Abstract
Mutant mouse models, genetically-engineered or spontaneous-mutations, serve as valuable tools for biomedical research. Genotyping of mutant mice is a critical requirement for maintaining the colony, to breed with other mutants and to match the phenotypic observations. The SCID mouse model has been extensively used as a common background-strain in many immunology and transplantation studies. Many different types of assays, including Restriction Fragment Length Polymorphism (RFLP), confronting two primer pairs PCR and end-point methods have been attempted for establishing a genotyping protocol for the SCID mutation. However, the best method that is thought to be reliable is sequencing, which requires additional time and resources to perform on a routine basis. In this report we describe a novel RFLP assay that is simple and reliable. The method is validated by sequencing analysis and this novel method can be adapted for routine genotyping of SCID model.
Keywords: SCID, NSG, Humanized mice, RFLP, Genotyping
Technical Note
Although genotyping is routinely performed in many labs, the genotyping strategies available for certain mutations are challenging (Han et al. 2006; McHugh et al. 2012). Inconclusive genotyping can often result in loosing of the precious strains, contamination of other mutations and also irreproducible results. A recent report indicated that over 15% of lines deposited to public repositories, like Jaxmice and MMRRC, did not carry the specified mutation by the depositor (Lloyd et al. 2015). Therefore it is of high importance to establish reliable genotyping protocols for every mutation. Further, the point mutations and short insertions and/or deletions can be difficult using PCR assay, the most common and quickest method used for thousands of genetically engineered mouse models. The SCID mouse model was first reported by Bosma, Custer, and Bosma (1983), and has been extensively used as a common background-strain in many immunology and transplantation studies (Rongvaux et al. 2014). The SCID genetic change is a recessive mutation in the Prkdc gene (Protein Kinase, DNA-Activated, Catalytic Polypeptide), which is a single base-pair substitution (T to A at codon 4045) that results in a premature stop codon (Blunt et al. 1996). The genotyping methods for identifying this single nucleotide change mutation have been challenging. We and other labs have experienced difficulty in genotyping this strain using the previously reported genotyping protocols. Therefore we sought to develop a reliable genotyping protocol for this strain.
Originally, this mutation was genotyped by using a PCR-RFLP (Restriction Fragment Length Polymorphism) technique in which the PCR product amplified from the target region is digested using AluI restriction enzyme (Blunt et al. 1996). The banding pattern of the RFLP assay was 241bp and 211bp bands for wild type and mutant bands respectively. However, because of the difficulty in resolving the bands, the reliability of this assay was questioned by Sealey, Hobbs, and Schmidt 2002 and they concluded that PCR-RFLP is unreliable and sequencing would be the best method to genotype the models accurately. Unlike PCR-RFLP method, which is quick and simple, sequencing takes more time to perform and is not a preferred method for routine genotyping. Subsequently, another RFLP method, with a different set of RFLP band sizes (mutant; 38bp, 28bp &11bp: het; 68bp, 38bp, 28bp & 11bp and wild type; 68bp & 11bp) was developed at Jax labs. The latter RFLP assay was also found to be challenging; it was discontinued and recently switched to an end-point analysis method (http://jaxmice.jax.org/strain/005557.html). The end-point analysis method is, again, not routinely used in many labs and it depends on the reliability of real time PCR, pipetting accuracy and also requires complex analysis for interpretation. In this report we developed a new RFLP assay with easily resolvable banding pattern (mutant; 162bp, 91bp & 30bp: het; 162bp, 121bp, 91bp & 30bp and wild type; 162bp & 121bp) that detects the mutant and het and wild type alleles very reliably. Further, we demonstrate that the new RFLP assay results match perfectly with the sequencing results.
