blue) labeling all nuclei. Representative high magnification images are shown and indicate there is no colocalization of BMP6 with endothelium. Scale bars = 50 μm.
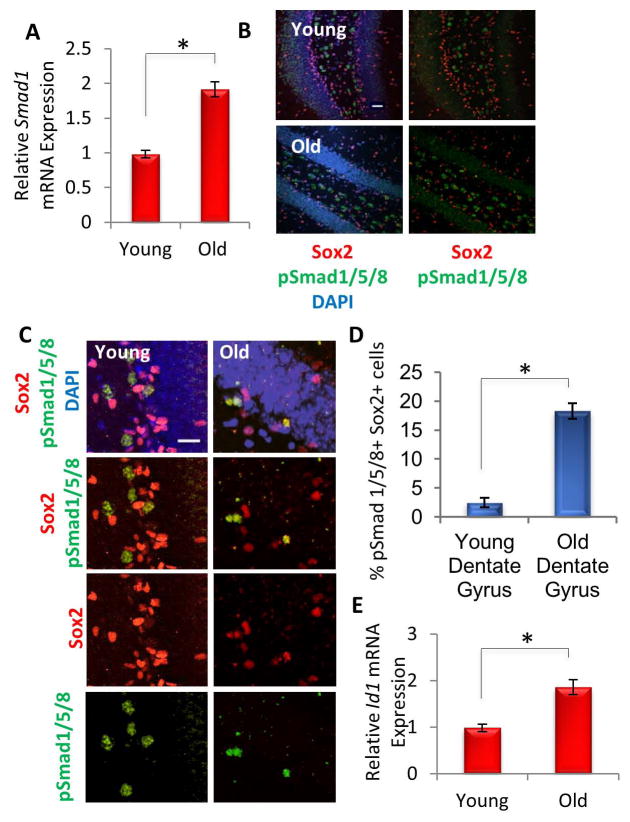
BMP 2, 4, and 6 signaling converge at the phosphorylation and activation of SMAD 1,5, and 8, and we thus analyzed downstream SMAD signaling in young versus aged neural stem cells in vivo. qRT-PCR revealed that in vivo hippocampal expression of total Smad1, a transcription factor activated by BMP signaling [21], was elevated with age (Figure 3A). In addition, while active, phosphorylated SMAD1/5/8 protein levels did not appear to change across the hippocampus as a whole with age (Figure 3B), the percentage of Sox2+ cells in the SGZ that were also pSmad1/5/8+ increased substantially to nearly 20% (Figure 3C, quantified in D). pSmad1 levels vary depending on the stage of stem cell division [34], so it is possible that even higher proportions of cells expressed pSmad1/5/8 overall. Furthermore, Id1, a known downstream target of the BMP/pSmad pathway [22, 35] that inhibits the function of proneural transcription factors and thereby represses neurogenic differentiation [35], was elevated with age (Figure 3E). These data are the first to demonstrate that in the hippocampus BMP transcript and protein levels and Id1 transcript levels increase with age, and that in hippocampal Sox2+ neural stem and progenitor cells SMAD1/5/8 phosphorylation is correspondingly elevated with age.
Figure 3. Downstream effectors of BMP Signaling increase with Age in Mice Hippocampi.
(A) qRT-PCR quantification of Smad1 mRNA expression in young and old hippocampi. The relative expression levels were normalized to GAPDH and presented relative to that of young hippocampi. Significant differences were identified by Student’s t-tests (*p<0.01), and error bars indicate the standard error of the mean (n=3). (B) Immunofluorescence was performed on young and old brain tissue sections (n=4) for pSmad1/5/8 (green) and Sox2 (red), with DAPI (blue) labeling all nuclei. Representative images are shown. Scale bar = 100 μM (C) Representative high magnification images of brain sections stained for pSmad1/5/8 (green) and Sox2 (red), with DAPI (blue) labeling all nuclei. Scale bar = 50 μM (D) Quantification of the percentage of pSmad1/5/8+ Sox2+ neural stem and progenitor cells in the denate gyrus demonstrates an increase with age in BMP signaling to NPCs. Significant differences were identified by Student’s t-tests (*p<0.01), and error bars indicate the standard deviation (n=4) (E) qRT-PCR quantification of Id1 mRNA levels in RNA extracted from young and old hippocampi. The relative expression levels were normalized to GAPDH and presented relative to that of young hippocampi. Significant differences were identified by Student’s t-tests (*p<0.02), and error bars indicate the standard error of the mean (n=4).
Partial rescue of neurogenesis in aged hippocampi by in vivo genetic inhibition of BMP/pSmad1 signaling
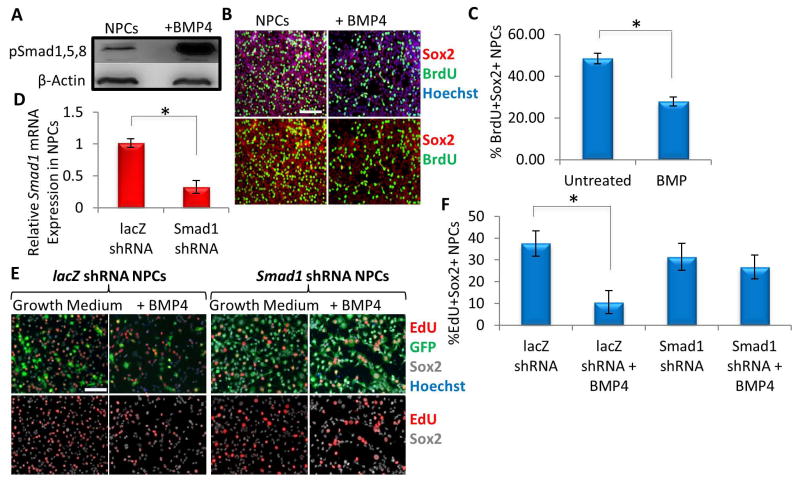
To probe whether an increase in BMP may be functionally relevant for neurogenesis, BMP4 was added to hippocampal-derived Sox2+ NPC cultures. After 30 minutes, phosphorylated Smad1/5/8 (Figure 4A) was upregulated, and after 24 hours cell proliferation was decreased, as assayed by reduced BrdU uptake (Figure 4B, quantified in C). To further assess the importance of downstream BMP signaling in regulating NPC proliferation, we generated a lentiviral vector encoding a shRNA against Smad1, plus a GFP reporter, and RNAi efficacy was confirmed in mouse neural progenitor cells in vitro via qRT-PCR (Figure 4D). As shown and quantified in Figures 4E and F, Smad1 shRNA but not control shRNA prevented the BMP4-induced decrease in proliferation. Collectively, these results indicate that BMP/pSmad1 signaling inhibits NPC proliferation.
Figure 4. BMP inhibits neural progenitor cell proliferation.
(A) Immunoblotting analysis shows that BMP4 (50 ng/mL) addition for 30 minutes activated its downstream effectors pSmad1/5/8 in rNPCs cultured in growth medium (DMF12 + N2 + 10 ng/mL FGF2). Protein loading was normalized to β-actin (B) Functional validation of increasing BMP4. Primary rNPCs were cultured in growth medium in the presence or absence of BMP4 (100 ng/mL) for 24 hrs. A 2 hour BrdU (10 μM) pulse was performed before cell fixation to label proliferating cells. Immunofluorescence was performed for BrdU (green) and Sox2 (red), with Hoechst (blue) labeling all nuclei. Representative images are shown. Scale bar = 100 μM (C) Proliferation of rNPCs were quantified by cell scoring in 25 random fields of each condition using an automated imager and MetaXpress cell scoring software. Results are displayed as the mean percent of BrdU+ proliferating rNPCs +/−SD, respectively. Significant differences were identified by Student’s t-tests (*p< 0.001), and error bars indicate the standard deviation (n=4). (D) Quantification of Smad1 mRNA expression by qRT-PCR was performed on RNA extracted from mNPCs transduced with shRNA to Smad1 or control virus and passaged for 2 weeks. The relative cDNA expression level was normalized to GAPDH and presented relative to that of mNPCs transduced with lacZ shRNA lentivirus. Significant differences were identified by Student’s t-tests (*p<0.05). Error bars indicate standard deviation (n=3). (E) rNPCs were transduced with control or Smad1 shRNA lentivirus. 72 hours post transduction, cells were cultured for 16 hours in growth medium (containing FGF2) in the presence or absence of BMP4 (200 ng/mL). A 4 hour EdU (30 μM) pulse was performed before cell fixation to label proliferating cells. Immunofluorescence was performed for GFP (green), EdU (red) and Sox2 (gray), with Hoechst (blue) labeling all nuclei. Representative images are shown. Scale bar = 100 μM. (F) Proliferation of rNPCs were quantified by cell scoring in 36 random fields of each condition using an automated imager and MetaXpress cell scoring software. Results are displayed as the mean percent of EdU+ proliferating rNPCs +/−SD, respectively. Significant differences were identified by Student’s t-tests (*p< 2 x10−34), and error bars indicate the standard deviation (n=36 technical replicates, with 4 biological replicates per condition).
