Abstract
People of African ancestry (Blacks) have increased risk of kidney failure due to numerous socioeconomic, environmental, and clinical factors. Two variants in the APOL1 gene are now thought to account for much of the racial disparity associated with hypertensive kidney failure in Blacks. However, this knowledge has not been translated into clinical care to help improve patient outcomes and address disparities. GUARDD is a randomized trial to evaluate the effects and challenges of incorporating genetic risk information into primary care. Hypertensive, non-diabetic, adults with self-reported African ancestry, without kidney dysfunction, are recruited from diverse clinical settings and randomized to undergo APOL1 genetic testing at baseline (intervention) or at one year (waitlist control). Providers are educated about genomics and APOL1. Guided by a genetic counselor, trained staff return APOL1 results to patients and provide low-literacy educational materials. Real-time clinical decision support tools alert clinicians of their patients’ APOL1 results and associated risk status at the point of care. Our academic-community-clinical partnership designed a study to generate information about the impact of genetic risk information on patient care (blood pressure and renal surveillance) and on patient and provider knowledge, attitudes, beliefs, and behaviors. GUARDD will help establish the effective implementation of APOL1 risk-informed management of hypertensive patients at high risk of CKD, and will provide a robust framework for future endeavors to implement genomic medicine in diverse clinical practices. It will also add to the important dialogue about factors that contribute to and may help eliminate racial disparities in kidney disease.
Keywords: Genetics, Chronic Kidney Disease, Disparities, African Ancestry, Race, Community-Based Research
INTRODUCTION
While a growing number of initiatives integrate genetic testing into clinical care, few focus on genetic risk factors for common chronic diseases, in part because there are few identified genetic variants that increase the risk of such illnesses. New knowledge of genetic variants that confer an increased risk of chronic kidney disease (CKD) has not been translated to clinical care. We describe the design and implementation of a randomized trial exploring the impact on patients, clinicians and clinical care, of incorporating genetic risk information for CKD into the treatment of hypertensive adults of African ancestry (Blacks) in diverse primary care settings. To our knowledge, this is the first genomic medicine program to integrate genetic risk for a common chronic disease into primary care.
CKD is commonly associated with hypertension (28%) and affects 26 million adults in the US. Blacks with hypertension have a 5-fold increased risk of end stage renal disease (ESRD) compared to Whites. Previous studies have implicated the APOL1 gene in increasing the risk for CKD and ESRD [1]. This increased risk is conferred by two variants (G1 and G2) in the last exon of APOL1 [2, 3]. The presence of two APOL1 risk variants confers a 5-fold increased risk for hypertensive CKD and a 7-fold increased risk for hypertension-attributed ESRD [1–4]. The added risk is lower among adults who also have diabetes [5, 6]. While one in seven Black adults carry two APOL1 risk variants [5, 6], these variants are nearly absent in other populations. This is likely because the G1 and G2 variants protect against infection with African trypanosomiasis (sleeping sickness) that is transmitted by tsetse flies in sub-Saharan Africa. As the disease is endemic to Africa, these variants are almost exclusively present in Blacks, in a manner similar to the high prevalence of sickle cell trait in regions exposed to malaria [7].
While it remains critical to recognize the contribution of multiple factors, particularly social determinants, to chronic disease disparities [8–15], genomic contributors also warrant careful evaluation [16]. High-risk APOL1 variants are thought to explain approximately 70% of the excess prevalence of CKD in Blacks [4]. As patients are not routinely screened for their APOL1 status, it is unknown whether patient or clinician knowledge of this genetic risk for CKD impacts patient care (i.e., renal surveillance, antihypertensive medication intensification), patient affect or behaviors such as anxiety and medication adherence, or patient outcomes such as blood pressure control and CKD.
GUARDD (Genetic testing to Understand and Address Renal Disease Disparities) is a randomized trial designed to determine the effects and challenges of incorporating APOL1 information into primary care management of Black adults with hypertension. Led by an academic-community-clinical partnership, GUARDD was designed to assess primary outcomes, including blood pressure reduction and renal surveillance, secondary psycho-behavioral outcomes, and best processes to improve adoption of genomic medicine.
METHODS
Study Overview
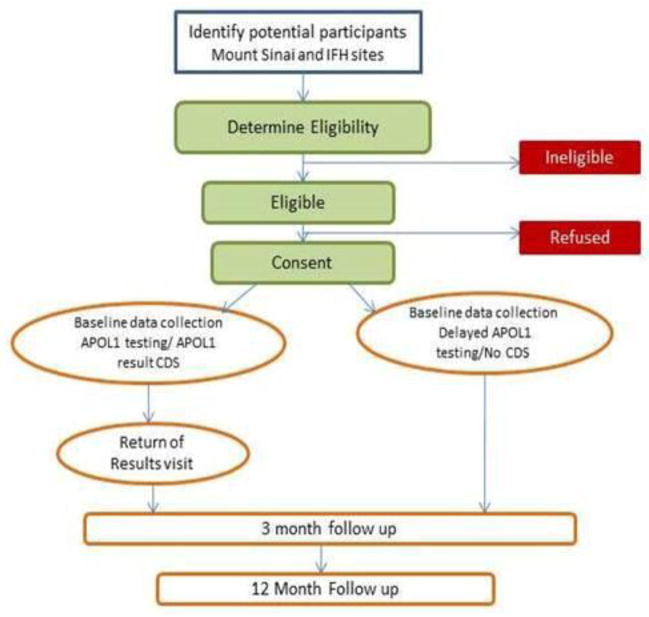
As shown in Figure 1, at participating clinical sites, study coordinators enroll eligible, interested, consented patients, collect a baseline survey and clinical measures, and randomize patients to immediate (intervention group) or delayed (control group) APOL1 testing. Patients in the intervention group receive their results from study coordinators, and the results are then sent to their primary care clinicians via an electronic health record (EHR) best practice alert. All patients are scheduled to return for 3 and 12-month follow up visits and control patients receive APOL1 testing at 12 months. The study received Institutional Review Board approval at all sites.
Figure 1.
