Abstract
Background
Trypanosoma (T.) evansi is a dyskinetoplastic variant of T. brucei that has gained the ability to be transmitted by all sorts of biting flies. T. evansi can be divided into type A, which is the most abundant and found in Africa, Asia and Latin America and type B, which has so far been isolated only from Kenyan dromedary camels. This study aimed at the isolation and the genetic and phenotypic characterisation of type A and B T. evansi stocks from camels in Northern Ethiopia.
Methodology/principal findings
T. evansi was isolated in mice by inoculation with the cryopreserved buffy coat of parasitologically confirmed animals. Fourteen stocks were thus isolated and subject to genotyping with PCRs targeting type-specific variant surface glycoprotein genes, mitochondrial minicircles and maxicircles, minisatellite markers and the F1-ATP synthase γ subunit gene. Nine stocks corresponded to type A, two stocks were type B and three stocks represented mixed infections between A and B, but not hybrids. One T. evansi type A stock was completely akinetoplastic. Five stocks were adapted to in vitro culture and subjected to a drug sensitivity assay with melarsomine dihydrochloride, diminazene diaceturate, isometamidium chloride and suramin. In vitro adaptation induced some loss of kinetoplasts within 60 days. No correlation between drug sensitivity and absence of the kinetoplast was observed. Sequencing the full coding sequence of the F1-ATP synthase γ subunit revealed new type-specific single nucleotide polymorphisms and deletions.
Conclusions/significance
This study addresses some limitations of current molecular markers for T. evansi genotyping. Polymorphism within the F1-ATP synthase γ subunit gene may provide new markers to identify the T. evansi type that do not rely on variant surface glycoprotein genes or kinetoplast DNA.
Author Summary
Trypanosoma (T.) evansi causes surra in various animal species in Africa, Latin America and Asia. Despite inducing important animal suffering, economic losses and being a World Animal Health Organisation (OIE) notifiable disease, surra is severely neglected in terms of awareness, control interventions and research into improved control tools. Most serological tests can only detect T. evansi type A, while molecular tests rely on detection of highly variable genes or on fragile kinetoplast DNA. Even more, the obscure T. evansi type B, first isolated decades ago in Kenya, totally escapes surveillance due to absence of reliable diagnostic tools. In the present study we isolated new type B stocks from Ethiopia, thus suggesting that this type of T. evansi is probably more widely distributed than previously thought. We further report on an alternative molecular marker for both types of T. evansi and present data on the drug sensitivity of the Ethiopian isolates.
Introduction
Surra, a wasting disease caused by Trypanosoma (T.) evansi, is one of the non tsetse-transmitted Animal African Trypanosomoses (AAT) occurring in Ethiopia. The disease imposes significant financial losses due to reduced fertility and mortality and is prohibiting the import of highly productive yet trypanosusceptible cattle breeds [1–3]. T. evansi belongs to the subgenus Trypanozoon, that also comprises T. brucei and T. equiperdum [4–6]. In terms of geographical distribution, Trypanosoma equiperdum and T. evansi, causing respectively dourine in horses and surra in livestock in Africa, Asia, and South America, have been far more successful than T. brucei, a parasite confined to sub-Saharan Africa where its vector, the tsetse fly, is present [7]. Recent phylogenetic studies suggest that T. evansi and T. equiperdum evolved from T. brucei on several occasions and from genetically distinct T. brucei strains and therefore could be considered as subspecies of T. brucei [8,9].
Trypanosomes are characterised by the presence of a structure called kinetoplast that corresponds with the DNA (kDNA) of their unique mitochondrion. T. brucei kDNA contains 20–50 copies of maxicircles (about 23 kb) and a highly diverse set of thousands of minicircles (about 1 kb). Maxicircles contain rRNA coding regions and genes coding for subunits of the respiratory chain complexes while minicircles code for guide RNAs required for editing [10].
T. equiperdum and T. evansi are dyskinetoplastic (kDNA-) since they lack part of the kDNA [8–11]. T. equiperdum typically has retained maxicircles, in some cases with substantial deletions, but has lost its minicircle diversity. T. evansi does not have maxicircles and either shows minicircle homogeneity or are akinetoplastic (kDNA) [10,12–14]. Based on their minicircle restriction digestion profile, T. evansi can be divided into type A and type B [15,16].
T. evansi type A is the most abundant and is found in Africa, South America and Asia. It is characterised by the presence of the gene for the variant surface glycoprotein (VSG) RoTat 1.2. This RoTat 1.2 VSG is expressed early during infections resulting in the detectability of anti-RoTat 1.2 antibodies in animals infected with T. evansi type A [17,18]. In contrast, T. evansi type B is far less common and has so far been isolated only from camels in Kenya [16,19]. More recently, serological and molecular evidence for the presence of T. evansi type B in Sudan, Ethiopia and Chad was published [20–24]. T. evansi type B lacks the RoTat 1.2 gene and as a consequence, infections with this type are not detected with serological and molecular tests based on RoTat 1.2 VSG, such as the CATT/T. evansi and RoTat 1.2 PCR [15,18,19,25]. So far, three molecular tests have been developed for the identification of T. evansi type B: the EVAB PCR, targeting a type B-specific minicircle DNA sequence, and a PCR and a LAMP targeting a type B-specific VSG JN 2118Hu [15,19,26]. T. equiperdum is the least known parasite of the Trypanozoon group, with very few isolates available for research, albeit new stocks were isolated from Ethiopian and Venezuelan horses recently [24,27].
Unlike T. brucei, T. evansi and T. equiperdum cannot develop in tsetse flies due to their inability to transform into the procyclic life stage. They can only survive in a mammalian host where they produce ATP exclusively through glycolysis. In contrast to bloodstream forms, ATP production in procyclic trypanosomes relies on oxidative phosphorylation and, therefore, on the capacity to express the full set of corresponding mitochondrial genes, including some which are encoded by the kDNA [10,28]. Bloodstream forms of T. evansi, T. equiperdum and laboratory-generated T. brucei strains that have lost all or critical parts of their kDNA, can survive without kDNA due to specific single amino acid mutations in the gamma (γ) subunit of the mitochondrial F1-ATP synthase [28]. Interestingly, the specific mutations/deletions in the C-terminal region of F1-ATP synthase γ subunit enable differentiation among the Trypanozoon strains [8]. Furthermore, when the F1-ATP synthase γ subunits of T. evansi type A (A281del), T. equiperdum (A273P) and the laboratory-generated T. brucei (L262P) strains are overexpressed in a T. brucei γ subunit knock out strain, the latter can survive after loss of its kinetoplast after treatment with DNA intercalating drugs such as acriflavin or ethidium bromide [28,29]. Once the genetically modified T. brucei are independent from kDNA maintenance and expression, they become multidrug resistant to the diamidine and phenanthridine class of drugs [30].
In T. evansi, drug resistance has been reported in several type A strains originating from Africa, Asia and Latin America [31–34]. Some Chinese strains appear to be innately resistant to the phenanthridine class of drugs [35]. In contrast, nothing is known on the drug susceptibly of the T. evansi type B strains. In a previous study, we reported that T. evansi infections are very common in camels, equines, cattle and small ruminants in Tigray and Afar provinces in Northern Ethiopia [20]. We also provided molecular and serological evidence that both T. evansi type A and type B occur in these provinces. In that study, of those dromedary camels that were parasitologically positive, buffy coat samples were collected and cryopreserved in liquid nitrogen for later isolation of the parasite. We here report on the isolation, adaptation to in vitro culture, genetic and phenotypic characterisation and in vitro drug sensitivity of T. evansi type A and B from Northern Ethiopia.
Materials and Methods
Ethics statement
The Animal Experimentation Ethics Committee (AEEC) of the Institute of Tropical Medicine (ITM) advised on the protocol for collection of blood samples from dromedary camels (EXT2012-1) and for the isolation of trypanosomes via inoculation of mice (EXT2012-2) at the College of Veterinary Medicine, Mekelle University. The study protocol for in vivo expansion of trypanosomes at ITM was approved by the AEEC (BM2013-1). Collecting blood from camels and experiments on mice were conducted according to the national guidelines of the Ethiopian Ministry of Livestock and Fishery Development and the Institutional Review Board of the Ministry of Science and Technology.
In vivo isolation of parasites from cryopreserved buffy coat in mice
Details on the collection and cryopreservation of buffy coat samples from dromedary camels that were parasitologically confirmed in the micro haematocrit centrifugation technique have been fully described elsewhere [20]. Two hundred μl of thawed buffy coat were inoculated intraperitoneally (IP) in two 25–30 g Swiss albino mice that were immunosuppressed with 0.16 μg kg-1 body weight dexamethasone (Shanghai Central Pharmaceutical, China) one day prior to inoculation [36]. Parasitaemia was checked in 5 μl of tail blood using the matching method [37], starting from day 7 post-infection and subsequently on every third day. As soon as trypanosomes were detected in at least one mouse, the animal was anaesthetised (the other kept as a backup), its blood was collected on heparin by heart puncture, diluted in an equal volume of phosphate buffered saline glucose (PSG; 7.5 g/l Na2HPO42H2O, 0.34 g/l NaH2 PO4H2O, 2.12 g/l NaCl, 10 g/l D-glucose, pH 8) and subinoculated into four naïve mice (200 μl each) which were monitored for parasitaemia as described above. Mice used as backup were euthanised when the newly infected mice became positive. When parasitemia reached about ± 107.8 cells ml−1 of blood, two of these parasitaemic mice were euthanised (the other two were kept as back up) and blood was taken for subinoculation into four other naïve mice. This protocol was repeated until the parasitaemia reached about 108.4 cells ml−1. At this stage the stock was considered in vivo adapted. All four mice were anaesthetised and exsanguinated by heart puncture in an equal volume of Triladyl-egg yolk-phosphate buffered saline glucose (TEP) cryomedium [38] for cryopreservation in 1 ml aliquots.
In vivo expansion and purification of parasite populations
Cryostabilates were thawed in a water bath at 37°C and diluted in PSG to 1 trypanosome per field (± 105.7 cells ml−1). Two-hundred μl volumes were injected IP in two naïve 20–30 g female OF-1 mice (Charles River, Belgium). Starting from three days post infection (DPI), parasitaemia was monitored daily and harvested at first peak parasitemia, typically at day 4 to 5 post-infection, as described above. Volumes of 0.5 ml of the blood were run over a mini Anion Exchange Centrifugation Technique (mAECT) column to separate the trypanosomes from the blood [39]. The trypanosomes eluted from the column were washed twice with 5 ml ice-cold PSG by centrifugation at 1500 g for 15 min. After the last centrifugation, the supernatant PSG was discarded and the trypanosome sediment was re-suspended in 100 μl of PSG. Part of this suspension was used for in vitro culture adaptation. The remainder was centrifuged at 1500 g for 5 min and the sediment was frozen at -80°C until DNA extraction. The isolates used for in vivo isolation and expansion and the corresponding T. evansi type A and B specific PCR result on their corresponding buffy coat DNA are indicated in Table 1.
