Abstract
For more than a decade, biometric moderation models have been used to examine whether genetic and environmental influences on individual differences might vary within the population. These quantitative gene × environment interaction (G×E) models not only have the potential to elucidate when genetic and environmental influences on a phenotype might differ, but why, as they provide an empirical test of several theoretical paradigms that serve as useful heuristics to explain etiology—diathesis-stress, bioecological, differential susceptibility, and social control. In the current manuscript, we review how these developmental theories align with different patterns of findings from statistical models of gene-environment interplay. We then describe the extant empirical evidence, using work by our own research group and others, to lay out genetically-informative plausible accounts of how phenotypes related to social inequality—physical health and cognition—might relate to these theoretical models.
For decades, biometric modeling of genetically informative family data was able to provide the relative magnitude of genetic and environmental influences on variables related to social inequality: constructs as diverse as mental health, physical health, well-being, family functioning and even income and morbidity. We know that almost any psychologically important variable related to social inequality that differs between people, like mental health or well-being, has a significant, non-zero heritability (proportion of variance in a phenotype that is due to genetic differences between individuals; Turkheimer, 2000). Interestingly, even putatively “environmental” variables, like education, employment, and wealth, have genetic influences (Kendler & Baker, 2007; Rowe, 1981). This explains why in recent years researchers have begun to search for the molecular genetic underpinnings of constructs like educational attainment (Martin et al., 2011; Rietveld et al., 2013b), economic and political preferences (Benjamin et al., 2012) and self-employment (van der Loos et al., 2013).
Potentially more interesting, however, than simply knowing whether there are genetic influences on indicators of social inequality would be to understand how the interplay between genes and environment shapes individual differences in social inequality over the lifespan. Biometric moderation models, introduced more than a decade ago, are able to provide estimates of genetic and environmental influences that are personalized to (i.e., dependent on) an individual's standing on a variable other than the phenotype (i.e., observed trait or behavior) of interest. Take for example, findings from our recent work on health; if we average across the population, we estimate the genetic influences on self-reported physical health at 16%; but, if we look at physical health as a function of marital relationship quality, we see that genetic influences are much higher among those with either very happy or very distressed relationships (South & Krueger, 2013). In the current manuscript, we review how these statistical models of gene-environment interplay align with existing theoretical models of development to lay out genetically-informative plausible accounts of how social inequality develops. We then describe how the existing evidence, including but not limited to work by our own research group, lines up with those accounts. We argue in this manuscript that these quantitative models provide a tool for testing long-standing theories about the development of individual differences. We conclude our review by summarizing what these findings imply about determining risk for indicators of social inequality based on a person's relative standing on important risk factors in the population, and we posit ways in which future research can build on this work to move toward investigating the interplay between measured genes and environments in the development of social inequality.
Behavior Genetic Methods: Classic Approaches and Models for Gene × Environment Interplay
Behavior genetic approaches were vital in demonstrating that almost every psychologically important individual difference variable, particularly personality and mental health, was a function of both genes and environment (that is, nature and nurture at play). An unexpected and important further contribution of this family of methods was to demonstrate that even putatively environmental or socio-cultural variables that are generally included under the larger umbrella of ‘social inequality’ had a heritable component; in other words the variance in things like income, wealth, education, and well-being could be explained, at least in part, by genetic differences within the population (Kendler & Baker, 2007). In this section, we first describe the classic behavior genetic approaches before moving on to newer models that allow for examination of quantitative Gene × Environment interaction (quantG×E).
Classic Approaches
Behavior genetic methods utilize genetically-informative data sets (e.g., twins, adopted children and parents) to estimate genetic and environmental variance in a phenotype (i.e. observed variable). In essence, these methods are able to estimate the contributions of nature and nurture to variance in the population. The well-known and frequently discussed heritability statistic is often used as evidence of “genes” determining a variable; however, the true definition of this quantity requires some precision. Heritability, by definition, is the proportion of total variance in a variable, in a specific sample drawn from a larger population, which is due to genetic variance in that sample. In other words, an individual difference variable differs along a spectrum in the population, from more to less, and heritability is an estimate of how much of that variance is due to genetic differences between people.