Study Flow
The study has two primary endpoints, comparing patients who are APOL1 positive (high risk) and APOL1 negative at three months after enrollment. The primary aim is a renal care endpoint, the correct utilization, by clinicians, of serum creatinine and/or urine albumin tests. The primary sub-aim is reduction of systolic blood pressure. Secondary outcomes include impact on primary outcomes at 12 months, psycho-behavioral differences of patients between groups and over time, clinician knowledge, attitudes and beliefs at baseline and 12 months, and differences in outcomes between those tested and not tested. We will also conduct focus groups with study participants after study completion to investigate their experiences in more depth.
Stakeholder Engagement
Our team engages several stakeholder groups who planned and developed this transdisciplinary, translational research program. Stakeholders include our core team of genomics, health equity, community-based, chronic disease, informatics, and primary care researchers, primary care providers, patients, community leaders, biostatisticians, and experts in clinical decision support (CDS), genetic counseling, hypertension, APOL1 and CKD. GUARDD has a Community Board including community leaders of African ancestry, clinicians and patients with the APOL1 risk variant. They participated in designing the study and conceptual framework, including eligibility criteria, consent procedures, patient and clinician educational materials, surveys, and recruitment strategies and materials. They meet on a monthly basis to discuss study status and challenges, using principles of community-based participatory research to guide their work and ensure meaningful participation [17, 18]. A Scientific Board, comprised of experts in APOL1, genomics, health disparities, genetic counseling, hypertension and CKD, reviews key study materials and decisions.
Recruitment sites
The study takes place in two large clinical entities. The Mount Sinai Health System is a large integrated health system serving approximately 3.5 million patients, with over 140 ambulatory practice locations in the New York City area. The Institute for Family Health (IFH), a network of federally qualified health centers, provides comprehensive, family practice-based care to over 100,000 patients yearly.
Development of Study Materials
Formative Research
To inform study design, surveys, and to pilot test consent forms, return of results procedures, and patient and clinician education materials, we conducted and analyzed semi-structured interviews with 15 primary care clinicians, and with 26 patients of African Ancestry before they underwent APOL1 testing and after receiving their test results from a genetic counselor.
Survey Development
The GUARDD team developed patient and provider surveys to ascertain knowledge, attitudes, beliefs and behaviors at study enrollment, at 3 and 12-month follow up for patients, and at enrollment and 12-month follow up for clinicians. Guided by literature review, results of the formative interviews and consultation with experts, the team built a conceptual framework (see Results section) and developed surveys, primarily using scales validated for use in diverse populations.
For the patient survey, we ask about general health [19], family history of hypertension and kidney disease, comorbidities [20], beliefs about hypertension and kidney disease [21], beliefs about medications [22], medication adherence [23], patient-provider relationship including communication and trust and satisfaction [24]. We also ask about perceived racism [25], patient activation [26], knowledge about genetics [27, 28], depression [29], general anxiety [30], perceived stress [31], life chaos [32], social support [33], health literacy [34], access to health care [35], diet and physical activity [36], health care utilization [37], and demographics [36]. We developed items related to additional domains including genetic testing, APOL1 testing and anticipated reaction toward testing. Follow-up surveys added items to assess reaction to the testing and results, test-related distress [38], satisfaction and decision regret around testing [39, 40].
For the clinician survey, we assess history of receiving genetics education, perceived knowledge of genetics and genomics [41–44], experience, comfort and concerns with genetic testing for chronic disease risk [43], experience with APOL1 genetic testing, connections between ancestry and genetic risk for common disease, utility of genomic medicine and CDS [41, 45], preferred type of CDS, and demographics. In the provider follow-up survey, we add items to assess perceived utility and use of genomic CDS to help care for patients who have APOL1 genetic testing.
In an iterative manner, the team conducted in-depth pilot testing of the surveys among Black community residents with hypertension, and with providers, and revised the surveys as needed. The final 166-item patient survey is approximately 30 minutes in length; the 45-item clinician survey takes about 5 minutes.
Clinical measures
At baseline, 3 and 12 month follow-up, trained study coordinators measure blood pressure digitally, taking three measurements, two minutes apart, discarding the first measurement and recording the second and the third measurements to calculate a mean blood pressure [46]. They measure weight with portable high capacity platform scales and height with portable stadiometers. Measures of renal testing and medications prescribed come directly from EHRs.
Testing and Return of Results
APOL1 genetic testing is performed by a clinically validated assay that interrogates the G1 (c.[1072A>G;1200T>G) and G2 (c.1212_1217del6) variants by multiplex allele-specific primer extension bead-based genotyping (Luminex, Austin, TX). Testing is conducted in a Clinical Laboratory Improvement Amendments (CLIA), New York State (NYS) approved,College of American Pathologists (CAP) accredited clinical laboratory. Coordinators return results under the guidance of the study’s genetic counselor who carefully trained and supervised them during their first return of results visits. Patients receive a “positive” result if they are homozygous or compound heterozygous for G1 and/or G2 variants, and a “negative” result if they are heterozygous G1 or G2 carriers or homozygous wild type. Patients with negative results have their results returned over the phone, which takes about 10 minutes, but are invited to meet with the study coordinator if desired. Patients with a positive result must come in for their results during an approximately 15 minute visit. All patients are offered the opportunity to speak or meet with the study’s genetic counselor, and all receive their genetic, blood pressure and body mass index results with lay explanations in person or by mail along with a letter for their provider. They receive an educational pamphlet with graphics. After returning results to the patient, coordinators activate a dataflow pipeline (see next section), which triggers a best practice alert containing the results that fire at the next clinician’s encounter with the patient. The team chose to have coordinators return all results to ensure that all patients would receive standardized information about their results in a timely manner. This entire process was informed by the formative interviews, developed in partnership with the Community Board and our experts, tailored by and for patients of African ancestry and for low-literate populations, and extensively piloted and revised.
