Abstract
Transcription of Bone Morphogenetic Proteins (BMPs) and their antagonists in precise spatiotemporal patterns is essential for proper skeletal development, maturation, maintenance, and repair. Nevertheless, transcriptional activity of these molecules in skeletal tissues beyond embryogenesis has not been well-characterized. In this study, we used several transgenic reporter mouse lines to define the transcriptional activity of two potent BMP ligands, Bmp2 and Bmp4, and their antagonist Noggin in the postnatal skeleton. At 3–4 weeks of age, Bmp4 and Noggin reporter activity was readily apparent in most cells of the osteogenic or chondrogenic lineages, respectively, while Bmp2 reporter activity was strongest in terminally differentiated cells of both lineages. By 5–6 months, activity of the reporters had generally abated; however, the Noggin and Bmp2 reporters remained remarkably active in articular chondrocytes and persisted there indefinitely. We further found that endogenous Bmp2, Bmp4, and Noggin transcript levels in postnatal bone and cartilage mirrored the activity of their respective reporters in these tissues. Finally, we found that the activity of the Bmp2, Bmp4, and Noggin reporters in bone and cartilage at 3–4 weeks could be recapitulated in both osteogenic and chondrogenic culture models. These results reveal that Bmp2, Bmp4, and Noggin transcription persists to varying degrees in skeletal tissues postnatally, with each gene exhibiting its own cell-type specific pattern of activity. Illuminating these patterns and their dynamics will guide future studies aimed at elucidating both the causes and consequences of aberrant BMP signaling in the postnatal skeleton.
Keywords: Bmp2, Bmp4, Noggin, bone, cartilage
Introduction
Tight control of Bone Morphogenetic Protein (BMP) signaling is required for proper skeletal development, maturation, maintenance, and repair (1, 2). A fundamental means by which control over BMP signaling is achieved is the transcription of individual ligands in precise spatiotemporal patterns (3). This is well illustrated by the prototypical BMP ligands Bmp2 and Bmp4, both of which are transcribed in widespread, yet distinct patterns during embryonic development. Their transcription, in turn, is governed by constellations of cis-regulatory sequences scattered across vast non-coding regions, also known as gene deserts (4). These regulatory sequences drive tissue-specific transcription of Bmp2 and Bmp4 in the skeleton, among other organs. Moreover, the consequences of their mis-regulation in skeletal tissues are severe (5–13).
Control of BMP signaling can also be achieved via extracellular ligand antagonists. Noggin is one such inhibitor, with specificity for Bmp2, Bmp4 and closely related BMP family members. Similar to these two ligands, Noggin is transcribed in many tissues during embryogenesis, including those of the skeleton. Comparatively little is known about the transcriptional control of Noggin; however, similar to Bmp2 and Bmp4, its mis-regulation in the skeleton also has devastating consequences (9, 12, 14–17).
Transgenic reporters are useful tools for characterizing the transcriptional output of specific genes in cellular detail over time. This is especially true of reporters that have been targeted to a native gene locus via homologous recombination in mouse embryonic stem cells. So-called “knock-in” reporters tend to faithfully recapitulate endogenous gene transcription in vivo, making it possible to follow cell-type specific transcription of a gene in any organ or tissue throughout development and beyond. Effective reporters for this purpose can also be generated through non-targeted insertion of modified bacterial artificial chromosomes (BACs). These typically include a reporter cassette in place of the gene of interest, along with up to several hundred kilobases of endogenous flanking sequence. In addition to highlighting transcriptional activity, BAC reporters are useful for mapping cis-regulatory sequences (4).
Knock-in or BAC-based transgenic reporters have been generated for several BMP ligands and their inhibitors, including Bmp2, Bmp4, and Noggin (14, 18–20). Their activity in the skeleton during embryogenesis has been well documented, providing important information regarding the spatial and temporal specifications of BMP signaling in early skeletal development, as well as the location of key cis-regulatory sequences. In contrast, activity of these same reporters in the postnatal skeleton has been characterized on a very limited basis (17, 21–25), despite the critical role of BMP signaling in skeletal maturation, maintenance, and repair, as well as the therapeutic benefits of manipulating BMP signaling in adults. We have assembled a collection of knock-in and BAC-based reporter transgenes for Bmp2, Bmp4, and Noggin, and here we characterize their activity in the postnatal skeleton.
Materials and Methods
Transgenic reporter mice
The Bmp2 and Bmp4 BAC reporter lines used in this study were generated in our lab (18, 19), while the Bmp4 and Noggin knock-in reporter lines were generated elsewhere (14, 20). All of the lines were maintained on mixed genetic backgrounds, typically by breeding a heterozygous male with a wild-type CD1 or B6/D2-F1 female. Test-crosses and X-gal staining of resulting embryos confirmed the continuity of previously-reported expression patterns with each successive generation (Supplemental Fig. 1A). Only male offspring were used for these experiments. All animal usage was in accordance with policies and protocols established by the Institutional Animal Care and Use Committee at Vanderbilt University Medical Center.
Isolation and culture of primary cells
All primary cells were harvested from femurs of male transgenic mice at 3–4 weeks of age. Chondrocytes were isolated from cartilage of the proximal epiphysis, which lacks a secondary ossification center (26). Briefly, the epiphysis was removed with a pair of tweezers and incubated in Ca2+- and Mg2+-free HBSS with 0.2% bacterial collagenase (Sigma, St. Louis) for 3–4 hours at 37°C to release chondrocytes from their extracellular matrix. BMSCs were isolated from femurs by cutting off both ends and flushing out the contents of the remaining diaphyseal shaft via brief high-speed centrifugation. Chondrocytes or BMSCs from multiple animals were pooled and plated at a density of 1 × 105 cells/cm2 in 24-well tissue-culture-treated plates with complete media consisting of DMEM and the following components: 10% FBS, 4.5mg/ml D-glucose, 100 units/ml penicillin, 100μg/ml streptomycin, 0.25μg/ml Fungizone (Life Technologies, Carlsbad), 2 mM L-alanyl-L-glutamine, and 2mM sodium pyruvate. Cultures were incubated at 37°C in a humidified chamber with 5% CO2 and media was replaced every 2–3 days. Beginning at confluence, the complete media was supplemented with 100μg/ml ascorbate-2-phosphate and 2mM sodium phosphate (pH 7.4) to support mineralization.