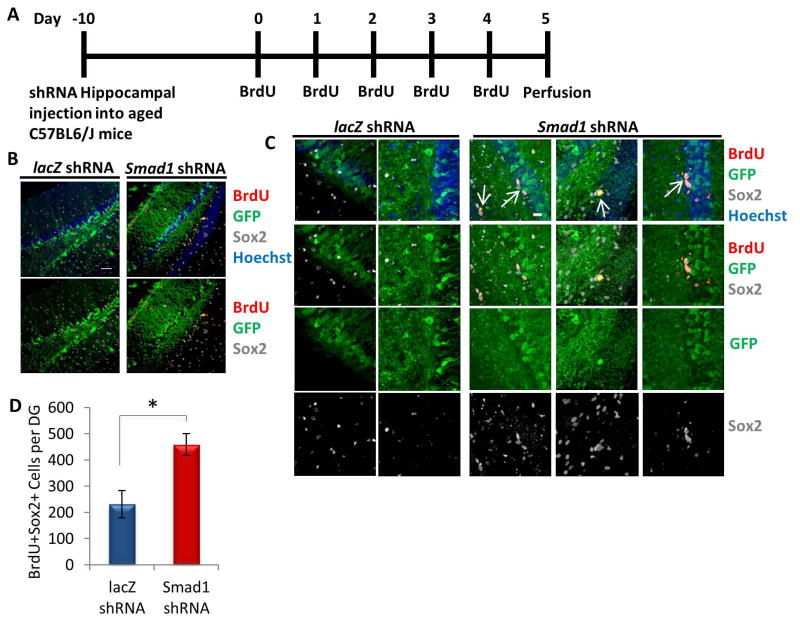
To further build upon these findings, the function of elevated BMP/pSmad1 signaling in the aged hippocampus was assessed in vivo via genetic perturbation of the pathway. After a single stereotaxic hippocampal injection of equal titers of either a lentiviral vector encoding GFP plus the shRNA against Smad1 – or a control shRNA against LacZ – into 18 month old mice, animals were allowed to recover for 10 days, followed by five consecutive days of BrdU administration (Figure 5A). As shown in Figure 5B–D, the numbers of Sox2+ proliferating cells (quantified in the GFP+ region of tissue sections throughout the entire hippocampus) were significantly increased after a single injection of shRNA to Smad1, compared to the control shRNA. Proliferation was thus significantly enhanced by the inhibition of pSmad1 signaling in the local niche of neural stem cells in 18 month old mice. This may contrast with young animals, in which physiological levels of BMP signaling are required for maintenance of the NSC pool [26]. Accordingly, over-inhibition of BMP signaling that was already at physiological young levels led to a transient increase in proliferating progenitors in the SGZ of young mice, but after a period of time this proliferation decreased due to a depletion of the stem cell pool [26].
Figure 5. Increase in NPC proliferation in aged hippocampi by in vivo genetic inhibition of pSmad1.
(A) Schematic of stereotaxic lentiviral injection experiment. Aged (18 month old) mice received stereotaxic injections into hippocampi (coordinates from bregma: AP: −2.12, ML: +/−1.5, VD: −1.55) of lentiviral vectors delivering either shRNA against Smad1 or shRNA against lacZ. The mice were allowed to recover for 10 days, followed by daily BrdU (50 mg/kg) intraperitonial injections for 5 days. Mice were perfused the morning after the last BrdU injection (Day 5). (B) Brain sections of lacZ or Smad1 shRNA injected mice (n=5 lacZ shRNA, 4 Smad1 shRNA) spanning the entire hippocampus were immunostained with GFP (green), BrdU (red), and Sox2 (gray), with Hoechst (blue) labeling cell nuclei. Representative images are shown. Scale bar = 50 μM. (C) Representative high magnification of immunostaining of the subgranular zone of the dentate gyrus.. Scale bar = 50 μM. Arrows indicate Sox2+BrdU+ proliferating NPCs in the SGZ of Smad1 shRNA injected mice. (D) Smad1 shRNA increases the number of BrdU+Sox2+ cells per GFP+ aged murine dentate gyrus. Significant differences were identified by Student’s t-tests (*p<0.04). Error bars indicate standard error of the mean (n=5 lacZ shRNA, 4 Smad1 shRNA brains)
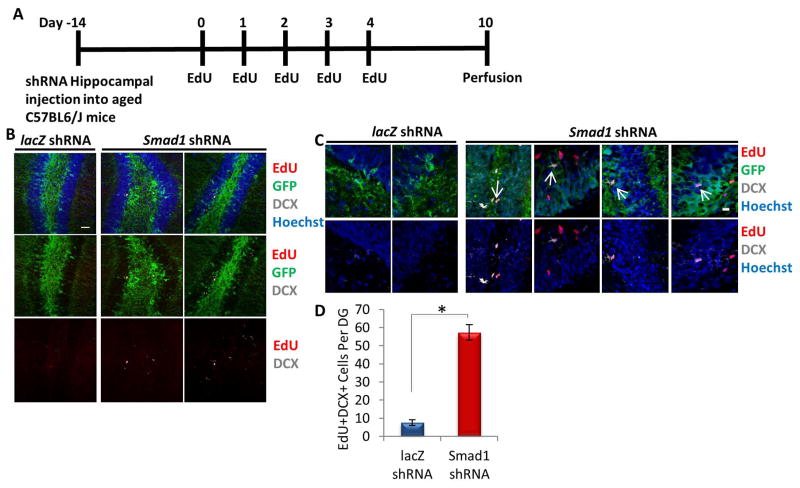
To analyze effects of BMP inhibition on downstream, type 2b doublecortin (DCX)+ transit amplifying cells, a second cohort of animals was injected with the shRNA lentiviral vectors, mitotically labeled 2 weeks later with EdU, and investigated 5 days after the last EdU injection to analyze cells that had progressed towards neuronal commitment [1] (Figure 6A). There was a substantial increase in the total number of EdU+DCX+GFP+ type 2b transit amplifying cells (Figure 6B–D). Thus, the in vivo delivery of lentiviral shRNA against Smad1 partially rejuvenated neural stem cell function and confirmed that canonical BMP/pSmad1 signaling is involved in the age-imposed inhibition of neurogenesis.
Figure 6. Increase of type 2b cell proliferation in aged hippocampi by in vivo genetic inhibition of Smad1.
(A) Schematic of injection experiment. Aged (18 month old) mice received stereotaxic injections into hippocampi (coordinates from bregma: AP: −2.12, ML: +/−1.5, VD: −1.55) of lentiviral vectors delivering either shRNA against Smad1 or shRNA against lacZ. The mice were allowed to recover for 14 days, followed by daily EdU (50 mg/kg) intraperitonial injections for 5 days. Five days after the last EdU injection, mice were saline and 4% PFA perfused. (B) Brain sections of lacZ or Smad1 shRNA injected mice (n=3 lacZ shRNA, 5 Smad1 shRNA) spanning the entire hippocampus were immunostained with GFP (green), EdU (red) and DCX (gray), with Hoechst (blue) labeling cell nuclei. Representative images are shown. Scale bar = 50 μM. (C) Representative high magnification images are shown. Scale bar = 50 μM. Arrows indicate DCX+BrdU+ proliferating neuronal precursors in the SGZ of Smad1 shRNA injected mice. (D) Smad1 shRNA increases the number of EdU+DCX+GFP+ cells in aged murine dentate gyri. Significant differences were identified by unpaired Student’s t-tests (*p<0.0002), and Mann Whitney value of *p<0.04. Error bars indicate standard error of the mean (n=3 lacZ shRNA, 5 Smad1 shRNA).