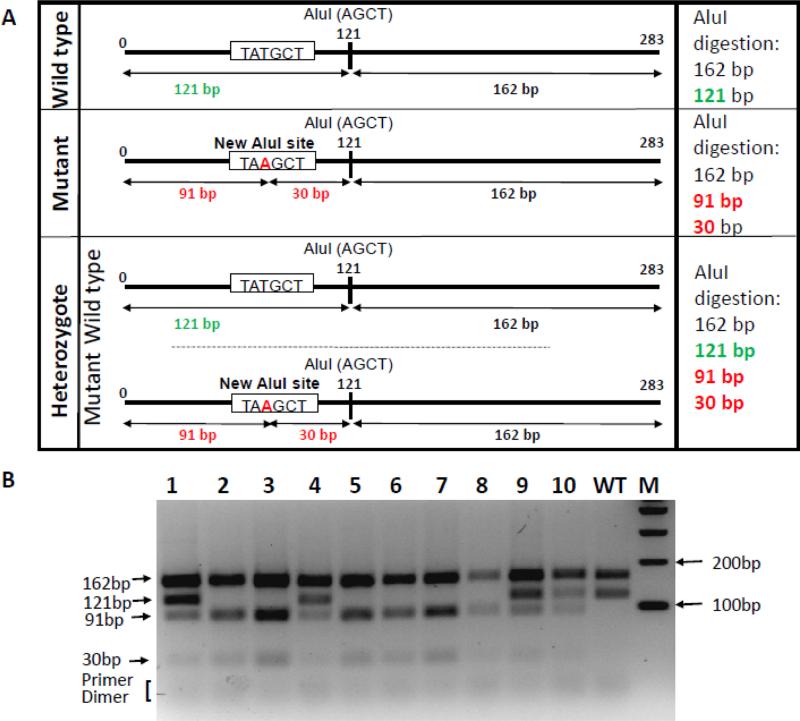
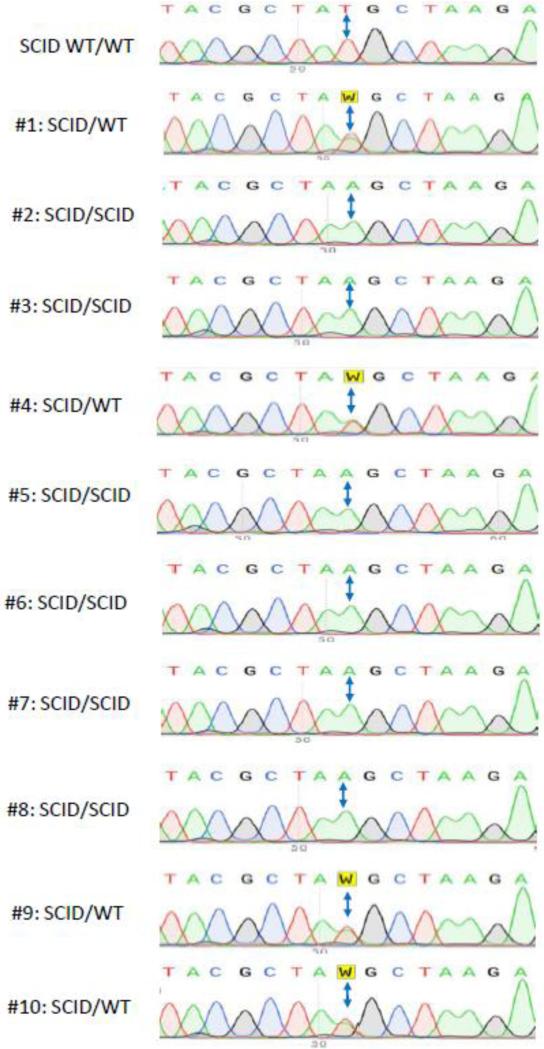
For the new RFLP assay, we designed two new primers (RQ-SCID-Gtp-F:GAGCAGACAATGCTGAGAAAAGGAGG and RQ-SCID-Gtp-R:GCCCAGTCCTTGCAGGCTGGGCTTGG), that bind 91 bp upstream and 192 bases downstream from the mutation site (TATGCT to TAAGCT: mutation nucleotide is bold faced and the AluI-RE site is underlined). The primers amplify a 283 bp band flanking the SCID mutation. The expected banding pattern of AluI digestion results are given in Figure 1A. The digested products were run in 3% Agarose gel. Genotyping results of the representative litter shown in Figure 1B is from a heterozygous SCID mutant male and a NSG female (homozygous for SCID); the samples are either heterozygotes or homozygotes. Figure 2 shows sequencing of all the samples from this litter including a wild type sample showing that PCR-RFLP and sequencing results agree with each other. Notably, the new RFLP strategy has been yielding reliable results, and is successfully used in our laboratory for over two years.
Figure 1.
(A) Schematic of SCID mutation genotyping by PCR-AluI-RFLP. Shown are the schematics of wild type, mutant and heterozygote SCID alleles and the expected band sizes from these alleles. (B) A representative 3% agarose gel image showing genotyping of a litter obtained by breeding of a homozygous and a heterozygous SCID mutant parents. As expected, the samples are either heterozygotes (lanes 1, 4,, 9, and 10) or homozygotes (lanes 2, 3, and 5-8). A wild type DNA was included as a control (lane: WT). The lane marked ‘M’ was loaded with 100 base pair marker showing only up to 400 bp bands
Figure 2.