Clinical Decision Support and Data Flow
GUARDD is designed to inform clinicians of their patients’ APOL1 results and associated risk of CKD at the point of care, without the need for explicit look up, and with recommendations pertaining to management of hypertension and monitoring of renal function. Our CDS delivers GUARDD best practice alerts that are crisp yet informative, and have one-click links to study-specific provider and patient information. There is privacy-protected, accurate dataflow between the EHR, genetic testing lab and study collection team. CLinical Implementation of Personalized Medicine through Electronic health Records and Genomics (CLIPMERGE), our independent CDS engine with bi-directional real-time communication with the EPIC EHR systems [47] receives data from participants’ EHRs and data collected by study personnel. CLIPMERGE is implemented in a Cloud instance with secure communications to the participating sites’ EPIC servers. The team built a Research Electronic Data Capture (REDCap) software database [48] to support the necessary workflow including study recruitment, randomization, collection of genetic test results, clinical measures and surveys. We customized this secure platform to include: (1) A list of eligible study participants, as per EPIC data queries; (2) Fields for participant demographics that can be supplemented by recruiters; (3) Set logic to determine subject eligibility based on screening questions by recruiters; (4) Randomization scheme, uploaded prior to the beginning of the study; (5) Follow-up information and scheduling using REDCap’s longitudinal module, calendar and scheduling functionalities; (6) Customizable report builder for coordinators to monitor and track their patients and build calendars to show all baseline, follow-up, return of results, patient intercept and reminder call activities; and (7) Complex report builder for the study leads to track and analyze study progress using R software [49–51].
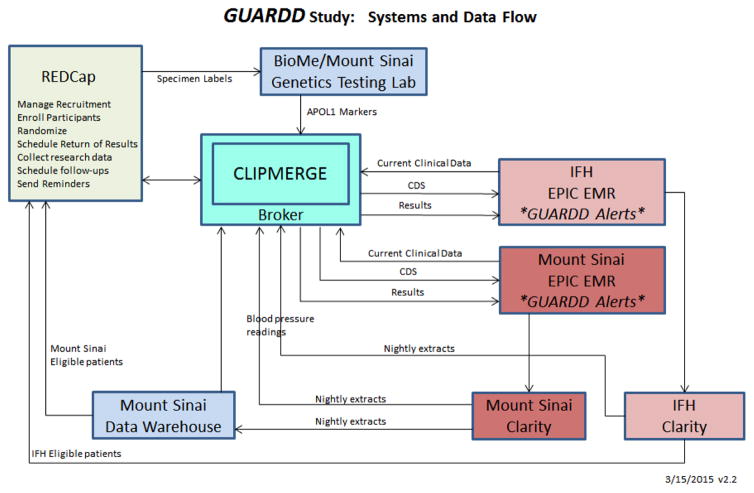
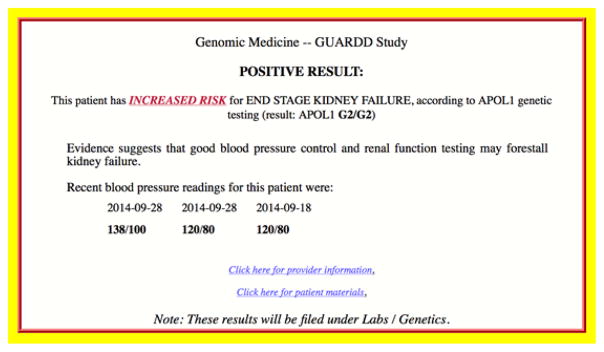
As shown in Figure 2, eligible patients are identified by custom EHR queries. Their contact information flows into REDCap, where a subject numbering scheme allows re-identification when necessary to support patient contact or data transmissions in and out of the EHR. When patients undergo the genetic test, their de-identified specimens are sent to the CLIA-certified Mount Sinai Genetic Testing Lab, which populates in REDCap via CLIPMERGE so that results can be returned by the study team. Once the study team returns the results to a participant, CLIPMERGE integrates this into the EHR. When a participating primary care provider opens an encounter, CLIPMERGE determines within one-half second whether an alert should be sent, and if so, formulates and returns a GUARDD alert to EPIC (Figure 3). Alerts were carefully developed for readability and ability to easily differentiate positive from negative results, accompanied by recent blood pressure readings. The alert includes links to information pages for providers and patients with one-click access and one-click printing. Once the alert is presented to a provider, CLIPMERGE files the result in EPIC under Lab Results in a Genetics folder, so clinicians can easily find the result at any time.
Figure 2.
Systems and Data Flow supporting the GUARDD Study
Figure 3.
GUARDD Best Practice Alert
Clinician Recruitment and Data Collection
Clinicians at all participating sites are informed about GUARDD and enrolled into the study during regularly scheduled group clinical meetings. A medical geneticist presents a brief education session about GUARDD study, including background information on hypertension, CKD, APOL1, and potential outcomes. Clinicians who consent to enroll their patients sign a letter, which the team sends to patients inviting them to join the study. Clinicians then fill out a baseline survey. Those who have not attended one of the information sessions are approached individually for education about GUARDD and enrollment into the study.
Participant Recruitment
We identify patients who meet our eligibility criteria through EHRs and confirm eligibility before enrollment. Inclusion criteria are: (1) Self-identified African American/Black or having African ancestry; (2) Age 18–70 years; (3) An ICD-9 diagnosis of hypertension (401.XX 402.XX AND 405.XX) and/or taking anti-hypertensive medications, and/or 2 systolic blood pressure readings >140 or 2 diastolic readings > 90 at least six months apart; and (4) Receiving primary care from one of the participating clinical sites. Exclusion criteria are: (1) Diabetes; (2) CKD; (3) Pregnancy; (4) Non-English speaking; (5) Planning on moving out of the area during the study period; (6) Having cognitive impairment; and (7) Not community dwelling. Coordinators mail potentially eligible patients a recruitment letter, call those who have not declined in response to the letter, and schedule enrollment. They call patients a minimum of 12 times, at least once in the evening and once on a weekend, and if not available, contact their listed next of kin.