Partial rescue of neurogenesis in aged hippocampi by in vivo transgenic inhibition of BMP signaling
To further investigate the finding that attenuation of BMP signaling enhances neurogenesis, we directly perturbed BMP signaling through Cre-mediated deletion of Bmpr1a in aged (18–22 month old) Bmpr1alox/lox Bmpr1b+/− Rosa26lacZ mice. It has been demonstrated that the majority of type 1 and 2a cells specifically express BMPR1A in the SGZ, whereas granule neurons express BMPR1B [26]. After a single stereotaxic hippocampal injection of equal titer of either a lentiviral vector encoding mCherry as well as Cre recombinase – or control vector encoding GFP only – into 18–22 month aged mice, animals were allowed to recover for 2 weeks, followed by five consecutive days of BrdU administration (Supplemental Figure 1A). Cre recombination was evident by β-galactosidase immunoreactivity in animals administered with the Cre, but not the control, virus (Supplemental Figure 1B). Furthermore, the number of Sox2+ proliferating cells (quantified in the mCherry+ or GFP+ region of tissue sections throughout the entire hippocampus) was significantly increased after a single injection of Cre recombinase, as compared with control GFP lentivirus (Supplemental Figure 1C–E). Proliferation was thus significantly enhanced by the inhibition of BMP signaling through the BMPR1A receptor in NSCs of 18–22 month old mice.
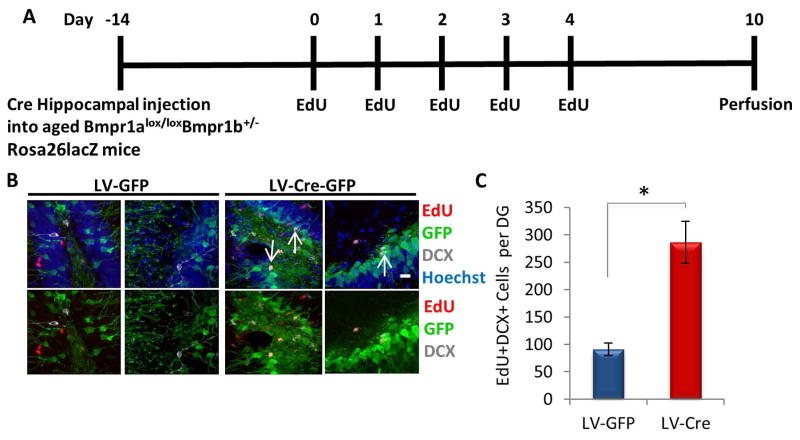
To analyze effects on downstream type 2b transit amplifying cells, additional aged, Bmpr1a floxed animals were injected with equal titer of a lentiviral vector encoding GFP and Cre, or control vector encoding GFP. Two weeks after the injection, cells were mitotically labeled with EdU, then investigated 5 days after the last EdU injection (Figure 7A). There was a substantial increase in the number of EdU+/DCX+ type 2b cells (quantified in the GFP+ region of tissue sections throughout the entire hippocampus) (Figure 7B–C). Thus, inhibition of BMP signaling via Cre-induced deletion of Bmpr1a in vivo further established that canonical BMP/pSmad signaling is involved in the age-imposed inhibition of neurogenesis.
Figure 7. Increase in type 2b cell proliferation in aged hippocampi by in vivo Cre-mediated Bmpr1a deletion.
(A) Schematic of stereotaxic lentiviral vector injection experiment. Aged (18–22 month old) Bmpr1alox/lox Bmpr1b+/− Rosa26lacZ mice received stereotaxic injections into hippocampi (coordinates from bregma: AP: −2.12, ML: +/−1.5, VD: −1.55) of lentiviral vectors encoding either Cre-GFP or GFP. The mice were allowed to recover for 14 days, followed by daily EdU (50 mg/kg) intraperitonial injections for 5 days. Mice were saline and PFA perfused 5 days after the last EdU injection (Day 10). (B) Brain sections of Cre or GFP injected mice (n=5) spanning the entire hippocampus were immunostained with GFP (green), EdU (red), and DCX (gray), with Hoechst (blue) labeling cell nuclei. Representative images are shown. Scale bar = 50 μM. Arrows indicate DCX+BrdU+ proliferating NPCs in the SGZ of Cre injected mice. (C) Cre-induced deletion of Bmpr1a increases the number of EdU+DCX+ cells in GFP+ aged murine dentate gyri. Significant differences were identified by Student’s t-tests (*p<0.03). Error bars indicate standard error of the mean (n=5 per group).
Discussion
Collectively, our data indicate that BMP signaling reversibly inhibits the capacity of stem cells in the aged brain to contribute to neurogenesis. Specifically, an age-associated increase of BMP/pSmad1,5,8 signaling in the neural stem cell niche was identified and shown to inhibit neural progenitor cell proliferation. Importantly, genetic inhibition of BMP signaling partially rescued neural stem and progenitor cell proliferation, as well as levels of neuronally committed transit amplifying type 2b cells, in aged mice. This ability to rapidly enhance neural precursor cell function via modulating the levels of signaling pathways in the stem cell microenvironment suggests promising strategies for combatting the loss of cognitive function and memory with aging.
In humans, there are 20 known BMP family members [36], and BMPs and many GDFs signal through SMAD1/5/8 [20]. Specifically, BMPs bind type 2 (BMPR2 or ACVR2) and type 1 receptors (BMPR1A (ALK3), BMPR1B (ALK6), ALK2, or ALK1) to induce downstream SMAD1/5/8 signaling. BMPs thereby regulate numerous downstream target genes, which vary as a function of microenvironmental, transcriptional, and epigenetic context during development and adulthood [20, 21]. Here, we show an age-associated increase in the expression of BMPs 2, 4, and 6, which bind BMPR2, BMPR1A, and BMPR1B and subsequently signal via SMAD1,5,8 [36]. We also found that SMAD inhibition partially rescued neurogenesis. It will be interesting to determine whether differences in the signaling properties of these three BMPs may differentially contribute to downstream consequences.
While pSmad1/5/8 levels across the hippocampus did not appear to change in young versus old dentate gyri, quantified across the full granule layer and hilus, the percentage of Sox2+ cells that coexpressed high levels of pSmad1/5/8 increased significantly with age (Figure 3). pSmad2/3 levels reportedly vary during different phases of the cell cycle within human embryonic stem cells, which in turn determines cell differentiation potential [37]. Furthermore, pSmad1/5 signaling levels can determine whether embryonic NSCs undergo symmetric versus asymmetric division in the CNS [34], suggesting an effect of BMP signaling on cell-fate determination. Additionally, in Sox2+ cells in the young dentate gyri, pSmad1 – which was primarily nuclearly localized – exhibited varying intensity [26]. It would thus be interesting to assess in future work whether cell cycle or other variation in SMAD1/5/8 signaling may also modulate multipotent adult stem cell behavior, and in particular whether our observed age-specific changes in the levels of SMAD1/5/8 activation in NSCs and NPCs influence their cell fate commitment.
Within the developing CNS, several downstream BMP targets act to inhibit embryonic neurogenesis [22]. For example, Id1 and Id3 inhibit the activity of transcription factors that promote neuronal differentiation, including Neurogenin, Ascl1, and NeuroD1 [22, 35]. BMPs also induce expression of Hes genes, which are known to inhibit neuronal differentiation [23]. It would be interesting to determine whether analogous downstream molecular events occur in adult neurogenesis and change with aging.
The increase in BMP signaling in the hippocampal stem cell niche results in an inhibitory microenvironment unsupportive of stem cell regenerative function. What is the source of such microenvironmental changes in protein expression? To address this question, we investigated the cellular localization of BMPs. Within the SVZ, BMP is reportedly secreted by neural stem and progenitor cells (i.e. resident radial glial-like cells and transient amplifying cells), while the ependymal cells in this neurogenic niche secrete Noggin, an inhibitor of various BMP ligands [23]. Different cells within the niche can thus push BMP signaling in different directions. Within the young murine hippocampus, BMP4 is also expressed [38], though the source(s) of its production is not well known. In general, hippocampal NSCs are known to interact with several cell types, including astrocytes, microglia, endothelial cells, and neurons [39]. Here, we established endothelial cells as a likely source of BMP4 production in the dentate gyrus (Figure 2A,B). Neural stem and progenitor cells are known to be closely associated with blood vessels in a vascular niche, where endothelial cells are important regulators of neurogenesis that have been reported to promote self-renewal and prevent differentiation of NSCs through secretion of soluble factors [40]. Vascular-derived BMPs may thus regulate neurogenesis. Interestingly, BMPs are produced by endothelial cells under stress, and are known to regulate angiogenesis and neurogenesis in young animals [26, 33, 41, 42].