Rough estimates of heritability can be calculated using the difference in twin correlations between identical (monozygotic, MZ) and fraternal (dizygotic, DZ) twins (i.e., h2= 2*(rMZ-rDZ); Falconer, 1965). It is also possible to decompose the total phenotypic variance of a phenotype using structural equation models. There are three sources of variance: additive genetic influences, usually abbreviated (A); influences siblings in the same family share in common and which make them more similar, abbreviated (C); and the unique environment, influences that siblings do not share and which make them different from other family members, abbreviated (E). Thus, the acronym “ACE model” is often used to describe biometric modeling of twin data.
Findings from classic biometric modeling had important implications for theoretical understandings of etiology; for instance, knowing that a phenotype has a significant non-zero heritability led to a large investment in the search for the molecular genetic underpinnings of social science constructs including subjective well-being (Rietveld et al., 2013a), self-employment (van der Loos et al., 2013) and educational attainment (Rietveld et al., 2013b). The limitation of the basic univariate ACE model is that it says nothing about a) what, specifically, those nonshared environmental influences were (Turkheimer & Waldron, 2000), or b) how genetic influences and environmental influences mediated relationships between variables. One attempt to answer these questions was to use extended ACE models that included multiple variables. The advantage of these multivariate models is that in addition to estimating genetic and environmental influences on each variable, it is possible to estimate the genetic and environmental overlap between two or more variables. For instance, the bivariate (Cholesky) decomposition can determine how much of the phenotypic correlation between two variables (e.g., well-being and education) is due to genetic influences shared in common between the two variables (i.e., bivariate heritability). Another way of looking at overlap is to estimate a genetic correlation. Like a phenotypic correlation, a genetic correlation (and commensurate estimates for shared and nonshared environmental correlations) range from -1 to +1 and indicate how much the genetic influences on one phenotype (e.g., well-being) overlap with the genetic influences on another phenotype (e.g., education). In this vein, Weiss et al. (2008) examined the relationships among subjective well-being and domains of the Five Factor Model/Big Five Model of personality using a Cholesky decomposition. They concluded that all of the genetic variance in subjective well-being was shared in common with genetic influences on Neuroticism, Agreeableness, Extraversion, Conscientiousness, and Openness.
These extended multivariate models did answer some questions. First, they were able to determine whether two variables that are phenotypically correlated share the same underlying genetic basis. For instance, finding that, among boys, intelligence was etiologically related to antisocial behavior through genetic overlap (Koenen, Caspi, Moffitt, Rijsdijk, & Taylor, 2006) suggests the same genes may affect both IQ and delinquent behavior. Second, these models also pointed toward possible sources of the nonshared environment, which are anonymous latent constructs in biometric modeling. The fact that the nonshared environmental influences on marital satisfaction overlap with wives' positive mental health implicates the husband as that very source of nonshared environment (Spotts et al., 2005). What this work could not inform, however, is the ways in which genetic influences and environmental contexts worked in tandem to produce a phenotypic outcome. For that, new statistical models were needed.
Quantitative Models for Gene × Environment Interplay
The heritability statistic and commensurate estimates of environmental influences (both between-family E influences and within-family C influences) are limited in two important ways. First, these estimates are presumed to be independent—that is, obtaining the relative magnitude of genetic vs. environmental influences on social inequality presumes that these two influences do not interact. These models say nothing about the interplay between genes and environment. Second, these estimates average across the entire (sample-specific) population. Many people, particularly lay individuals, misunderstand the concept of heritability, interpreting a 50% estimate to mean that 50% of their depressive tendency, for instance, is attributed to genetics. In fact, this type of individual-level heritability is not possible with quantitative modeling of twin data. The correct interpretation of heritability, in that case, is to say that 50% of the variation in depression, in a specific sample drawn from one population, is due to genetic differences between individuals in that sample. But, again, this heritability value tells us nothing about how different environments may affect that 50% estimate.