Sequencing of offspring genotyped in Figure 1. chromatograms showing the SCID mutation double headed arrow).
One of the reasons for the unreliable outcomes of the previous two RFLP methods was that they relied on bands of very small sizes (as small as 11 bp) and small difference in sizes (as less as 17 bp difference). Such small sized bands and short size differences makes quite challenging to resolve using agarose gel electrophoresis. In comparison, the differences and sizes in the banding-pattern of new RFLP assay (smallest band as 30 bp and the difference between the closely running bands is 61 bp) are quite easy to resolve in a standard agarose gel electrophoresis (Figure1A). Although another strategy using Confronting Two-Pair primers PCR was available for genotyping the SCID mutation (Maruyama et al. 2002), this method was not suggested on the Jaxmice website, instead a previously developed PCR-RFLP assay was followed until 2014.
In summary, we developed a new RFLP method that can be used in many labs for routine genotyping of SCID mutation. Considering that many genetically engineered mutations are bred routinely with the SCID mutation, the assay described here will come as a very handy tool for many researchers.
Acknowledgements
This work was partially supported by the National Institute of General Medical Sciences of the National Institutes of Health under Grant no. P20GM103471 and the Center for Humanized Mice from ORIP/DPCPSI/NIH/1R24OD018546-01. We acknowledge Santhi Gorantla for reading the manuscript, Donald W Harms, Jaclyn S. Knibbe and Poonam S. Joshi for their help in mice breeding. RQ and CBG acknowledge the Nebraska Research Initiative and UNMC Vice-Chancellor for Research Office for supporting the mouse genome engineering core facility.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blunt T, Gell D, Fox M, Taccioli GE, Lehmann AR, Jackson SP, Jeggo PA. Identification of a Nonsense Mutation in the Carboxyl-Terminal Region of DNA- Dependent Protein Kinase Catalytic Subunit in the Scid Mouse. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(19):10285–90. doi: 10.1073/pnas.93.19.10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosma GC, Custer RP, Bosma MJ. A Severe Combined Immunodeficiency Mutation in the Mouse. Nature. 1983;301(5900):527–30. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- Han Dawn D., Chen Rong, Hill Erik R., Tilley Michael R., Gu Howard H. Cause and Solutions to the Polymerase Chain Reaction Smear Problem in Genotyping. Analytical Biochemistry. 2006;353(2):296–98. doi: 10.1016/j.ab.2006.03.041. doi:10.1016/j.ab.2006.03.041. [DOI] [PubMed] [Google Scholar]
- Lloyd Kent, Franklin Craig, Lutz Cat, Magnuson Terry. Reproducibility: Use Mouse Biobanks or Lose Them. Nature. 2015;522(7555):151–53. doi: 10.1038/522151a. doi:10.1038/522151a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama Chika, Suemizu Hiroshi, Tamamushi Shojiro, Kimoto Shigenobu, Tamaoki Norikazu, Ohnishi Yasuyuki. Genotyping the Mouse Severe Combined Immunodeficiency Mutation Using the Polymerase Chain Reaction with Confronting Two-Pair Primers (PCR-CTPP). Experimental Animals / Japanese Association for Laboratory Animal Science. 2002;51(4):391–93. doi: 10.1538/expanim.51.391. [DOI] [PubMed] [Google Scholar]
- McHugh Donal, O'Connor Tracy, Bremer Juliane, Aguzzi Adriano. Tang Ya-Ping., editor. ZyFISH: A Simple, Rapid and Reliable Zygosity Assay for Transgenic Mice. PLoS ONE. 2012;7(5):e37881. doi: 10.1371/journal.pone.0037881. doi:10.1371/journal.pone.0037881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongvaux Anthony, Willinger Tim, Martinek Jan, Strowig Till, Gearty Sofia V, Teichmann Lino L, Saito Yasuyuki, et al. Development and Function of Human Innate Immune Cells in a Humanized Mouse Model. Nature Biotechnology. 2014;32(4):364–72. doi: 10.1038/nbt.2858. doi:10.1038/nbt.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sealey Amy L, Hobbs Nicole K, Schmidt Edward E. Molecular Genotyping of the Mouse Scid Allele. Journal of Immunological Methods. 2002;260(1-2):303–4. doi: 10.1016/s0022-1759(01)00524-5. doi:10.1016/S0022-1759(01)00524-5. [DOI] [PubMed] [Google Scholar]