In addition, high levels of BMP6 are present in microglia in young and aged brains (Figure 2C, D). Transcriptional profiling of microglia in the developing brain revealed Bmp2 as highly expressed in reactive microglia [43], though expression of BMP family members in adult microglia has not been validated. In addition, microglia are known not only to increase in number in the aged brain but also to develop a more inflammatory phenotype with higher expression of several cytokines, including BMP family member TGF-β1 [44, 45]. However, the role of BMP signaling in microglial function and interaction with neural precursor cells is not well validated and would be interesting to explore. NSCs have been shown to regulate microglia proliferation and activation in part through secretion of VEGF [46], and it would thus be interesting to assess whether BMP signaling is involved in microglia-NSC interactions, potentially through cross-talk with VEGF. Finally, BMP signaling through SMAD1/5/8 and induction of Id2 reportedly promotes the conversion of microglia into neurons and astrocytes [47], though this proposed function needs further investigation.
While the role of BMPs in the aging of the hippocampal stem cell niche and decline of neurogenesis has not previously been elucidated, downstream effects of BMPs have been implicated in age-associated CNS pathologies. Specifically, an increase in BMP6 has been shown in human Alzheimer’s Disease patients and in corresponding mouse models, is associated with decreased neurogenesis in both patients and mice models [31]. Furthermore, BMP4 was also shown to increase in Alzheimer’s mouse models and inhibit hippocampal cell proliferation [48]. Interestingly, hippocampal neurogenesis is rescued in AD models by inhibiting BMP signaling with Noggin [49]. It has also been demonstrated that increased BMP signaling results in cognitive impairments, whereas its inhibition improves cognition in hippocampus-dependent learning in young mice [28, 29].
In general, it may prove beneficial to inhibit BMP signaling in the elderly to enhance neurogenesis and potentially reverse age-induced brain pathologies, for example through systemic administration of small molecule inhibitors, though the role of systemic BMP inhibition on various aged tissues will need consideration. Furthermore, while inhibiting excessive BMP signaling may promote neurogenesis in the aged, it may prove important to tune the level of signaling in old to that seen in the young, in order to rejuvenate the stem cell niche while preserving the crucial role of BMP signaling in maintaining the stem cell pool. For example, over-inhibition of BMP signaling that was already at physiological young levels led to a transient increase in proliferating progenitors in the SGZ of young mice, but after a period of time this proliferation decreased due to a depletion of the stem cell pool [26].
Several studies have demonstrated that not just one signaling pathway, but a network of highly interactive morphogenic signaling pathways become affected by the aging process in various tissue stem cell compartments, including the brain [15, 17, 50, 51]. While inhibition of BMP signaling through both genetic and transgenic approaches led to a two-fold increase in proliferating NPCs and immature neurons (Figures 5–7 and Supplemental Figure 2), it did not rescue these phenotypes close to the level seen in young mice (Figure 1A) [8]. Thus, in addition to BMP signaling, it may be necessary to retune additional signaling pathways to fully rescue aged neurogenesis. Hippocampal neurogenesis is regulated at different stages by multiple signaling pathways, including TGF-β/BMP, Notch, Shh, Wnt, and ephrin-B signaling [52–54]. Of these pathways, Wnt/β-catenin signaling, important for neurogenic differentiation [54, 55], has been demonstrated to decrease with aging in the hippocampus [15–17]. When upstream or downstream Wnt pathway regulators were modulated in aged mice, there was a 2 to 3 fold increase in neurogenesis [17], similar to the level of increase found in this study upon inhibiting BMP signaling. Therefore, it may be beneficial to simultaneously modulate multiple signaling pathways that are deregulated with age in order to obtain an enhancement of neurogenesis to levels seen in the young.
In summary, this work improves our understanding of the aging of neural stem cells by revealing a key signaling pathway, BMP that becomes deregulated with age. Partial rescue of neurogenesis through acute inhibition of this pathway demonstrates that decline in functionality of neuronal stem cells with age is reversible and can be rescued by the modulation of regulatory signaling in the stem cell microenvironment.
Materials and Methods
Animals
Young (2–3 month old) and old (18–24 month old) C57BL6/J male mice were purchased from the Jackson Laboratory and the NIH. A breeding pair of Bmpr1alox/lox Bmpr1b+/− Rosa26lacZ were obtained as a gift from the laboratory of Professor John Ngai, UC Berkeley. The mice were bred and aged in house, in accordance with the Guide for Care and Use of Laboratory Animals of the National Institutes of Health.
For each experiment on aged (18–24 month old) mice, an n of at least 7 per group was used initially. If aged mice were lost due to surgery or age-related health issues, a minimum of n=3 remaining mice were analyzed and assessed for statistical significance (described in statistical analysis below). Mice of the same genetic strain and age were assigned to groups at random. The animal experimental procedures were performed in accordance with the Guide for Care and Use of Laboratory Animals of the National Institutes of Health, and approved by the Office of Laboratory Animal Care, UC Berkeley.
Cell Culture
Primary rat neural progenitor cells isolated from the hippocampi of 6-week-old female Fisher 344 rats (Charles River) were cultured in growth medium (DMEM/F12 (Life Technologies) containing N2 supplement (Life Technologies) and 10 ng/mL FGF-2 (PeproTech)) on laminin (Roche) and polyornithine (Sigma) coated tissue culture plates, with subculturing on reaching 80% confluency using Accutase (Phoenix Flow Systems), as previously described [54].
Primary mouse neural progenitor cells were isolated form C57BL6/J mice (Charles River) as previously described [56]. Cells were cultures in growth medium (Neurobasal A (Gibco) containing B27 supplement (Gibco), Glutamax-1 supplement (Gibco), 20 ng/mL FGF-2 (Peprotech), and 20 ng/mL EGF (PeproTech)) on Poly-d-Lysine (Sigma) and Laminin (Roche) coated tissue culture plate, with subculturing on reaching 80% confluency using Accutase (Phoenix Flow Systems). Progenitor cells were tested for mycoplasma contamination at the UC Berkeley Stem Cell Core Facility and using Hoechst DNA stain.
In vitro validation of BMP Signaling and Proliferation Assay
Rat NPCs were cultured in growth medium (DMF12 + N2 + 10 ng/mL FGF-2) as described above. Cells were cultured at a density of 200,000 cells per well of a 6-well culture slide in the presence/absence of BMP4 (50 ng/mL) (R&D) for 30 minutes, followed by a PBS wash and cell scraping into RIPA buffer for Western blot analysis as described below. Rat hippocampal-derived NPCs were also cultured at a density of 80,000 cells per well of an 8-well chamber slide in growth medium plus the presence/absence of BMP4 (100 ng/mL) for 24 hours. NPCs were pulsed for 2 hours with 10 μM BrdU (Sigma Aldrich) before cell fixation with 70% cold ethanol for immunocytochemistry analysis as described below.
Dissection and Preparation of Murine Hippocampi for RNA or Protein Analysis
Young or old mice were anesthetized and perfused with 20 mL saline, followed by dissection and isolation of hippocampi. For RNA extraction, tissue was placed in 1 mL Trizol (Life Technologies) and homogenized, followed by chloroform extraction as previously described [54]. For protein extraction, hippocampi tissue was homogenized in RIPA buffer (50 mM Tris, 150 mM NaCl, 1% NP40, 0.25% sodium deoxycholate and 1 mM EDTA, pH 7.4) containing 1X protease inhibitor (Roche), 1 mM Phenylmethylsulfonyl fluoride (PMSF), 1 mM sodium fluoride, and 1 mM sodium orthovanadate. Lysates were spun at 10K rpm for 5 min at 4°C to remove debris, and supernatant containing protein extract was snap frozen with dry ice.
RNA extraction, RT-PCR and real-time PCR
Total RNA was extracted from primary neural progenitor cells or young and old murine hippocampi using Trizol reagent (Life Technologies) according to manufacturer’s instructions. 1 ug of total RNA was used for cDNA synthesis with oligo dT primers (Life Technologies). For real-time PCR amplification and quantification of genes of interest, an initial amplification using specific primers to each gene of interest (realtimeprimers.com) was conducted with a denaturation step at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 1 min, primer annealing at 55°C for 30 s, and primer extension at 72°C for 30 s. Real-time PCR was performed using SYBR and an ABI PRISM 7500 Sequence Detection System (Applied Biosystems). Reactions were run in triplicate in three independent experiments. The geometric mean of housekeeping gene GAPDH was used as an internal control to normalize the variability in expression levels, which were analyzed using the 2−ΔΔCT method described [57].
ELISA
The concentration of active BMP4 in hippocampal tissue protein lysate was determined using enzyme-linked immunoabsorbent assay (ELISA)-based cytokine antibody array (R&D), according to manufacturer instructions.