For over a decade now, however, modeling of biometric moderation in a structural modeling framework has made it possible to differentiate estimates of genetic and environmental influences dependent on a person's standing within the population (Purcell, 2002; Rathouz, Van Hulle, Rodgers, Waldman, & Lahey, 2008; van der Sluis, Posthuma, & Dolan, 2012; van Hulle, Lahey, & Rathouz, 2013). Biometric moderation models allow for different ACE estimates depending on a person's standing within the population on a “moderator” variable—hence the term moderation model. That is, instead of estimating one heritability coefficient that averages across all differences within a sample drawn from a population, biometric moderation models allow heritability to differ within the population. This is not a completely new concept; biometric sex limitation models, for instance, provide for a statistical test of whether genetic and environmental influences on a phenotype are the same for men and women. While still not able to individualize to the etiology of one specific person, we are now able to get closer and closer to understanding how certain risk factors can change the etiology of important outcomes for people within the population. To use one illustrative example, would the heritability of health differ depending on a person's socioeconomic status? The short answer is yes—at low levels of income, genetic influences on physical health are greater than at high levels of income (Johnson & Krueger, 2005a).
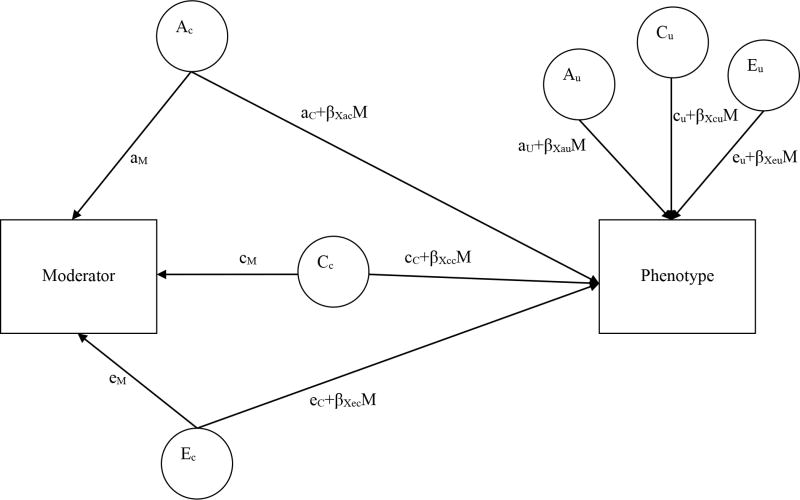
A path model of the bivariate moderation model (Purcell, 2002) is shown in Figure 1. This model is an extension of a bivariate decomposition, in which the variance in two variables, and the covariance between them, is partitioned into three sources as noted above (i.e., genetic effects (A), common or shared environmental effects (C), and nonshared environmental effects (E). In the moderation model, the six paths leading to the downstream variable include extra parameters (aC+βXacM) that allow for estimation of genetic and environmental variance at different levels of the moderator variable. This parameterization of the model makes it possible to estimate ACE influences at any possible level of the moderator (M). Another advantage of the model in Figure 1 is that it is also possible to estimate the genetic and environmental overlap between the two variables in the form of genetic and environmental correlations, ranging from -1 to +1 in the usual way, and these correlations will also differ depending on the level of the moderator. To utilize this model, however, the moderator must be a variable that is unique to each twin (e.g., income as adults, marital status, relationship satisfaction, etc.). If the moderator is a variable that is necessarily shared between twins (e.g., neighborhood SES as children), then it is necessary to use an alternative, univariate model that “controls” for gene-environment correlation by regressing out the effect of the moderator on the outcome but still allows for separate ACE estimates at different levels of the moderator (Purcell, 2002; van der Sluis et al., 2012).
Figure 1.
Bivariate moderation model.
The model is shown for only one member of a twin pair. Genetic and environmental influences on the Outcome Variable vary by level of the Moderator Variable. A=additive genetics, C= shared environmental influences, and E=non-shared environmental influences. AC, CC, and EC are variance components underlying the Moderator that also influence the Outcome (i.e., “common components”), and AU, CU, and EU represent residual (“unique”) variance in the Outcome after accounting for the variance in common with the Moderator. β coefficients index the direction and magnitude of moderation. When all β coefficients are set to zero, this represents no moderation effects. Total phenotypic variance in the Outcome can be calculated by squaring and summing all of the paths leading to it: Var(Outcome) = (aC+βacM)2 + (aU+βauM)2 + (cC+βccM)2 + (cU+βcuM)2 + (eC+βecM)2 + (eU+βeuM)2.