Table 1. List of Ethiopian T. evansi isolates with data on origin and results in RoTat 1.2 PCR and EVAB PCR performed on DNA extracted from the buffy coat specimens from the infected camels.
pos: positive, neg: negative.
| Stabilate code | Region | District | Station | RoTat 1.2 PCR | EVAB PCR | In vivo subpassages before first cryostabilate | In vivo expansion at ITM |
|---|---|---|---|---|---|---|---|
| MCAM/ET/2013/MU/01 | Afar | Megalle | Adahara | pos | neg | 3 | yes |
| MCAM/ET/2013/MU/02 | Tigray | Raya-Azebo | Chercher | pos | neg | 5 | yes |
| MCAM/ET/2013/MU/03 | Tigray | Raya-Azebo | Kukufto | pos | neg | 5 | no |
| MCAM/ET/2013/MU/04 | Tigray | Raya-Azebo | Chercher | pos | neg | 3 | yes |
| MCAM/ET/2013/MU/05 | Tigray | Raya-Azebo | Balla | pos | neg | 4 | yes |
| MCAM/ET/2013/MU/06 | Tigray | Raya-Azebo | Balla | pos | neg | 3 | yes |
| MCAM/ET/2013/MU/07 | Afar | Yallo | Gubidera | pos | neg | 2 | yes |
| MCAM/ET/2013/MU/08 | Afar | Golina | Ullel-ella | pos | neg | 2 | yes |
| MCAM/ET/2013/MU/09 | Tigray | Raya-Azebo | Kukufto | pos | neg | 3 | yes |
| MCAM/ET/2013/MU/10 | Afar | Awash Fentale | Alibete | neg | pos | 2 | yes |
| MCAM/ET/2013/MU/11 | Afar | Megalle | Adahara | pos | neg | 3 | yes |
| MCAM/ET/2013/MU/12 | Afar | Yallo | Gubidera | pos | neg | 3 | no |
| MCAM/ET/2013/MU/13 | Afar | Golina | Ullel-ella | pos | neg | 3 | yes |
| MCAM/ET/2013/MU/14 | Afar | Awash Fentale | Alibete | neg | pos | 3 | yes |
| MCAM/ET/2013/MU/15 | Afar | Awash Fentale | Dihoon | pos | neg | 2 | yes |
| MCAM/ET/2013/MU/16 | Afar | Golina | Ullel-ella | pos | neg | 2 | no |
| MCAM/ET/2013/MU/17 | Afar | Awash Fentale | Dihoon | pos | neg | 2 | yes |
| MCAM/ET/2013/MU/18 | Afar | Megalle | Adahara | pos | neg | 2 | no |
| MCAM/ET/2013/MU/19 | Afar | Megalle | Adahara | pos | neg | 3 | no |
| MCAM/ET/2013/MU/20 | Afar | Golina | Ullel-ella | pos | neg | 2 | no |
| MCAM/ET/2013/MU/21 | Afar | Megalle | Adahara | pos | neg | 3 | no |
| MCAM/ET/2013/MU/22 | Afar | Megalle | Adahara | pos | neg | 3 | no |
In vitro adaptation in HMI-9 medium with horse serum
The highly concentrated trypanosome suspension in PSG was diluted to 2 x 105 cells ml−1 in Hirumi’s modified Iscove’s medium 9 (HMI-9), complemented with 15% (v/v) heat-inactivated foetal bovine serum (Gibco, Belgium) and 5% (v/v) heat-inactivated horse serum (Gibco, Belgium) (abbreviated as HMI-9 (HS)) [40,41]. Parasites were seeded at 2 x 104, 2 x 103 and 2 x 102 cells ml−1, in a total volume of 500 μl in a 48-well plate (Nunc, Denmark) and incubated at 37°C and 5% CO2. After 72 hours, a well, where trypanosome density had increased above 2 x 105 cells ml−1, was used for further subpassage in 500 μl of HMI-9 (HS). The well with the highest density of viable parasites was then further maintained in HMI-9 without horse serum [40]. When possible, log phase growing in vitro cultures were scaled up in flasks (Nunc, Denmark) to obtain larger numbers of parasites for cryostabilisation, DNA extraction and in vitro drug sensitivity testing [42]. The in vitro growth curves of the different stocks were generated by seeding cells at 1 x 104 cells ml−1 in 500 μl of HMI-9 in three replicate wells that were counted every 24 h. The doubling times (Td) were calculated from the exponential part of the curve using non-linear regression fitted with an exponential equation in GraphPad Prism 6 (GraphPad, version 6, USA).
Molecular characterisation of parasite populations
DNA extraction of trypanosome sediments prepared from the in vivo expanded and the in vitro adapted populations was performed with DNA Isolation Kit (Roche Diagnostics, Germany) following the protocol recommended for isolation of DNA from mammalian tissue. From T.b. brucei AnTat 1.1E, T.b. gambiense LiTat 1.3, T.b. gambiense type II ABBA and T. equiperdum Dodola 940, DNA was extracted using the Maxwell 16 Tissue DNA Purification kit on a Maxwell 16 instrument according to the manufacturer's instructions (Promega, Belgium). DNA concentrations were measured using the Nanodrop ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies, USA) and adjusted to 10 ng μl-1. A set of PCRs targeting VSG genes (RoTat 1.2 and JN 2118Hu), maxicircle genes (ND4, ND5, ND7 and A6), class A minicircles (miniA PCR) and class B minicircles (EVAB PCR) minisatellites (MORF-2REP), P2 adenosine transporter (AT1) and the F1-ATP synthase γ subunit were adopted to characterise the studied parasite populations [4,15,19,28,43–45]. Where applicable, the published PCR protocols were adjusted to the requirements of the HotStarTaq Plus DNA polymerase (Qiagen, Germany). Primer sequences, reaction mixture contents, cycling conditions and expected amplicon size are described and referenced in Table 2. All PCR amplifications were carried out in 200 μl thin-wall PCR tubes (ABgene, UK) in a T3 thermocycler 48 (Biometra, Germany). Ten μl of amplified products were electrophoresed in 1 to 2% agarose gel at 135 V for 30 min and afterwards stained with ethidium bromide for visualization under UV light. For direct sequencing, PCR was performed in 50–100 μl volumes and amplicons were cleaned up and concentrated using a PCR cleanup kit (QIAquick PCR Purification Kit, Qiagen, Germany) and sent out for bidirectional direct sequencing at the Genetic Sequencing Facility (VIB, Belgium) using the described PCR primers.
Table 2. PCRs used in the present study with target sequence, primer name and sequences, length of expected amplicon, reaction mixtures and cycling conditions.
Reaction mixture 1: 25 μl containing 25 ng DNA, 1X CoralLoad buffer, 1.5 mM of MgCl2, 200 μM of dNTPs, 0.5 μM of each primer, 0.5 U of HotStar TaqPlus. Reaction mixture 2: 25 μl containing 25 ng DNA, 1X CoralLoad buffer, 1.5 mM of MgCl2, 200 μM of dNTPs, 1 μM of each primer, 0.5 U of HotStar TaqPlus. Reaction mixture 3: 25 μl containing 25 ng DNA, 1X CloneAmp HiFi PCR premix and 0.25 μM of each primer. bp: base pair, P: Plus DNA strand, M = Minus DNA strand.
| Target sequence | Primers | Primer sequences | Amplicon length | Reaction mixture | Cycling conditions | Adapted from |
|---|---|---|---|---|---|---|
| VSG RoTat 1.2 | ILO7957 | 5′-GCC ACC ACG GCG AAA GAC-3′ | 488 bp | 1 | 95°C for 5 min and 35 cycles of 30 sec at 94°C, 30 sec at 58°C, 30 sec at 72°C and final extension for 5 min at 72°C | [43] |
| ILO8091 | 5′-TAA TCA GTG TGG TGT GC-3′ | [43] | ||||
| VSG JN 2118Hu | Forward | 5′-TTCTACCAACTGACGGAGCG-3′ | 273 bp | 1 | 95°C for 5 min and 35 cycles of 30 sec at 94°C, 30 sec at 55°C, 30 sec at 72°C and final extension for 5 min at 72°C | [19] |
| Reverse | 5′-TAGCTCCGGATGCATCGGT-3′ | [19] | ||||
| Maxicircle A6 | Forward | 5′-AAAAATAAGTATTTTGATATTATTAAAG-3′ | 381 bp | 2 | 95°C for 5 min and 30 cycles of 94°C for 1 min, 54°C for 1 min, and 72°C for 30 s followed by a final elongation step at 72°C for 8 min | [44] |
| Reverse | 5′-TATTATTAACTTATTTGATC-3′ | [44] | ||||
| Maxicircle ND4 | Forward | 5′-TGTGTGACTACCAGAGAT-3′ | 256 bp | 2 | Idem as above | [44] |
| Reverse | 5′-ATCCTATACCCGTGTGTA-3′ | [44] | ||||
| Maxicircle ND5 | Forward | 5′-TGGGTTTATATCAGGTTCATTTATG-3 | 400 bp | 2 | Idem as above | [28] |
| Reverse | 5′-CCCTAATAATCTCATCCGCAGTACG-3′ | [28] | ||||
| Maxicircle ND7 | Forward | 5′-ATGACTACATGATAAGTA-3 | 167 bp | 2 | Idem as above | [44] |
| Reverse | 5′-CGGAAGACATTGTTCTACAC-3′ | [44] | ||||
| Minicircle class A | MiniA | 5′-GGGTTTTTTAGGTCCGAG-3′ | 1000 bp | 1 | 95°C for 5 min and 35 cycles of 30 sec at 94°C, 30 sec at 58°C, 30 sec at 72°C and final extension for 5 min at 72°C | [15] |
| Reverse MiniB | 5′-CCGAAAATAGCACGTG-3’ | [15] | ||||
| Minicircle class B | EVAB1 | 5’-CACAGTCCGAGAGATAGAG-3’ | 436 bp | 1 | 95°C for 5 min and 30 cycles of 30 sec at 94°C, 30 sec at 60°C, 60 sec at 72°C and final extension for 10 min at 72°C | [15] |
| EVAB2 | 5’-CTGTACTCTACATCTACCTC-3’ | [15] | ||||
| Minisatellite MORF2-REP | P | 5’TGCATGGCAATAGCGATGGGC-3’ | repeated 102 bp sequence | 1 | 95°C for 5 min and 30 cycles of denaturing at 94°C for 30 s, annealing at 60°C for 30 sec and extension at 72°C for 3 min. Elongation was continued for 72°C for 5 min | [4] |
| M | 5’ATCGTCACCTGGTGTACTTCTC-3’ | [4] | ||||
| TeAT1 | TbAT1-F | 5’-GAAATCCCCGTCTTTTCTCAC-3’ | 1600 bp | 1 | 95°C for 5 min, 24 cycles of 1 min at 95°C followed by 1 min at 56°C and 2 min at 72°C, and final extension at 72°C for 5 min | [45] |
| TbAT1-R- | 5’-ATGTGCTGAGCCTTTTTCCTT-3’ | [45] | ||||
| F1-ATP synthase γ subunit | F1-ATP-F | 5’-AACTGCAACGAAGCTTATGTCGGGCAAGCTTCGTC-3’ | 918 bp | 3 | 98°C for 30 sand 35 cycles of 98°C for 15 s, 59.4°C for 15 s, 72°C for 20 s and 72°C for 5 min followed by cool down to 4°C | |
| F1-ATP-R | 5’-TAAATGGGCAGGATCCCTACTTGGTTACTGCCCCTTC-3’ |
The full length sequence of the F1-ATP synthase γ subunit was cloned into a BamHI and HindIII double digested pHD309 vector using the In-Fusion Cloning kit (Clontech, Japan). Primers contained a F1-ATP synthase γ subunit specific sequence based on the T. evansi sequence of STIB 810 (EU185797) and a 5′ extension of 15 bp specific to the place of integration in pHD309, containing the restriction sites and sequence overlap with the vector, as required for the In-Fusion Cloning reaction. Proofreading-PCR was performed using the Clone-Amp HiFi PCR premix (Clontech, Japan). Amplicons were cleaned up (QIAquick PCR Purification Kit, Qiagen, Germany) before use in the In-Fusion protocol. The reaction products were transformed in Stellar competent cells according to the manufacturer's recommendations (Clontech, Japan). Transformant clones were checked for the presence of insert using colony PCR, cultured in LB medium, plasmid purified (QIAprep Spin Miniprep Kit, Qiagen, Germany) and at least 7 to 12 clones per transformation were bidirectionally sequenced at the Genetic Sequencing Facility (VIB, Belgium) using primers binding to pHD309.
