Abstract
Yes-associated protein 1 (YAP1) is a transcriptional coactivator in the Hippo signaling pathway. Increased YAP1- activity promotes the growth of tumors, including that of colorectal cancer (CRC). Verteporfin, a drug that enhances phototherapy to treat neovascular macular degeneration, is an inhibitor of YAP1. Here, we found that verteporfin inhibited tumor growth independently of its effects on YAP1 or the related protein TAZ in genetic or chemical-induced mouse models of CRC, in patient-derived xenografts and in enteroid models of CRC. Instead, verteporfin exhibited in vivo selectivity for killing tumor cells in part by impairing the global clearance of high molecular weight oligomerized proteins, particularly p62 (a sequestrome involved in autophagy) and STAT3 (a transcription factor). Verteporfin inhibited cytokine-induced STAT3 activity and cell proliferation and reduced the viabilty of cultured CRC cells. Although verteporfin accumulated to a greater extent in normal cells than in tumor cells in vivo, experiments with cultured cells indicated that the normal cells efficiently cleared verteporfin-induced protein oligomers through autophagic and proteasomal pathways. Culturing CRC cells in hypoxic or nutrient-deprived conditions (modeling a typical CRC microenvironment) impaired the clearance of protein oligomers and resulted in cell death; whereas culturing cells in normoxic or glucose-replete conditions protected cell viability and proliferation in the presence of verteporfin. Furthermore, verteporfin suppressed the proliferation of other cancer cell lines even in the absence of YAP1, suggesting that verteporfin may be effective against multiple types of solid cancers.
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer type and the third leading cause of cancer deaths for both men and women in the United States (1). A key contributing event to the pathogenesis of upwards of 80% of sporadic CRCs is inactivation of the APC (adenomatous polyposis coli) tumor suppressor gene. APC inactivation appears to be an early genetic event in the multi-step pathway to CRC, and mutations affecting KRAS and TP53 are observed in a significant proportion of CRC cases (2). Whereas the bulk of CRCs are termed sporadic and are not associated with any clear predisposing factors, a small subset of CRCs arise in patients with inflammatory bowel disease (IBD), a chronic relapsing inflammatory disease of the intestine. CRCs arising in IBD patients are often termed as colitis-associated colon cancers (CACs) (3). Recent work has demonstrated that the growth-promoting role of the transcription co-activator Yes-associated protein 1 (YAP1) is critical for the proliferative response observed during intestinal inflammation as well as in sporadic CRC (4–6).
YAP1 is a downstream effector in the Hippo signaling pathway, which functions as an essential regulator of proliferation, organ size and cell differentiation (7, 8). In response to a decrease in Hippo signaling, YAP1 translocates into the nucleus and acts as transcriptional co-activator to TEA domain (TEAD) family members (TEAD1–4). TEADs regulate various genes the encode proteins with critical roles in proliferation and inhibition of apoptosis (9–11). Activation of YAP1, through disruption of upstream kinases, leads to hyperproliferation and spontaneous tumor formation in certain tissues, including the intestine (6, 12). Recently, it has been shown that YAP1 is an essential component of the β-catenin degradation complex. Upon activation of Wnt signaling, both β-catenin and YAP1 signaling are activated, and YAP1 plays a key role in the hyperproliferative response upon disruption of Apc or activation of β-catenin (13–15). Because YAP1 is a critical component of the β-catenin degradation complex, disruption of YAP1 leads to enhanced β-catenin activation and increased proliferation in the intestine (16). Thus, through its scaffolding role in β-catenin regulation, YAP1 can function as a tumor suppressor. On the other hand, through its transcriptional co-activator role, YAP1 can function as an oncogene (17). Therefore, approaches that specifically inhibit YAP1 transcriptional activity without affecting YAP1’s role in β-catenin turnover, could have a therapeutic role in CRC. Several inhibitors that interfere with the YAP1-TEAD complex have been identified (18), including selected porphyrin-containing drugs that potently inhibit YAP1 transcriptional activity (18). Verteporfin (VP), an FDA approved drug used as a photosensitizer for photodynamic therapy in patients with age-related macular degeneration (AMD) potently inhibits YAP1 transcriptional activity, independent of light activation (18, 19). Currently, VP has been the main pharmacological tool to understand the role of YAP1 in various cancers (18, 20–26).
Here we investigated the mechanism of action of VP in CRC using CRC-derived cell lines, mouse models and patient-derived adenoma and adenocarcinoma enteroid models. Unexpectedly, we uncovered a tumor suppressive function of VP that was independent of its effects on YAP1.
RESULTS
YAP1 signaling is activated in colon-derived cell lines and CRC
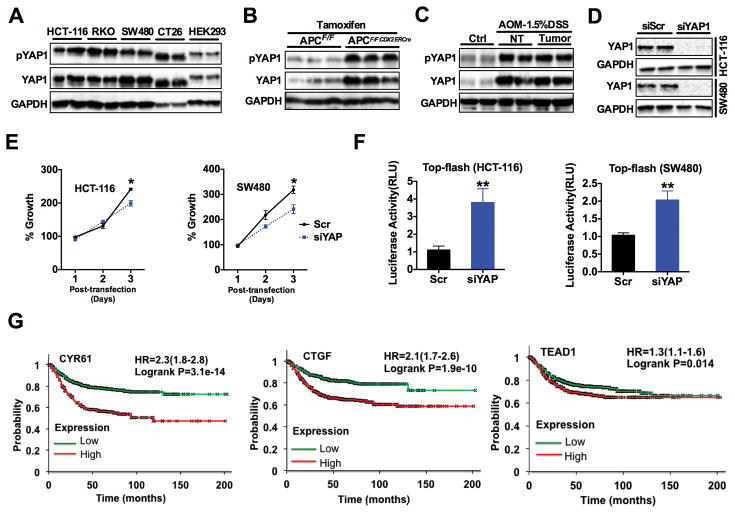
To explore the role of YAP1 signaling in CRC cells, we assessed the abundance of total and phosphorylated YAP1 in various CRC-derived cell lines as well as in the HEK293 embryonic kidney-derived cell line, which is known to express active YAP1. All the CRC cell lines studied had high abundance of total and phosphorylated YAP1 (Fig. 1A and fig. S1A). YAP1 was also assessed in dysplastic and hyperproliferative colon tissues from mice where both Apc alleles were inactivated by a colon epithelial-specific tamoxifen inducible Cre transgene (ApcF/F:CDX2ERCre) (27). Seven days after colon-specific disruption of Apc, the abundance of total and phosphorylated YAP1 increased in the colon of ApcF/F:CDX2ERCre mice compared to littermate controls (Fig. 1B and fig. S1B). In addition, the abundance of total and phosphorylated YAP1 was increased in colon tumor epithelium of a mouse colitis-associated cancer (CAC) model (Fig. 1C and fig. S1C). To further evaluate the function of YAP1 in CRC, we first explored the effects on CRC cell growth in vitro after siRNA-mediated knockdown of YAP1 (Fig. 1D and fig. S1D). Inhibition of YAP1 resulted in a modest decrease in the proliferation of CRC cells, perhaps because antagonism of YAP1 function in Hippo signaling was offset in part by further activation of β-catenin signaling in the cells, due to loss of YAP1-dependent degradation of β-catenin (Fig. 1E–F) (13, 16). Our findings are consistent with the complex and sometimes contradictory data on YAP1 function in CRC (4, 6, 16). To address the potential relevance of YAP1 transcriptional activity in CRC clinical specimens, Kaplan-Meier survival curves were generated and stratified by well-characterized YAP1 transcriptional target genes using datasets published under the GEO accession numbers GSE12945, GSE14333, GSE17538, GSE31595, GSE33114, GSE37892, GSE39582, and GSE41258. Increased expression of YAP1-regulated target genes such as CYR61, CTGF, RUNX2, ITGB2, LSP1, PMEPA1, EPHA2 and FLI1 or TEAD1, all predicted worse patient survival (Fig. 1G and fig. S1E). Collectively, these data suggested that further studies on the effects of inhibiting YAP1 transcriptional function in CRC are of potential interest.
Fig. 1. YAP1 signaling is activated in colon derived cell lines and colon cancer.
(A to C) Western blot analysis of total and phosphorylatedYAP1 (pYAP1) in (A) human colon cancer cell lines and HEK293A cells, (B) in the colons of ApcF/F and ApcF/F:CDXERCre mice 9 days after treatment with tamoxifen (n=3 mice/group), and (C) in tumor and adjacent normal colon tissues of AOM/DSS treated mice (n=3 mice/group). (D) Western analysis of YAP1 in HCT-116 and SW480 cells transfected with either control siRNA (Scr) or a siRNA targeting YAP1 (siYAP). (E) MTT assay performed in HCT-116 and SW480 cells after knockdown of YAP1. Data are mean ± S.E.M. (F) Luciferase analysis using Top-flash promoter after knockdown of YAP1 in HCT-116 or SW480 cells. Luciferase values were normalized to β-Gal. Data are mean ± SD. * p < 0.05, ** p<0.01 vs Scr. All cell line experiments were done in triplicates and repeated at least three times. (G) Kaplan–Meier survival plots with hazard rates (HR) and confidence intervals computed using data of 1,332 patients stratified by differential expression of YAP1 target genes CYR61, CTGF and TEAD1.