Because these models estimate whether the total latent (unmeasured but assumed) genetic influences on a phenotype differ depending on a person's standing on a second, putatively environmental, variable, they are commonly referred to as quantitative estimates of Gene × Environment interaction (quantG×E, to distinguish from measured gene × measured environment interactions, discussed below). Theoretically, the presence of G×E could mean that the genetic effects on a phenotype only become apparent in the right environment; or, conversely, environmental effects on a phenotype are dependent on a person having the right genotype. Earlier attempts to statistically model G×E using twin data would stratify the sample by level of the moderator variable and examine genetic and environmental influences as a function of these subgroups (Cleveland, 2003). For instance Heath and colleagues (1989) found that genetic influences on alcohol consumption varied from 31% in young, married women to 76% in older, unmarried women, demonstrating that an important sociodemographic variable, marital status, can have a moderating effect on the etiology of alcohol use. The newer biometric moderation models have several advantages over previous methods to examine G×E in genetically informative family data (e.g., stratification of twin correlations). First, it is possible to formally test the presence vs. absence of moderation using a variety of well-validated fit indices. Second, both the univariate and bivariate versions model the main effect of the moderator on the phenotype, either by including a direct path or by decomposing the effect of the moderator on the outcome (thus accounting for gene-environment correlation). Finally, it is possible to obtain ACE estimates of the phenotype along the full spectrum of the moderator. For these reasons, the popularity of this model has led to a growth in quantG×E studies over the last decade. We next turn to discussing how these results fit with work on measured gene × measured environment interactions.
Gene × Environment Interaction: Measured vs. Latent Genetic Influences
Gene × environment interaction has long been theorized for complex human behavioral phenotypes, including personality, psychopathology, and cognition. If found, G×E would imply that the effect of the environment on a phenotype would depend on a person's genotype; or alternatively that the expression of a genotype would depend on the right environmental context. The idea of G×E lines up nicely with many developmental theories. For instance, the diathesis-stress model of psychopathology posits that a diathesis (possibly genetic) for mental illness will only be trigged in the right environmental context (Monroe & Simons, 1991), a topic we return to again, below. Empirical studies of G×E, however, have only begun in earnest in this century, following recent statistical and methodological advances. We first discuss candidate molecular G×E before returning to consider quantG×E.
Measured Gene × Measured Environment Interaction
Candidate G×E (or cG×E) studies are distinguished from quantG×E by the use of both a measured environment and a measured gene. In the most common use of this technique, a candidate gene is chosen based on a known or suspected mechanism of action and an environmental context is ideally selected based on evidence that it elicits variable responses among different individuals and affects a neurobiological system underlying the trait of interest (Moffitt, Caspi, & Rutter, 2006). Genes are specific protein coding segments of DNA and can be anywhere from a few hundred to thousands of base pairs in length. Genes at the same location on the genome (locus) can differ in specific physical ways, between persons. For example, a cG×E interaction could be posited such that an environmental risk factor only has its effect on the phenotype in the presence of the greatest genetic risk (e.g., in persons with two copies—homozygous—of the risk allele).
The increase in cG×E research followed from the rather disappointing lack of significant and replicable main effects for specific genetic polymorphisms on individual differences and major mental disorders. Normative personality traits, for example, have approximately 50% heritability (Bouchard & Loehlin, 2001) but candidate gene studies failed to find any significant, replicable loci with an appreciable effect size (Munafo et al., 2003). Explanations for the failure of candidate gene studies include generally underpowered studies and the fact that many relevant polymorphisms may not be in the protein coding region of the genome, but instead outside of well-characterized genes that were hypothesized as potential candidates (Duncan, Pollastri, & Smoller, 2014). A huge technological advance in gene-hunting techniques, genome-wide association studies (GWAS) were able to search across first thousands and currently millions of single nucleotide polymorphisms (genetic variants). GWAS led to much more successful replication, but the total percentage of variance explained by this technique was still much lower than the total heritability of phenotypes as estimated by twin and family methods (Manolio et al., 2009). Several explanations have been proposed to explain this “missing heritability” (for a review, see Manuck & McCaffery, 2014). First, the molecular genetic architecture of most studied phenotypes could be due to a larger number of common variants of much smaller effect size than previously thought, requiring larger than anticipated samples to identify these variants. Second, complex phenotypes (e.g, psychiatric disorders) may be due to structural variants (e.g., copy number variants, such as deletions or insertions) or rare variants (alleles present in <1% of the population) of large effect size that are not well captured by GWAS, which tends to include DNA markers that are fairly common in the population. Third, the accuracy of heritability estimates could have been affected by nonadditive genetic effects (dominance or epistasis).