X-gal staining and histology
Bacterial β-galactosidase (LacZ) activity was detected in postnatal femurs and cultured cells via X-gal staining. Briefly, the cells or femurs were pre-fixed with 10% neutral-buffered formalin (NBF), equilibrated in wash buffer (100mM sodium phosphate, pH 7.4; 2mM magnesium chloride; 1% sodium deoxycholate; 1% Igepal CA630), and incubated with staining solution (wash buffer plus 0.5mg/ml 5-Bromo-4-chloro-3-indolyl-β-D-galactoside, 4mM potassium ferricyanide, and 4mM potassium ferrocyanide). Cultured cells were pre-fixed at room temperature for 10 minutes and stained overnight at 37°C, while femurs were pre-fixed for 1 hour at 4°C and stained overnight at room temperature or 4°C to increase the sensitivity or specificity of the assay, respectively.
Stained cultured cells and femurs were rinsed with PBS to remove all traces of staining solution and post-fixed. Cultured cells were fixed in 10% NBF for one hour at room temperature and femurs were fixed at 4°C for 48 hours in either 10% NBF or 100% ethanol. Femurs fixed in NBF were subsequently decalcified for two weeks in 10% EDTA, while those fixed in ethanol were cleared for two weeks in 2% potassium hydroxide. Decalcified femurs were embedded in paraffin, sectioned at 10μm increments, and counter-stained with Nuclear Fast Red. Cultured cells were counter-stained in some cases with 0.1% Alizarin Red solution (pH 4.2) for 10 minutes at room temperature to highlight calcified nodules. Cells and cleared femurs were transferred to glycerol for imaging and permanent storage.
Nucleic acid isolation and RT-qPCR
Total RNA and genomic DNA (gDNA) were collected from femurs and tails, respectively, of up to 10 male CD1 mice at 3–4 weeks or 5–6 months of age. Briefly, femurs were dissected in cold PBS, after which the proximal epiphyses were removed with a pair of forceps, pooled, and set aside. Next, the ends of the femurs were removed with a pair of scissors, the marrow was flushed out by high-speed centrifugation, and the remaining cortical bone pieces were pooled. The pooled cartilage and bone samples were then transferred to TriZol (Life Technologies, Carlsbad) and homogenized on ice using a rotor-stator homogenizer. After phase separation with chloroform, the crude RNA was further purified using the RNeasy Micro kit (Qiagen, Valencia). Genomic DNA was isolated from tail snips using the DNeasy Blood & Tissue kit (Qiagen). Quality and quantity of the nucleic acids were assessed via NanoDrop (Thermo Scientific, Waltham) and agarose gel electrophoresis. For each sample, 100 nanograms of total RNA were reverse-transcribed into cDNA using SuperScript VILO MasterMix (Life Technologies), according to the manufacturer’s instructions. Matching no-RT control samples were generated in parallel. The cDNA samples, their corresponding controls, and approximately 100ng of gDNA were then each diluted 10-fold into 200μl of H20.
Quantitative PCR was carried out using SsoAdvanced SYBR Green Supermix on a CFX96 Real-Time PCR Detection System (BioRad, Hercules) according to the manufacturer’s instructions. Primers for Bmp2, Bmp4, Noggin, LacZ, Polr1a, and Polr3b (Supplemental Table 1) were designed using Primer3 (27) and validated on an 8-point standard curve generated with two-fold serial dilutions of gDNA. All primer sets yielded R2 values greater than 0.9, with amplification efficiencies in the range of 95–105%; moreover, they all gave rise to a single amplicon as indicated by their melt curves. Each primer pair was used to amplify 5μl of gDNA, cDNA, and no-RT control from each sample in duplicate. Relative normalized expression was calculated from the resulting Ct values as follows. First, the average Ct value for each target in each cDNA sample was transformed via the 2ΔCt method into a relative quantity using the average Ct value from the corresponding gDNA sample as a reference. This step relates expression to genomic copy number, facilitating direct comparisons between different targets in the same cDNA sample. Next, the relative quantities of Bmp2, Bmp4, Noggin, and LacZ were normalized to the geometric mean of the relative quantities of Polr1a and Polr3b via the 2ΔΔCt method, permitting comparisons across samples. All calculations, including standard deviation, were performed using the CFX Manager software (BioRad). Expression of a gene in a given cDNA sample was considered undetectable by our methods if the relative quantity did not differ by more than 10-fold from that of its corresponding no-RT control.
Results
We assessed the transcription of Bmp2, Bmp4, and Noggin in the postnatal skeleton using six reporter transgenes (Fig. 1). Four of these were BAC-based constructs and two were targeted knock-in alleles. The BAC reporters contain the complete Bmp2 or Bmp4 gene, along with approximately 200 kilobases of non-coding flanking sequence in primarily the 5′ or the 3′ direction. This design captures the activities of various distant non-coding enhancers around either Bmp2 or Bmp4 (18, 19). The knock-in reporters for Bmp4 and Noggin have been previously described (14, 20) and can be studied in heterozygous animals, which are viable in both cases and exhibit no overt skeletal abnormalities. For each reporter, the coding sequence of the endogenous gene has been disrupted by a bacterial LacZ cassette so that beta-galactosidase is expressed in place of functional Bmp2, Bmp4, or Noggin.