YAP1 transcriptional inhibitor verteporfin inhibits CRC growth in vivo
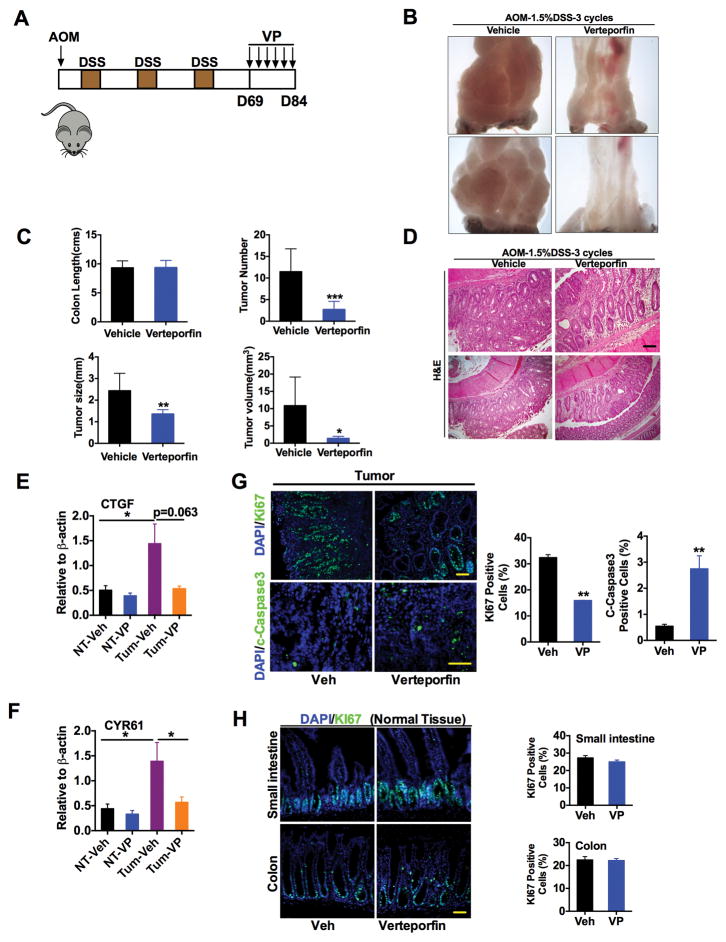
VP, previously identified as a transcriptional inhibitor of YAP1 (18) was assessed in an azoxymethane (AOM)/dextran sulfate sodium (DSS)-induced model of colon cancer. After three cycles of DSS, colon tumors were observed in all treated C57BL/6 mice. Treatment was then initiated with 100mg/kg VP injected intraperitoneally at 3-day intervals, and mice were assessed after six doses of VP (Fig. 2A). VP treatment significantly and rapidly reduced tumor multiplicity and tumor size compared to vehicle treatment (Fig. 2B and C). Histological analyses demonstrated that the tumors present in the VP-treated mice were minimally dysplastic adenomas, compared to more advanced adenomas in vehicle-treated mice (Fig. 2D). A significant decrease in the expression of YAP1-target genes (CTGF and CYR61) and in cell proliferation and an increase in apoptosis (assessed by cleaved caspase-3 abundance) were observed in tumors from VP-treated mice (Fig. 2E–G). However, VP treatment did not affect normal intestinal homeostasis as assessed by histological examination and Ki67 staining (Fig. 2H). In addition, kidney, liver, small intestine and colon tissues did not exhibit any apparent toxic effects upon VP treatment (fig. S2).
Fig. 2. Verteporfin treatment decreases colon tumors in a mouse model of colon cancer.
(A) Schematic representation of AOM/DSS CAC mouse model that were treated with 100 mg/kg verteporfin (VP) twice a week for three-weeks. (B to D) Representative image of tumors (B), colon length and number, size and volume of colon tumors (C), and H&E staining (D) of representative tumors from mice treated with vehicle (Veh) (n=9 mice/group) or VP (n=8). Scale bar, 50μm. (E and F) qPCR analysis of CTGF and CYR61 expression in normal and tumor colon tissues from vehicle- or VP-treated mice, normalized to mRNA encoding β-actin. Data are mean ± S.E.M (n=4 mice/group). (G and H) Immunostaining for the Ki67 or c-Caspase3 in (G) tumor and (H) normal colon tissues from mice treated with vehicle or VP. Data are mean ± S.E.M. *p < 0.05; **p<0.01; ***p<0.001 vs Veh. Scale bars, 50μm.
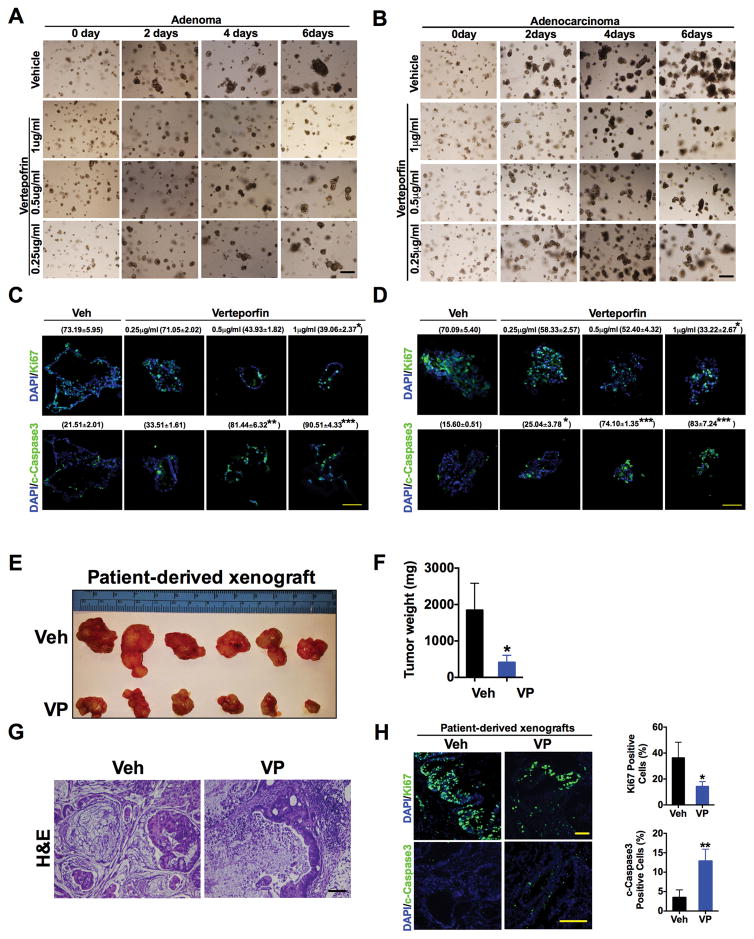
We further assessed the efficacy of VP to decrease cell proliferation in patient-derived colon adenoma and adenocarcinoma enteroid cultures and patient-derived xenograft models. Three-dimensional (3D) patient-derived enteroid culture systems recapitulate many relevant aspects of colon tumor growth, in addition to reflecting the phenotypic and genetic changes seen in primary patient tumors compared to cultured cell lines. Moreover, the enteroid cultures are enriched in presumptive tumor-initiating cells, which have been suggested to be resistant to chemotherapy- and radiation-based treatments (28). The growth of both adenoma and adenocarcinoma enteroid cultures decreased upon exposure to VP in a dose- and time-dependent manner (Fig. 3A–D). VP was further assessed in CRC patient-derived xenograft model. When tumors reached an average size of 150mm2 the mice were injected at 3-day intervals for 3 weeks with vehicle or VP. A significant decrease in tumor size, tumor weight, necrosis, and proliferation, and an increase in apoptosis were observed in xenografts from VP-treated mice (Fig. 3E–H). These results were consistent with the effects seen in CRC-derived cell lines and an additional xenograft model using the mouse CRC-derived CT-26 cells (fig. S3A–D). VP had a more potent response in activating apoptosis, as assessed by the abundance of cleaved PARP and cleaved caspase-3, compared to the effects on cell proliferation as assessed by MTT assay and analysis of PCNA and Ki67 in tumor enteroids and CRC-derived cell lines (Fig. 3C and D and fig. S3E–G). Together, the data suggest that the use of VP has substantial anti-tumor effects and minimal toxicity in vivo.
Fig. 3. Verteporfin decreases cell growth in patient-derived enteroid and xenograft models.
(A and B) Representative images of (A) adenoma or (B) adenocarcinoma patient-derived enteroids treated with verteporfin (VP) for the indicated doses and times. (C and D) Immunofluorescence staining and quantification (% positive staining ± S.E.M, n=3 enteroids of Ki67 and cleaved caspase3 in patient-derived (C) adenoma or (D) adenocarcinoma enteroids in the presence of vehicle (Veh) or VP for 6 days. (E to H) Size (E), weight (F), H&E staining (G), and proliferative and apoptotic analysis (H) of CRC patient-derived xenografts in mice treated with vehicle (n=6) or VP (n=8). Data are mean ± S.E.M of at least three independent experiments. *p < 0.05; **p<0.01; ***p<0.001 vs Veh. Scale bars, 50μm.