In vitro drug sensitivity testing
Melarsomine dihydrochloride (Cymelarsan, Sanofi Aventis, France) and isometamidium hydrochloride (Veridium, Ceva Santé Animale, Belgium) were prepared as 10 mg ml−1 stock solutions in distilled water. Dophanil powder (Dophanil, Docpharma, Belgium), containing 445 mg diminazene diaceturate and 555 mg antipyrine per gram, was concentrated to a 10 mg ml−1 diminazene diaceturate solution in DMSO (Sigma, Belgium). Suramin (Germanin, Bayer, Germany) was prepared as a 100 mg ml−1 in DMSO. A method to measure the IC50 values of compounds in 96-well plates was performed as described elsewhere [46]. Briefly, 2 × 104 cells ml−1 from in vitro adapted stocks were exposed to seven threefold drug dilutions, ranging from 5000 to 7 ng ml−1 for suramin, 500 to 0.7 ng ml−1 for diminazene diaceturate and from 250 to 0.35 ng ml−1 for melarsomine dihydrochloride and isometamidium hydrochloride, in a total volume of 200 μl of HMI-9 medium. Next, the plate was incubated for 72 hours at 37°C with 5% CO2 followed by addition of 20 μl of resazurin (Sigma, Belgium; 12.5 mg in 100 ml PBS) for measuring trypanosomes viability. After a further 24 h incubation at 37°C and 5% CO2, fluorescence was measured (excitation λ = 560 nm; emission λ = 590 nm) with a VictorX3 multimodal plate reader using top reading (Perkin Elmer, Belgium) [42]. The results were expressed as the percent reduction in parasite viability compared to the parasite viability in control wells without drugs. The 50% inhibitory concentration (IC50) was calculated using non-linear regression fitted with a (log) inhibitor versus normalised response (variable slope) equation (GraphPad, version 6, USA). The IC50 values obtained from day 30 and day 60 in vitro cultures were compared using t-tests corrected for multiple testing according to the Holm-Sidak method (α = 0.05) (GraphPad, version 6, USA).
Microscopic examination for presence of a kinetoplast in trypanosomes
Trypanosome populations at different stages of in vivo and in vitro expansion were examined for the presence of the kinetoplast using 4',6-diamidino-2-phenylindole (DAPI) staining. Briefly, live trypanosomes in PSG or in vitro culture medium were washed in PBS by centrifugation, deposited onto microscope slides, air dried and fixed with methanol for 30 min. Subsequently, the slides were rehydrated in PBS and mounted in 87% glycerol containing 1 μg ml-1 DAPI (Sigma, Belgium) [28]. Images were captured with an epifluorescence microscope (Olympus BX41, Olympus, Japan) equipped with a NU fluorescent cube (excitation: 360–370 nm and emission > 420 nm)) and Cell˄D software (Olympus, Japan). DAPI stained trypanosomes were grouped according to the number of kinetoplasts (K) and nuclei (N) present within each cell. The percentage of kinetoplastic cells in a DAPI stained slide was calculated on the basis of on average 300 examined trypanosomes, by dividing the sum of 1K1N + 2K1N + 2K2N cells by the sum of 1K1N + 2K1N + 2K2N + 0K1N + 0K2N cells. A two-tailed Spearman correlation matrix (using a confidence interval of 95%) was used to find the correlation between the percentage of kinetoplastic cells at day 30 and day 60 of in vitro culture and the respective IC50 value for a particular drug (GraphPad, version 6, USA).
In vivo infectivity check
To check the in vivo infectivity of trypanosome populations that were cryostabilised after continuous propagation in vitro for 60 days, 5 x 106 cells in 300 μl were inoculated in a single OF-1 mouse where after parasitaemia was checked as described above.
Results
Isolation of Ethiopian T. evansi stocks
Thirty cryopreserved buffy coat specimens from parasitologically positive dromedary camels were inoculated in immunosuppressed Swiss albino mice. In total, 22 parasite stocks originating from 22 different animals could be isolated and cryopreserved after 2 to 5 subpassages in mice. They were labelled as MCAM/ET/2013/MU/01 to MCAM/ET/2013/MU/22. Based on positivity in RoTat 1.2 PCR and EVAB PCR of the corresponding cryopreserved buffy coats, 20 of these stocks are T. evansi type A and 2 are T. evansi type B (Table 1) [20]. Copy cryovials of these primary isolates were brought to ITM, Antwerp and 14 were selected for further expansion in mice. The selection was based on their geographical origin and subtype: 12 type A stocks originated from different sampling stations in Afar and Tigray (MCAM/ET/2013/MU/01, 02, 04, 05, 06, 07, 08, 09, 11, 13, 15, 17) and two type B stocks (MCAM/ET/2013/MU/10 and 14) were from Awash Fentale in Afar. At peak parasitaemia, between 4 to 7 DPI, parasites were harvested, purified from blood using a mAECT column, washed with PSG and pelleted for DNA extraction and for in vitro culture adaptation.
Molecular typing based on specific VSG sequences of in vivo expanded stocks
DNA extracts of in vivo expanded stocks were subjected to RoTat 1.2 PCR and JN 2118Hu PCR to identify the T. evansi type based on type-specific VSG sequences. In addition, the specificity of these PCRs was tested on DNA of other Trypanozoon strains (T.b. brucei AnTat 1.1E, T.b. gambiense LiTat 1.3, T.b. gambiense type II ABBA, T. evansi type A RoTat 1.2, T. evansi type B KETRI 2479 and T. equiperdum Dodola 940). Results are represented in Table 3. All the in vivo expanded stocks that originated from RoTat 1.2 PCR positive buffy coats, were also positive in RoTat 1.2 PCR (MCAM/ET/2013/MU/01, 02, 04, 05, 06, 07, 08, 09, 11, 13, 15 and 17). Direct sequencing of the 488 bp amplicons from these putative T. evansi type A stocks and the T. evansi RoTat 1.2 strain revealed 100% identity (in a 350 bp sequenced fragment) with the published RoTat 1.2 VSG sequence (AF317914), thus identifying them as T. evansi type A. Only one synonymous polymorphism (C699A) was found in MCAM/2013/ET/MU/04. The gel with the RoTat 1.2 PCR products from the purified trypanosomes showed a faint band of about 400 bp amplified in T. evansi KETRI 2479 and in MCAM/ET/2013/MU/10 and 14. Direct sequencing of these 400 bp amplicons failed. The PCR targeting the T. evansi type B specific VSG JN 2118Hu generated the expected amplicon in T. evansi type B KETRI 2479 and in MCAM/ET/2013/MU/10 and 14. Additionally, an amplicon was generated from MCAM/ET/2013/MU/15. Also for T.b. brucei AnTat 1.1E and T.b. gambiense type II ABBA, amplicons of 273 bp were produced in the JN 2118Hu PCR. Direct sequencing of these amplicons revealed that the Ethiopian T. evansi type B MCAM/ET/2013/MU/10 and 14, T. evansi type B KETRI 2479 and T.b. brucei AnTat 1.1E were 100% identical (in a 190 bp sequenced fragment) to the corresponding sequence of JN 2118Hu VSG (AJ870486). In T.b. gambiense type II ABBA, one synonymous mutation (G300A) was found.
Table 3. Genetic characteristics of the trypanosome populations studied.
pos = positive, neg = negative, seq = sequence identity, n.a. = not applicable, n.d. = not done, (f) = faint, * amplification failed may be due to restricted elongation time in PCR protocol or probably high number of repeats present.