To enhance recruitment, the team implements additional strategies, all developed in partnership with our community board, including study flyers, large poster boards and TV monitor advertisements in clinic waiting areas. We created a profile of the study on Team4Cure, a patient-centric web-based application (app) for patient recruitment into research trials [52]. These strategies also allow the patients with no listed race in our databases to self-identify as having African ancestry and enroll. We receive daily lists of clinic appointments so recruiters can intercept patients, assess their interest and eligibility, and enroll them. We also ask providers to refer eligible patients.
Recruiter training
Study procedures, protocols, and recruitment and return of results scripts are documented in a recruitment manual, which was the basis of study coordinator training. Coordinators are either of African ancestry or from the communities where we recruit patients, are trained in all aspects of human subjects protection, recruitment, consent, survey administration and clinical measures including genetic testing. As part of their training, coordinators learn procedures proven effective in recruiting minority patients and interview mock patients to receive feedback [53–60]. We hold biweekly recruiter meetings to discuss recruitment progress, highlight successful techniques, and brainstorm ways to handle difficult situations that arise.
Data Collection and Randomization
Study coordinators enroll patients and collect all study data, entering it directly into REDCap. Enrollment visits are scheduled around patients’ availability, including evenings and weekends. Study coordinators send reminder letters, and place reminder calls or text messages the day before the visit. During initial 1.5 hour visits, coordinators obtain informed consent, administer the survey, obtain clinical measures, and collect venous blood (or if not possible, saliva [62]) for genetic testing if the participant is randomized to the intervention. Randomization is stratified by the clinical site to create balanced in-group assignment and eliminate the risk of cross-contamination between facilities. The randomization schedule using block design is generated using SAS 9.3 software [63]. Coordinators receive the assignment for each participant via REDCap after baseline data collection is complete, so study assignment does not impact participants’ responses to the survey. They obtain alternate contact numbers and desired method of communication (phone, mail, text, e-mail) for each participant. Participants receive a form with their biological measurements, dates for their return of results contact, 3-and 12-month follow-up visits, and $40 in gift cards to a local retailer of their choice.
Retention/Follow-Up Visits
To help maintain high retention rates, we facilitate relationship building as coordinators are assigned to specific participants, mail them personalized birthday cards, and call them at 9 months after enrollment to check in. For those difficult to reach for follow-up visits, coordinators intercept patients when they come for clinical care and/or send a return receipt letter asking patients to call back.
Primary Endpoints and Sample Size
Outcome measures will be compared among three arms of the GUARDD study, the APOL1-positive and APOL1-negative intervention groups, and the control group. The study was designed to randomize 2050 patients to intervention or control in a 7:1 ratio. Of the 1800 intervention patients, approximately 250 will test positive. We will thus have approximately 250 high risk, 250 control, and 1550 low risk patients. The sample size for the study was calculated assuming a 10% improvement in practice guideline-appropriate renal function test ordering in the APOL1-positive group (40% estimated) vs. APOL1-negative group (30% estimated) to yield 87% power to detect the difference of interest using a two-sided chi-square test. The blood pressure sub-aim, 5mmHg improvement in systolic blood pressure at 3-month follow up in APOL1-positive compared with APOL1-negative group, can be detected with 95% power.
RESULTS
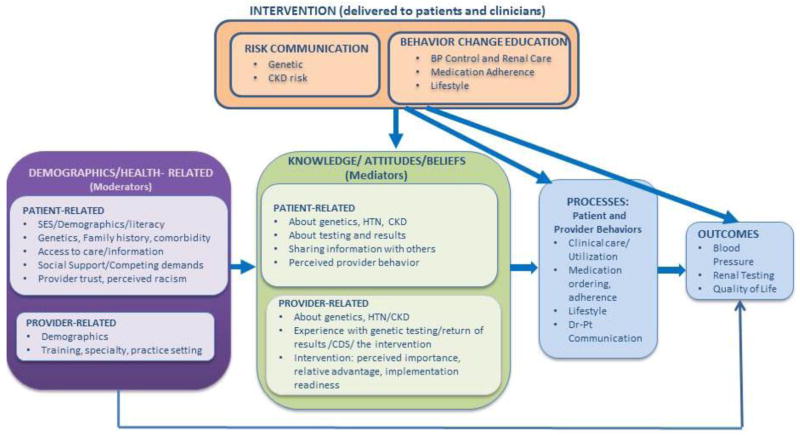
Formative research and piloting revealed several important factors that guided our study design and materials, and could inform similar studies. Study development also led to construction of a conceptual framework (Figure 4) to guide instrument development and our approach to patients and clinicians. The framework was based on the Common Sense Model [64, 65], developed in close collaboration with our advisory board and experts in genomic risk communication. It shows that patient and provider demographics and other health-related characteristics influence mediators such as knowledge, attitudes and beliefs about genetics, hypertension, and CKD. It also recognizes that these characteristics may independently impact outcomes. Mediators in turn impact patient and provider behaviors (processes) that are needed to impact outcomes. GUARDD, which is delivered to both patients and clinicians, focuses on risk communication and behavior change education targeting the listed patient and provider mediators, processes and outcomes.
Figure 4.
Conceptual Framework
Through study development, our advisory boards and formative research, we also uncovered important patient and clinician preferences for study content and conduct. We found that patients had several strong preferences. First, they asked for written results and information about hypertension, CKD, and APOL1; what they could do to prevent CKD, and if they are at high-risk, a glossary of terms; ways to talk to their doctor with this new information; and a blood pressure log. Second, they asked that we focus on specific details on what the test would mean for them. Third, they wanted the information from a trusted source, which could be their clinician, a genetic counselor, or a trained person from their background or community.
We also uncovered important clinician preferences. Clinicians asked us to provide a quick alert on their computers with the test result, and links to what genetic testing and APOL1 mean, and to materials they could give patients. Second, they preferred to receive very brief trainings on this information (minutes, not hours). Finally, they wanted the study team to make sure all patients received the results, as we were ordering the tests (the clinicians were not), but that we provide clinicians with the results as soon as their patients have received them.