Verteporfin inhibits STAT3 signaling in colon tumors and colon tumor-derived cell lines
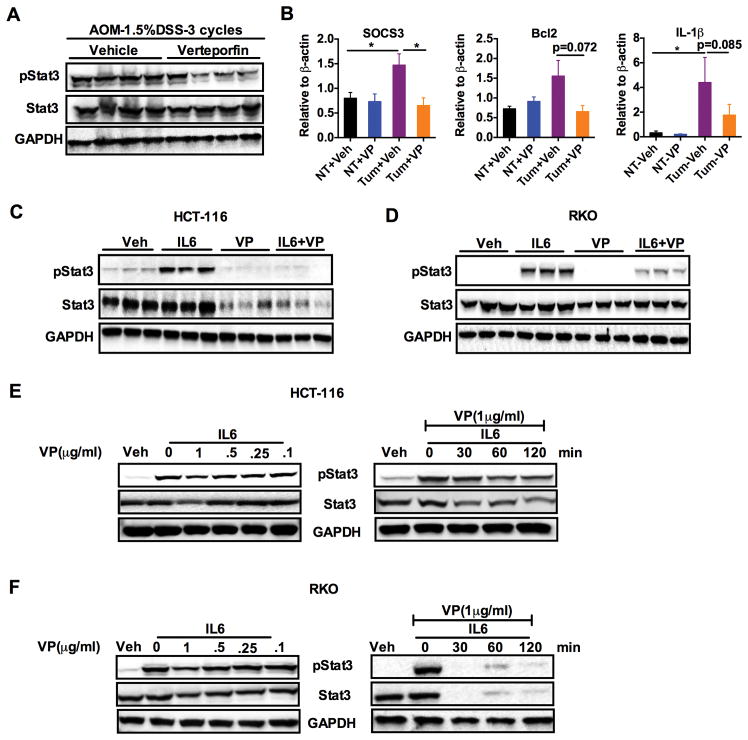
YAP1-dependent transcriptional activation is critical for colon tumor progression; therefore, we investigated what downstream mechanisms might be involved. Interestingly, we found that the phosphorylation of the pro-proliferative kinases such as AKT and ERK were significantly increased in tumors from AOM-DSS mice that were treated with VP (Fig 2A and fig. S4A). Decreased phosphorylation of STAT3 after VP treatment was also observed (Fig. 4A and fig. S4B). This is consistent with previous data demonstrating that increased nuclear abundance of YAP1 increases STAT3 phosphorylation (6). STAT3 signaling plays an important role in the survival and proliferation of intestinal epithelial cells in CRC (29, 30). Moreover, cross-talk between the IL-6 co-receptor gp130 and YAP1 was recently identified as a major mechanism leading to epithelial cell proliferation in the colon in response to inflammation (5, 31). Expression of STAT3-target genes Socs3, Bcl2 and Il-1β were decreased in tumor tissues after VP treatment (Fig. 4B). Furthermore IL-6-stimulated STAT3 phosphorylation was ablated after VP exposure in a panel of CRC-derived cells (Fig. 4, C and D, and fig. S4, C–E). In addition, VP also significantly decreased total STAT3 abundance in the cell lines studied (Fig. 4, C and D, and fig. S4, C–E), and a dose- and time- dependent decrease in IL-6-induced STAT3 signaling (Fig. 4, E and F, and fig. S4, F–H). Furthermore, VP significantly decreased both IL-6-mediated induction of a luciferase construct containing a STAT3 response element as well as induction of Socs3 in HCT-116 cells (fig. S4, I and J). The collective data suggest that VP may antagonize colon tumorigenesis by inhibiting STAT3 signaling.
Fig. 4. Verteporfin inhibits STAT3 signaling in colon tumors and colon-derived cell lines.
(A) Western analysis for phosphorylated and total STAT3 in the tumors of AOM-DSS treated mice from the vehicle and verteporfin group (n=4 mice/group). (B) qPCR analysis of Socs3, Bcl2 and Il-1β in normal tissue (NT) or tumor tissue (Tum) from vehicle (Veh) and verteporfin (VP)-treated AOM/DSS mice in. Expression was normalized to mRNA encoding β-actin. *p < 0.05 vs Veh. (C and D) Western analysis of phosphorylated and total STAT3 in (C) HCT-116 or (D) RKO that were pre-treated with vehicle or VP (1μg/ml) for 2 hours then treated with IL-6 (10ng/ml) for 20 min. (E and F) Time- and dose-dependent effects of VP on IL-6 induced STAT3 phosphorylation in (E) HCT-116 or (F) RKO cells. All cell line experiments were done in triplicates and repeated at least three times.
Verteporfin inhibits cell proliferation through YAP1-independent mechanisms
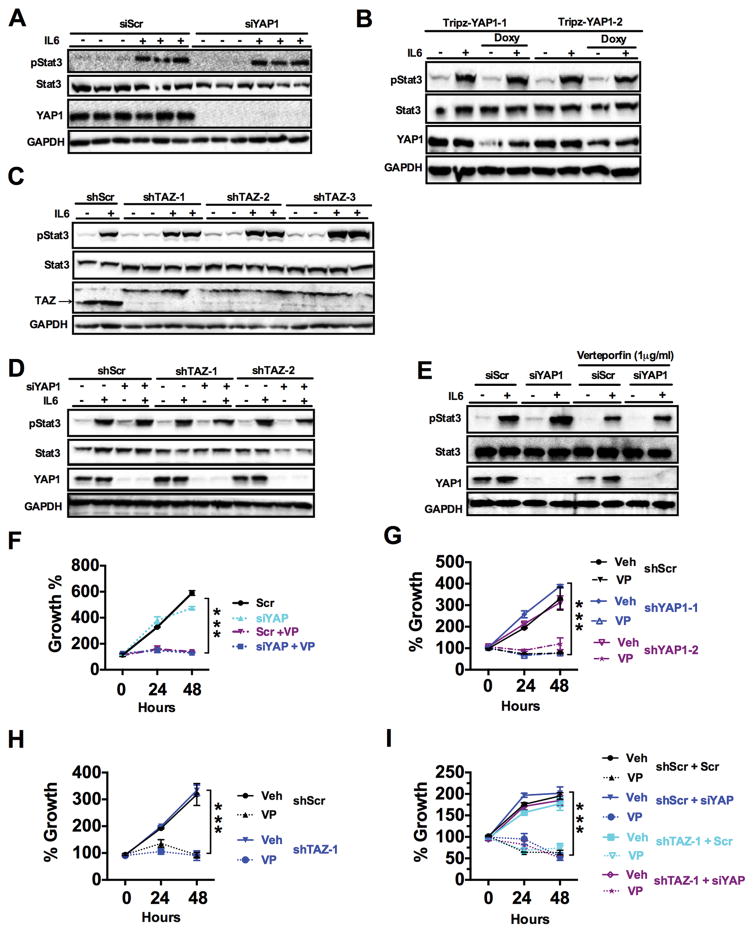
The anti-tumor role of VP has been previously reported to be through its inhibition of YAP1 signaling, independent of light activation (18, 26). Given the association of YAP1 and STAT3 (18, 26), this mechanism may occur through its inhibitory effects on YAP1. However, knockdown of YAP using siRNA or stable cells expressing two different doxycycline inducible shRNAs against YAP (TripZ-YAP1-1 and TripZ-YAP1-2), did not affect IL-6-induced phosphorylation of STAT3 in HCT-116 cells (Fig. 5, A and B, and fig. S5, A and B). It is possible that VP could also inhibit TAZ, a close homolog with redundant functions, which also binds to TEAD (13, 14). However, expression of shRNA against TAZ alone or with siRNA against YAP did not affect IL-6 induced STAT3 phosphorylation (Fig. 5, C and D, and fig. S5, C and D). Furthermore, VP repressed the IL-6-mediated phosphorylation of STAT3 even in the absence of YAP1 (Fig. 5E and fig. S5E). These data clearly demonstrate that inhibition of STAT3 signaling by VP is independent of YAP1. Further analysis revealed that VP could decrease cell proliferation in cells treated with a STAT3 inhibitor (fig. S5F), suggesting that VP does not inhibit cell proliferation solely by repressing STAT3 signaling. These data prompted us to further assess if the growth inhibitory role of VP was independent of YAP1. Surprisingly and in contrast to what has been previously reported (18, 25, 26, 32), VP decreased cell proliferation in colon cancer-derived cell lines in the absence of YAP1, TAZ, or YAP1 and TAZ (Fig. 5, F–I, and fig. S5G). Moreover, the growth inhibitory effects of VP were independent of YAP1 in breast, cervical, bone and liver cancer-derived cell lines (fig. S5H). Therefore, our data demonstrate VP reduces cell growth in cancer cells through a YAP1-independent mechanism.
Fig. 5. Verteporfin inhibits growth of CRC-derived cells independent of YAP1.