Another possibility is that genes influencing complex diseases/disorders might only be expressed in the right environmental circumstances. In a seminal paper, Caspi and colleagues (2002) showed that the likelihood of depression and number of depressive symptoms was greatest among those with both a positive history of environmental risk and the risky (two copies of the short version of the allele) version of the serotonin gene 5-HTTLPR. Since that study was published, the cost of genotyping has decreased substantially, allowing researchers to add molecular genetic data to everything from highly focal lab-based studies (Burt, 2009) to larger, ongoing studies like the Health and Retirement Study (The Health and Retirement Study: A Longitudinal Study of Health). Over the past decade, there has been a wealth of published cG×E papers, examining the likelihood of an outcome as a function of risk allele and environmental risk factor (for recent reviews, see Duncan et al., 2014; Manuck & McCaffery, 2014). Unfortunately, the problems with replicability found for candidate gene, linkage, and association studies are also an issue for cG×E. Indeed, numerous replication attempts have been made of the original Caspi et al. (2002) findings on 5-HTTLPR and life stress on depression; as reviewed elsewhere (Duncan et al., 2014), two meta-analyses have failed to support the original finding (Munafò, Durrant, Lewis, & Flint, 2009; Risch et al., 2009), and the one meta-analysis in support of the interaction (Karg, Burmeister, Shedden, & Sen, 2011) has been criticized on methodological grounds (Duncan & Keller, 2011). A recent review found 103 cG×E studies in the first decade of research; only 6 had two or more replication attempts, and of these none had unequivocal support for the initial finding (Duncan et al., 2014). Like candidate gene studies, cG×E studies may be underpowered. Further, one report suggests publication bias in favor of positive cG×E for reports of novel findings as compared to much lower rates for replication attempts (Duncan & Keller, 2011). The replication failures of both candidate gene and cG×E studies led the editor of Behavior Genetics to establish strict new guidelines for consideration of publication, including: submission of a well-powered replication study, or a novel finding with adequate power; exploratory or novel finding meeting statistical criteria for genome wide significance; or meta-analysis of the same genetic variant and/or environmental variable and behavioral outcome (Hewitt, 2012).
Quantitative Gene × Measured Environment Interaction
Like measured G×E, quantG×E work also includes an environmental “moderator” but estimates genetic risk from known degrees of similarity among different types of relatives. As with classic behavior genetic approaches, quantG×E does not identify which genes are involved at a molecular level. Instead, biometric modeling suggests the presence of quantG×E when genetic influences differ as a function of the moderator; we then infer that the effect of additive genetic influence across all genes that directly impact the phenotype (most likely many loci of very small effect size) will depend on the level of the moderator. Even though quantG×E is not testing the moderating influence of a specific gene, however, there are advantages to quantG×E over cG×E.
First, quantG×E allows for estimation of aggregate genetic and environmental effects, instead of focusing on the environmental interaction with a specific candidate gene that may turn out to be spurious. Findings from genome-wide association studies (GWAS) suggest that the genetic architecture of complex human behavioral phenotypes, from mental illness to educational attainment, consists of hundreds if not thousands of genes of very small effect size (Chabris et al., 2013). If the main effects of genetic influences are the result of so many different individual genes, it stands to reason that when G×E occurs, it is the expression of many genes that is being moderated by the environmental context. Thus, quantG×E, which identifies aggregate moderation of all additive genetic influences, has a statistical and methodological advantage over cG×E, which by definition will only examine the interaction at one gene.
Second, quantG×E tests for not only variation in heritability but in environmental influences as well. For example, some quantG×E studies have found non-zero estimates of shared environment at extreme ends of the moderator variable (e.g., Krueger et al., 2008; South & Krueger, 2008), and shared environmental influences are notoriously difficult to find in “classic” twin modeling. Identifying contexts in which the shared or unique environmental influences on a phenotype are notable influences on etiology has important implications for designing and implementing new forms of prevention or intervention.