DISCUSSION
The GUARDD study is designed to generate essential insights for sustainable adoption and large-scale dissemination of genomic medicine in diverse clinical settings providing care for common adult-onset diseases in general, and for underserved African Ancestry populations with large excess burden of non-diabetic kidney diseases specifically. GUARDD is a randomized controlled trial that tests patients of African ancestry for genetic variants that increase their risk of kidney failure, returns results to patients and providers (through EHRs), and evaluates impact on patients, providers, and clinical care. The study was developed in close partnership with patients, advocates and clinicians of diverse socioeconomic status, in diverse clinical settings, and uses mixed methods to plan the study and to evaluate impact.
For patients, the study is designed for and with people of African ancestry, to test the impact of sharing genetic information in the management of hypertension. In doing so, we are providing this priority population access to advances in science that may ameliorate the disproportionate prevalence and burden of chronic disease it faces. GUARDD’s procedures and materials are developed for and with diverse individuals of diverse educational and socioeconomic backgrounds. The team of recruiters, from the background and communities of the patients, employs low-literacy, engaging, easy to understand materials, flexible recruitment times and places, multiple methods of contact, including in-person, phone, text and via an app. Through carefully designed qualitative interviews and quantitative surveys, we will be able to determine the impact of genetic testing and return of results on patients’ attitudes, beliefs, behaviors and outcomes. Will patients, knowing they have a genetic variant, feel empowered or stigmatized? Will they engage in care to control their blood pressure, of feel they are doomed to renal disease and become less adherent? GUARDD should answer these questions. Race is a social construct, but ancestry has important biological implications, and GUARDD explicitly acknowledges and studies both.
For primary care providers, GUARDD is an opportunity to incorporate genomic medicine into their everyday practice and to do so in the context of a common chronic illness. Providers practicing today have had little if any training in discussing and addressing genetic and genomic risks with patients [66, 67], yet they will need to advise patients about the genetic and genomic aspects of disease. Will such testing fit neatly into the arsenal of other lab tests, such as renal function, so that clinicians will merely share patients’ risks with little explanation as to what kind of test it is? Will clinicians need to engage patients in discussions about genetics and genomics as part of testing? Providers need to be prepared to work with patients, and the medical community will need information on how best to efficiently and effectively weave this work into their busy practices, information we aim to provide through this study. Are clinicians prepared for genomics in primary care? GUARDD was built with clinical partners so that these questions and challenges can be asked and addressed.
For health systems, we developed a CDS system that exceeds the capabilities typically available within commercial EHRs. This includes storing actionable genetic results, holding back the filing of these genetic results in the EHR (while allowing other clinical genetics results to file), and providing an external CDS engine that can support complex decision and timing rules. We developed the content and format of GUARDD alerts based on user feedback during the formative stage of the project, and did so alongside front-line providers and clinical leaders of large, busy practices, so that their use could easily continue after the research is completed.
There is a paucity of research that adequately includes Blacks and adults with limited resources [68–70]. Yet, these populations have the poorest outcomes from common, chronic illnesses such as CKD, and are least likely to benefit from diagnostic and therapeutic advances [71, 72]. Translational genomic research also should take into account real-world primary care operations, and the need for real-time, point-of-care integration of test results with the EHR using CDS tools [73–75]. Hypertension-associated CKD and APOL1 genetic risk in Black populations is a highly relevant and well-suited opportunity for a ‘prototype’ genomic medicine demonstration project for common chronic illnesses managed in primary care settings. The approaches described to develop, conduct, and analyze this study provide a framework to implement genomic medicine in diverse clinical settings.
Supplementary Material
Acknowledgments
This study is supported by NHGRI (5U01HG007278, U01HG006380) and NCATS (UL1TR000067). Neither NHGRI nor NCATS had any involvement in the study design, collection, analysis or interpretation of data, writing of this article or decision to submit it for publication. The authors would also like to thank the GUARDD team of academic, community, clinical partners, study coordinators, and staff at study sites, and their partners in the IGNITE Network, a consortium of genomic medicine pilot demonstration projects funded and guided by the NHGRI, for their valuable contributions to this project. We would like to thank Linda Cameron, Richard Cooper, Howard Leventhal, Gbenga Ogedegbe, Martin Pollack, Saskia Sanderson and Stuart Scott for their input.
Footnotes
Trial Registration: ClinicalTrials.gov identifier NCT02234063
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
CR Horowitz, Email: carol.horowitz@mssm.edu.
NS Abul-Husn, Email: noura.abul-husn@mssm.edu.
S Ellis, Email: steve.ellis@mountsinai.org.
MA Ramos, Email: michelle.ramos@mountsinai.org.
R Negron, Email: rennie.negron@yale.edu.
M Suprun, Email: maria.suprun@mountsinai.org.
RE Zinberg, Email: randi.zinberg@mssm.edu.
T Sabin, Email: tatiana.sabin@mountsinai.org.
D Hauser, Email: dhauser@institute.org.
N Calman, Email: ncalman@institute.org.
E Bagiella, Email: emilia.bagiella@mssm.edu.
EP Bottinger, Email: erwin.bottinger@mssm.edu.