Immunocytochemistry
Mice were anesthetized and perfused with saline and 4% PFA. Brains were collected and placed in 4% PFA overnight for post-fixation, followed by dehydration in 30% sucrose/PBS at 4°C for 2 days. Brains were then sectioned at 40 μM using a sliding microtome and stored in a glycerol-based cryoprotectant at −20°C until further analysis by immunostaining.
For BrdU in vivo labeling of mitotic cells, mice were intraperitoneally injected with BrdU (50 mg/kg of body weight, Sigma Aldrich) dissolved in saline to label mitotic cells, as previously described [58]. Sections were incubated in SSC/formamide at 65°C water for 2 hours, washed in TBS, followed by 2N HCl for 15 minutes. They were then placed in 2X Saline-Sodium Citrate (SSC) for 30 minutes, .1 mM Borate Buffer for 15 minutes, followed by 6, 15 minute washes in TBS, then blocked in a permeabilization/staining buffer, TBS++ (3% Donkey Serum and .25% Triton-X-100) for 2 hours. Sections were then incubated with αRat-BrdU (Abcam Inc. ab6326) and other antibodies (see Antibodies below) in TBS++ at 4°C for 72 hours. Secondary staining was done as described below.
For EdU in vivo labeling of mitotic cells, mice were intraperitoneally injected with EdU (50 mg/kg of body weight, Life Technologies) dissolved in phosphate-buffered saline. Brain sections were post-fixed with 4% PFA for 30 minutes after primary and secondary staining, and treated for EdU visualization using the Click-iT EdU kit (Life Technologies), as per the manual’s instructions.
For non-BrdU/EdU staining, sections were washed 3 times for 15 minutes in TBS, followed by one hour blocking in a permeabilization/staining buffer, TBS++ (3% Donkey Serum and .25% Triton-X-100 in TBS), then incubated with primary antibodies of interest (see Antibodies) for 72 hours. For secondary staining, sections were washed 3 times, 15 minutes each in TBS, followed by 2 hour incubation in donkey raised, fluorophore-conjugated, species-specific secondary antibodies (Jackson Immunoresearch) at 1:250 dilution in TBS++. Following secondary staining, sections underwent 3, 15 minute washes in TBS, with 4 μM Hoechst in the second wash. Finally, the sections were mounted on positively charged frosted slides, dried overnight and imaged with a prairie confocal microscope.
Antibodies
αRabbit-pSmad1/5/8 (Cell Signaling #9511), αRabbit-βactin (Cell Signaling #4967), αMouse-BMP6 (Chemicon International MAB1048), αRabbit-BMP4 (Abcam ab39973), αMouse-β-galactosidase (MP Biomedicals 08633651), αMouse-mCherry (Novus Biologicals 1C51), EdU Click-it kit (Life Technologies C10337 and C10338), αGuineyPig-DCX (Millipore AB2253), αGoat-Sox2 (Santa Cruz SC-17320), αChicken-GFP (Abcam ab13970), αRat-BrdU (Abcam ab6326), BMP4 ELISA kit (R&D DY314), αGoat-Iba1 (Abcam ab5076), αRabbit-GFAP (Abcam ab7260), αRat-CD31 (BD Biosciences 550274)
Western Blot Analysis
Neural progenitor cells were lysed in RIPA buffer (50 mM Tris, 150 mM NaCl, 1% NP40, 0.25% sodium deoxycholate and 1 mM EDTA, pH 7.4) containing 1X protease inhibitor (Roche), 1 mM Phenylmethylsulfonyl fluoride (PMSF), 1 mM sodium fluoride and 1 mM sodium orthovanadate. The protein concentration was determined by a Bradford assay (Bio-Rad). Lysates were resuspended in 1X Laemmli buffer (Bio-Rad), boiled for 5 minutes, and separated on precast 7.5% or 4–15% TGX gels (Bio-Rad). Primary antibodies were diluted in 5% non-fat milk in TBS + 0.1% Tween-20, and nitrocellulose membranes were incubated with antibody mixtures overnight at 4°C. HRP-conjugated secondary antibodies (Santa Cruz Biotech) were diluted 1:500 in 5% non-fat milk in TBS + 0.1% Tween-20 and incubated for 1 hour at room temperature. Blots were developed using Western Lightning ECL reagent (Perkin Elmer), and analyzed with Bio-Rad Gel Doc/Chemi Doc Imaging System and Quantity One software. Results of multiple assays were quantified using Applied Biosystems of Image J software. Pixel intensity of bands of interest were normalized with the corresponding pixel intensities of glyceraldehydes-3-phosphate dehydrogenase or β-actin.
Lentiviral Vector Construction, Packaging, and Purification
A DNA cassette encoding human U6 promoter-driven expression of shRNA against mouse SMAD1 (Gene ID: 17125) was constructed by PCR with flanking PacI sites and, following restriction digestion and phenol/chloroform purification, ligated into the PacI site of the pFUGW lentiviral vector [59]. Five candidate sequences were tested for knockdown efficiency, and the most effective sequence (shSMAD 1.3 in Supplemental Table 1) was selected for experimental studies. Sequences for all shRNAs tested are provided in Supplemental Table 1. The control shRNA vector against LacZ was constructed previously [54]. PCR was performed with Phusion DNA Polymerase (New England Biolabs) under the following conditions: 98°C for 2 min, 30 cycles of 12 s at 95°C, 30 s at 65°C, and 25 s at 72°C, with a final extension step of 2 min at 72°C. Lentiviral and retroviral vectors were packaged and purified using standard methods as described [60, 61].
Cre lentiviral plasmids were obtained (Addgene http://www.addgene.org/20781/ and http://www.addgene.org/27546/), packaged, and purified as previously described [60, 61]. The control GFP vector was constructed previously [59].
In vitro validation of Smad1 shRNA Vector
Mouse neural progenitor cells (NPCs) were plated at 200,000 cells per well of a 6 well tissue culture plate in growth medium, transduced with lentivirus encoding shRNA to Smad1 or lacZ at a multiplicity of infection (MOI) of 5, and cultured for two weeks. RNA was extracted with Trizol (Life Technologies), followed by qPCR to assess levels of Smad1 (see Supplemental Table 1 for sequences).
To assess shRNA functionality, rNPCs were plated at 200,000 cells per well of a 6 well tissue culture plate in growth medium, transduced with lentivirus encoding shRNA to Smad1 or lacZ at a multiplicity of infection (MOI) of 10, and cultured for 72 hours. Cells were then plated at 105 cells per well of 8 well chamber slides and cultured for 16 hours in growth medium in the presence or absence of 200 ng/mL BMP4. A 4 hour EdU (30 μM) pulse was performed before cell fixation in 4% PFA to label proliferating cells. Quantification described below.
In vivo loss of function via lentiviral vector stereotaxic injections to hippocampus
Aged (18 month) C57BL6/J male mice received lateral intrahippocampal injections of equal volume and similar titer of lentiviral solutions (1 μl of LV-shRNA-Smad1-GFP or LV-shRNA-lacZ-GFP, 1–3 × 10^8 IU/mL) in PBS on day −10, on the right hemisphere hippocampus, at 0.25 uL per minute. The injection coordinates with respect to bregma were −2.12 mm anteriorposterior, −1.55 mm dorsoventral (from dura), and 1.5 mm mediolateral (refer to Figure 5a). Mice were allowed to recover 10 days, followed by BrdU intraperitoneal (IP) injections (50 mg/kg bodyweight) 1 times daily for 5 days. One day after receiving the fifth BrdU IP injection, mice were saline and 4% PFA perfused. Immunostaining and quantification described in immunocytochemistry section and quantification and statistical analysis section.
Aged (18 month) C57BL6/J male mice received lateral intrahippocampal injections of equal volume and similar titer of lentiviral solutions (1 μl of LV-shRNA-Smad1-GFP or LV-shRNA-lacZ-GFP, 1–3 × 10^8 IU/mL) in PBS on day −14, on the right hemisphere hippocampus, at 0.25 uL per minute. The injection coordinates with respect to bregma were −2.12 mm anteriorposterior, −1.55 mm dorsoventral (from dura), and 1.5 mm mediolateral (refer to Figure 6a). Mice were allowed to recover 14 days, followed by EdU IP injections (50 mg/kg bodyweight) 1 times daily for 5 days. Five days after receiving the fifth IP injection, mice were saline and 4% PFA perfused. Immunostaining and quantification described in quantification and statistical analysis section.