| RoTat 1.2 | JN 2118Hu | Maxicircle PCR | Minicircle class | Fraction of kinetoplastic cells in vivo | Minisatellite profile | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trypanosome stock or strain | PCR | seq | PCR | seq | ND4 | ND5 | ND7 | A6 | A | B | % | MORF2-REP |
| T.b. brucei AnTat 1.1E | neg | n.a. | pos | identical | pos | pos | pos | pos | pos (f) | neg | n.d. | neg*. |
| T.b. gambiense LiTat 1.3 | neg | n.a. | neg | n.a. | pos | pos | pos | pos | neg | neg | n.d. | 7,11 (f) |
| T. b. gambiense ABBA | neg | n.a. | pos | G300A | pos | pos | pos | pos | neg | neg | n.d. | 3 |
| T. evansi RoTat 1.2 | pos | identical | neg | n.a. | neg | neg | neg | neg | pos | neg | 97 | 4,6 |
| T. evansi KETRI 2479 | neg | n.a. | pos | identical | neg | neg | neg | neg | neg | pos | 98 | 3,5 |
| T. equiperdum Dodola 940 | neg | n.a. | neg | n.a. | pos | pos | pos | pos | neg | neg | n.d. | 11(f) |
| MCAM/ET/2013/MU/001 | pos | identical | neg | n.a. | neg | neg | neg | neg | pos | neg | 97 | 7 |
| MCAM/ET/2013/MU/002 | pos | identical | neg | n.a. | neg | neg | neg | neg | pos | neg | 98 | 6,7 |
| MCAM/ET/2013/MU/004 | pos | C699A | neg | n.a. | neg | neg | neg | neg | pos | neg | 99 | 6,7 |
| MCAM/ET/2013/MU/005 | pos | identical | neg | n.a. | neg | neg | neg | neg | pos | neg | 98 | 6,7 |
| MCAM/ET/2013/MU/006 | pos | identical | neg | n.a. | neg | neg | neg | neg | pos | neg | 99 | 6,7 |
| MCAM/ET/2013/MU/007 | pos | identical | neg | n.a. | neg | neg | neg | neg | pos | neg | 100 | 6,7 |
| MCAM/ET/2013/MU/008 | pos | identical | neg | n.a. | neg | neg | neg | neg | pos | neg | 98 | 6,7 |
| MCAM/ET/2013/MU/009 | pos | identical | neg | n.a. | neg | neg | neg | neg | neg | neg | 0% | 6,7 |
| MCAM/ET/2013/MU/010 | neg | n.a. | pos | identical | neg | neg | neg | neg | neg | pos | 98 | 3,4 |
| MCAM/ET/2013/MU/011 | pos | identical | neg | n.a. | neg | neg | neg | neg | pos | pos (f) | 98 | 7 |
| MCAM/ET/2013/MU/013 | pos | identical | neg | n.a. | neg | neg | neg | neg | pos | neg | 99 | 6,7 |
| MCAM/ET/2013/MU/014 | neg | n.a. | pos | identical | neg | neg | neg | neg | neg | pos | 99 | 3,4 |
| MCAM/ET/2013/MU/015 | pos | n.d. | pos | n.d. | neg | neg | neg | neg | pos | pos | 98 | 3,4,6,7 |
| MCAM/ET/2013/MU/017 | pos | identical | neg | n.a. | neg | neg | neg | neg | pos | pos (f) | 99 | 6,7 |
Morphological and genotypic kDNA status of the in vivo expanded stocks
Four PCRs that target maxicircle DNAs, of which three NADH-dehydrogenase subunits (ND4, ND5, ND7) and the ATPase subunit 6 (A6), and two PCRs that target class-specific minicircle sequences (miniA PCR and EVAB PCR) were run on DNA extracts of the purified trypanosomes (Table 3). All Ethiopian T. evansi stocks and T. evansi type A RoTat 1.2 and T. evansi type B KETRI 2479 were negative for all four maxicircle genes, while T.b. brucei AnTat 1.1E, T.b. gambiense LiTat 1.3, T.b. gambiense type II ABBA and T. equiperdum Dodola 940 were positive for all four maxicircle genes.
All stocks that contain RoTat 1.2 VSG, except MCAM/ET/2013/MU/09, were positive in miniA PCR. Additionally, weak amplification was seen in T.b. brucei AnTat 1.1E. MCAM/ET/2013/MU/10 and 14 were positive in EVAB PCR, confirming their identification as T. evansi type B as observed on their corresponding buffy coat specimens (Table 1). Additionally, EVAB PCR amplicons were detected in 3 stocks that were also positive for RoTat 1.2 VSG PCR suggesting a mixed infection with type A and B: a strong amplification was present in MCAM/ET/2013/MU/15, while a weak amplification was visible in MCAM/ET/2013/MU/11 and 17. The presence of kinetoplasts in the trypanosome cells was demonstrated using fluorescence microscopy with DAPI staining on ex vivo isolated trypanosomes (Table 3). T. evansi RoTat 1.2, T. evansi KETRI 2479 and all but one Ethiopian T. evansi stocks show a kinetoplast in > 96% of the cells. Stock MCAM/ET/2013/MU/09 was found to be akinetoplastic since only the nucleus of the trypanosomes was visible with DAPI.
MORF2-REP minisatellite profile of the in vivo expanded stocks
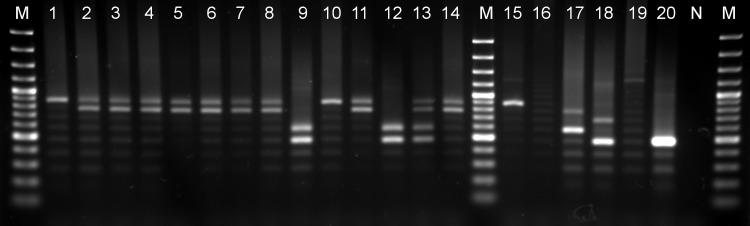
In T. evansi RoTat 1.2, the MORF2-REP locus consists of 4 and 6 repeats, while in T. evansi KETRI 2479, 3 and 5 repeats were found (Table 3). In vivo expanded Ethiopian stocks of type A had either 1 allele (7 repeats) or 2 alleles (6 and 7 repeats), thus displaying a different pattern than T. evansi type A RoTat 1.2. The Ethiopian type B stocks MCAM/ET/2013/MU/10 and 14 contain 3 and 4 repeats, and thus have a pattern different from T. evansi type B KETRI 2479. MCAM/ET/2013/MU/15 showed a clear pattern of the Ethiopian type B (3 and 4 repeats), and double allele pattern of the Ethiopian type A (6 and 7 repeats). The other presumed mixed type A and type B stocks MCAM/ET/2013/MU/11 and 17 showed only the Ethiopian type A T. evansi pattern (Fig 1). DNA extracted from the buffy coats revealed similar MORF2-REP patterns as the in vivo expanded trypanosomes except for the buffy coat of MCAM/ET/2013/MU/15 that revealed only the Ethiopian type A MORF2-REP pattern. The other Trypanozoon strains showed the following patterns: T. b. gambiense LiTat 1.3 had 7 and 11 repeats, T.b. gambiense type II ABBA had 3 repeats, T. equiperdum Dodola 940 had 11 repeats, while no amplicons were generated from T.b. brucei AnTat 1.1E under the giving PCR conditions.
Fig 1. MORF2-REP profiles of Ethiopian T. evansi stocks and T. evansi and T. brucei reference strains.
1.5% agarose gel showing MORF2-REP minisatellite PCR amplicons. Lane M: 100 bp plus marker, lanes 1 to 14: Ethiopian T. evansi stocks MCAM/ET/2013/MU/01-02-04-05-06-07-08-09-10-11-13-14-15-17, lane 15: T.b. gambiense LiTat 1.3, lane 16: T.b. brucei AnTat 1.1E lane 17: T. evansi type A (RoTat 1.2), lane 18: T. evansi type B (KETRI 2479), lane 19: T. equiperdum Dodola 940, lane 20: T. b. gambiense ABBA, lane N: negative control
F1-ATP synthase γ subunit genotyping
Sequence analysis of in total 136 clones of the full length F1-ATP synthase γ subunit, amplified from DNA of the in vivo expanded Ethiopian stocks MCAM/ET/2013/MU/04, 06, 09, 10, 11, 13, 14, 15 and of T.b. brucei AnTat 1.1E, T.b. gambiense LiTat 1.3, T. evansi RoTat 1.2, T. evansi KETRI 2479, T. b. gambiense type II ABBA and T. equiperdum Dodola 940 revealed diverse homozygous and heterozygous nucleotide polymorphisms spread over the entire coding sequence (Table 4).
Table 4. F1-ATP synthase γ subunit single nucleotide polymorphism (SNP) observed within the studied trypanosome stocks and strains or retrieved from GenBank.
Some SNPs were only present in T.b. b. TREU927 (G6A, C9T,C572G), T.b.b 29–13 (C149G, A168C, C866T) and T.b.b. STIB 920 (G738C) and are not represented in the table. del = deletion, GAN: GenBank accession number, * identical to all Ethiopian T. evansi type A stocks, ** identical to all Ethiopian T. evansi type B stocks. Blank spaces indicate no change and–indicates missing sequence information.
| Stock/strain | A93G | C142T | C194T | A198G | G294A | T321C | A356T | T654C | T663C | G801T | T807C | G817C | 841–843 GCT | A844T | T867G | A882G | T892C | GAN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T.b.g. DAL972 | G | G | T | G | Tbg972.10.90 | |||||||||||||
| T.b.g. LiTat 1.3 | G | G | T | G | KT934830 | |||||||||||||
| T.b.g. ABBA | G | G | KT934831 | |||||||||||||||
| T. ev. RoTat 1.2* | T | T | KT934832 | |||||||||||||||
| T. ev. RoTat 1.2* | T | del | KT934833 | |||||||||||||||
| T. ev. STIB 810 | T | EU185797 | ||||||||||||||||
| T. ev. STIB 810 | T | del | EU185798 | |||||||||||||||
| T. ev. KETRI 2479** | C | C | G | KT934834 | ||||||||||||||
| T. ev. KETRI 2479** | T | KT934835 | ||||||||||||||||
| T. ev. KETRI 2479 | - | - | - | - | - | - | - | - | - | C | T | G | EU185794 | |||||
| T. eq BoTat 1.1 | C | C | C | EU185793 | ||||||||||||||
| T. eq. STIB 841 | Y | R | C | W | Y | Y | G | C | EU185792 | |||||||||
| T. eq. Dodola 940 | T | C | C | G | KT934836 | |||||||||||||
| T.b.b. AnTat 1.1E | T | C | C | G | KT934837 | |||||||||||||
| T.b.b. AnTat 1.1E | A | C | KT934838 | |||||||||||||||
| T.b.b. STIB 920 | C | C | C | G | EU185791 | |||||||||||||
| T.b.b. 29–13 | - | T | C | C | G | C | EU185790 | |||||||||||
| T.b.b. TREU927 | G | A | C | T | C | Tb927.10.180 |
The F1-ATP synthase γ subunit of T.b. gambiense LiTat 1.3 (KT934830) appeared homozygous and identical to the T.b. gambiense DAL972 sequence (Tbg972.10.90). T.b. gambiense type II ABBA (KT934831) appeared homozygous and differed in only 2 SNPs (G801T and A882G) from the T.b. gambiense sequence. T. evansi RoTat 1.2 and the Ethiopian stocks MCAM/ET/2013/MU/04, 06, 09,11 and 13 were heterozygous and revealed in one allele (KT934833), identical to the published full length T. evansi STIB 810 (EU185798) sequence, the deletion of nucleotides A841-843del. The second allele contained a C142T polymorphism (KT934832), that is not present in the wild-type T. evansi STIB 810 sequence (EU185797), but that could be identified in the genome sequence of the Chinese akinetoplastic T. evansi STIB 805 strain [9]. For T. evansi KETRI 2479 and the Ethiopian stocks MCAM/ET/2013/MU/10 and 14 we obtained heterozygous alleles, different from the partial sequence of T. evansi KETRI 2479 (EU185794). The first allele had the unique A844T polymorphism (KT934835), and differed from the second allele in 3 additional SNPs (T321C, T807C, T867G) that were also found in some T.b. brucei and T. equiperdum. Interestingly, the in vivo expanded stock of MCAM/ET/2013/MU/15 revealed alleles that belonged to T. evansi type A and type B. In contrast, when the original buffy coat of this stock was tested, only alleles of T. evansi type A were found. Finally, T. equiperdum Dodola 940 (KT934836) appeared homozygous and its single allele was identical to one of the two alleles found in T.b. brucei AnTat 1.1E (KT934837), but differed in 5 SNPs with the sequence from T. equiperdum BoTat 1.1 (EU185793) and in 6 SNPs with T. equiperdum STIB 841 (EU185792). However, for the T. equiperdum STIB 841 strain, 5 of the 6 SNPs were ambiguous polymorphisms that do not rule out similarity to T. equiperdum Dodola 940.