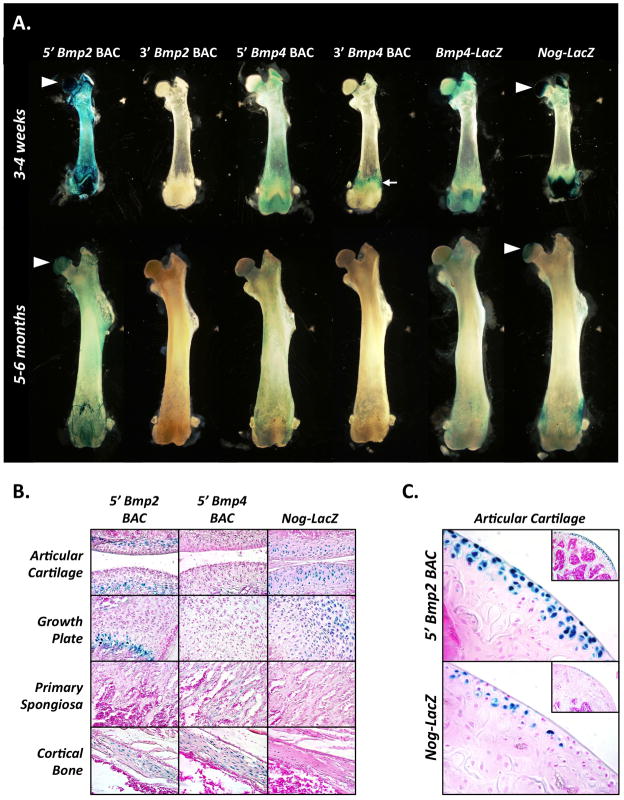
Figure 1. Bmp2, Bmp4, and Noggin transgenic reporters.
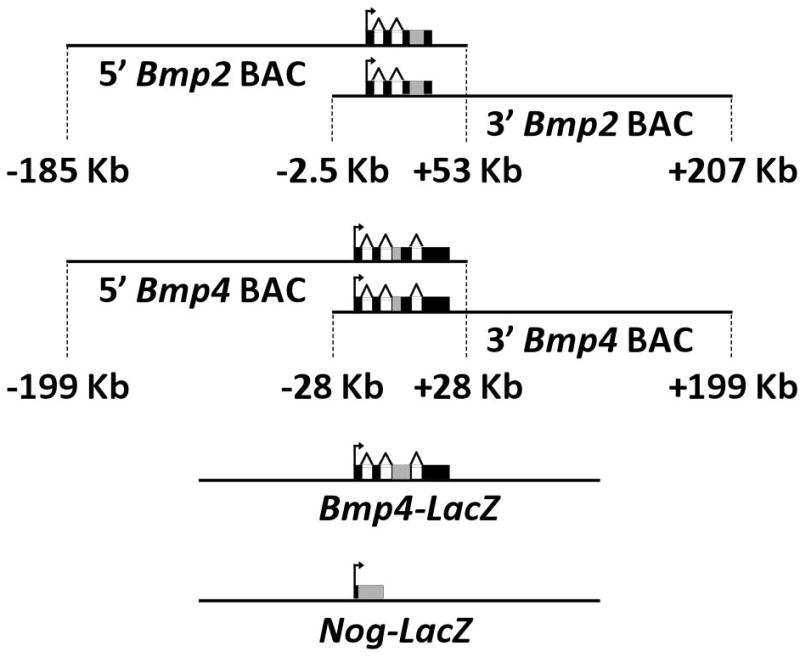
Schematic diagrams of the six reporter transgenes used in this study are shown, including the location of the LacZ cassette (gray rectangle) with respect to the endogenous exons (black rectangles). For the BAC reporters, relative amounts of flanking non-coding sequence contained by the constructs are indicated.
Bmp2, Bmp4, and Noggin are persistently transcribed in the postnatal skeleton
To characterize reporter activity on a gross level, we performed whole-mount X-gal staining on various skeletal elements from transgenic mice at 3–4 weeks and 5–6 months of age (Fig. 2A and Supplemental Fig. 1B). At 3–4 weeks, the 5′ Bmp2 BAC was strongly active in both bone and cartilage, while activity of the 3′ Bmp2 BAC was completely undetectable in either tissue. The 5′ Bmp4 BAC was likewise much more active postnatally than its 3′ counterpart, albeit with some important differences. Specifically, activity of the 5′ Bmp4 BAC was restricted to bone (Fig. 2A), while activity of the 3′ Bmp4 BAC was detectable in tendons up to their sites of insertion (Fig 2A and Supplemental Figure 1B). In agreement with the 5′ Bmp4 BAC, the Bmp4-LacZ knock-in reporter was active in bone, but not cartilage. The Nog-LacZ reporter, by contrast, was highly active in cartilage compared to bone.
Figure 2. Bmp2, Bmp4, and Noggin are dynamically transcribed in distinct cell-type specific patterns in the postnatal skeleton.
(A) Whole-mount X-gal-stained femurs from each of the six transgenic lines at points of peak postnatal growth (3–4 weeks) and maturity (5–6 months) are shown. Arrowheads indicate intensely-stained femoral heads of the 5′ Bmp2 BAC and Nog-LacZ reporter lines. Tendon insertion staining in the 3′ Bmp4 BAC reporter line is indicated by a white arrow. (B) Sections taken from whole-mount X-gal stained femurs of 5′ Bmp2 BAC, 5′ Bmp4 BAC, or Nog-LacZ mice at 3–4 weeks are shown with key skeletal cell populations highlighted, including chondrocytes (articular cartilage, growth plate), osteoblasts (primary spongiosa), and osteocytes (cortical bone). (C) Activity of the 5′ Bmp2 BAC and the Nog-LacZ reporters in articular cartilage of the proximal femur at 5–6 months (shown at low magnification in insets).