References
- 1.Lipkowitz MS, Freedman BI, Langefeld CD, Comeau ME, Bowden DW, Kao WH, Astor BC, Bottinger EP, Iyengar SK, Klotman PE, et al. Apolipoprotein L1 gene variants associate with hypertension-attributed nephropathy and the rate of kidney function decline in African Americans. Kidney Int. 2013;83(1):114–120. doi: 10.1038/ki.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329(5993):841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tzur S, Rosset S, Shemer R, Yudkovsky G, Selig S, Tarekegn A, Bekele E, Bradman N, Wasser WG, Behar DM, et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet. 2010;128(3):345–350. doi: 10.1007/s00439-010-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsa A, Kao WH, Xie D, Astor BC, Li M, Hsu CY, Feldman HI, Parekh RS, Kusek JW, Greene TH, et al. APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med. 2013;369(23):2183–2196. doi: 10.1056/NEJMoa1310345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster MC, Coresh J, Fornage M, Astor BC, Grams M, Franceschini N, Boerwinkle E, Parekh RS, Kao WH. APOL1 variants associate with increased risk of CKD among African Americans. J Am Soc Nephrol. 2013;24(9):1484–1491. doi: 10.1681/ASN.2013010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman DJ, Kozlitina J, Genovese G, Jog P, Pollak MR. Population-based risk assessment of APOL1 on renal disease. J Am Soc Nephrol. 2011;22(11):2098–2105. doi: 10.1681/ASN.2011050519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanhamme L, Paturiaux-Hanocq F, Poelvoorde P, Nolan DP, Lins L, Van Den Abbeele J, Pays A, Tebabi P, Van Xong H, Jacquet A, et al. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature. 2003;422(6927):83–87. doi: 10.1038/nature01461. [DOI] [PubMed] [Google Scholar]
- 8.Gao SW, Oliver DK, Das N, Hurst FP, Lentine KL, Agodoa LY, Sawyers ES, Abbott KC. Assessment of racial disparities in chronic kidney disease stage 3 and 4 care in the department of defense health system. Clin J Am Soc Nephrol. 2008;3(2):442–449. doi: 10.2215/CJN.03940907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu CY, Lin F, Vittinghoff E, Shlipak MG. Racial differences in the progression from chronic renal insufficiency to end-stage renal disease in the United States. J Am Soc Nephrol. 2003;14(11):2902–2907. doi: 10.1097/01.asn.0000091586.46532.b4. [DOI] [PubMed] [Google Scholar]
- 10.Johns TS, Estrella MM, Crews DC, Appel LJ, Anderson CA, Ephraim PL, Cook C, Boulware LE. Neighborhood socioeconomic status, race, and mortality in young adult dialysis patients. J Am Soc Nephrol. 2014;25(11):2649–2657. doi: 10.1681/ASN.2013111207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keith D, Ashby VB, Port FK, Leichtman AB. Insurance type and minority status associated with large disparities in prelisting dialysis among candidates for kidney transplantation. Clin J Am Soc Nephrol. 2008;3(2):463–470. doi: 10.2215/CJN.02220507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norris KC, Agodoa LY. Unraveling the racial disparities associated with kidney disease. Kidney Int. 2005;68(3):914–924. doi: 10.1111/j.1523-1755.2005.00485.x. [DOI] [PubMed] [Google Scholar]
- 13.Peralta CA, Katz R, DeBoer I, Ix J, Sarnak M, Kramer H, Siscovick D, Shea S, Szklo M, Shlipak M. Racial and ethnic differences in kidney function decline among persons without chronic kidney disease. J Am Soc Nephrol. 2011;22(7):1327–1334. doi: 10.1681/ASN.2010090960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powe NR. To have and have not: Health and health care disparities in chronic kidney disease. Kidney Int. 2003;64(2):763–772. doi: 10.1046/j.1523-1755.2003.00138.x. [DOI] [PubMed] [Google Scholar]
- 15.Vargas RB, Norris KC. Kidney disease progression and screening cost-effectiveness among African Americans. J Am Soc Nephrol. 2012;23(12):1915–1916. doi: 10.1681/ASN.2012101028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fullerton SM, Knerr S, Burke W. Finding a place for genomics in health disparities research. Public Health Genomics. 2012;15(3–4):156–163. doi: 10.1159/000334717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horowitz CR, Robinson M, Seifer S. Community-based participatory research from the margin to the mainstream: are researchers prepared? Circulation. 2009;119(19):2633–2642. doi: 10.1161/CIRCULATIONAHA.107.729863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Israel BA, Schulz AJ, Parker EA, Becker AB. Review of community-based research: assessing partnership approaches to improve public health. Annu Rev Public Health. 1998;19:173–202. doi: 10.1146/annurev.publhealth.19.1.173. [DOI] [PubMed] [Google Scholar]
- 19.Stewart AL, Hays RD, Ware JE., Jr The MOS short-form general health survey. Reliability and validity in a patient population. Med Care. 1988;26(7):724–735. doi: 10.1097/00005650-198807000-00007. [DOI] [PubMed] [Google Scholar]
- 20.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40(5):373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 21.Broadbent E, Petrie KJ, Main J, Weinman J. The brief illness perception questionnaire. J Psychosom Res. 2006;60(6):631–637. doi: 10.1016/j.jpsychores.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 22.Svarstad BL, Chewning BA, Sleath BL, Claesson C. The Brief Medication Questionnaire: a tool for screening patient adherence and barriers to adherence. Patient Educ Couns. 1999;37(2):113–124. doi: 10.1016/s0738-3991(98)00107-4. [DOI] [PubMed] [Google Scholar]
- 23.Voils CI, Maciejewski ML, Hoyle RH, Reeve BB, Gallagher P, Bryson CL, Yancy WS., Jr Initial validation of a self-report measure of the extent of and reasons for medication nonadherence. Med Care. 2012;50(12):1013–1019. doi: 10.1097/MLR.0b013e318269e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Safran DG, Kosinski M, Tarlov AR, Rogers WH, Taira DH, Lieberman N, Ware JE. The Primary Care Assessment Survey: tests of data quality and measurement performance. Med Care. 1998;36(5):728–739. doi: 10.1097/00005650-199805000-00012. [DOI] [PubMed] [Google Scholar]