Third, there are many existing twin databases that have many nuanced and careful measures of the environment, have very large sample sizes, and could be used to conduct quantG×E studies; cG×E studies are often underpowered (Duncan & Keller, 2011), make use of existing genetic information in a database instead of collecting genes based on a priori hypotheses of biological mechanisms (Young-Wolff, Enoch, & Prescott, 2011), or require returning to an already-existing sample to collect new genetic data. That is not to say that issues of power and sample size are straightforward when it comes to quantG×E. For instance, there has been a healthy debate in the literature surrounding the different quantG×E models and how they relate to false positives (van der Sluis et al., 2012; van Hulle et al., 2013). van der Sluis and colleagues suggest that the extended univariate moderation model has greater power to detect significant moderation compared to the bivariate moderation model, as long as the moderation is not on the covariance common to the moderator and the outcome. Further, some warn that what looks like significant moderation may actually be masking nonlinear effects of the moderator on the outcome (van Hulle et al., 2013). Van Hulle et al. articulated procedures for formally testing quantG×E moderation against nonlinear main effects and conducted simulations with samples of 500 pairs and 2000 pairs. For example, rejecting a quantG×E model with nonlinear main effects in favor of the bivariate moderation model with moderation only on A would take a sample of up to 910 twin pairs for 90% power; much smaller sample sizes were required for omnibus tests of moderation on A, C, and E, which is what is often done and reported in the literature. They reported that with samples of 2000 twin pairs it was generally possible to reject a model that was not the “true” model (whether it was the Purcell moderation model or not), but concluded that tests for GxM are generally underpowered. The model simulations reported by Van Hulle et al. are complex, as they considered several competing models (with or without G×E, with or without nonlinear main effects) and moderation on all, some or none of the ACE parameters. We agree with their recommendation for always reporting parameter estimates from quantG×E studies as part of any research report; the size of the moderation effect in different samples drawn from different populations will aid in issues such as determining power and sample size for model fit indices (e.g., likelihood ratio tests and BIC).
Having elucidated the differences between quantG×E and cG×E, we can now turn to describing how, instead of seeing these methods as competing, they are really best thought of as complimentary. From a practical standpoint, quantG×E is a way of determining where to direct the time and money necessary for molecular genetic inquiry. Studies now show that the heritability of alcohol use is greater among adolescents with peers who use alcohol (Dick et al., 2007), in urban areas versus rural ones (Legrand et al., 2008; Rose et al., 2001), among girls with less parental closeness (Miles et al., 2005), in women without a religious upbringing versus women with such an upbringing (Koopmans et al., 1999), and in unmarried women versus married women (Heath et al., 1989). Our own work has shown that genetic influences on alcohol use are higher among individuals with low levels of SES (Hamdi, Krueger, & South, in press) or with distressed marriages (Jarnecke & South, 2014). These studies of quantG×E point toward particular segments of the population which may prove to be more fruitful for gene-finding efforts. Further, establishing a replicable effect using quantG×E may then reduce the chances of obtaining a null effect with cG×E. As noted above, cG×E are notorious for difficulties with replication. To our knowledge, there has been no comprehensive meta-analysis of all quantG×E studies conducted since Purcell's (2002) model was published. A review of quantG×E for alcohol and related phenotypes found significant quantG×E for almost all studies reviewed (14 of 16), and for all studies that used twin data to examine qunatG×E, the pattern was in the same direction (greater genetic influences in more permissive environments). As we review below, replication of one of the first studies in this area, moderation of heritability for IQ by SES (Turkheimer, Haley, Waldron, D'Onofrio, & Gottesman, 2003), has been less straightforward. However, the existence of similar phenotypes and environmental measures across different already existing twin studies suggests that it would be feasible to attempt numerous replications using quantG×E before moving to the more expensive step of molecular genetic work.