At 5–6 months of age, activity of the 5′ Bmp2 BAC and Nog-LacZ reporters in cartilage was still remarkably strong (Fig. 2A and Supplemental Fig. 1C). In contrast, activity of the 5′ Bmp2 BAC, the 5′ Bmp4 BAC, and the Bmp4-LacZ reporters in bone was diminished. The 3′ Bmp2 and Bmp4 BACs continued to exhibit no activity in either skeletal tissue, although the 3′ Bmp4 BAC remained highly active in tendons (Supplemental Fig. 1C). These patterns of reporter activity persisted in mice up to 1–2 years of age (data not shown). Taken together, these results reveal that Bmp2, Bmp4, and Noggin remain transcriptionally active in the skeleton beyond embryogenesis, with Bmp2 and Noggin transcription persisting indefinitely in adult cartilage. Moreover, the inactivity of the 3′ Bmp2 BAC – despite inclusion of several kilobases of promoter – indicates that Bmp2 transcription in the postnatal skeleton requires distant upstream regulatory sequences present only in the 5′ BAC.
Bmp2, Bmp4, and Noggin are transcribed in distinct cell-type specific patterns postnatally
To pinpoint the precise cell populations in which reporters for Bmp2, Bmp4, and Noggin are most active postnatally, we examined sections from whole-mount X-gal stained femurs. At 3–4 weeks of age, the Bmp2, Bmp4, and Noggin reporters each had cell-type-specific patterns of activity with varying degrees of overlap (Fig. 2B). The 5′ Bmp2 BAC was active in hypertrophic sub-populations of chondrocytes in both growth plate and articular cartilage (Fig. 2B, top row, left two panels). The 5′ Bmp2 BAC was also active in osteocytes embedded in cortical bone, but not in osteoblasts within the primary spongiosa (Fig. 2B, top row, right two panels). The 5′ Bmp4 BAC, in contrast, was active in both osteocytes and osteoblasts, but not chondrocytes (Fig. 2B, middle row). Finally, the Noggin-LacZ reporter was active in chondrocytes at various stages of maturation in both growth plate and articular cartilage, but not in osteoblasts or osteocytes (Fig. 2B, bottom row). These results indicate that transcription of Bmp4 and Noggin in the skeleton during postnatal development is lineage-dependent, with Bmp4 primarily transcribed by cells of the osteogenic lineage and Noggin primarily transcribed in cells of the chondrogenic lineage. In contrast, Bmp2 transcription is linked to maturational status, with cells of both lineages exhibiting increased transcriptional activity as they progress towards terminal differentiation.
At 5–6 months of age, activity of the 5′ Bmp2 BAC was restricted to chondrocytes at the articular surface (Fig. 2C), a sub-population from which it was excluded during earlier postnatal development (Fig. 2B, top row, left panel). This shift in activity from the deeper to the more superficial, permanent chondrocyte layers occurred between two and three months of age (data not shown), coinciding with the onset of skeletal maturity. The Nog-LacZ reporter was also active in this same cell population, albeit to a lesser extent (Fig. 2C). These patterns of activity persisted in mice up to 1–2 years of age with little alteration (data not shown). Thus, Bmp2 and Noggin are transcribed throughout adulthood in a sub-population of articular chondrocytes distinct from that in which Bmp2 is transcribed during postnatal development.
Bmp2, Bmp4, and Noggin mRNA levels mirror activity of their reporters in postnatal skeletal tissues
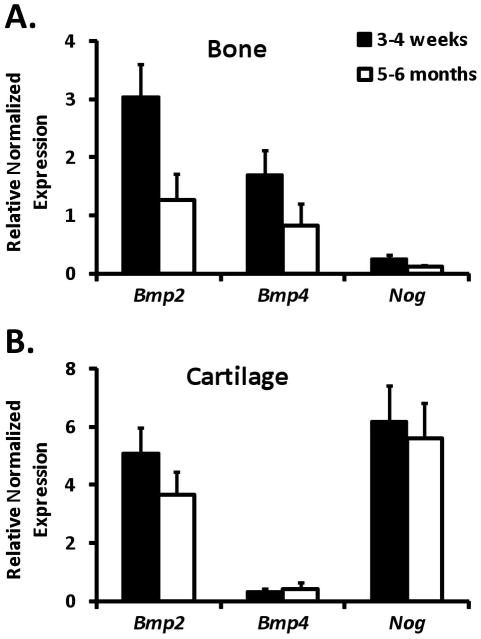
Next, we examined endogenous Bmp2, Bmp4, and Noggin expression in postnatal skeletal tissues. Using total RNA isolated from epiphyseal cartilage or cortical bone derived from femurs of 3–4 week-old or 5–6 month-old wild-type mice, we measured relative transcript levels of the three genes by RT-qPCR. To facilitate direct comparisons between different transcripts, we normalized RT-qPCR data for each gene to qPCR results obtained with the same primers on a pool of genomic DNA isolated from the same mice, in which the 3 genes were present in equimolar amounts. The most abundant transcript in bone at 3–4 weeks of age was Bmp2, followed by Bmp4, which in turn was nearly an order of magnitude higher than Noggin (Fig. 3A). By 5–6 months, the transcript levels of all 3 genes had decreased by about 2-fold (Fig 3A). In cartilage at 3–4 weeks of age, Bmp2 and Noggin transcripts were present in roughly equal amounts and were far more abundant than Bmp4 transcripts, which were barely detectable (Fig. 3B). Transcript levels of all three genes remained about the same in cartilage between 3–4 weeks and 5–6 months of age (Fig. 3B). Collectively, these results confirm that Bmp2, Bmp4, and Noggin transcript levels mirror the activity of their reporters in the postnatal skeleton.