(A and B) Western blot analysis in HCT-116 cells transfected with (A) scrambled (siScr) or siRNA against YAP1 (siYAP1), (B) HCT-116 cells stably expressing doxycycline (Doxy)-inducible shRNA against YAP1 (clone 1 and 2). Cells were untreated or exposed to IL-6 (10 ng/ml, 20 min). (C and D) Western blotting in lysates from HCT-116 cells stably expressing (C) control (shScr) or shRNA against TAZ (shTAZ clone 1-3) or (D) shTAZ (clone 1 and 2) and either siScr or siYAP and treated with IL-6 (10 ng/ml, 20 min). (E) Western blot analysis in HCT-116 cells transfected with siScr or siYAP and treated with 1μg/ml verteporfin (VP) then 10 ng/ml IL-6 for 20 min. (F) MTT assay in HCT-116 cells transfected with siScr or siYAP and treated with vehicle or VP (1μg/ml). (G to I) MTT assay in 1μg/ml VP-treated HCT-116 cells stably expressing (G) doxycycline-inducible shYAP, (H) shTAZ, or (I) shTAZ and siScr or siYAP. Data are mean ± S.E.M of at least three independent experiments, each done in triplicate. ***p<0.001 vs Veh.
Verteporfin increases apoptosis by inducing high molecular weight oligomerization of proteins
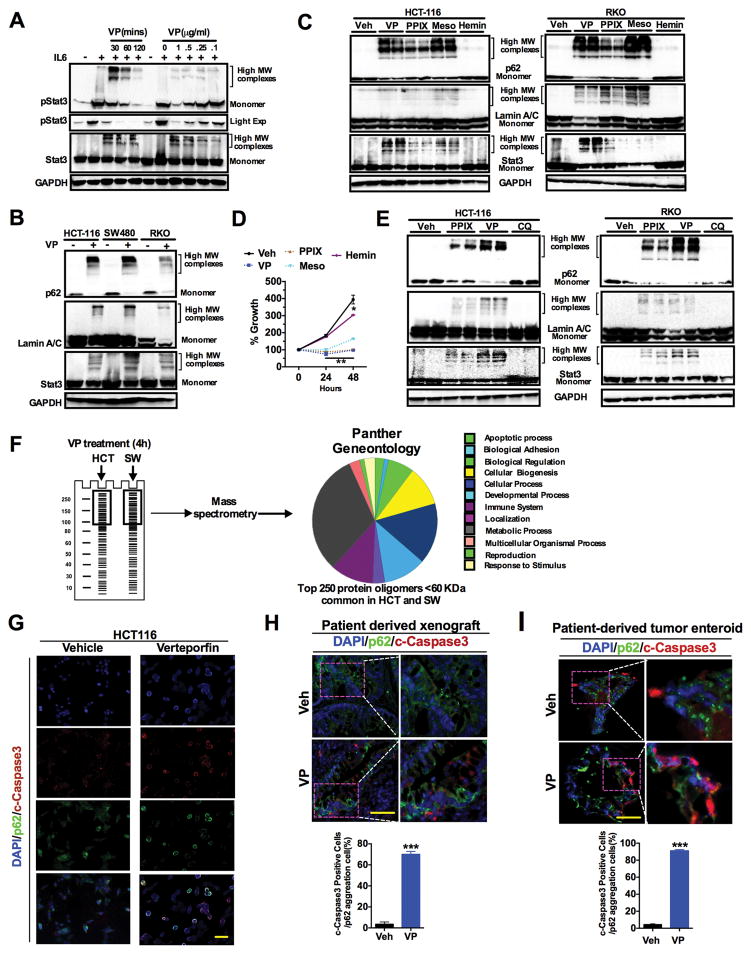
Besides its reported effects on YAP1, VP has been shown to inhibit autophagy (32, 33). Autophagy is a critical mechanism in many tumors for nutrient recycling and growth, and autophagy inhibitors have been shown to decrease CRC growth (34–36). To assess whether VP decreases CRC cell growth through inhibition of autophagy, the well-characterized autophagy inhibitors bafilomycin A1 or chloroquine were used alone or in conjunction with VP in cultures of HCT-116 cells. Treatment with bafilomycin A1 or chloroquine along with VP did not significantly alter growth compared to VP alone (fig. S6, A and B). Structurally, VP belongs to the porphyrin class of drugs. Intracellular porphyrins can cross-link proteins, leading to formation of high-molecular weight oligomers of proteins (33, 37). Therefore, to assess whether the loss of STAT3 signaling by VP was due to oligomerization of STAT3, HCT-116 cells were treated with VP in a dose- and time-dependent manner. VP treatment led to a shift in the molecular weights of both phosphorylated and total STAT3 toward less monomer abundance at the higher VP concentrations or incubation periods (Fig. 6A and fig. S6C). Similarly, proteins such as p62 and LaminA/C that oligomerize upon porphyrin treatment were also found to form protein oligomers after VP treatment (Fig. 6B and fig. S6D). Although YAP1 protein decreased by VP treatment, we could not detect high molecular oligomers of YAP1, possibly because the YAP1 epitope was masked among the protein oligomers. In addition, we also observed oligomerization of p62, STAT3 and LaminA/C in the lysates from VP treated patient-derived xenografts (fig. S6E). The VP structural analogues protoporphyrin IX (PPIX) and mesoporphyrin (Meso) also caused protein oligomerization, whereas another analogue hemin did not induce protein oligomers (Fig. 6C and fig. S6F). Protein oligomerization strongly correlated inhibition of cell proliferation, because exposure to VP, PPIX or Meso, but not hemin, resulted in significantly decreased cell proliferation (Fig. 6D). Protein oligomerization by VP was associated with induction of endoplasmic reticulum (ER) stress markers in HCT-116 cells in a time- and dose-dependent manner (fig. S6G). However, pretreatment with tauroursodeoxycholic acid (TUDCA), a chemical chaperone that reduces ER stress (38), did not protect HCT-116 cells from VP-mediated decrease in cell proliferation (fig. S6H). Inhibition of autophagy by chloroquine did not induce the formation of protein oligomers, demonstrating that the oligomerization of proteins by VP is independent of its function as an autophagy inhibitor, but rather due to cross-linking of protein (Fig. 6E and fig. S6I) (32, 33, 35).
Fig. 6. Verteporfin-induced protein oligomerizations causes apoptotic cell death.
(A) Abundance of monomers and high molecular weight (MW) oligomers of phosphorylated and total STAT3 in response to 10 ng/ml IL-6 in HCT-116 cells pretreated with VP (left, 1μg/ml VP for indicated times; right, treatment for 2 hours). (B) Abundance of monomers and high molecular weight (MW) oligomers of p62, Lamin A/C and STAT3 in CRC cell lines treated with 1μg/ml VP for 2 hours. (C and D) HCT-116 and RKO cells were exposed for 2 hours to vehicle (Veh), 1μg/ml verteporfin (VP), 5 μM protoporphyrin IX (PPIX), 5 μM mesoporphyrin (Meso), or 30 μM hemin. Both cell lines were then assessed for high MW oligomers or monomers by Western blot (C) and HCT-116 cells were additionally assessed for proliferation (D). (E) As in (C), also in response to 2 hours exposure to 100 μM chloroquine (CQ). (F) Identification and classification of VP-induced oligomers in HCT-116 (HCT) and SW480 (SW) by mass-spectrometry and Panther Gene Ontology. (G to I) Co-immunostaining for cleaved caspase 3 and p62 in (G) HCT-116 cells treated with 1μg/ml VP for 4 hours, (H) patient-derived colon tumors from Fig. 3E,, and (I) patient-derived enteroids exposed to 1μg/ml VP for 8 hours. Scale bar, 50μm. Data are mean value ± S.E.M of at least three independent experiments, each performed in triplicate. Blots and images are representative of three independent experiments. *p < 0.05; **p<0.01 ***p<0.001 vs Veh.
To further understand the mechanism by which VP decreases cell growth, a proteomic approach to globally assess VP-induced high-molecular weight oligomers in HCT-116 and SW480 cells was carried out. High-molecular weight proteins (>100kD) were isolated, and proteins that were identified with the predicted size less than 60 kD were considered as VP-induced high-molecular weight oligomerized proteins (Fig. 6F). The common proteins identified in both HCT-116 cells and SW480 are listed in table S1. p62 and LaminA/C were identified in the proteomic approach, thus increasing the confidence that this method could identify novel VP-induced oligomers. Interestingly, numerous proteins involved in various cellular processes such as apoptosis, signaling, bioenergetics, structural proteins and cellular biogenesis were present in the high-molecular weight fraction upon treatment of HCT-116 and SW480 cells with VP (Fig. 6F). To assess whether cells with increased VP-induced oligomerized protein could be identified by immunofluorescence, p62 was visualized after VP treatment in HCT-116 cells. In untreated cells, p62 diffusely stained throughout the cytosol, whereas p62 protein was highly increased in the perinuclear region in response to VP treatment (fig. S6J). To assess whether VP-induced apoptosis was caused specifically by protein oligomerization, dual staining for cleaved caspase3 (a marker of apoptosis) and p62 (a marker of oligomerization) was performed in HCT-116 cells, human enteroid cultures, and patient-derived xenografts (Fig. 6, G–I). Over 70% of the cells were co-stained for p62 oligomerization and cleaved caspase3 in VP-treated colon tumor mouse models. Together the data suggests that protein oligomerization by VP is associated with apoptotic cell death.