In vitro adaptation of Ethiopian T. evansi stocks
Fourteen Ethiopian T. evansi stocks, T. evansi RoTat 1.2 and T. evansi KETRI 2479 were expanded in mice and purified from blood at peak parasitaemia to initiate primary in vitro cultures in HMI-9 (HS) medium. After 96 hours, the initial 2x104 cells ml−1 inoculum reached concentrations above 2x105 cells ml−1 for all the different stocks. These cells were used for further in vitro propagation by subpassage in fresh medium. Over the next 72 hours, only MCAM/ET/2013/MU/09, 14 and 15, and T. evansi RoTat 1.2 and T. evansi KETRI 2479 showed proliferation. In contrast, slightly increased cell densities were observed for MCAM/ET/2013/MU/01, 04, 06 and 10. For all other strains not a single inoculum proliferated and longer incubation led to growth cessation.
Because the HMI-9 (HS) medium did not support sufficient in vitro culture growth for most of the Ethiopian T. evansi stocks, it was abandoned and replaced with HMI-9 without horse serum. In vitro adapted strains of T.b. brucei AnTat 1.1E and T.b. gambiense LiTat 1.3 were cultured in HMI-9 in parallel. In vitro cultures were only considered adapted to HMI-9 medium when it was possible to maintain the parasites in continuous proliferation. To this extent, dense parasite cultures, containing 2–5 x 105 cells ml−1, were subpassaged into new wells using serial fivefold dilutions in fresh medium. When these subpassages reached densities above 2 x 105 cells ml−1 within a 48–96 hours period, the stock was considered adapted. The five stocks that already grew well in the HMI-9 (HS) medium continued proliferating when inoculated from the dense cultures at serial fivefold dilutions in HMI-9. These five stocks were considered to be in vitro adapted after 15 days of in vitro culture. Out of the four remaining stocks, only MCAM/ET/2013/MU/04 and 10 slowly regained the ability to proliferate in HMI-9 at a reduced subpassaging scheme using serial twofold dilutions. MCAM/ET/2013/MU/04 required 25 days to adapt, while MCAM/ET/2013/MU/10 was only fully adapted after day 35 of in vitro culture. Gradually increasing the culture volume allowed to obtain sufficient parasites from the adapted cultures for in vitro drug testing, DNA extraction, and cryostabilisation at day 30 (all, except MCAM/ET/2013/MU/10) and at day 60 of in vitro culture (all stocks).
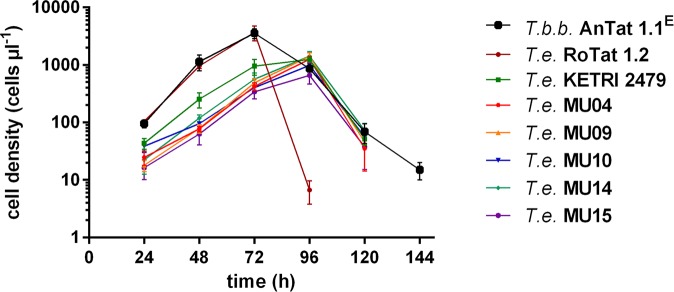
DNA of the in vitro adapted stocks was subjected to RoTat 1.2 PCR, EVAB PCR and MORF2-REP PCR. All in vitro stocks had similar molecular profiles as their corresponding in vivo expanded parental stocks, except MCAM/ET/2013/MU/15. While the in vivo expanded stock of the latter was identified as a mixed infection of T. evansi type A and type B, the in vitro adapted stock (at day 30 and day 60 in vitro culture) was identified as pure T. evansi type B with the above mentioned PCRs and confirmed by cloning and sequencing of the F1-ATP synthase γ subunit. Thus, beside T. evansi RoTat 1.2 and T. evansi KETRI 2479, we achieved the in vitro adaptation of 2 Ethiopian type A stocks, 2 Ethiopian type B stocks and additionally ended up with a pure T. evansi type B in vitro adapted stock originating from a mixed type A and type B in vivo adapted stock. Growth curves were generated for T.b. brucei AnTat 1.1E and all seven in vitro adapted stocks (Fig 2). T.b. brucei AnTat 1.1E and T. evansi RoTat 1.2 had the shortest Td, 7.5 ± 0.3 h-1 and 7.7 ± 0.2 h-1 respectively, and reached the highest maximum population density (MPD) of ± 3–4 x 106 cells ml-1, while T. evansi KETRI 2479 had a longer Td, 10.8 ± 0.2 h-1, and a lower MPD of ± 1 x 106 cells ml-1. The Ethiopian type A stocks MCAM/ET/2013/MU04 and MU09 had a Td of 11.2 ± 0.4 and 11.3 ± 0.4 respectively, and a MPD of ± 1 x 106 cells ml-1. Similarly, the Ethiopian type B stocks MCAM/ET/2013/MU10, 14 and 15 had a Td of 12.9 ± 0.5, 11.3 0.5 and 12.1 ± 0.6 respectively, and a MPD of ± 0.7–1 x 106 cells ml-1 (Fig 2).
Fig 2. In vitro growth curve of trypanosome stocks and strains.
T.b.b. = T.b. brucei, T.e. = T. evansi, MU = MCAM/ET/2013/MU.
In vitro drug sensitivity and relation to kDNA
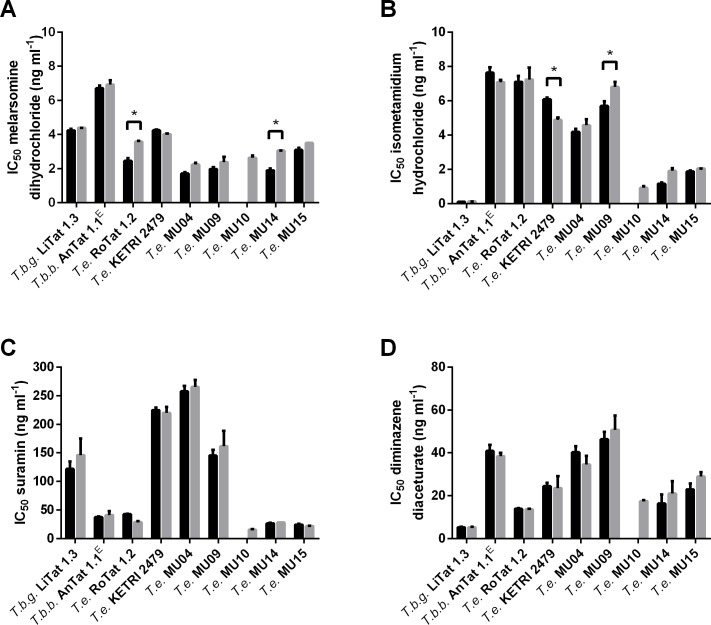
After day 30 and 60 of in vitro culture, IC50 values were determined for melarsomine dihydrochloride (Cymelarsan) (Fig 3A), isometamidium hydrochloride (Veridium) (Fig 3B), diminazene diaceturate (Dophanil) (Fig 3C) and suramin (Germanin) (Fig 3D). In general, non-significant differences (p > 0.05) were found between IC50 values recorded at day 30 and day 60 of in vitro culture, except for the melarsomine dihydrochloride IC50 values of T. evansi RoTat 1.2 and T. evansi MCAM/ET/2013/MU/14 and for the isometamidium hydrochloride IC50 values of T. evansi KETRI 2479 and T. evansi MCAM/ET/2013/MU/09 (p < 0.05). For comparison between the different stocks, the IC50 values of day 30 and day 60 of in vitro cultures were averaged. All Ethiopian T. evansi stocks had IC50 values for melarsomine dihydrochloride (IC50 1.9–3.3 ng ml-1) that were similar to those of T.b. gambiense LiTat 1.3 (IC50 4.3 ng ml-1), T.b. brucei AnTat 1.1E (IC50 6.8 ng ml-1), T. evansi RoTat 1.2 (IC50 3.0 ng ml-1) and T. evansi KETRI 2479 (IC50 4.1 ng ml-1). For isometamidium hydrochloride, the IC50 values of the Ethiopian T. evansi (IC50 0.6–6.2 ng ml-1) fall within the range of T.b. gambiense LiTat 1.3 (IC50 0.1 ng ml-1), T.b. brucei AnTat 1.1E (IC50 7.3 ng ml-1), T. evansi RoTat 1.2 (IC50 7.1 ng ml-1) and T. evansi KETRI 2479 (IC50 5.5 ng ml-1). However, the two Ethiopian T. evansi type A stocks (IC50 4.3–6.2 ng ml-1) appear to be threefold less sensitive that the three type B stocks (IC50 0.6–1.9 ng ml-1). For suramin, large differences in IC50 values were found among the Ethiopian T. evansi (IC50 15.9–261.5 ng ml-1) stocks and among the other strains: T.b. brucei AnTat 1.1E (IC50 39.5 ng ml-1) and T. evansi RoTat 1.2 (IC50 35.8 ng ml-1) appear highly susceptible, while T.b. gambiense LiTat 1.3 (IC50 134.0 ng ml-1) and T. evansi KETRI 2479 (IC50 222.4 ng ml-1) are less susceptible. The two Ethiopian T. evansi type A (IC50 153.5–261.5 ng ml-1) appear to be tenfold less sensitive than the three type B (IC50 15.9–27.6 ng ml-1). For diminazene diaceturate, the IC50 values of all Ethiopian T. evansi (IC50 17.5–48.5 ng ml-1) are higher than those of T.b. gambiense LiTat 1.3 (IC50 5.2 ng ml-1) and T. evansi RoTat 1.2 (IC50 13.8 ng ml-1), but similar to T.b. brucei AnTat 1.1E (IC50 39.6 ng ml-1) and T. evansi KETRI 2479 (IC50 24.0 ng ml-1). The two Ethiopian T. evansi type A (IC50 37.4–48.5 ng ml-1) appear to be twofold less sensitive than the three type B (IC50 17.5–25.9 ng ml-1). Direct sequencing of the full length TeAT1 PCR amplicons of MCAM/ET/2013/MU/04, 09, 10, 14, and 15, T. evansi type A RoTat 1.2 and T. evansi Type B KETRI 2479 revealed no polmorphisms to the wild-type TeAT1 sequence (AB124588).
Fig 3. In vitro drug sensitivity of T. evansi.
IC50 values for different drugs, with standard deviation error bars, obtained with different Trypanosoma sp. stocks and strains after 30 (black bars) and 60 days (grey bars) in vitro culture. Significant differences between IC50 values of 30 and 60 days in vitro cultures are indicated by an asterisk. T.b.g. = T.b. gambiense, T.b.b. = T.b. brucei, T.e. = T. evansi, MU = MCAM/ET/2013/MU. Panel A: IC50 values for melarsomine dihydrochloride. Panel B: IC50 values for isometamidium hydrochloride. Panel C. IC50 values for suramin and Panel D: IC50 values for diminazene diaceturate.