Figure 3. Expression of endogenous Bmp2, Bmp4, and Noggin mRNA in postnatal skeletal tissues.
Relative transcript levels of Bmp2, Bmp4, and Noggin were measured by RT-qPCR using total RNA isolated from cortical bone (A) or epiphyseal cartilage (B) of femurs from wild-type mice at 3–4 weeks (black bars) or 5–6 months (white bars) of age. Graphed values represent the mean (± SD, n=3) expression level calculated for each transcript via the 2ΔΔCt method, with genomic DNA isolated from the same mice serving as reference samples and Polr1a and Polr3b serving as internal reference genes.
Bmp2, Bmp4, and Noggin are transcribed in cultured skeletal lineage cells and their precursors
Finally, we assessed Bmp2, Bmp4, and Noggin reporter activity in primary cultures of committed skeletal-lineage cells and their precursors. To this end, we isolated epiphyseal chondrocytes and bone marrow stromal cells (BMSCs) from femurs of 3–4 week-old mice harboring the 5′ Bmp2 BAC, 5′ Bmp4 BAC, or Nog-LacZ reporters. We then cultured these for up to three weeks in media supplemented with ascorbate and inorganic phosphate. Both cell types secrete and mineralize a thick extracellular matrix under these conditions and express markers generally consistent with terminal osteogenic or chondrogenic differentiation (28, 29), (Supplemental Figure 2). Thus, these systems provide us with an opportunity to follow the dynamics of reporter activity in differentiating cells of both lineages.
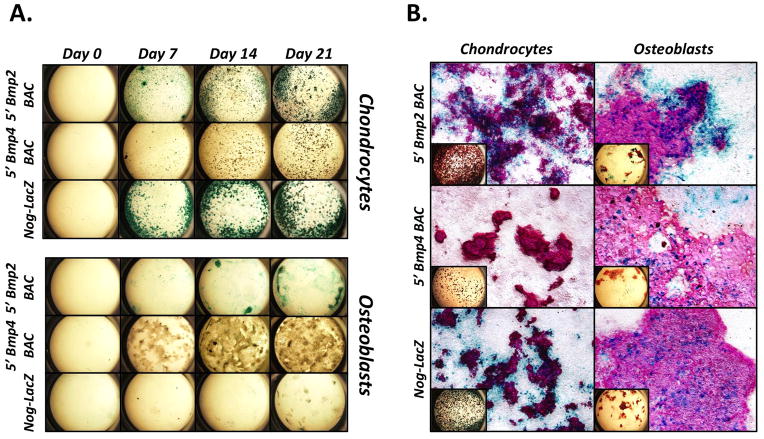
When assayed via X-gal staining, activity of the 5′ Bmp2 BAC, 5′ Bmp4 BAC, and Nog-LacZ reporters was undetectable in both culture models prior to supplementation (Fig. 4A, Day 0). After supplementation, and concomitant with mineralization, the 5′ Bmp2 BAC and Nog-LacZ reporters both exhibited robust activity in chondrocyte cultures, while only the 5′ Bmp2 BAC reporter appeared active in BMSCs (Fig. 4A, Days 7–21). Closer inspection revealed that both the Nog-LacZ and 5′ Bmp4 BAC reporters were indeed active in BMSCs at modest levels (Fig. 4B, middle and bottom panels on right); however, there was no detectable activity in 5′ Bmp4 BAC transgenic chondrocyte cultures (Fig. 4B, middle panel on left). Interestingly, activity of the Nog-LacZ and 5′ Bmp4 BAC reporters was located primarily within mineralized nodules of the BMSC cultures, whereas activity of the 5′ Bmp2 BAC was also present in flattened cells at the periphery of the nodules. A similar pattern of activity was seen in chondrocyte cultures derived from 5′ Bmp2 BAC mice. Finally, Nog-LacZ reporter activity localized to the periphery of mineralized nodules in chondrocyte cultures, in complete contrast to its pattern in BMSCs.
Figure 4. Characterization of Bmp2, Bmp4, and Noggin transcription in cultured skeletal lineage cells and their precursors.
(A and B) Epiphyseal chondrocytes or BMSCs (osteoblasts) were isolated from the femurs of 3–4 week-old 5′ Bmp2 BAC, 5′ Bmp4 BAC, or Nog-LacZ mice, cultured with ascorbate and inorganic phosphate for up to 3 weeks, and stained with X-gal. (A) Time course showing a week-by-week progression of staining, beginning at the point when mineralization-supporting media was first added to the cultures (Day 0). (B) Day 21 cultures counter-stained with Alizarin Red to highlight mineralized nodules (shown at low magnification in insets).
These results reveal that Bmp2, Bmp4, and Noggin are transcribed in cells harvested directly from the postnatal skeleton and cultured in monolayers under mineralization-inducing conditions. In general, their patterns of transcriptional activity are reminiscent of those seen in skeletal lineage cells in vivo. The chondrocyte cultures, in particular, exhibit robust Bmp2 and Noggin–but not Bmp4–transcriptional activity. BMSCs, on the other hand, transcribe all three genes at more modest levels, suggesting they may be a mixture of both chondrogenic and osteogenic cells.
Discussion
The presence of Bmp2 and Bmp4 in the postnatal skeleton has been appreciated since their discovery more than two decades ago (30). Subsequent immunohistochemical studies have shed some light on their expression patterns (13, 31–34), (Supplemental Fig. 3). To our knowledge, this is the first study to characterize their transcriptional activity in skeletal tissues beyond embryogenesis. Our results reveal that while both genes are transcribed by osteocytes embedded in cortical bone, their transcription patterns in the growth plate and trabecular bone compartments are completely different. This striking divergence may help explain why limb-specific deletions of Bmp2 or Bmp4 result in different skeletal phenotypes (11, 35), despite their extremely high homology and generally similar signaling abilities (36).