Tumors exhibit impaired clearance of VP-induced protein oligomers resulting in apoptotic cell death
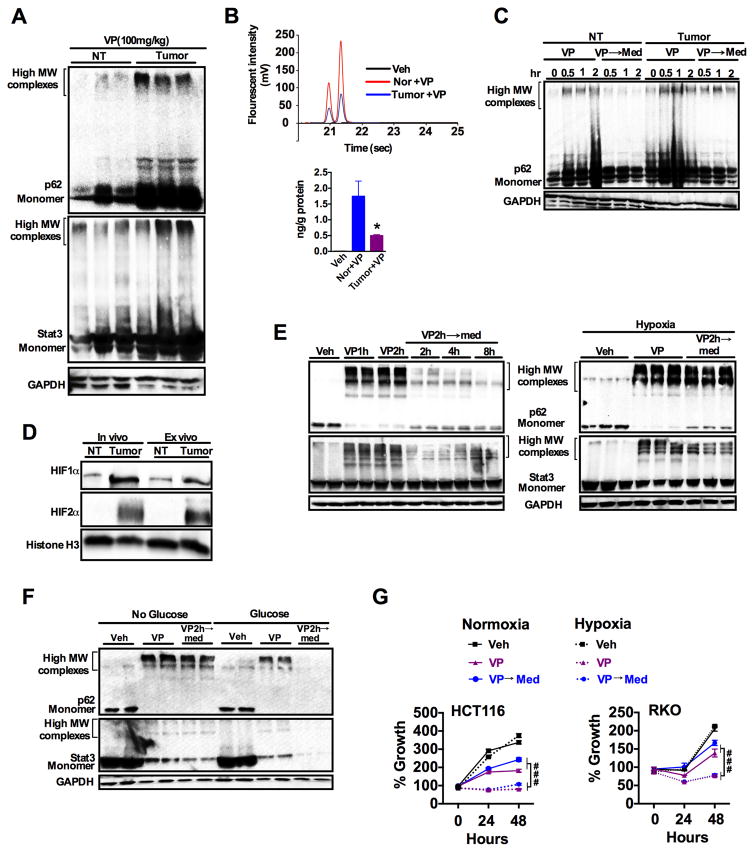
VP has very low toxicity, and in vivo treatment resulted in decreased tumor growth without affecting normal tissues (39, 40). To understand the mechanism by which VP elicits tumor-specific growth inhibition, mice with AOM/DSS-induced colon tumors were treated with VP then assessed for protein oligomerization. Notably, VP treatment induced significantly greater oligomerization of p62 and STAT3 in the tumor tissue compared to adjacent normal tissue (Fig. 7A and fig. S7A). Previous work has demonstrated that oxidative stress after exposure to porphyrins could lead to increased protein oligomerization (41). However, protein oligomers induced by VP was only slightly attenuated by the antioxidants N-acetyl cysteine (NAC), butylated hydroxyanisole (BHA) and sodium azide (NaN3) (fig. S7, B and C). To assess whether there was a difference in VP uptake between the tumor and normal adjacent tissue, free intracellular VP was assessed by HPLC. Verteporfin has two regioisomers, which eluted at 21 and 22 seconds, respectively. Surprisingly, the abundance of the free form of VP was significantly less in tumors than in adjacent normal tissue (Fig. 7B). This may be explained by increased VP in oligomerized proteins in tumors, but clearly demonstrates that normal tissues take up VP. We then assessed whether the protein oligomer clearance mechanism is impaired in tumors using explants of the tumor and adjacent normal tissue that were then treated with VP ex vivo, followed by replacing the media without VP (washout period). Western blot analysis revealed effective clearance of high-molecular p62 protein oligomers in normal tissue but not in tumors (Fig. 7C and fig. S7D). Interestingly, unlike in vivo tumors, colon-cancer derived cell lines were able to effectively clear the high molecular weight oligomers (fig. S7E). However, inhibition of proteasome (MG-132) and autophagy pathways using early autophagy inhibitor (Wortmannin) or late autophagy inhibitor (Bafilomycin) impaired the clearance of protein oligomers after a washout period (fig. S7E).
Fig. 7. Tumor selective apoptosis by verteporfin is associated impaired clearance of protein oligomers.
(A) Western blot analysis of p62 and STAT3 in normal colon tissue (NT) and colon tumor tissue from mice treated intraperitoneally with verteporfin (VP, 100 mg/kg) for 4 hours (n=3 mice/group). (B) Intracellular amount of VP in tumor and adjacent normal colon tissues after 4 hours VP treatment (n=3 samples/group). *p < 0.05 vs normal. (C) Western blot analysis of p62 in the normal and tumor colon tissues treated ex vivo with VP (1μg/ml), followed by refreshing with DMEM media (→ Med) for indicated time (hr, hours). (D) Western blot analysis for HIF1α and HIF2α in normal (NT) and tumor tissues either freshly isolated from mice (in vivo) or after culturing for 4 hours (ex vivo). (E and F) Abundance of p62 and STAT3 high-molecular weight oligomers in HCT-116 cells exposed to VP (1μg/ml; h, hours) followed by a washout period in (E) normoxic (21% O2) or hypoxic (1% O2) culture conditions or (F) in the presence or absence of glucose. (G) MTT assays in HCT-116 and RKO cells exposed to VP in normoxic or hypoxic culture conditions. Data are mean ± S.E.M.; ###p<0.001 vs hypoxic cells. All cell line experiments were done in triplicates and repeated at least three times.
In vivo, tumors grow in nutrient-deprived and hypoxic environments; ex vivo, although grown in nutrient-rich media, tumors still exhibit significant hypoxia as assessed by increased abundance of the hypoxic transcription factors hypoxia-inducible factor (HIF)1α and HIF2α (Fig. 7D and fig. S7F). Tumors in nutrient-deprived or hypoxic conditions rely heavily on recycled and scavenged nutrients and metabolites through proteasomal and autophagic turnover (34–36, 42). The role of hypoxia and nutrient deprivation in the clearance of protein oligomers was assessed in cultured cells. HCT-116 cells were treated with VP for 2 hours followed by a washout period for up to 8 hours, under normoxic or hypoxic conditions. Normoxic HCT-116 cells cleared the protein oligomers rapidly compared to hypoxic cells (Fig. 7E and fig. S7G). Similarly, glucose deprivation decreased the clearance of VP-induced protein oligomers (Fig. 7F and fig. S7H). Consistent with the clearance of high molecular weight oligomers, the washout period partially restored cell proliferation in HCT-116 and RKO rectal carcinoma cells (Fig. 7G). However, hypoxic cells were highly susceptible to a decrease in cell proliferation after exposure to VP (Fig. 7G). Replacing the cells in normal media without VP did not restore their proliferation in hypoxic conditions (Fig. 7G). Similar studies could not be performed in glucose-deprived media, because the cells could not be cultured for time points amenable for growth assays.
Protein oligomerization is a critical mechanism in VP-induced growth inhibition
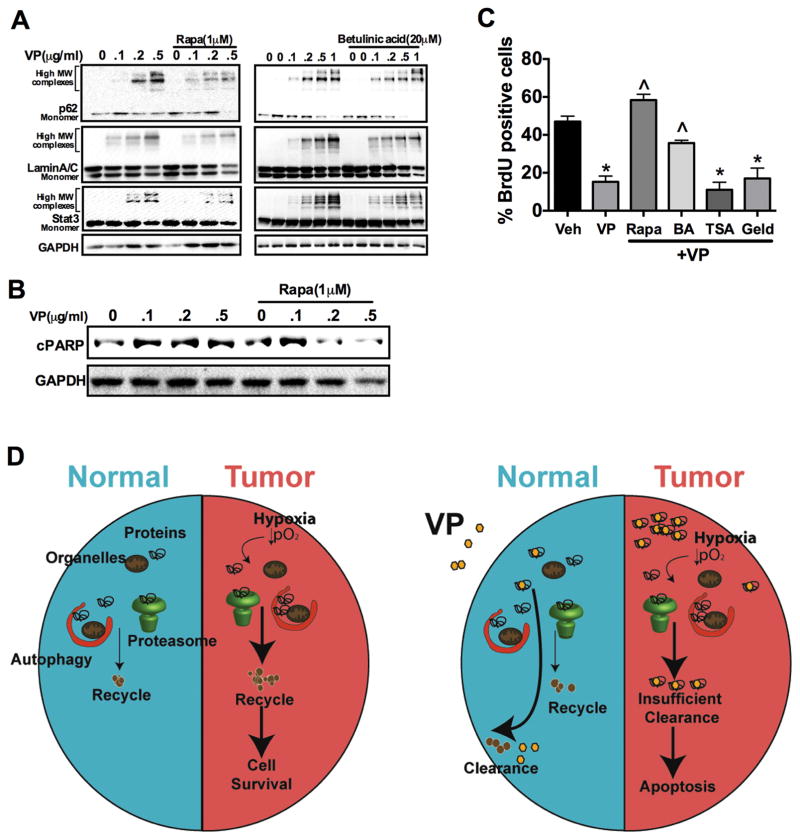
To further establish a cause-effect relationship between protein oligomerization and growth inhibition by VP, we determined whether decreasing VP-induced protein oligomers could protect cells from VP-mediated inhibition of proliferation. Pretreating HCT-116 cells with the autophagy activator (rapamycin) or proteasome activator (betulinic acid) decreased the abundance of protein oligomers after cells were exposed to VP (Fig. 8A and fig. S8A). However, rapamycin and betulinic acid could not resolve VP-induced oligomers after longer-term VP treatment (fig. S8B), suggesting that VP oligmerization may eventually saturate the clearance pathways. Moreover, treatment with proteasomal regulators such as trichostatin A (a histone deacetylase inhibitor) or geldanamycin (a heat shock protein 90 inhibitor) did not prevent VP-induced protein oligomerization (fig S8C), possibly because of the weak regulation of proteasome activity by these regulators compared to that of betulinic acid. Because activation of autophagy and proteasome pathways resolved acute VP-induced oligermization, we assessed early parameters of cell proliferation to confirm that protein oligomerization is a major mechanism by which VP decreases cell proliferation. Decreasing the abundance of protein oligomers using rapamycin or betulinic acid prevented the effects of VP on apoptosis (assessed by cleaved PARP abundance) and cell proliferation (Fig. 8, B and C, and fig. S8D). Collectively, the data suggests that normoxic cells in a nutrient-rich environment could efficiently use the autophagy and proteasomal machinery to clear VP cross-linked proteins. However, hypoxic or nutrient-deprived tumor cells, which depend on autophagic and proteasomal pathways for nutrient supply, fail to clear VP cross-linked protein oligomers resulting in apoptotic cell death and cancer regression (Fig. 8D).