DAPI staining was performed on in vivo and in vitro propagated stocks (Fig 4). In vitro culture did not change the percentage of kinetoplastic cells in T.b. gambiense LiTat 1.3 (99%), T.b. brucei AnTat 1.1E (99%) and MCAM/ET/2013/MU/09 (0%). On the other hand, already after 30 days in vitro culture a decrease in the percentage of kinetoplastic cells was observed in T. evansi RoTat 1.2 (89%), T. evansi KETRI 2479 (81%), MCAM/ET/2013/MU/04 (97%), 14 (93%) and 15 (94%) compared to non-in vitro adapted trypanosomes. After 60 days of in vitro culture, the percentage of kinetoplastic cells dropped even further for T. evansi KETRI 2479 (64%), MCAM/ET/2013/MU/04 (89%) and 10 (35%). No significant correlation was observed between the percentage of kinetoplastid cells of all in vitro adapted T. evansi stocks (including day 30 and day 60) and their IC50 values for melarsomine dihydrochloride (ρ = -0.13, p = 0. 67), isometamidium hydrochloride (ρ = -0.324, p = 0.278), suramin (ρ = -0.097, p = 0.752) and diminazene diacetureate (ρ = -0.355, p = 0.233). These data suggest that among the in vitro adapted Ethiopian T. evansi stocks there is no relation between the drug sensitivity and the presence of kinetoplast DNA. Furthermore, their loss of kDNA does not seem to influence rodent infectivity since all cryostabilates made from day 60 in vitro cultures remained infective for mice with detectable parasitaemia at 4–5 DPI.
Fig 4. Percentage of kinetoplastic cells within T. evansi populations.
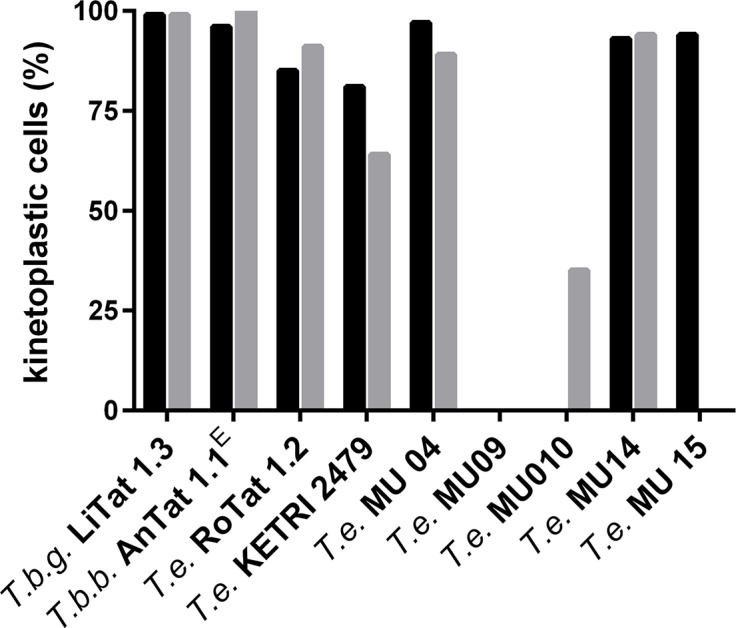
Percentage of kinetoplastic cells visualised after DAPI staining and fluorescence microscopy within populations after 30 (black bars) and 60 days (grey bars) in vitro propagation. T.b.g. = T.b. gambiense, T.b.b. = T.b. brucei, T.e = T. evansi, MU = MCAM/ET/2013/MU.
Discussion
Previous molecular and serological studies revealed that trypanosome infections in camels from Northern Ethiopia are caused by either RoTat 1.2 PCR or EVAB PCR positive parasites. In some instances amplicons of both PCRs were detected within the same buffy coat extract, suggesting the occurrence of mixed infections [20]. The present study was undertaken to isolate the trypanosomes from camels carrying apparent single infections through inoculation of their buffy coats in immunosuppressed mice. The in vivo inoculation led to the successful isolation of 22 stocks, out of which 14 were selected on the basis of their geographical origins for further investigations (5 stocks from Tigray and 9 stocks from Afar). Next, we performed an in-depth comparative molecular analysis on DNA extracts from the isolated parasite stocks using diverse PCRs. Furthermore, we analysed the specificity of each of these PCRs on a collection of Trypanozoon strains.
The RoTat 1.2 VSG sequence can be used to characterise T. evansi type A [25,43]. In our collection, all buffy coats positive in RoTat 1.2 PCR yielded in vivo isolated stocks that were RoTat 1.2 PCR positive but that were negative in the maxicircle gene targeting PCRs. Furthermore, with the exception of the akinetoplastic stock MCAM/ET/2013/MU/09, all these strains had type A minicircles. MCAM/ET/2013/MU/09 may be naturally akinetoplastic since the DNA extracted from the original buffy coat was negative in all PCRs targeting kinetoplast DNA. The occurrence of naturally akinetoplastic strains was previously documented in Latin America and China [12–14,47]. One stock (MCAM/ET/2013/MU/04) contained a SNP in its RoTat 1.2 VSG PCR amplicon. SNPs in RoTat 1.2 amplicons were previously reported in Egypt but do not necessarily lead to a negative result in RoTat 1.2 based antibody detection tests. This was also the case for the camel from which MCAM/ET/2013/MU/04 was isolated [48,49].
Initially defined by minicircle class B, identification of T. evansi type B is possible with EVAB PCR that amplifies a fragment of this minicircle [15]. Additionally, it was proposed that the VSG JN 2118Hu, first described in a Kenyan T. evansi strain, is a specific marker for T. evansi type B [19].
In our collection, 2 buffy coat extracts that were positive in EVAB PCR yielded in vivo isolated stocks that were EVAB PCR positive as well. Interestingly, an EVAB PCR amplicon was also detected in three additional in vivo expanded stocks that were RoTat 1.2 PCR positive but for which the corresponding buffy coats were EVAB PCR negative. These three stocks might be mixed infections. JN 2118Hu VSG PCR appeared to be less sensitive because it detected only 3 out of 5 EVAB PCR positive isolated stocks. Furthermore, the JN 2118Hu VSG PCR appeared to be less specific since T.b. brucei AnTat 1.1E and T.b. gambiense type II ABBA were also positive in this PCR. None of the EVAB PCR positive isolated stocks contained maxicircle DNA and they were all negative in miniA PCR, except for the three mixed infections. Therefore, we conclude that we isolated at least two “pure” T. evansi type B stocks from Ethiopian camels, decades after the initial isolation of T. evansi type B from camels in Kenya [15].
We used the minisatellite locus MORF2-REP to verify whether both putative mixed stocks, that were positive in RoTat 1.2 PCR and EVAB PCR, were real mixed infections or hybrids between T. evansi type A and B. The Ethiopian isolates clustered in two classes of T. evansi type A, of which one with a previously described heterozygous profile (6 and 7 repeats) and one with a homozygous profile (7 repeats). The Ethiopian T. evansi type B stocks had a heterozygous profile (3 and 4 repeats) differing from the only known profile described for Kenyan type B isolates (3 and 5 repeats) [50]. In one of the mixed infections we observed a profile that can be interpreted as a mixture of Ethiopian type A and type B, while the others only revealed the Ethiopian type A pattern. These results prove that we are dealing with mixed infections and not with hybrids between T. evansi type A and type B. To exclude that these apparent mixed infections represent cross-contamination with genetic material, we attempted in vitro cultivation of the in vivo expanded stocks.
Previously we have shown that addition of 1,1% methylcellulose to HMI-9 greatly helps the in vitro adaptation of Trypanozoon strains, including T.b. gambiense and T. evansi RoTat 1.2 [40]. However, to avoid the use of this highly viscous medium we preferred the use of horse serum to adapt T. evansi stocks as is suggested in previous reports [51–53]. While this approach proved to be successful for all type B stocks, only two out of nine Ethiopian T. evansi type A could be adapted. Interestingly, in the case of mixed stock MCAM/ET/2013/MU/15, this medium selected T. evansi type B out of the mixed population. While only the type A infection was detected in the buffy coat DNA extract, both types could be detected in the in vivo expanded stock DNA, but eventually only type B was detected in the in vitro adapted stock.
Gillingwater and colleagues reported on the drug sensitivity profiles of a panel of T. evansi and T. equiperdum strains where they considered T. evansi STIB 806K to be a reference sensitive strain for suramin (IC50 70.4 ng ml-1), diminazene diaceturate (IC50 4.5 ng ml-1) and melarsomine dihydrochloride (IC50 1.4 ng ml-1). They reported drug resistance in two T. evansi stocks with an IC50 for suramin > 10000 ng ml-1 (STIB 780 and STIB 781), and in the T. equiperdum OVI strain, with an IC50 for diminazene diaceturate of 302 ng ml-1 and an IC50 for melarsomine dihydrochloride of 17.6 ng ml-1 [46]. The only strain that is shared between their panel and our collection is T. evansi RoTat 1.2, which despite different approaches in the experimental testing, yielded corresponding IC50 values, especially for diminazene diaceturate and melarsomine dihydrochloride, thus facilitating comparison between both studies. In our Ethiopian T. evansi collection, no resistance against melarsomine dihydrochloride was found. However, some stocks appeared to have raised IC50 values for suramin (> 200 ng ml-1) and diminazene diaceturate (> 50 ng ml-1). The IC50 values that we observe for T.b. gambiense LiTat 1.3 and the Ethiopian T. evansi type B are similar to the in vitro IC50 value of 0.82 ng ml-1 found by Sahin and coworkers for T. congolense IL3000 which is sensitive to isometamidium (Veridium) in vivo [54]. In the same study, an in vitro IC50 of 11.06 ng ml-1 is reported for T.b. brucei AnTat 1.1 strain, which is slightly higher than the value that we obtained in experiments with our T.b. brucei AnTat 1.1 strain and the other T. evansi stocks [54]. Nevertheless, defining our T. evansi stocks as either sensitive or resistant based solely on the in vitro drug sensitivity results may be too audacious, given the fact that IC50 values were determined in only one assay, the resazurin viability assay [55–57]. Therefore, an in vivo drug sensitivity profile of all our Trypanozoon strains against the commonly used trypanocides remains to be elucidated. Interestingly, both Ethiopian T. evansi type A stocks appear to be less susceptible to suramin, diminazene diaceturate and isometamidium hydrochloride than the three type B stocks. In T.b. brucei, resistance against suramin and isometamidum hydrochloride has been linked to several proteins [58,59], while resistance to diamidine and melaminophenyl classes of drugs is attributed to the transporter protein TbAT1 and the aquaporin AQP2 [60–62]. The lower sensitivity to diminazene diaceturate was not caused by mutations in the T. evansi TeAT1 [63].