Aged (18 month) Bmpr1alox/lox Bmpr1b+/− Rosa26lacZ male mice received lateral intrahippocampal injections of equal volume and similar titer of lentiviral solutions (1 μl of LV-Cre-mCherry or LV-FUGW (LV-GFP), 1–3 × 10^7 IU/mL) in PBS on day −14, on the right hemisphere hippocampus, at 0.25 uL per minute. The injection coordinates with respect to bregma were −2.12 mm anteriorposterior, −1.55 mm dorsoventral (from dura), and 1.5 mm mediolateral (refer to Supplemental Figure 1a). Mice were allowed to recover 14 days, followed by BrdU IP injections (50 mg/kg bodyweight) 1 times daily for 5 days. One day after receiving the fifth BrdU IP injection, mice were saline and 4% PFA perfused. Immunostaining and Quantification described elsewhere.
Aged (18 month) C57BL6/J male mice received lateral intrahippocampal injections of equal volume and similar titer of lentiviral solutions (1 μl of LV-Cre-GFP or LV-FUGW (LV-GFP), 1–3 × 10^7 IU/mL) in PBS on day −14, on the right hemisphere hippocampus, at 0.25 uL per minute. The injection coordinates with respect to bregma were −2.12 mm anteriorposterior, −1.55 mm dorsoventral (from dura), and 1.5 mm mediolateral (refer to Figure 7a). Mice were allowed to recover 14 days, followed by EdU IP injections (50 mg/kg bodyweight) 1 times daily for 5 days. Five days after receiving the fifth EdU IP injection, mice were saline and 4% PFA perfused. Immunostaining and quantification described in Quantification and Statistical Analysis section.
Quantification and Statistical Analysis
For quantification of in vitro immunofluorescent images for BrdU or EdU incorporation, 25 20x images per replicate were taken on the Molecular Devices ImageXpress Micro automated epifluorescence imager, followed by automated cell quantification using the multiwavelength cell scoring module within the MetaXpress analysis software. Data was analyzed using an unpaired Student’s t-test or Mann Whitney nonparametric U test, and p values equal or lower than 0.05 were considered statistically significant. Sample sizes of n=3 or greater were used for each experiment based on preliminary results of pilot experiments in combination with standard power analysis to assess for the minimum number of animals needed to achieve statistical significance, assuming an effect size of 0.5 or greater, α = 0.05 and a power of 0.8, adhering to NIH and IACUC standards for using the minimum number of animals, and previously published experimental group numbers [16, 54]. Significance was assessed based on p values and heteroscedastic variance between groups that are statistically compared. For pixel intensity quantifications of immunofluorescent images, ImageJ was used to determine integrated pixel intensity of thresholded images, with n=3–4 young or old mice brains, 3 coronal or sagittal tissue sections per brain, and 2 images (right and left hippocampi or hemisphere) per section assessed.
For quantification of the number of BrdU+Sox2+ or EdU+DCX+ cells in shRNA or Cre injected mice, confocal stacks of 8 coronal GFP+ or mCherry+ brain sections spanning the hippocampus (40 microns thick, 200 microns apart) were acquired on a Prairie confocal microscope, and cells within the dentate granule cell layer were counted. Cell numbers were normalized to the volume of the DG granule cell layer using ImageJ as previously described [17, 62]. Briefly, volume was calculated based on a threshold of the granule layer of each image as determined with Hoechst staining, then calculating the volumetric fraction based on the thickness of the brain slice (40 μM) and the interval at which hippocampi sections were analyzed (every 6th section). Cell densities in the dentate gyrus were calculated as the number of immunopositive cells divided by the volume of the GCL and expressed as the number per dentate granule cell layer total volume.
Only aged mice that were lost during the study due to health reasons were excluded from analysis. For automated cell counting using MetaXpress analysis software, only sites that were blurry with indistinguishable colors (such as occasional areas on the slide with bubbles or rare images acquired with incorrect focus by the Molecular Devices ImageXpress Micro automated epifluorescence imager), or in the case of cell culture occasional areas with large cell clumps, were excluded from the cell quantification analysis. These criteria were pre-established.
When performing in vivo experiments, there was no blinding as to experimental groups. For quantification and analysis, researchers were blinded to the group allocation when performing cell counts.
Supplementary Material
Acknowledgments
This work was supported by California Institute for Regenerative Medicine grant RT2-02022 and NIH R01 NS074831 to DVS, grants from the National Institutes of Health R01 AG02725201 and California Institute for Regenerative Medicine RN1-00532-1 to IMC, and a NSF Graduate Fellowship to HY. We thank Mary West and the CIRM/QB3 Shared Stem Cell Facility at UC Berkeley for use of the Molecular Devices ImageXpress and Prairie 2P/Confocal microscope and training on these instruments. We thank Drs. John Ngai and Russell Fletcher at UC Berkeley for providing a breeding pair of Bmpr1alox/lox Bmpr1b+/− Rosa26lacZ mice, Mustafa Alkhouli for animal care and assistance with brain staining and image processing, Hikaru Mamiya and Matthew Zeiderman for assistance in in vitro NPC staining and qPCRs, Manika Paul for her assistance with brain staining and image processing, George Sun for assistance with qPCRs, and Ashutosh Shrestha for assistance with cell staining and imaging.
Footnotes
Author Contributions
HY produced and assessed all viral particles, designed, performed and analyzed the experiments for Figures 1–7 and Supplemental Figures 1 and 2, interpreted these data and co-wrote the manuscript; AM performed and analyzed experiments for Figures 5–7 and Supplemental Figure 2; CS performed and analyzed experiments for Figures 5, 7, and Supplemental Figure 2; LB cloned Smad1 shRNA constructs; IMC participated in the design of the experiments, interpreted the data, and edited the manuscript; DVS designed, directed and integrated the study, interpreted the data, and co-wrote the manuscript.
There are no conflict of interests.
References
- 1.Ming G, Song H. Adult neurogenesis in the mammalian central nervous system. [Accessed October 17, 2013];ANNU REV NEUROSCI. 2005 28:223–50. doi: 10.1146/annurev.neuro.28.051804.101459. Available at: http://www.ncbi.nlm.nih.gov/pubmed/16022595. [DOI] [PubMed] [Google Scholar]
- 2.Deng W, Aimone JB, Gage FH. New neurons and new memories: how does adult hippocampal neurogenesis affect learning and memory? NAT REV NEUROSCI. 2010;11(5):339–350. doi: 10.1038/nrn2822. Available at: http://dx.doi.org/10.1038/nrn2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng W, Saxe MD, Gallina IS, et al. Adult-born hippocampal dentate granule cells undergoing maturation modulate learning and memory in the brain. [Accessed October 20, 2013];J NEUROSCI. 2009 29(43):13532–42. doi: 10.1523/JNEUROSCI.3362-09.2009. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2787190&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oboti L, Savalli G, Giachino C, et al. Integration and sensory experience-dependent survival of newly-generated neurons in the accessory olfactory bulb of female mice. EUR J NEUROSCI. 2009;29(4):679–692. doi: 10.1111/j.1460-9568.2009.06614.x. Available at: http://dx.doi.org/10.1111/j.1460-9568.2009.06614.x. [DOI] [PubMed] [Google Scholar]
- 5.Maslov AY, Barone Ta, Plunkett RJ, et al. Neural stem cell detection, characterization, and age-related changes in the subventricular zone of mice. [Accessed October 26, 2013];J NEUROSCI. 2004 24(7):1726–33. doi: 10.1523/JNEUROSCI.4608-03.2004. Available at: http://www.ncbi.nlm.nih.gov/pubmed/14973255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olariu ANA, Cleaver KM, Cameron HA. Decreased Neurogenesis in Aged Rats Results from Loss of Granule. Cell Cell Cycle. 2007;667(October 2006):659–667. doi: 10.1002/cne.21268. [DOI] [PubMed] [Google Scholar]