Fig. 8. Schematic representations summarizing the tumor-specific cytotoxicity induced by verteporfin.
(A) Western blot analysis of p62, LaminA/C and STAT3 in HCT-116 cells that were pretreated with 1μM rapamycin (right) or 20μM betulinic acid (left) for 16 hours then verteporfin (VP) for 90 min. (B) Western blot analysis of cleaved PARP (cPARP) in HCT-116 cells pretreated with 1μM rapamycin (Rapa) for 16 hours then VP for 6 hours. (C) Cell proliferation assessed by BrdU staining in HCT-116 cells pretreated with vehicle, 1μM Rapa, 20μM betulinic acid (BA), 1μM TrichostatinA (TSA) or 1μM geldanamycin (Geld) for 16 hours followed by treatment with VP for 90 min. Experiments were done in triplicate and repeated twice. *p < 0.05 vs Veh; ^p < 0.05 vs VP alone. (D) In normal cells, VP-induced protein oligomers are efficiently cleared through autophagy and proteasomal degradation. However, hypoxic tumor cells which highly rely on autophagy and proteasomal machinery for nutrients and biomolecules fail to clear the protein oligomers resulting in apoptotic cell death.
DISCUSSION
VP photodynamic therapy is frequently used as a first line therapy for AMD, and a meta-analysis has demonstrated that VP photodynamic therapy is an efficacious treatment for preventing vision loss in patients (39). Currently anti-angiogenic treatments targeting vascular endothelial growth factors are the most common therapy for neovascular AMD (43). However, the characterization of VP as a YAP1 transcriptional inhibitor has revived interest in VP’s potential use as a potential therapy for cancer. VP in many different tumor models potently limits tumor growth (18, 25, 26). Consistent with these reports, the data presented here expand these findings to CRC. Using CRC-derived cell lines, patient-derived enteroids and xenografts, and a mouse model of CRC, we demonstrate that VP exerts its tumor inhibitory effects via inhibition of cell proliferation and induction of apoptosis. Importantly, our studies demonstrate that growth inhibition after VP treatment was independent of YAP1 or the related Hippo effector TAZ. The present work identifies a VP-induced mechanism that is toxic specifically to tumors, initiating cell death by inducing high-molecular weight oligomerization of numerous proteins involved in key cellular pathways.
A potential generalized mechanism for VP inhibition of CRC growth through high molecular protein oligomers
VP and other related porphyrin molecules induce high molecular oligomers (33, 37). However, hemin, which also inhibits YAP1, could not form protein oligomers (18). We show that the VP-induced cell death correlates with the ability of porphyrins to induce high-molecular weight protein oligomers. In addition, cells with cytosolic protein oligomers in colon-cell lines, CRC enteroids and xenografts exhibited an increase in markers of apoptosis and decreasing oligomers by activating autophagy and proteasome pathways further support protein oligomer-induced proteotoxicity as an important mechanism by which VP induces cell death. Porphyrin-related molecules induce oligomerization through a direct non-enzymatic crosslinking of proteins forming monomeric high-weight oligomers potentiated by oxidative stress (33, 37). Through proteomic analysis, we identified VP to oligomerize nearly 250 proteins involved in various biological pathways in two different colon cancer-derived cell lines. VP-induced protein oligomerization appears to be a selective phenomenon observed with specific proteins such as p62, STAT3 and LaminA/C. Further efforts will be needed to better understand protein features and possible motifs that are critical in initiating oligomerization and to identify specific proteins and mechanisms that are essential for proteotoxicity-induced apoptosis.
Impaired clearance of protein oligomers contributes to tumor-selective cell death in CRC
Colon tumors are exquisitely sensitive to VP-induced cell death. Cancer-derived cell lines in culture and normal tissues are relatively less energetically stressed, and do not rely on autophagic or proteasomal recycling for their key nutrients. Therefore, they can efficiently utilize these pathways to remove high-molecular weight protein oligomers. However, when colon cancer cell lines are energetically stressed with hypoxia or glucose deprivation, mimicking the conditions of tumors in vivo, high-molecular weight protein oligomers are not cleared efficiently. Rescuing cell growth by decreasing protein oligomers through activation of autophagy or proteasome in cells treated with VP suggests that VP inhibits cell growth partly by inducing high molecular oligomerization of key proteins involved in various biological processes. The inability to clear protein oligomers correlates to increased cell death. Cancer cells heavily depend on the autophagic and proteasomal pathways to scavenge nutrients that are essential for their growth and survival (35, 36, 42). Therefore, re-routing of autophagy and proteasomal pathways to promote clearance of oligomers results in nutrient deprivation of cancer cells. On the other hand, failure to clear the protein oligomers results in persistent accumulation of toxic protein oligomers. Thus, the compromised cellular function both energetically and in the clearance of protein oligomers results in tumor-specific increase in apoptotic cell death by VP (Fig. 9).
VP is a critical tool for understanding the role of YAP1 signaling in cancer (18, 26). The present data clearly identifies a novel mechanism of action for VP in inhibiting cancer growth that is independent of its previously known photoactivation or YAP1 inhibition. Our data demonstrating that VP treatment leads to tumor specific proteotoxicity provides an alternative treatment strategy for CRC. Moreover, the present work provides evidence that VP might have potential for treatment approaches in some patients diagnosed with CRC.
Material and Methods
Animals and treatments
C57BL/6 mice were used and maintained in standard cages in a light- and temperature-controlled room, and were fed with standard chow and water ad libitum. For the azoxymethane (AOM)/dextran sulfate sodium (DSS) tumor model, six-week-old mice were injected intraperitoneally with AOM (10 mg/kg body weight). One week after AOM injection, mice were placed on 1.5% DSS in drinking water for 7-days (inflammatory phase) and were given regular drinking water for 14-days (recovery phase) and repeated three times. Mice were assessed four-weeks after the last cycle of DSS treatment. For VP treatment (USP reference standard, Rockville, MD), VP dissolved in DMSO was diluted using PBS and injected intraperitoneally at 100mg/kg as described in the figure legends. Control mice were injected with DMSO diluted with PBS. All the animal studies were carried out in accordance with the Institute of Laboratory Animal Resources guidelines and approved by the University Committee on the Use and Care of Animals at the University of Michigan.
Cell culture and proliferation assays
Cells were maintained in DMEM medium (supplemented with 10% fetal bovine serum and 1% antibiotic/antimycotic agent) at 37°C in 5% CO2 and 21% O2. For hypoxia experiments, cells were incubated in a hypoxia chamber (5% CO2 and 1% O2) at 37°C. For glucose-deprivation, cells were cultured in DMEM without glucose, supplemented with 0.1% BSA and 1% antibiotic/antimycotic. For inhibitory RNA (RNAi) experiments, HCT-116 cells were transfected with 25nM scrambled siRNA or siRNA against YAP1 (Dharmacon, Lafayette, CO), using lipofectamine 2000 (Invitrogen), or stable shRNA-expressing cells were generated using the doxycycline inducible pTRIPZ vector in which scrambled, or two shRNAs against YAP1 were cloned (kind gifts from Dr. Kun-Liang Guan) (24). Stable cells for TAZ knockdown were generated using pGIPZ vector cloned with scrambled or three independent shRNAs against TAZ. Growth assays were performed using 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) reagent.