In contrast to Bmp2 and Bmp4, reports on Noggin protein localization in the postnatal skeleton are scarce. Our results indicate it is robustly transcribed by most cells of the chondrogenic lineage. This, in turn, suggests that cartilage is the primary source of Noggin in the postnatal skeleton, and that Noggin-mediated antagonism of BMP signaling is required in this tissue. Interestingly, conditional knock-out of Noggin in the skeleton using a human Osteocalcin-Cre driver results in mild postnatal osteopenia and may reflect the importance of its expression by cells of the osteogenic lineage, albeit at low levels (15). Future studies aimed at determining the relative importance of Noggin expression by chondrogenic or osteogenic cells in the postnatal skeleton are thus warranted.
The robust and persistent transcription of Bmp2 by adult articular chondrocytes is consistent with the requirement of BMP signaling for articular cartilage maintenance (37). Moreover, a cell-autonomous role for Bmp2 in promoting cartilage anabolism is well-supported by in vitro studies (38). On the other hand, the concurrent transcriptional activity of Noggin suggests that BMP signaling is antagonized in articular cartilage. Interestingly, immunohistochemical studies have detected Noggin (39), but not Bmp2 protein in normal adult articular cartilage (40–42). Absence of the latter may be due to a repressive post-transcriptional regulatory mechanism (43, 44). Taken together, these results suggest that the potent anabolic activity of Bmp2 must be carefully balanced in adult articular cartilage. Indeed, joint pathologies such as osteoarthritis may reflect a disruption of this delicate balance (45).
Our previous work demonstrated that the 3′ Bmp2 BAC is active in bone during embryogenesis, mediated by a distant downstream enhancer termed ECR1 (19). Surprisingly, we were unable to detect activity of the 3′ Bmp2 BAC reporter in the skeleton from 3–4 weeks of age and beyond, or in cells cultured from postnatal skeletal tissues (Fig. 2A and data not shown). Thus, the influence of ECR1 on Bmp2 transcription in bone appears to be a transient embryonic phenomenon. Moreover, it is not recapitulated in primary osteoblast culture models derived from postnatal skeletal tissues. In contrast, the 5′ Bmp2 BAC exhibits robust and persistent activity in chondrocytes during embryogenesis and throughout postnatal life. It is also active in osteogenic cells during postnatal development. Finally, the 5′ Bmp2 BAC is active in differentiating cells of both lineages in vitro, thus making it amenable to studies aimed at dissecting distant upstream transcriptional regulatory mechanisms.
In conclusion, we have used some of the best available reporters for Bmp2, Bmp4, and Noggin to illuminate both the dynamics and cellular localization of their transcription in the postnatal skeleton. By pinpointing specific cell populations in which these three genes are transcribed, our results will guide future studies aimed at further elucidating the causes and consequences of BMP signaling in the skeleton during postnatal development and beyond.
Supplementary Material
Acknowledgments
This work was supported by grants F32AR057649 (S.K.P.), R01HL114751 (D.P.M.), and P30CA068485 (Vanderbilt University Translational Pathology Shared Resource). The authors would like to acknowledge Emily Poulin, Xiangli Yang, and Matt Stewart for their assistance in revising the manuscript, as well as Yun Ma, Brigid Hogan, and Chin Chiang for providing BMSC cDNA, Bmp4-LacZ mice, and Nog-LacZ mice, respectively.
Footnotes
Disclosures
The authors state that they have no conflicts of interest.
Authors’ roles
Study design and conduct: SKP. Data collection and analysis: SKP. Data interpretation: SKP and DPM. Drafting of manuscript: SKP. Revising and approving final version of manuscript: SKP and DPM. SKP and DPM share responsibility for the integrity of the data analysis.
This article includes supplemental methods, a supplemental table, and three supplemental figures.
References
- 1.Gazzerro E, Canalis E. Bone morphogenetic proteins and their antagonists. Rev Endocr Metab Disord. 2006 Jun;7(1–2):51–65. doi: 10.1007/s11154-006-9000-6. [DOI] [PubMed] [Google Scholar]
- 2.Kamiya N, Mishina Y. New insights on the roles of BMP signaling in bone-A review of recent mouse genetic studies. Biofactors. 2011 Mar-Apr;37(2):75–82. doi: 10.1002/biof.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kingsley DM. What do BMPs do in mammals? Clues from the mouse short-ear mutation. Trends Genet. 1994 Jan;10(1):16–21. doi: 10.1016/0168-9525(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 4.Pregizer S, Mortlock DP. Control of BMP gene expression by long-range regulatory elements. Cytokine Growth Factor Rev. 2009 Oct-Dec;20(5–6):509–15. doi: 10.1016/j.cytogfr.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bandyopadhyay A, Tsuji K, Cox K, Harfe BD, Rosen V, Tabin CJ. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006 Dec;2(12):e216. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonilla-Claudio M, Wang J, Bai Y, Klysik E, Selever J, Martin JF. Bmp signaling regulates a dose-dependent transcriptional program to control facial skeletal development. Development. 