- 7.Walter J, Keiner S, Witte OW, et al. Age-related effects on hippocampal precursor cell subpopulations and neurogenesis. [Accessed October 26, 2013];NEUROBIOL AGING. 2011 32(10):1906–14. doi: 10.1016/j.neurobiolaging.2009.11.011. Available at: http://www.ncbi.nlm.nih.gov/pubmed/20006411. [DOI] [PubMed] [Google Scholar]
- 8.Kuhn HG, Dickinson-Anson H, Gage FH. Neurogenesis in the Dentate Gyrus of the Adult Decrease of Neuronal Progenitor Proliferation Rat : Age-Related. 1996;76(6):2027–2033. doi: 10.1523/JNEUROSCI.16-06-02027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hattiangady B, Shetty AK. Aging does not alter the number or phenotype of putative stem/progenitor cells in the neurogenic region of the hippocampus. NEUROBIOL AGING. 2008;29(1):129–147. doi: 10.1016/j.neurobiolaging.2006.09.015. Available at: http://www.neurobiologyofaging.org/article/S0197-4580(06)00340-X/abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aizawa K, Ageyama N, Yokoyama C, et al. Age-dependent alteration in hippocampal neurogenesis correlates with learning performance of macaque monkeys. EXP ANIM. 2009;58(4):403–7. doi: 10.1538/expanim.58.403. Available at: http://www.ncbi.nlm.nih.gov/pubmed/19654438. [DOI] [PubMed] [Google Scholar]
- 11.Aizawa K, Ageyama N, Terao K, et al. Primate-specific alterations in neural stem/progenitor cells in the aged hippocampus. [Accessed July 15, 2014];NEUROBIOL AGING. 2011 32(1):140–50. doi: 10.1016/j.neurobiolaging.2008.12.011. Available at: http://www.sciencedirect.com/science/article/pii/S0197458008004272. [DOI] [PubMed] [Google Scholar]
- 12.Eriksson PS, Perfilieva E, Björk-Eriksson T, et al. Neurogenesis in the adult human hippocampus. NAT MED. 1998;4(11):1313–7. doi: 10.1038/3305. Available at: http://www.ncbi.nlm.nih.gov/pubmed/19714567. [DOI] [PubMed] [Google Scholar]
- 13.Spalding KL, Bergmann O, Alkass K, et al. Dynamics of Hippocampal Neurogenesis in Adult Humans. CELL. 2013;153(6):1219–1227. doi: 10.1016/j.cell.2013.05.002. Available at: http://linkinghub.elsevier.com/retrieve/pii/S0092867413005333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vukovic J, Colditz MJ, Blackmore DG, et al. Microglia modulate hippocampal neural precursor activity in response to exercise and aging. [Accessed October 25, 2013];J NEUROSCI. 2012 32(19):6435–43. doi: 10.1523/JNEUROSCI.5925-11.2012. Available at: http://www.ncbi.nlm.nih.gov/pubmed/22573666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamoto M, Inoue K, Iwamura H, et al. Reduction in paracrine Wnt3 factors during aging causes impaired adult neurogenesis. [Accessed October 26, 2013];FASEB J. 2011 25(10):3570–82. doi: 10.1096/fj.11-184697. Available at: http://www.ncbi.nlm.nih.gov/pubmed/21746862. [DOI] [PubMed] [Google Scholar]
- 16.Miranda CJ, Braun L, Jiang Y, et al. Aging brain microenvironment decreases hippocampal neurogenesis through Wnt-mediated survivin signaling. [Accessed October 26, 2013];AGING CELL. 2012 11(3):542–52. doi: 10.1111/j.1474-9726.2012.00816.x. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3350615&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seib DRM, Corsini NS, Ellwanger K, et al. Loss of Dickkopf-1 restores neurogenesis in old age and counteracts cognitive decline. [Accessed October 24, 2013];CELL STEM CELL. 2013 12(2):204–14. doi: 10.1016/j.stem.2012.11.010. Available at: http://www.ncbi.nlm.nih.gov/pubmed/23395445. [DOI] [PubMed] [Google Scholar]
- 18.Villeda Sa, Luo J, Mosher KI, et al. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. [Accessed October 20, 2013];NATURE. 2011 477(7362):90–4. doi: 10.1038/nature10357. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3170097&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pineda JR, Daynac M, Chicheportiche A, et al. Vascular-derived TGF-β increases in the stem cell niche and perturbs neurogenesis during aging and following irradiation in the adult mouse brain. [Accessed November 10, 2013];EMBO MOL MED. 2013 5(4):548–62. doi: 10.1002/emmm.201202197. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3628106&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oshimori N, Fuchs E. The harmonies played by TGF-β in stem cell biology. [Accessed October 26, 2013];CELL STEM CELL. 2012 11(6):751–64. doi: 10.1016/j.stem.2012.11.001. Available at: http://www.ncbi.nlm.nih.gov/pubmed/23217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massagué J. TGFβ signalling in context. [Accessed October 17, 2013];NAT REV MOL CELL BIOL. 2012 13(10):616–30. doi: 10.1038/nrm3434. Available at: http://www.ncbi.nlm.nih.gov/pubmed/22992590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakashima K, Takizawa T, Ochiai W, et al. BMP2-mediated alteration in the developmental pathway of fetal mouse brain cells from neurogenesis to astrocytogenesis. PROC NATL ACAD SCI U S A. 2001;98(10):5868–73. doi: 10.1073/pnas.101109698. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=33305&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bond AM, Bhalala OG, Kessler Ja. The dynamic role of bone morphogenetic proteins in neural stem cell fate and maturation. [Accessed October 22, 2013];DEV NEUROBIOL. 2012 72(7):1068–84. doi: 10.1002/dneu.22022. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3773925&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim DA, Tramontin AD, Trevejo JM, et al. Noggin Antagonizes BMP Signaling to Create a Niche for Adult Neurogenesis. NEURON. 2000;28(3):713–726. doi: 10.1016/s0896-6273(00)00148-3. Available at: http://linkinghub.elsevier.com/retrieve/pii/S0896627300001483. [DOI] [PubMed] [Google Scholar]
- 25.Porlan E, Morante-Redolat JM, Marques-Torrejon MA, et al. Transcriptional repression of Bmp2 by p21Waf1/Cip1 links quiescence to neural stem cell maintenance. NAT NEUROSCI. 2013;16(11):1567–1575. doi: 10.1038/nn.3545. Available at: http://dx.doi.org/10.1038/nn.3545. [DOI] [PubMed] [Google Scholar]
- 26.Mira H, Andreu Z, Suh H, et al. Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. [Accessed October 25, 2013];CELL STEM CELL. 2010 7(1):78–89. doi: 10.1016/j.stem.2010.04.016. Available at: http://www.ncbi.nlm.nih.gov/pubmed/20621052. [DOI] [PubMed] [Google Scholar]
- 27.Bonaguidi Ma, Peng C-Y, McGuire T, et al. Noggin expands neural stem cells in the adult hippocampus. [Accessed October 28, 2013];J NEUROSCI. 2008 28(37):9194–204. doi: 10.1523/JNEUROSCI.3314-07.2008. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3651371&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gobeske KT, Das S, Bonaguidi Ma, et al. BMP signaling mediates effects of exercise on hippocampal neurogenesis and cognition in mice. [Accessed October 24, 2013];PLOS ONE. 2009 4(10):e7506. doi: 10.1371/journal.pone.0007506. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2759555&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan X-T, Cai W-Q, Yang Z, et al. Effect of antisense oligonucleotide of noggin on spatial learning and memory of rats. ACTA PHARMACOL SIN. 2003;24(5):394–7. Available at: http://www.ncbi.nlm.nih.gov/pubmed/12740172. [PubMed] [Google Scholar]
- 30.Keyes BE, Segal JP, Heller E, et al. Nfatc1 orchestrates aging in hair follicle stem cells. PROC NATL ACAD SCI U S A. 2013;110(51):E4950–E4959. doi: 10.1073/pnas.1320301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crews L, Adame A, Patrick C, et al. Increased BMP6 levels in the brains of Alzheimer’s disease patients and APP transgenic mice are accompanied by impaired neurogenesis. [Accessed October 29, 2013];J NEUROSCI. 2010 30(37):12252–62. doi: 10.1523/JNEUROSCI.1305-10.2010. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2978735&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shetty Ga, Hattiangady B, Shetty AK. Neural stem cell- and neurogenesis-related gene expression profiles in the young and aged dentate gyrus. [Accessed March 24, 2014];AGE (DORDR) 2013 35(6):2165–76. doi: 10.1007/s11357-012-9507-6. Available at: http://www.ncbi.nlm.nih.gov/pubmed/23322452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sorescu GP, Sykes M, Weiss D, et al. Bone morphogenic protein 4 produced in endothelial cells by oscillatory shear stress stimulates an inflammatory response. [Accessed April 16, 2014];J BIOL CHEM. 2003 278(33):31128–35. doi: 10.1074/jbc.M300703200. Available at: http://www.ncbi.nlm.nih.gov/pubmed/12766166. [DOI] [PubMed] [Google Scholar]