- 25.LaVeist TA, Nickerson KJ, Bowie JV. Attitudes about racism, medical mistrust, and satisfaction with care among African American and white cardiac patients. Med Care Res Rev. 2000;57(Suppl 1):146–161. doi: 10.1177/1077558700057001S07. [DOI] [PubMed] [Google Scholar]
- 26.Hibbard JH, Stockard J, Mahoney ER, Tusler M. Development of the Patient Activation Measure (PAM): conceptualizing and measuring activation in patients and consumers. Health Serv Res. 2004;39(4 Pt 1):1005–1026. doi: 10.1111/j.1475-6773.2004.00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henneman L, Timmermans DR, van der Wal G. Public experiences, knowledge and expectations about medical genetics and the use of genetic information. Community Genet. 2004;7(1):33–43. doi: 10.1159/000080302. [DOI] [PubMed] [Google Scholar]
- 28.Sanderson SC, Linderman MD, Suckiel SA, Diaz GA, Zinberg RE, Ferryman K, Wasserstein M, Kasarskis A, Schadt EE. Motivations, concerns and preferences of personal genome sequencing research participants: Baseline findings from the HealthSeq project. Eur J Hum Genet. 2015 [Google Scholar]
- 29.Kroenke K, Strine TW, Spitzer RL, Williams JB, Berry JT, Mokdad AH. The PHQ-8 as a measure of current depression in the general population. J Affect Disord. 2009;114(1–3):163–173. doi: 10.1016/j.jad.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 30.Kroenke K, Spitzer RL, Williams JB, Monahan PO, Lowe B. Anxiety disorders in primary care: prevalence, impairment, comorbidity, and detection. Ann Intern Med. 2007;146(5):317–325. doi: 10.7326/0003-4819-146-5-200703060-00004. [DOI] [PubMed] [Google Scholar]
- 31.Cohen S, Kamarck T, Mermelstein R. A global measure of perceived stress. J Health Soc Behav. 1983;24(4):385–396. [PubMed] [Google Scholar]
- 32.Wong MD, Sarkisian CA, Davis C, Kinsler J, Cunningham WE. The association between life chaos, health care use, and health status among HIV-infected persons. J Gen Intern Med. 2007;22(9):1286–1291. doi: 10.1007/s11606-007-0265-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherbourne CD, Stewart AL. The MOS social support survey. Soc Sci Med. 1991;32(6):705–714. doi: 10.1016/0277-9536(91)90150-b. [DOI] [PubMed] [Google Scholar]
- 34.Wallace LS, Rogers ES, Roskos SE, Holiday DB, Weiss BD. Brief report: screening items to identify patients with limited health literacy skills. J Gen Intern Med. 2006;21(8):874–877. doi: 10.1111/j.1525-1497.2006.00532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bindman AB, Grumbach K, Osmond D, Komaromy M, Vranizan K, Lurie N, Billings J, Stewart A. Preventable hospitalizations and access to health care. JAMA. 1995;274(4):305–311. [PubMed] [Google Scholar]
- 36.National Health and Nutrition Examination Survey Questionnaire. [ http://www.cdc.gov/nchs/nhanes/nhanes2013-2014/questionnaires13_14.htm]
- 37.Bhandari A, Wagner T. Self-reported utilization of health care services: improving measurement and accuracy. Med Care Res Rev. 2006;63(2):217–235. doi: 10.1177/1077558705285298. [DOI] [PubMed] [Google Scholar]
- 38.Cella D, Hughes C, Peterman A, Chang CH, Peshkin BN, Schwartz MD, Wenzel L, Lemke A, Marcus AC, Lerman C. A brief assessment of concerns associated with genetic testing for cancer: the Multidimensional Impact of Cancer Risk Assessment (MICRA) questionnaire. Health Psychol. 2002;21(6):564–572. [PubMed] [Google Scholar]
- 39.Brehaut JC, O’Connor AM, Wood TJ, Hack TF, Siminoff L, Gordon E, Feldman-Stewart D. Validation of a decision regret scale. Med Decis Making. 2003;23(4):281–292. doi: 10.1177/0272989X03256005. [DOI] [PubMed] [Google Scholar]
- 40.Holmes-Rovner M, Kroll J, Schmitt N, Rovner DR, Breer ML, Rothert ML, Padonu G, Talarczyk G. Patient satisfaction with health care decisions: the satisfaction with decision scale. Med Decis Making. 1996;16(1):58–64. doi: 10.1177/0272989X9601600114. [DOI] [PubMed] [Google Scholar]
- 41.Bernhardt BA, Zayac C, Gordon ES, Wawak L, Pyeritz RE, Gollust SE. Incorporating direct-to-consumer genomic information into patient care: attitudes and experiences of primary care physicians. Per Med. 2012;9(7):683–692. doi: 10.2217/pme.12.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonham VL, Sellers SL, Woolford S. Physicians’ knowledge, beliefs, and use of race and human genetic variation: new measures and insights. BMC Health Serv Res. 2014;14:456. doi: 10.1186/1472-6963-14-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selkirk CG, Weissman SM, Anderson A, Hulick PJ. Physicians’ preparedness for integration of genomic and pharmacogenetic testing into practice within a major healthcare system. Genet Test Mol Biomarkers. 2013;17(3):219–225. doi: 10.1089/gtmb.2012.0165. [DOI] [PubMed] [Google Scholar]
- 44.van Langen IM, Birnie E, Leschot NJ, Bonsel GJ, Wilde AA. Genetic knowledge and counselling skills of Dutch cardiologists: sufficient for the genomics era? Eur Heart J. 2003;24(6):560–566. doi: 10.1016/s0195-668x(02)00522-5. [DOI] [PubMed] [Google Scholar]
- 45.Overby CL, Erwin AL, Abul-Husn NS, Ellis SB, Scott SA, Obeng AO, Kannry JL, Hripcsak G, Bottinger EP, Gottesman O. Physician Attitudes toward Adopting Genome-Guided Prescribing through Clinical Decision Support. J Pers Med. 2014;4(1):35–49. doi: 10.3390/jpm4010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pickering TG, Hall JE, Appel LJ, Falkner BE, Graves J, Hill MN, Jones DW, Kurtz T, Sheps SG, Roccella EJ, et al. Recommendations for blood pressure measurement in humans and experimental animals: Part 1: blood pressure measurement in humans: a statement for professionals from the Subcommittee of Professional and Public Education of the American Heart Association Council on High Blood Pressure Research. Hypertension. 2005;45(1):142–161. doi: 10.1161/01.HYP.0000150859.47929.8e. [DOI] [PubMed] [Google Scholar]