2012 Feb;139(4):709–19. doi: 10.1242/dev.073197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maatouk DM, Choi KS, Bouldin CM, Harfe BD. In the limb AER Bmp2 and Bmp4 are required for dorsal-ventral patterning and interdigital cell death but not limb outgrowth. Dev Biol. 2009 Mar 15;327(2):516–23. doi: 10.1016/j.ydbio.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Mi M, Jin H, Wang B, Yukata K, Sheu TJ, Ke QH, et al. Chondrocyte BMP2 signaling plays an essential role in bone fracture healing. Gene. 2013 Jan 10;512(2):211–8. doi: 10.1016/j.gene.2012.09.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okamoto M, Murai J, Yoshikawa H, Tsumaki N. Bone morphogenetic proteins in bone stimulate osteoclasts and osteoblasts during bone development. J Bone Miner Res. 2006 Jul;21(7):1022–33. doi: 10.1359/jbmr.060411. [DOI] [PubMed] [Google Scholar]
- 10.Shu B, Zhang M, Xie R, Wang M, Jin H, Hou W, et al. BMP2, but not BMP4, is crucial for chondrocyte proliferation and maturation during endochondral bone development. J Cell Sci. 2011 Oct 15;124(Pt 20):3428–40. doi: 10.1242/jcs.083659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsuji K, Bandyopadhyay A, Harfe BD, Cox K, Kakar S, Gerstenfeld L, et al. BMP2 activity, although dispensable for bone formation, is required for the initiation of fracture healing. Nat Genet. 2006 Dec;38(12):1424–9. doi: 10.1038/ng1916. [DOI] [PubMed] [Google Scholar]
- 12.Tsumaki N, Nakase T, Miyaji T, Kakiuchi M, Kimura T, Ochi T, et al. Bone morphogenetic protein signals are required for cartilage formation and differently regulate joint development during skeletogenesis. J Bone Miner Res. 2002 May;17(5):898–906. doi: 10.1359/jbmr.2002.17.5.898. [DOI] [PubMed] [Google Scholar]
- 13.Yang W, Guo D, Harris MA, Cui Y, Gluhak-Heinrich J, Wu J, et al. Bmp2 in osteoblasts of periosteum and trabecular bone links bone formation to vascularization and mesenchymal stem cells. J Cell Sci. 2013 Sep 15;126(Pt 18):4085–98. doi: 10.1242/jcs.118596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunet LJ, McMahon JA, McMahon AP, Harland RM. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science. 1998 May 29;280(5368):1455–7. doi: 10.1126/science.280.5368.1455. [DOI] [PubMed] [Google Scholar]
- 15.Canalis E, Brunet LJ, Parker K, Zanotti S. Conditional inactivation of noggin in the postnatal skeleton causes osteopenia. Endocrinology. 2012 Apr;153(4):1616–26. doi: 10.1210/en.2011-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devlin RD, Du Z, Pereira RC, Kimble RB, Economides AN, Jorgetti V, et al. Skeletal overexpression of noggin results in osteopenia and reduced bone formation. Endocrinology. 2003 May;144(5):1972–8. doi: 10.1210/en.2002-220918. [DOI] [PubMed] [Google Scholar]
- 17.Wu XB, Li Y, Schneider A, Yu W, Rajendren G, Iqbal J, et al. Impaired osteoblastic differentiation, reduced bone formation, and severe osteoporosis in noggin-overexpressing mice. J Clin Invest. 2003 Sep;112(6):924–34. doi: 10.1172/JCI15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandler KJ, Chandler RL, Mortlock DP. Identification of an ancient Bmp4 mesoderm enhancer located 46 kb from the promoter. Dev Biol. 2009 Mar 15;327(2):590–602. doi: 10.1016/j.ydbio.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandler RL, Chandler KJ, McFarland KA, Mortlock DP. Bmp2 transcription in osteoblast progenitors is regulated by a distant 3′ enhancer located 156.3 kilobases from the promoter. Mol Cell Biol. 2007 Apr;27(8):2934–51. doi: 10.1128/MCB.01609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawson KA, Dunn NR, Roelen BA, Zeinstra LM, Davis AM, Wright CV, et al. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999 Feb 15;13(4):424–36. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Granero-Molto F, Weis JA, Miga MI, Landis B, Myers TJ, O’Rear L, et al. Regenerative effects of transplanted mesenchymal stem cells in fracture healing. Stem Cells. 2009 Aug;27(8):1887–98. doi: 10.1002/stem.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamiya N, Shafer S, Oxendine I, Mortlock DP, Chandler RL, Oxburgh L, et al. Acute BMP2 upregulation following induction of ischemic osteonecrosis in immature femoral head. Bone. 2013 Mar;53(1):239–47. doi: 10.1016/j.bone.2012.11.023. [DOI] [PubMed] [Google Scholar]
- 23.Marsell R, Steen B, Bais MV, Mortlock DP, Einhorn TA, Gerstenfeld LC. Skeletal trauma generates systemic BMP2 activation that is temporally related to the mobilization of CD73+ cells. J Orthop Res. 2013 Sep 9; doi: 10.1002/jor.22487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsubara H, Hogan DE, Morgan EF, Mortlock DP, Einhorn TA, Gerstenfeld LC. Vascular tissues are a primary source of BMP2 expression during bone formation induced by distraction osteogenesis. Bone. 2012 Jul;51(1):168–80. doi: 10.1016/j.bone.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warren SM, Brunet LJ, Harland RM, Economides AN, Longaker MT. The BMP antagonist noggin regulates cranial suture fusion. Nature. 2003 Apr 10;422(6932):625–9. doi: 10.1038/nature01545. [DOI] [PubMed] [Google Scholar]