- 34.Le Dréau G, Saade M, Gutiérrez-Vallejo I, et al. The strength of SMAD1/5 activity determines the mode of stem cell division in the developing spinal cord. [Accessed February 12, 2014];J CELL BIOL. 2014 204(4):591–605. doi: 10.1083/jcb.201307031. Available at: http://www.ncbi.nlm.nih.gov/pubmed/24515346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ross SE, Greenberg ME, Stiles CD. Basic Helix-Loop-Helix Factors in Cortical Development. NEURON. 2003;39(1):13–25. doi: 10.1016/s0896-6273(03)00365-9. Available at: http://linkinghub.elsevier.com/retrieve/pii/S0896627303003659. [DOI] [PubMed] [Google Scholar]
- 36.Chen D, Zhao M, Mundy GR. Bone Morphogenetic Proteins. GROWTH FACTORS. 2004;22(4):233–241. doi: 10.1080/08977190412331279890. Available at: http://dx.doi.org/10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- 37.Pauklin S, Vallier L. The Cell-Cycle State of Stem Cells Determines Cell Fate Propensity. CELL. 2013;155(1):135–147. doi: 10.1016/j.cell.2013.08.031. Available at: http://linkinghub.elsevier.com/retrieve/pii/S0092867413010258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lein ES, Hawrylycz MJ, Ao N, et al. Genome-wide atlas of gene expression in the adult mouse brain. NATURE. 2007;445(7124):168–176. doi: 10.1038/nature05453. Available at: http://dx.doi.org/10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 39.Basak O, Taylor V. Stem cells of the adult mammalian brain and their niche. [Accessed December 1, 2013];CELL MOL LIFE SCI. 2009 66(6):1057–72. doi: 10.1007/s00018-008-8544-x. Available at: http://www.ncbi.nlm.nih.gov/pubmed/19011753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen Q, Goderie SK, Jin L, et al. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. [Accessed January 21, 2014];SCIENCE. 2004 304(5675):1338–40. doi: 10.1126/science.1095505. Available at: http://www.ncbi.nlm.nih.gov/pubmed/15060285. [DOI] [PubMed] [Google Scholar]
- 41.Bouletreau P, Warren S, Spector J, et al. Hypoxia and VEGF up-regulate BMP-2 mRNA and protein expression in microvascular endothelial cells: implications for fracture healing. PLAST RECONSTR SURG. 2002;109(7):2384–2397. doi: 10.1097/00006534-200206000-00033. [DOI] [PubMed] [Google Scholar]
- 42.Beets K, Huylebroeck D, Moya IM, et al. Robustness in angiogenesis: Notch and BMP shaping waves. TRENDS GENET. 2013;29(3):140–149. doi: 10.1016/j.tig.2012.11.008. Available at: http://linkinghub.elsevier.com/retrieve/pii/S0168952512001928. [DOI] [PubMed] [Google Scholar]
- 43.Parakalan R, Jiang B, Nimmi B, et al. Transcriptome analysis of amoeboid and ramified microglia isolated from the corpus callosum of rat brain. BMC NEUROSCI. 2012;13(1):1. doi: 10.1186/1471-2202-13-64. Available at: BMC Neuroscience. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mouton PR, Long JM, Lei D-L, et al. Age and gender effects on microglia and astrocyte numbers in brains of mice. BRAIN RES. 2002;956(1):30–35. doi: 10.1016/s0006-8993(02)03475-3. Available at: http://www.sciencedirect.com/science/article/pii/S0006899302034753. [DOI] [PubMed] [Google Scholar]
- 45.Norden DM, Godbout JP. Review: Microglia of the aged brain: primed to be activated and resistant to regulation. NEUROPATHOL APPL NEUROBIOL. 2013;39(1):19–34. doi: 10.1111/j.1365-2990.2012.01306.x. Available at: http://dx.doi.org/10.1111/j.1365-2990.2012.01306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mosher KI, Andres RH, Fukuhara T, et al. Neural progenitor cells regulate microglia functions and activity. [Accessed January 27, 2014];NAT NEUROSCI. 2012 15(11):1485–7. doi: 10.1038/nn.3233. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3495979&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niidome T, Matsuda S, Nonaka H, et al. A molecular pathway involved in the generation of microtubule-associated protein 2-positive cells from microglia. BIOCHEM BIOPHYS RES COMMUN. 2008;370:184–188. doi: 10.1016/j.bbrc.2008.03.068. [DOI] [PubMed] [Google Scholar]
- 48.Li D, Tang J, Xu H, et al. Decreased hippocampal cell proliferation correlates with increased expression of BMP4 in the APPswe/PS1ΔE9 mouse model of Alzheimer’s disease. HIPPOCAMPUS. 2008;18(7):692–698. doi: 10.1002/hipo.20428. Available at: http://dx.doi.org/10.1002/hipo.20428. [DOI] [PubMed] [Google Scholar]
- 49.Tang J, Song M, Wang Y, et al. Noggin and BMP4 co-modulate adult hippocampal neurogenesis in the APPswe/PS1ΔE9 transgenic mouse model of Alzheimer’s disease. BIOCHEM BIOPHYS RES COMMUN. 2009;385(3):341–345. doi: 10.1016/j.bbrc.2009.05.067. Available at: http://www.sciencedirect.com/science/article/pii/S0006291X09009991. [DOI] [PubMed] [Google Scholar]
- 50.Conboy IM, Rando TA. Heterochronic parabiosis for the study of the effects of aging on stem cells and their niche. CELL CYCLE. 2012;11(12):2260–2267. doi: 10.4161/cc.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silva H, Conboy IM. Aging and stem cell renewal. [Accessed October 29, 2013];StemBook. 2008 :1–14. Available at: http://www.stembook.org/node/459. [PubMed]
- 52.Suh H, Deng W, Gage FH. Signaling in adult neurogenesis. [Accessed October 29, 2013];ANNU REV CELL DEV BIOL. 2009 25:253–75. doi: 10.1146/annurev.cellbio.042308.113256. Available at: http://www.ncbi.nlm.nih.gov/pubmed/19575663. [DOI] [PubMed] [Google Scholar]
- 53.Schwarz TJ, Ebert B, Lie DC. Stem cell maintenance in the adult mammalian hippocampus: a matter of signal integration? DEV. [Accessed October 29, 2013];NEUROBIOL. 2012 72(7):1006–15. doi: 10.1002/dneu.22026. Available at: http://www.ncbi.nlm.nih.gov/pubmed/22488809. [DOI] [PubMed] [Google Scholar]
- 54.Ashton RS, Conway A, Pangarkar C, et al. Astrocytes regulate adult hippocampal neurogenesis through ephrin-B signaling. [Accessed October 22, 2013];NAT NEUROSCI. 2012 15(10):1399–406. doi: 10.1038/nn.3212. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3458152&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lie D-C, Colamarino SA, Song H-J, et al. Wnt signalling regulates adult hippocampal neurogenesis. NAT PUBL GR. 2005;437(7063):1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- 56.Babu H, Claasen J-H, Kannan S, et al. A protocol for isolation and enriched monolayer cultivation of neural precursor cells from mouse dentate gyrus. [Accessed October 28, 2013];FRONT NEUROSCI. 2011 5(July):89. doi: 10.3389/fnins.2011.00089. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3140691&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. METHODS. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. Available at: http://www.sciencedirect.com/science/article/pii/S1046202301912629. [DOI] [PubMed] [Google Scholar]
- 58.Lai K, Kaspar BK, Gage FH, et al. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. [Accessed October 28, 2013];NAT NEUROSCI. 2003 6(1):21–7. doi: 10.1038/nn983. Available at: http://www.ncbi.nlm.nih.gov/pubmed/12469128. [DOI] [PubMed] [Google Scholar]
- 59.Lois C, Hong EJ, Pease S, et al. Germline Transmission and Tissue-Specific Expression of Transgenes Delivered by Lentiviral Vectors. SCI. 2002;295(5556):868–872. doi: 10.1126/science.1067081. Available at: http://www.sciencemag.org/content/295/5556/868.abstract. [DOI] [PubMed] [Google Scholar]
- 60.Yu JH, Schaffer DV. High-throughput, library-based selection of a murine leukemia virus variant to infect nondividing cells. [Accessed October 28, 2013];J VIROL. 2006 80(18):8981–8. doi: 10.1128/JVI.00615-06. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1563944&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peltier J, Schaffer D. Viral Packaging and Transduction of Adult Hippocampal Neural Progenitors. In: Conboy IM, Schaffer DV, Barcellos-Hoff MH, et al., editors. Protocols for Adult Stem Cells SE - 7. Vol 621. Methods in Molecular Biology™. Humana Press; 2010. pp. 103–116. Available at: http://dx.doi.org/10.1007/978-1-60761-063-2_7. [DOI] [PubMed] [Google Scholar]
- 62.He Y, Zhang H, Yung A, et al. ALK5-dependent TGF-[beta] signaling is a major determinant of late-stage adult neurogenesis. NAT NEUROSCI. 2014;17(7):943–952. doi: 10.1038/nn.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.