Interestingly, DAPI staining of the trypanosomes indicated slight to severe loss of the kDNA in all in vitro adapted T. evansi stocks, when compared to in vivo adapted stocks. The loss of kDNA in in vitro cultured T. evansi is a phenomenon that has been known for a long time [10,15,55,64]. Non-vital loss of the kinetoplast is made possible by mutations in the F1-ATP synthase γ subunit of T. evansi allowing to uncouple from the Fo subunit and effectively circumventing the requirement for mitochondrial gene expression [65]. Furthermore, it has been shown that the expression of certain T. evansi F1-ATP synthase γ subunit coding sequences in T. brucei allows this species to survive loss of its kDNA after chemical treatment [28]. Moreover, in such genetically modified T. brucei, independence of kDNA maintenance and expression is associated with multidrug resistance [30]. In our collection of T. evansi stocks we did not observe differences in drug sensitivity between populations that were partially or completely akinetoplast confirming earlier evidence that the presence or absence of kDNA is irrelevant within this context [30,55].
Recently, Carnes et al. showed that SNPs in the F1- ATP γ subunit could be used to genotypically support the multiple origins of at least 4 dyskinetoplastic T. evansi/T. equiperdum lineages: one major group of RoTat 1.2 VSG positive T. evansi/T. equiperdum type A, and three very small groups each represented by only a single strain: T. evansi type B KETRI 2479, T. equiperdum BoTat and T. equiperdum OVI [9]. All Ethiopian T. evansi type A had the corresponding mutation of the type A group. The Ethiopian type B T. evansi shared a similar profile as KETRI 2479. Finally, the Ethiopian T. equiperdum strain Dodola, which had some maxicircle genes but was negative for both type A and type B markers revealed an F1-ATP synthase sequence similar to T.b. brucei AnTat 1.1E strain, thus likely belongs to the same dyskinetoplastic group as T. equiperdum OVI [9,28].
In conclusion, our study shows that the apparent T. evansi type that is detected in a buffy coat of an infected camel does not necessarily represent the full diversity that is present in the infected animal. Moreover, the fact that 5 out of 22 new T. evansi isolates from camel in Ethiopia contain T. evansi type B may be an indication that is more widespread than currently known. The inoculation of the trypanosomes in immunosuppressed mice may allow the propagation of mixed populations. In contrast, in vitro cultivation seems to reduce the diversity by selecting for only one particular type, in our study T. evansi type B. Secondly, our study addresses some drawbacks of current molecular markers for T. evansi genotyping. To rely solely on VSG markers or kDNA markers for the molecular identification of T. evansi may be misleading due to possible recombinations occurring in VSG genes and to the presence of akinetoplastic T. evansi stocks. In this regard, we confirm that the F1-ATP synthase γ subunit gene, that is not related to the VSG repertoire nor to the presence of kDNA, may become an interesting target for genotyping T. evansi stocks in areas where both types overlap and where mixed infections can occur. Nevertheless, it is not possible to separate the Ethiopian T. equiperdum from T. brucei on the basis of this target gene. Thirdly, no evidence of in vitro drug resistance was found in our collection of T. evansi type A and type B stocks. The presence or partial absence of kDNA in the in vitro adapted T. evansi stocks did not correspond with the drug sensitivity phenotype.
Acknowledgments
The pHD309 vector was a kind gift from George Cross from the Rockefeller University. The authors would like to acknowledge for Bart Cuypers, Nicolas Bebronne, Jeroen Swiers, Kidane Weldu, Kumachew Belay and Alemu Tola for their help in the laboratory and shipment of trypanosome stabilates from Ethiopia to Belgium.
Data Availability
All F1 ATP synthase gamma subunit sequence files are available from the GenBank database (accession numbers KT934830, KT934831, KT934832, KT934833, KT934834, KT934835, KT934836, KT934837, KT934838).
Funding Statement
The PhD fellowship of BH was financed by the Belgian Directorate General for Development Cooperation (contract number, 0410057701). NVR is supported by “Les Amis de l'Institut Pasteur de Bruxelles” a.s.b.l. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript
References
- 1.Dargantes AP, Mercado RT, Dobson RJ, Reid SA (2009) Estimating the impact of Trypanosoma evansi infection (surra) on buffalo population dynamics in southern Philippines using data from cross-sectional surveys. Int J Parasitol 39: 1109–1114. 10.1016/j.ijpara.2009.02.012 [DOI] [PubMed] [Google Scholar]
- 2.Dobson RJ, Dargantes AP, Mercado RT, Reid SA (2009) Models for Trypanosoma evansi (surra), its control and economic impact on small-hold livestock owners in the Philippines. Int J Parasitol 39: 1115–1123. 10.1016/j.ijpara.2009.02.013 [DOI] [PubMed] [Google Scholar]
- 3.Reid SA (2002) Trypanosoma evansi control and containment in Australasia. Trends Parasitol 18: 219–224. [DOI] [PubMed] [Google Scholar]
- 4.Biteau N, Bringaud F, Gibson W, Truc P, Baltz T (2000) Characterization of Trypanozoon isolates using a repeated coding sequence and microsatellite markers. Mol Biochem Parasitol 105: 185–201. [PubMed] [Google Scholar]
- 5.Hide G, Cattand P, Le Ray D, Barry JD, Tait A (1990) The identification of Trypanosoma brucei subspecies using repetitive DNA sequences. Mol Biochem Parasitol 39: 213–225. [DOI] [PubMed] [Google Scholar]
- 6.Hoare C. A. (1972) The trypanosomes of mamals A zoological monograph. Oxford, UK: Blackwell Scientific Publications. [Google Scholar]
- 7.Desquesnes M, Holzmüller P, Lai DH, Dargantes A, Lun ZR et al. (2013) Trypanosoma evansi and surra: a review and perspectives on origin, history, distribution, taxonomy, morphology, hosts, and pathogenic effects. Biomed Res Int 2013: ID 194176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai DH, Hashimi H, Lun ZR, Ayala FJ, Lukes J (2008) Adaptations of Trypanosoma brucei to gradual loss of kinetoplast DNA: Trypanosoma equiperdum and Trypanosoma evansi are petite mutants of T. brucei. Proc Natl Acad Sci U S A 105: 1999–2004. 10.1073/pnas.0711799105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carnes J, Anupama A, Balmer O, Jackson A, Lewis M et al. (2015) Genome and phylogenetic analyses of Trypanosoma evansi reveal extensive similarity to T. brucei and multiple independent origins for dyskinetoplasty. PLoS Negl Trop Dis 9: e3404 10.1371/journal.pntd.0003404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schnaufer A, Domingo GJ, Stuart K (2002) Natural and induced dyskinetoplastic trypanosomatids: how to live without mitochondrial DNA. Int J Parasitol 32: 1071–1084. [DOI] [PubMed] [Google Scholar]
- 11.Claes F, Büscher P, Touratier L, Goddeeris BM (2005) Trypanosoma equiperdum: master of disguise or historical mistake? Trends Parasitol 21: 316–321. [DOI] [PubMed] [Google Scholar]
- 12.Ou YC, Giroud C, Baltz T (1991) Kinetoplast DNA analysis of four Trypanosoma evansi strains. Mol Biochem Parasitol 46: 97–102. [DOI] [PubMed] [Google Scholar]
- 13.Lun ZR, Vickerman K (1991) Multinuclear forms in a dyskinetoplastic strain of Trypanosoma evansi in mice. Ann Parasitol Hum Comp 66: 51–53. [DOI] [PubMed] [Google Scholar]
- 14.Ventura RM, Takata CS, Silva RA, Nunes VL, Takeda GF et al. (2000) Molecular and morphological studies of Brazilian Trypanosoma evansi stocks: the total absence of kDNA in trypanosomes from both laboratory stocks and naturally infected domestic and wild mammals. J Parasitol 86: 1289–1298. [DOI] [PubMed] [Google Scholar]
- 15.Njiru ZK, Constantine CC, Masiga DK, Reid SA, Thompson RC et al. (2006) Characterization of Trypanosoma evansi type B. Infect Genet Evol 6: 292–300. [DOI] [PubMed] [Google Scholar]
- 16.Borst P, Fase-Fowler F, Gibson WC (1987) Kinetoplast DNA of Trypanosoma evansi. Mol Biochem Parasitol 23: 31–38. [DOI] [PubMed] [Google Scholar]
- 17.Verloo D, Magnus E, Büscher P (2001) General expression of RoTat 1.2 variable antigen type in Trypanosoma evansi isolates from different origin. Vet Parasitol 97: 183–189. [DOI] [PubMed] [Google Scholar]
- 18.Bajyana Songa E, Hamers R (1988) A card agglutination test (CATT) for veterinary use based on an early VAT RoTat 1/2 of Trypanosoma evansi. Ann Soc Belg Med Trop 68: 233–240. [PubMed] [Google Scholar]
- 19.Ngaira JM, Olembo NK, Njagi EN, Ngeranwa JJ (2005) The detection of non-RoTat 1.2 Trypanosoma evansi. Exp Parasitol 110: 30–38. [DOI] [PubMed] [Google Scholar]
- 20.Birhanu H, Fikru R, Mussa S, Weldu K, Tadesse G et al. (2015) Epidemiology of Trypanosoma evansi and Trypanosoma vivax in domestic animals from selected districts of Tigray and Afar regions, Northern Ethiopia. Parasit Vectors 8: 212 10.1186/s13071-015-0818-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagos A, Yilkal K, Esayass T, Alemu T, Fikru R et al. (2009) Parasitological and serological survey on trypanosomosis (surra) in camels in dry and wet areas of Bale Zone, Oromyia Region, Ethiopia. Rev Méd Vét 160: 569–573. [Google Scholar]
- 22.Salim B, Bakheit MA, Kamau J, Nakamura I, Sugimoto C (2011) Molecular epidemiology of camel trypanosomiasis based on ITS1 rDNA and RoTat 1.2 VSG gene in the Sudan. Parasit Vectors 4: 31 10.1186/1756-3305-4-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boid R (1988) Isoenzyme characterisation of 15 stocks of Trypanosoma evansi isolated from camels in the Sudan. Trop Med Parasitol 39: 45–50. [PubMed] [Google Scholar]