- 26.Cole HA, Yuasa M, Hawley G, Cates JM, Nyman JS, Schoenecker JG. Differential development of the distal and proximal femoral epiphysis and physis in mice. Bone. 2013 Jan;52(1):337–46. doi: 10.1016/j.bone.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012 Aug;40(15):e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garimella R, Bi X, Camacho N, Sipe JB, Anderson HC. Primary culture of rat growth plate chondrocytes: an in vitro model of growth plate histotype, matrix vesicle biogenesis and mineralization. Bone. 2004 Jun;34(6):961–70. doi: 10.1016/j.bone.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 29.Hughes FJ, Aubin JE. Culture of Cells of the Osteoblast Lineage. In: Arnett T, Henderson B, editors. Methods in Bone Biology. London, U.K: Chapman & Hall; 1998. pp. 1–49. [Google Scholar]
- 30.Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, et al. Novel regulators of bone formation: molecular clones and activities. Science. 1988 Dec 16;242(4885):1528–34. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- 31.Anderson HC, Hodges PT, Aguilera XM, Missana L, Moylan PE. Bone morphogenetic protein (BMP) localization in developing human and rat growth plate, metaphysis, epiphysis, and articular cartilage. J Histochem Cytochem. 2000 Nov;48(11):1493–502. doi: 10.1177/002215540004801106. [DOI] [PubMed] [Google Scholar]
- 32.Ngo TQ, Scherer MA, Zhou FH, Foster BK, Xian CJ. Expression of bone morphogenic proteins and receptors at the injured growth plate cartilage in young rats. J Histochem Cytochem. 2006 Aug;54(8):945–54. doi: 10.1369/jhc.6A6939.2006. [DOI] [PubMed] [Google Scholar]
- 33.Yazaki Y, Matsunaga S, Onishi T, Nagamine T, Origuchi N, Yamamoto T, et al. Immunohistochemical localization of bone morphogenetic proteins and the receptors in epiphyseal growth plate. Anticancer Res. 1998 Jul-Aug;18(4A):2339–44. [PubMed] [Google Scholar]
- 34.Santos A, Bakker AD, Willems HM, Bravenboer N, Bronckers AL, Klein-Nulend J. Mechanical loading stimulates BMP7, but not BMP2, production by osteocytes. Calcif Tissue Int. 2011 Oct;89(4):318–26. doi: 10.1007/s00223-011-9521-1. [DOI] [PubMed] [Google Scholar]
- 35.Tsuji K, Cox K, Bandyopadhyay A, Harfe BD, Tabin CJ, Rosen V. BMP4 is dispensable for skeletogenesis and fracture-healing in the limb. J Bone Joint Surg Am. 2008 Feb;90(Suppl 1):14–8. doi: 10.2106/JBJS.G.01109. [DOI] [PubMed] [Google Scholar]
- 36.Lavery K, Swain P, Falb D, Alaoui-Ismaili MH. BMP-2/4 and BMP-6/7 differentially utilize cell surface receptors to induce osteoblastic differentiation of human bone marrow-derived mesenchymal stem cells. J Biol Chem. 2008 Jul 25;283(30):20948–58. doi: 10.1074/jbc.M800850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rountree RB, Schoor M, Chen H, Marks ME, Harley V, Mishina Y, et al. BMP receptor signaling is required for postnatal maintenance of articular cartilage. PLoS Biol. 2004 Nov;2(11):e355. doi: 10.1371/journal.pbio.0020355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oshin AO, Caporali E, Byron CR, Stewart AA, Stewart MC. Phenotypic maintenance of articular chondrocytes in vitro requires BMP activity. Vet Comp Orthop Traumatol. 2007;20(3):185–91. doi: 10.1160/vcot-06-07-0061. [DOI] [PubMed] [Google Scholar]
- 39.Lories RJ, Daans M, Derese I, Matthys P, Kasran A, Tylzanowski P, et al. Noggin haploinsufficiency differentially affects tissue responses in destructive and remodeling arthritis. Arthritis Rheum. 2006 Jun;54(6):1736–46. doi: 10.1002/art.21897. [DOI] [PubMed] [Google Scholar]
- 40.Blaney Davidson EN, Vitters EL, van der Kraan PM, van den Berg WB. Expression of transforming growth factor-beta (TGFbeta) and the TGFbeta signalling molecule SMAD-2P in spontaneous and instability-induced osteoarthritis: role in cartilage degradation, chondrogenesis and osteophyte formation. Ann Rheum Dis. 2006 Nov;65(11):1414–21. doi: 10.1136/ard.2005.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukui N, Zhu Y, Maloney WJ, Clohisy J, Sandell LJ. Stimulation of BMP-2 expression by pro-inflammatory cytokines IL-1 and TNF-alpha in normal and osteoarthritic chondrocytes. J Bone Joint Surg Am. 2003;85-A(Suppl 3):59–66. doi: 10.2106/00004623-200300003-00011. [DOI] [PubMed] [Google Scholar]
- 42.Nakase T, Miyaji T, Tomita T, Kaneko M, Kuriyama K, Myoui A, et al. Localization of bone morphogenetic protein-2 in human osteoarthritic cartilage and osteophyte. Osteoarthritis Cartilage. 2003 Apr;11(4):278–84. doi: 10.1016/s1063-4584(03)00004-9. [DOI] [PubMed] [Google Scholar]
- 43.Fukui N, Ikeda Y, Ohnuki T, Hikita A, Tanaka S, Yamane S, et al. Pro-inflammatory cytokine tumor necrosis factor-alpha induces bone morphogenetic protein-2 in chondrocytes via mRNA stabilization and transcriptional up-regulation. J Biol Chem. 2006 Sep 15;281(37):27229–41. doi: 10.1074/jbc.M603385200. [DOI] [PubMed] [Google Scholar]
- 44.Kruithof BP, Fritz DT, Liu Y, Garsetti DE, Frank DB, Pregizer SK, et al. An autonomous BMP2 regulatory element in mesenchymal cells. J Cell Biochem. 2011 Feb;112(2):666–74. doi: 10.1002/jcb.22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lories RJ, Luyten FP. Bone morphogenetic protein signaling and arthritis. Cytokine Growth Factor Rev. 2009 Oct-Dec;20(5–6):467–73. doi: 10.1016/j.cytogfr.2009.10.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.