Western blot analysis
Western blot analyses were performed as previously described (44). All antibodies used were from Cell Signaling Technology Inc. (Danvers, MA) except for LaminA/C (Active Motif, Carlsbad, CA), HIF-2 (Novus Biologicals, Littleton, CO), HIF-1, p62 and GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA),
Meta-analysis of CRC samples
CRC gene expression datasets along with the patient survival were identified in GEO using the search keywords “colon”, “cancer”, and “microarray” (http://www.ncbi.nlm.nih.gov/geo/). Only publications providing raw data, clinical survival information, and containing at least 30 patients were included. The gene chips were MAS 5.0 normalized in the R statistical environment (http://www.R-project.org) using the Bioconductor package Affy (http://www.bioconductor.org). Survival analysis using Cox proportional hazards regression was performed as described previously (45). The most reliable probe sets for each gene were selected using Jetset. Kaplan-Meier survival plots were generated using WinStat for Excel (Robert K Fitch Software, Germany).
Proteomic analysis of oligomers
Cells were treated with 1μg/ml VP for 2 hours, and protein lysates were separated on SDS-PAGE. The proteins that resolved at molecular weight higher than 100 kD were sequenced using the Proteomic/Mass-spectrometry core at the University of Michigan. Mass spec data were searched against Uniprot human protein database using Proteome Discoverer (v 1.4) with the following search parameters: Precursor and product ion tolerance window was set at 10ppm and 0.8Da. Carbamidomethylation of cysteine was considered a fixed modification. Oxidation of methionine, phosphorylation of serine, threonine and tyrosine, and ubiquitination of lysine were considered as dynamic modifications. Percolator was used to filter the data at <1% false discovery rate. The identified proteins that are smaller than 60 kD molecular weight but present in the high-molecular weight fraction were analyzed using Panther Geneontology tool. Data are available via ProteomeXchange with identifier PXD002793.
Patient-derived CRC enteroid cultures
Enteroids derived from a large adenoma and an adenocarcinoma were established as described before (46). Two-days after plating, enteroids were treated with VP and the growth was monitored.
Mouse and patient-derived xenograft models
The patient-derived colon xenograft model UM-CRC 13-1333 was established from a biospecimen procured from a patient undergoing resection of primary disease located in the sigmoid colon. The UM-CRC 13-1333 model has both a KRAS (G12D) as well as a PIK3CA (Q546L) mutation (J. S. Sebolt-Leopold, personal correspondence). Efficacy studies with VP were carried out in female NCR nude mice (CrTac:NCr-Foxn1nu 5-6-weeks of age from Taconic) implanted subcutaneously. Once tumors reached 150mm2 (30-days), 100mg/kg VP was injected intraperitoneally at 3-day intervals until the vehicle-treated tumors doubled in size (21-days). Colon cancer CT-26 cells were injected subcutaneously at 6 x 106 cells in aythmic nude mice and once tumors reached 100mm2 (7 days), 100mg/kg VP was injected intraperitoneally daily until the vehicle-treated tumors doubled in size (7 days). In both xenograft models, mice were euthanized 4 hours after the last injection.
High Performance Liquid Chromatography (HPLC) analysis
VP was measured at 430 nm (excitation) and 690 nm (emission). The column (equilibrated for 10min) was eluted at a flow rate of 1.0 ml/min with linear gradients of solvents A and B (A, 0.05mM monobasic sodium phosphate, pH3.5 in water; B, methanol). The solvent gradient was as follows: 0 to 4 min, 50–65% B; 4 to 8 min, 65–85% B; 8 to 22 min, 85–99% B; 22 to 27 min, 99–10% B; 27 to 30 min, 10–50% B. Cell/tissue lysates were diluted 10 folds in HPLC injection solvent (50% methanol, 50% 0.05mM monobasic sodium phosphate, pH3.5) and injected. Concentration of VP in the experimental samples were calculated from the standard curve of commercially obtained VP and normalized to the protein content of the tissue.
Histology and immunofluorescence
Formalin fixed sections were stained with hematoxylin and eosin (H&E) and frozen sections were used for immunofluorescence analysis as previously described (44). Primary antibodies against KI67 (1:100; Vector Laboratories. Inc., Burlingame, CA), cleaved caspase3 (1:200, Cell Signaling Technology), and p62 (1:100, Santa Cruz) were used.
Quantitative real-time RT-PCR
RNA extraction, reverse transcription and quantitative PCR (qPCR) were described previously (44). The primers used in the study are listed in Table S2.
Statistical analysis
Results are expressed as mean ± SD or mean ± SEM. Significance between two groups were calculated by Independent t-test and significance among different groups were tested using one-way analysis of variance followed by Dunnett’s post-hoc comparisons.
Supplementary Material
Fig. S1: Yap signaling is activated in human colon cancer cell lines and human colon cancer tissues
Fig. S2: Verteporfin treatment does not induce toxicity in normal tissues
Fig. S3: Verteporfin treatment decreases growth in CT-26 xenograft model and colon cancer cell lines
Fig. S4: Verteporfin treatment inhibits STAT3 signaling
Fig. S5: Growth inhibitory effect of verteporfin is YAP independent
Fig. S6: Growth inhibitor effect of verteporfin is associated with oligomerization of proteins
Fig. S7: Hypoxia and nutrient deprivation impairs clearance of VP induced protein oligomers
Fig. S8: Activating autophagy and proteasomal pathways partially clears protein oligomers
Table S1: Proteins that oligomerized in response to verteporfin
Table S2: List of primers.
Acknowledgments
We thank Dr. Kun-Liang Guan for sharing the doxycycline-inducible YAP1 shRNA constructs and Dr. Venkatesha Basrur for assistance with the mass spectrometry analysis. Organoid cultures of colonic neoplasia were provided by the Enteroid Core, a component of the GI SPORE Molecular Pathology and Biosample Core.
Funding: This work was supported by NIH grants: CA148828 and DK095201 to Y.M.S; CA181855-01 to J.V.; CA046592-26S3 to M.S.W.; DK52951 to M.B.O.; the University of Michigan Gastrointestinal Peptide Research Center DK034933; and GI SPORE CA130810. B.G. was supported by the OTKA K108655 grant. S.K.R was supported by a postdoctoral fellowship from the American Heart Association (15POST22650034).
Footnotes
This manuscript has been accepted for publication in Science Signaling. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencesignaling.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References and Notes
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Fearon ER. Molecular genetics of colorectal cancer. Annual review of pathology. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 3.Itzkowitz SH, Yio X. Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. American journal of physiology Gastrointestinal and liver physiology. 2004;287:G7–17. doi: 10.1152/ajpgi.00079.2004. [DOI] [PubMed] [Google Scholar]
- 4.Avruch J, Zhou D, Bardeesy N. YAP oncogene overexpression supercharges colon cancer proliferation. Cell cycle. 2012;11:1090–1096. doi: 10.4161/cc.11.6.19453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai J, Zhang N, Zheng Y, de Wilde RF, Maitra A, Pan D. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes & development. 2010;24:2383–2388. doi: 10.1101/gad.1978810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou D, Zhang Y, Wu H, Barry E, Yin Y, Lawrence E, Dawson D, Willis JE, Markowitz SD, Camargo FD, Avruch J. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E1312–1320. doi: 10.1073/pnas.1110428108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- 8.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes & development. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Current biology: CB. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 10.Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Developmental cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Developmental cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, Bardeesy N. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azzolin L, Panciera T, Soligo S, Enzo E, Bicciato S, Dupont S, Bresolin S, Frasson C, Basso G, Guzzardo V, Fassina A, Cordenonsi M, Piccolo S. YAP/TAZ incorporation in the beta-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158:157–170. doi: 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 14.Azzolin L, Zanconato F, Bresolin S, Forcato M, Basso G, Bicciato S, Cordenonsi M, Piccolo S. Role of TAZ as mediator of Wnt signaling. Cell. 2012;151:1443–1456. doi: 10.1016/j.cell.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 15.Rosenbluh J, Nijhawan D, Cox AG, Li X, Neal JT, Schafer EJ, Zack TI, Wang X, Tsherniak A, Schinzel AC, Shao DD, Schumacher SE, Weir BA, Vazquez F, Cowley GS, Root DE, Mesirov JP, Beroukhim R, Kuo CJ, Goessling W, Hahn WC. beta-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell. 2012;151:1457–1473. doi: 10.1016/j.cell.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barry ER, Morikawa T, Butler BL, Shrestha K, de la Rosa R, Yan KS, Fuchs CS, Magness ST, Smits R, Ogino S, Kuo CJ, Camargo FD. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2013;493:106–110. doi: 10.1038/nature11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li VS, Clevers H. Intestinal regeneration: YAP-tumor suppressor and oncoprotein? Current biology: CB. 2013;23:R110–112. doi: 10.1016/j.cub.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 18.Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, Liu JO, Pan D. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes & development. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziemssen F, Heimann H. Evaluation of verteporfin pharmakokinetics--redefining the need of photosensitizers in ophthalmology. Expert opinion on drug metabolism & toxicology. 2012;8:1023–1041. doi: 10.1517/17425255.2012.701617. [DOI] [PubMed] [Google Scholar]