- 24.Sánchez E, Perrone T, Recchimuzzi G, Cardozo I, Biteau N et al. (2015) Molecular characterization and classification of Trypanosoma spp. Venezuelan isolates based on microsatellite markers and kinetoplast maxicircle genes. Parasit Vectors 8: 536 10.1186/s13071-015-1129-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Claes F, Radwanska M, Urakawa T, Majiwa PA, Goddeeris B et al. (2004) Variable Surface Glycoprotein RoTat 1.2 PCR as a specific diagnostic tool for the detection of Trypanosoma evansi infections. Kinetoplastid Biol Dis 3: 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Njiru ZK, Ouma JO, Enyaru JC, Dargantes AP (2010) Loop-mediated Isothermal Amplification (LAMP) test for detection of Trypanosoma evansi strain B. Exp Parasitol 125: 196–201. 10.1016/j.exppara.2010.01.017 [DOI] [PubMed] [Google Scholar]
- 27.Hagos A, Goddeeris BM, Yilkal K, Alemu T, Fikru R et al. (2010) Efficacy of Cymelarsan and Diminasan against Trypanosoma equiperdum infections in mice and horses. Vet Parasitol 171: 200–206. 10.1016/j.vetpar.2010.03.041 [DOI] [PubMed] [Google Scholar]
- 28.Dean S, Gould MK, Dewar CE, Schnaufer AC (2013) Single point mutations in ATP synthase compensate for mitochondrial genome loss in trypanosomes. Proc Natl Acad Sci U S A 110: 14741–14746. 10.1073/pnas.1305404110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schnaufer A (2010) Evolution of dyskinetoplastic trypanosomes: how, and how often? Trends Parasitol 26: 557–558. 10.1016/j.pt.2010.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gould MK, Schnaufer A (2014) Independence from Kinetoplast DNA maintenance and expression is associated with multidrug resistance in Trypanosoma brucei in vitro. Antimicrob Agents Chemother 58: 2925–2928. 10.1128/AAC.00122-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El Rayah IE, Kaminsky R, Schmid C, El Malik KH (1999) Drug resistance in Sudanese Trypanosoma evansi. Vet Parasitol 80: 281–287. [DOI] [PubMed] [Google Scholar]
- 32.Payne RC, Sukanto IP, Partoutomo S, Jones TW, Luckins AG et al. (1994) Efficacy of Cymelarsan in Friesian Holstein calves infected with Trypanosoma evansi. Trop Anim Health Prod 26: 219–226. [DOI] [PubMed] [Google Scholar]
- 33.Boid R, Jones TW, Payne RC (1989) Malic enzyme type VII isoenzyme as an indicator of suramin resistance in Trypanosoma evansi. Exp Parasitol 69: 317–323. [DOI] [PubMed] [Google Scholar]
- 34.Zhou J, Shen J, Liao D, Zhou Y, Lin J (2004) Resistance to drug by different isolates Trypanosoma evansi in China. Acta Trop 90: 271–275. [DOI] [PubMed] [Google Scholar]
- 35.Brun R, Lun ZR (1994) Drug sensitivity of Chinese Trypanosoma evansi and Trypanosoma equiperdum isolates. Vet Parasitol 52: 37–46. [DOI] [PubMed] [Google Scholar]
- 36.Sultana K (1996) The effects of dexamethasone sodium phosphate on weight of bone (tibia) in mice. Pak J Pharm Sci 9: 298–303. [PubMed] [Google Scholar]
- 37.Herbert WJ, Lumsden WHR (1976) Trypanosoma brucei: A rapid ''matching'' method for estimating the host's parasitemia. Exp Parasitol 40: 427–431. [DOI] [PubMed] [Google Scholar]
- 38.Pyana Pati P, Ngay Lukusa I, Mumba Ngoyi D, Van Reet N, Kaiser M et al. (2011) Isolation of Trypanosoma brucei gambiense from cured and relapsed sleeping sickness patients and adaptation to laboratory mice. PLoS Negl Trop Dis 5: e1025 10.1371/journal.pntd.0001025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Büscher P, Mumba ND, Kabore J, Lejon V, Robays J et al. (2009) Improved models of mini Anion Exchange Centrifugation Technique (mAECT) and Modified Single Centrifugation (MSC) for sleeping sickness diagnosis and staging. PLoS Negl Trop Dis 3: e471 10.1371/journal.pntd.0000471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Reet N, Pyana PP, Deborggraeve S, Büscher P, Claes F (2011) Trypanosoma brucei gambiense: HMI-9 medium containing methylcellulose and human serum supports the continuous axenic in vitro propagation of the bloodstream form. Exp Parasitol 128: 285–290. 10.1016/j.exppara.2011.02.018 [DOI] [PubMed] [Google Scholar]
- 41.Hirumi H, Hirumi K (1989) Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J Parasitol 75: 985–989. [PubMed] [Google Scholar]
- 42.Van Reet N, Van de Vyer H, Pyana PP, Van der Linden AM, Büscher P (2014) A panel of Trypanosoma brucei strains tagged with blue and red-shifted luciferases for bioluminescent imaging in murine infection models. PLoS Negl Trop Dis 8: e3054 10.1371/journal.pntd.0003054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urakawa T, Verloo D, Moens L, Büscher P, Majiwa PA (2001) Trypanosoma evansi: cloning and expression in Spodoptera frugiperda insect cells of the diagnostic antigen RoTat1.2. Exp Parasitol 99: 181–189. [DOI] [PubMed] [Google Scholar]
- 44.Domingo GJ, Palazzo SS, Wang B, Pannicucci B, Salavati R et al. (2003) Dyskinetoplastic Trypanosoma brucei contains functional editing complexes. Eukaryot Cell 2: 569–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graf FE, Ludin P, Wenzler T, Kaiser M, Brun R et al. (2013) Aquaporin 2 mutations in Trypanosoma brucei gambiense field isolates correlate with decreased susceptibility to pentamidine and melarsoprol. PLoS Negl Trop Dis 7: e2475 10.1371/journal.pntd.0002475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gillingwater K, Büscher P, Brun R (2007) Establishment of a panel of reference Trypanosoma evansi and Trypanosoma equiperdum strains for drug screening. Vet Parasitol 148: 114–121. [DOI] [PubMed] [Google Scholar]
- 47.Stevens JR, Nunes VL, Lanham SM, Oshiro ET (1989) Isoenzyme characterization of Trypanosoma evansi isolated from capybaras and dogs in Brazil. Acta Trop 46: 213–222. [DOI] [PubMed] [Google Scholar]
- 48.Elhaig MM, Youssef AI, El-Gayar AK (2013) Molecular and parasitological detection of Trypanosoma evansi in camels in Ismailia, Egypt. Vet Parasitol 198: 214–218. 10.1016/j.vetpar.2013.08.003 [DOI] [PubMed] [Google Scholar]
- 49.Amer S, Ryu O, Tada C, Fukuda Y, Inoue N et al. (2011) Molecular identification and phylogenetic analysis of Trypanosoma evansi from dromedary camels (Camelus dromedarius) in Egypt, a pilot study. Acta Trop 117: 39–46. 10.1016/j.actatropica.2010.09.010 [DOI] [PubMed] [Google Scholar]
- 50.Masiga DK, Ndung'u K, Tweedie A, Tait A, Turner CM (2006) Trypanosoma evansi: genetic variability detected using amplified restriction fragment length polymorphism (AFLP) and random amplified polymorphic DNA (RAPD) analysis of Kenyan isolates. Exp Parasitol 114: 147–153. [DOI] [PubMed] [Google Scholar]
- 51.Hirumi H, Martin S, Hirumi K, Inoue N, Kanbara H et al. (1997) Cultivation of bloodstream forms of Trypanosoma brucei and T. evansi in a serum-free medium. Trop Med Int Health 2: 240–244. [DOI] [PubMed] [Google Scholar]
- 52.Kaminsky R, Brun R (1998) In vitro and in vivo activities of trybizine hydrochloride against various pathogenic trypanosome species. Antimicrob Agents Chemother 42: 2858–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zweygarth E, Rottcher D (1986) In vitro cultivation of African stocks of Trypanosoma (Trypanozoon) brucei evansi. Ann Soc Belg Med Trop 66: 145–151. [PubMed] [Google Scholar]
- 54.Sahin A, Asencio C, Izotte J, Pillay D, Coustou V et al. (2014) The susceptibility of Trypanosoma congolense and Trypanosoma brucei to isometamidium chloride and its synthetic impurities. Vet Parasitol 203: 270–275. 10.1016/j.vetpar.2014.04.002 [DOI] [PubMed] [Google Scholar]
- 55.Kaminsky R, Schmid C, Lun ZR (1997) Susceptibility of dyskinetoplastic Trypanosoma evansi and T. equiperdum to isometamidium chloride. Parasitol Res 83: 816–818. [DOI] [PubMed] [Google Scholar]
- 56.Raz B, Iten M, Grether-Buhler Y, Kaminsky R, Brun R (1997) The Alamar Blue assay to determine drug sensitivity of African trypanosomes (T.b. rhodesiense and T.b. gambiense) in vitro. Acta Trop 68: 139–147. [DOI] [PubMed] [Google Scholar]
- 57.Van Reet N., Pyana P, Rogé S, Claes F, Büscher P (2013) Luminescent multiplex viability assay for Trypanosoma brucei gambiense. Parasit Vectors 6: 207 10.1186/1756-3305-6-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alsford S, Eckert S, Baker N, Glover L, Sanchez-Flores A et al. (2012) High-throughput decoding of antitrypanosomal drug efficacy and resistance. Nature 482: 232–236. 10.1038/nature10771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baker N, Hamilton G, Wilkes JM, Hutchinson S, Barrett MP et al. (2015) Vacuolar ATPase depletion affects mitochondrial ATPase function, kinetoplast dependency, and drug sensitivity in trypanosomes. Proc Natl Acad Sci U S A 112: 9112–9117. 10.1073/pnas.1505411112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Munday JC, Settimo L, de Koning HP (2015) Transport proteins determine drug sensitivity and resistance in a protozoan parasite, Trypanosoma brucei. Front Pharmacol 6: 32 10.3389/fphar.2015.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Munday JC, Tagoe DN, Eze AA, Krezdorn JA, Rojas Lopez KE et al. (2015) Functional analysis of drug resistance-associated mutations in the Trypanosoma brucei adenosine transporter 1 (TbAT1) and the proposal of a structural model for the protein. Mol Microbiol 96: 887–900. 10.1111/mmi.12979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Munday JC, Eze AA, Baker N, Glover L, Clucas C et al. (2014) Trypanosoma brucei aquaglyceroporin 2 is a high-affinity transporter for pentamidine and melaminophenyl arsenic drugs and the main genetic determinant of resistance to these drugs. J Antimicrob Chemother 69: 651–663. 10.1093/jac/dkt442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Witola WH, Inoue N, Ohashi K, Onuma M (2004) RNA-interference silencing of the adenosine transporter-1 gene in Trypanosoma evansi confers resistance to diminazene aceturate. Exp Parasitol 107: 47–57. [DOI] [PubMed] [Google Scholar]
- 64.Zweygarth E, Kaminsky R, Webster P (1990) Trypanosoma brucei evansi: dyskinetoplasia and loss of infectivity after long-term in vitro cultivation. Acta Trop 48: 95–99. [DOI] [PubMed] [Google Scholar]
- 65.Schnaufer A, Clark-Walker GD, Steinberg AG, Stuart K (2005) The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J 24: 4029–4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All F1 ATP synthase gamma subunit sequence files are available from the GenBank database (accession numbers KT934830, KT934831, KT934832, KT934833, KT934834, KT934835, KT934836, KT934837, KT934838).