- 47.Gottesman O, Scott SA, Ellis SB, Overby CL, Ludtke A, Hulot JS, Hall J, Chatani K, Myers K, Kannry JL, et al. The CLIPMERGE PGx Program: clinical implementation of personalized medicine through electronic health records and genomics-pharmacogenomics. Clin Pharmacol Ther. 2013;94(2):214–217. doi: 10.1038/clpt.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R. A language and environment for statistical computing. [ http://www.R-project.org/]
- 50.REDCapR. Interaction between R and REDCap. [ http://CRAN.R-project.org/package=REDCapR]
- 51.redcapAPI. Accessing data from REDCap projects using the API. [ https://github.com/nutterb/redcapAPI/wiki]
- 52.Team4Cure - Home. [ http://www.team4cure.org/]
- 53.Corbie-Smith G, Thomas SB, Williams MV, Moody-Ayers S. Attitudes and beliefs of African Americans toward participation in medical research. J Gen Intern Med. 1999;14(9):537–546. doi: 10.1046/j.1525-1497.1999.07048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goldfinger JZ, Kronish IM, Fei K, Graciani A, Rosenfeld P, Lorig K, Horowitz CR. Peer education for secondary stroke prevention in inner-city minorities: design and methods of the prevent recurrence of all inner-city strokes through education randomized controlled trial. Contemp Clin Trials. 2012;33(5):1065–1073. doi: 10.1016/j.cct.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gorelick PB, Harris Y, Burnett B, Bonecutter FJ. The recruitment triangle: reasons why African Americans enroll, refuse to enroll, or voluntarily withdraw from a clinical trial. An interim report from the African-American Antiplatelet Stroke Prevention Study (AAASPS) J Natl Med Assoc. 1998;90(3):141–145. [PMC free article] [PubMed] [Google Scholar]
- 56.Horowitz CR, Eckhardt S, Talavera S, Goytia C, Lorig K. Effectively translating diabetes prevention: a successful model in a historically underserved community. Transl Behav Med. 2011;1(3):443–452. doi: 10.1007/s13142-011-0067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roberson NL. Clinical trial participation. Viewpoints from racial/ethnic groups. Cancer. 1994;74(9 Suppl):2687–2691. doi: 10.1002/1097-0142(19941101)74:9+<2687::aid-cncr2820741817>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 58.Shavers-Hornaday VL, Lynch CF, Burmeister LF, Torner JC. Why are African Americans under-represented in medical research studies? Impediments to participation. Ethn Health. 1997;2(1–2):31–45. doi: 10.1080/13557858.1997.9961813. [DOI] [PubMed] [Google Scholar]
- 59.Sisk JE, Horowitz CR, Wang JJ, McLaughlin MA, Hebert PL, Tuzzio L. The success of recruiting minorities, women, and elderly into a randomized controlled effectiveness trial. Mt Sinai J Med. 2008;75(1):37–43. doi: 10.1002/msj.20014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swanson GM, Ward AJ. Recruiting minorities into clinical trials: toward a participant-friendly system. J Natl Cancer Inst. 1995;87(23):1747–1759. doi: 10.1093/jnci/87.23.1747. [DOI] [PubMed] [Google Scholar]
- 61.Oragene Dx DNA collection device gets FDA clearance. [ http://www.medgadget.com/2011/12/oragene-dx-dna-collection-device-gets-fda-clearance.html]
- 62.Birnboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Generating randomization schedules using SAS programming. [ http://www2.sas.com/proceedings/sugi27/p267-27.pdf]
- 64.Leventhal H, Diefenbach M. Illness cognition: Using common sense to understand treatment adherence and affect cognition interactions. Cognitive Therapy and Research. 1992;16(2):143–163. [Google Scholar]
- 65.Leventhal H, Diefenbach M. The active side of illness cognition. In: Skelton JA, Croyle RT, editors. Mental representation in health and illness. New York: Springer-Verlag; 1991. pp. 247–272. [Google Scholar]
- 66.Klitzman R, Chung W, Marder K, Shanmugham A, Chin LJ, Stark M, Leu CS, Appelbaum PS. Attitudes and practices among internists concerning genetic testing. J Genet Couns. 2013;22(1):90–100. doi: 10.1007/s10897-012-9504-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Patay BA, Topol EJ. The unmet need of education in genomic medicine. The American journal of medicine. 2012;125(1):5–6. doi: 10.1016/j.amjmed.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 68.Alyass A, Turcotte M, Meyre D. From big data analysis to personalized medicine for all: challenges and opportunities. BMC Med Genomics. 2015;8:33. doi: 10.1186/s12920-015-0108-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haga SB. Impact of limited population diversity of genome-wide association studies. Genet Med. 2010;12(2):81–84. doi: 10.1097/GIM.0b013e3181ca2bbf. [DOI] [PubMed] [Google Scholar]
- 70.Moonesinghe R, Jones W, Honore PA, Truman BI, Graham G. Genomic medicine and racial/ethnic health disparities: promises, perils, and the challenges for health care and public health policy. Ethn Dis. 2009;19(4):473–478. [PubMed] [Google Scholar]
- 71.Smedley BD, Stith AY, Nelson AR, editors. Unequal Treatment: Confronting Racial and Ethnic Disparities in Health Care. Washington (DC): 2003. [PubMed] [Google Scholar]
- 72.Williams DR, Wyatt R. Racial Bias in Health Care and Health: Challenges and Opportunities. JAMA. 2015;314(6):555–556. doi: 10.1001/jama.2015.9260. [DOI] [PubMed] [Google Scholar]
- 73.Manolio TA, Chisholm RL, Ozenberger B, Roden DM, Williams MS, Wilson R, Bick D, Bottinger EP, Brilliant MH, Eng C, et al. Implementing genomic medicine in the clinic: the future is here. Genet Med. 2013;15(4):258–267. doi: 10.1038/gim.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Osheroff JA, Teich JM, Middleton B, Steen EB, Wright A, Detmer DE. A roadmap for national action on clinical decision support. J Am Med Inform Assoc. 2007;14(2):141–145. doi: 10.1197/jamia.M2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shirts BH, Salama JS, Aronson SJ, Chung WK, Gray SW, Hindorff LA, Jarvik GP, Plon SE, Stoffel EM, Tarczy-Hornoch PZ, et al. CSER and eMERGE: current and potential state of the display of genetic information in the electronic health record. J Am Med Inform Assoc. 2015 doi: 10.1093/jamia/ocv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.