- 20.Feng X, Degese MS, Iglesias-Bartolome R, Vaque JP, Molinolo AA, Rodrigues M, Zaidi MR, Ksander BR, Merlino G, Sodhi A, Chen Q, Gutkind JS. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer cell. 2014;25:831–845. doi: 10.1016/j.ccr.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang N, Hjorth-Jensen K, Hekmat O, Iglesias-Gato D, Kruse T, Wang C, Wei W, Ke B, Yan B, Niu Y, Olsen JV, Flores-Morales A. In vivo quantitative phosphoproteomic profiling identifies novel regulators of castration-resistant prostate cancer growth. Oncogene. 2014 doi: 10.1038/onc.2014.206. [DOI] [PubMed] [Google Scholar]
- 22.Perra A, Kowalik MA, Ghiso E, Ledda-Columbano GM, Di Tommaso L, Angioni MM, Raschioni C, Testore E, Roncalli M, Giordano S, Columbano A. YAP activation is an early event and a potential therapeutic target in liver cancer development. Journal of hepatology. 2014;61:1088–1096. doi: 10.1016/j.jhep.2014.06.033. [DOI] [PubMed] [Google Scholar]
- 23.Song S, Ajani JA, Honjo S, Maru DM, Chen Q, Scott AW, Heallen TR, Xiao L, Hofstetter WL, Weston B, Lee JH, Wadhwa R, Sudo K, Stroehlein JR, Martin JF, Hung MC, Johnson RL. Hippo coactivator YAP1 upregulates SOX9 and endows esophageal cancer cells with stem-like properties. Cancer research. 2014;74:4170–4182. doi: 10.1158/0008-5472.CAN-13-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu FX, Luo J, Mo JS, Liu G, Kim YC, Meng Z, Zhao L, Peyman G, Ouyang H, Jiang W, Zhao J, Chen X, Zhang L, Wang CY, Bastian BC, Zhang K, Guan KL. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer cell. 2014;25:822–830. doi: 10.1016/j.ccr.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huggett MT, Jermyn M, Gillams A, Illing R, Mosse S, Novelli M, Kent E, Bown SG, Hasan T, Pogue BW, Pereira SP. Phase I/II study of verteporfin photodynamic therapy in locally advanced pancreatic cancer. British journal of cancer. 2014;110:1698–1704. doi: 10.1038/bjc.2014.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brodowska K, Al-Moujahed A, Marmalidou A, Meyer Zu Horste M, Cichy J, Miller JW, Gragoudas E, Vavvas DG. The clinically used photosensitizer Verteporfin (VP) inhibits YAP-TEAD and human retinoblastoma cell growth in vitro without light activation. Exp Eye Res. 2014;124:67–73. doi: 10.1016/j.exer.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng Y, Sentani K, Wiese A, Sands E, Green M, Bommer GT, Cho KR, Fearon ER. Sox9 induction, ectopic Paneth cells, and mitotic spindle axis defects in mouse colon adenomatous epithelium arising from conditional biallelic Apc inactivation. The American journal of pathology. 2013;183:493–503. doi: 10.1016/j.ajpath.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dame MK, Bhagavathula N, Mankey C, DaSilva M, Paruchuri T, Aslam MN, Varani J. Human colon tissue in organ culture: preservation of normal and neoplastic characteristics. In vitro cellular & developmental biology. Animal. 2010;46:114–122. doi: 10.1007/s11626-009-9247-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bollrath J, Phesse TJ, von Burstin VA, Putoczki T, Bennecke M, Bateman T, Nebelsiek T, Lundgren-May T, Canli O, Schwitalla S, Matthews V, Schmid RM, Kirchner T, Arkan MC, Ernst M, Greten FR. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer cell. 2009;15:91–102. doi: 10.1016/j.ccr.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 30.Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, Karin M. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taniguchi K, Wu LW, Grivennikov SI, de Jong PR, Lian I, Yu FX, Wang K, Ho SB, Boland BS, Chang JT, Sandborn WJ, Hardiman G, Raz E, Maehara Y, Yoshimura A, Zucman-Rossi J, Guan KL, Karin M. A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature. 2015;519:57–62. doi: 10.1038/nature14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donohue E, Tovey A, Vogl AW, Arns S, Sternberg E, Young RN, Roberge M. Inhibition of autophagosome formation by the benzoporphyrin derivative verteporfin. The Journal of biological chemistry. 2011;286:7290–7300. doi: 10.1074/jbc.M110.139915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donohue E, Balgi AD, Komatsu M, Roberge M. Induction of Covalently Crosslinked p62 Oligomers with Reduced Binding to Polyubiquitinated Proteins by the Autophagy Inhibitor Verteporfin. PloS one. 2014;9:e114964. doi: 10.1371/journal.pone.0114964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nature reviews Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamark T, Johansen T. Aggrephagy: selective disposal of protein aggregates by macroautophagy. International journal of cell biology. 2012;2012:736905. doi: 10.1155/2012/736905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu YL, DeLay M, Jahangiri A, Molinaro AM, Rose SD, Carbonell WS, Aghi MK. Hypoxia-induced autophagy promotes tumor cell survival and adaptation to antiangiogenic treatment in glioblastoma. Cancer research. 2012;72:1773–1783. doi: 10.1158/0008-5472.CAN-11-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singla A, Griggs NW, Kwan R, Snider NT, Maitra D, Ernst SA, Herrmann H, Omary MB. Lamin aggregation is an early sensor of porphyria-induced liver injury. Journal of cell science. 2013;126:3105–3112. doi: 10.1242/jcs.123026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ozcan U, Yilmaz E, Ozcan L, Furuhashi M, Vaillancourt E, Smith RO, Gorgun CZ, Hotamisligil GS. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azab M, Benchaboune M, Blinder KJ, Bressler NM, Bressler SB, Gragoudas ES, Fish GE, Hao Y, Haynes L, Lim JI, Menchini U, Miller JW, Mones J, Potter MJ, Reaves A, Rosenfeld PJ, Strong A, Su XY, Slakter JS, Schmidt-Erfurth U, Sorenson JA G. Treatment of Age-Related Macular Degeneration with Photodynamic Therapy Study, G. Verteporfin in Photodynamic Therapy Study. Verteporfin therapy of subfoveal choroidal neovascularization in age-related macular degeneration: meta-analysis of 2-year safety results in three randomized clinical trials: Treatment Of Age-Related Macular Degeneration With Photodynamic Therapy and Verteporfin In Photodynamic Therapy Study Report no. 4. Retina. 2004;24:1–12. doi: 10.1097/00006982-200402000-00001. [DOI] [PubMed] [Google Scholar]
- 40.Miller JW, Schmidt-Erfurth U, Sickenberg M, Pournaras CJ, Laqua H, Barbazetto I, Zografos L, Piguet B, Donati G, Lane AM, Birngruber R, van den Berg H, Strong A, Manjuris U, Gray T, Fsadni M, Bressler NM, Gragoudas ES. Photodynamic therapy with verteporfin for choroidal neovascularization caused by age-related macular degeneration: results of a single treatment in a phase 1 and 2 study. Archives of ophthalmology. 1999;117:1161–1173. doi: 10.1001/archopht.117.9.1161. [DOI] [PubMed] [Google Scholar]
- 41.Singla A, Moons DS, Snider NT, Wagenmaker ER, Jayasundera VB, Omary MB. Oxidative stress, Nrf2 and keratin up-regulation associate with Mallory-Denk body formation in mouse erythropoietic protoporphyria. Hepatology. 2012;56:322–331. doi: 10.1002/hep.25664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mani A, Gelmann EP. The ubiquitin-proteasome pathway and its role in cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23:4776–4789. doi: 10.1200/JCO.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 43.Barakat MR, Kaiser PK. VEGF inhibitors for the treatment of neovascular age-related macular degeneration. Expert opinion on investigational drugs. 2009;18:637–646. doi: 10.1517/13543780902855316. [DOI] [PubMed] [Google Scholar]
- 44.Xue X, Ramakrishnan S, Anderson E, Taylor M, Zimmermann EM, Spence JR, Huang S, Greenson JK, Shah YM. Endothelial PAS domain protein 1 activates the inflammatory response in the intestinal epithelium to promote colitis in mice. Gastroenterology. 2013;145:831–841. doi: 10.1053/j.gastro.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PloS one. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dame MK, Jiang Y, Appelman HD, Copley KD, McClintock SD, Aslam MN, Attili D, Elmunzer BJ, Brenner DE, Varani J, Turgeon DK. Human colonic crypts in culture: segregation of immunochemical markers in normal versus adenoma-derived. Lab Invest. 2014;94:222–234. doi: 10.1038/labinvest.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Rios D, Dianes JA, Sun Z, Farrah T, Bandeira N, Binz PA, Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkley RJ, Kraus HJ, Albar JP, Martinez-Bartolome S, Apweiler R, Omenn GS, Martens L, Jones AR, Hermjakob H. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1: Yap signaling is activated in human colon cancer cell lines and human colon cancer tissues
Fig. S2: Verteporfin treatment does not induce toxicity in normal tissues
Fig. S3: Verteporfin treatment decreases growth in CT-26 xenograft model and colon cancer cell lines
Fig. S4: Verteporfin treatment inhibits STAT3 signaling
Fig. S5: Growth inhibitory effect of verteporfin is YAP independent
Fig. S6: Growth inhibitor effect of verteporfin is associated with oligomerization of proteins
Fig. S7: Hypoxia and nutrient deprivation impairs clearance of VP induced protein oligomers
Fig. S8: Activating autophagy and proteasomal pathways partially clears protein oligomers
Table S1: Proteins that oligomerized in response to verteporfin
Table S2: List of primers.