We use the example of physical activity and genetic influences on body mass index to show how quantG×E can effectively direct molecular genetics research. Classic biometric modeling approaches have shown that BMI is robustly heritable, with heritability estimates ranging from .45 to .84 in men and .64 to .85 in women in a sample of 37,000 twin pairs ranging from age 20 to 39 (Schousboe et al., 2003). Using biometric moderation to examine quantG×E, studies have shown that heritability of BMI is moderated by certain environmental factors, including physical activity (Mustelin, Silventoinen, Pietilainen, Rissanen, & Kaprio, 2009). Recently, a new study demonstrated that genetic risk for BMI, as measured by a risk score across 12 SNPs in obesity-susceptibility loci, was moderated by physical activity, such that the association between genetic risk score and BMI was greater in sedentary individuals (Li et al., 2010). We see this as a blueprint for future work that combines quantG×E and cG×E; the wealth of data from existing twin and family studies alone on measured environmental variables and phenotypes of interest to social scientists could be used to help narrow the scope of focus for molecular genetics researchers.
Conceptual Models and Corresponding Quantitative G×E Model
In this section, we describe four broad theoretical models that have been posited to explain myriad phenotypes related to social inequality (e.g., personality, psychopathology, health). For each, we describe how such a theoretical model would be supported empirically by biometric moderation modeling. We then illustrate how work to this point has, or has not, supported each theory. We focus on variables of particular importance to social inequality—cognition and physical health. Table 1 provides a brief summary of each of the four theories as well as a theoretical origin paper and an empirical example of quantG×E. Note that even though we present these models as distinct, it is entirely possible that more than one model will be necessary to explain the pattern of G×E found for a specific combination of moderator and phenotype. That is, these models are best thought of as useful heuristics that can be used to interpret the effects found in these quantG×E studies, but the findings from any one specific study may suggest the plausibility of more than one model.
Table 1. Theories Guiding the Study of Quantitative Gene × Environment Interaction Studies.
| Name of the Theory | Brief Description of the Theory | Paper Describing the Theoretical Background | Empirical Paper Demonstrating Evidence of quant G×E |
|---|---|---|---|
| Diathesis-Stress | A predisposition for the phenotype (i.e., a diathesis), in the form of premorbid risk factors that can be genetic, cognitive, affective, etc., lies dormant until it is triggered by some sort of stressor. | (Monroe & Simons, 1991) | (South & Krueger, 2008) |
| Bioecological Model | Genetic influences are maximized in stable and adaptive environments that permit positive and enduring interactions—proximal processes—between individuals and their immediate surroundings, which enable them to actualize their genetic potentials. | (Bronfenbrenner & Ceci, 1994) | (Turkheimer et al., 2003) |
| Differential Susceptibility | Plasticity to the environment is an individual difference, with some people being far more susceptible to (that is, genetically influenced by) the effect of both positive and negative environments. | (Belsky & Pluess, 2009) | (South & Krueger, 2013) |
| Social Control and Social Compensation | Genetic influences are dampened in certain environmental contexts; for social control, structural process/social norms impose constraints, and for social compensation, the environment lacks stress or possesses enriching properties. | (Shanahan & Hofer, 2005) | (Dick et al., 2007) |
Diathesis-Stress
The diathesis-stress model is particularly well known in the field of psychopathology, but has been broadened to include outcomes as diverse as subjective well-being (Burns & Machin, 2013), academic achievement (Jaekel, Pluess, Belsky, & Wolke, 2014), and even chronic pain (Turk, 2002). This model posits that a diathesis, or predisposition in the form of premorbid risk factors, for the phenotype (generally an undesirable outcome) lies dormant until it is triggered by some sort of stressor. The diathesis can be genetic, biological, or even cognitive, and the stressor can range from major, acute life event to minor, chronic daily hassles (Monroe & Simons, 1991). This fits well within the context of G×E, if we think of additive genetic influences as a “distal” diathesis and a measured environment as a stressor which triggers the expression of those genetic influences; indeed, Shanahan and Hofer (2005) have referred to this as contextual triggering. It is important to remember, however, that the diathesis itself may have an effect on whether the environment is experienced at all. This is known as gene-environment correlation (rGE), of which there are three types: active rGE, in which a person's genetically influenced characteristics lead them to choose certain environments; evocative rGE, in which those same traits evoke a reaction from others in the environment; or passive rGE, in which the genetically-influenced characteristics of one's parents influence the environment that a person experiences growing up, such that the family environment and the genotype one inherits are correlated. Gene-environment correlation cannot be directly tested in classic twin study approaches or the univariate moderation model, although the extended Purcell (2002) moderation model does estimate genetic and environmental overlap between the moderator and outcome, which can be used to infer rGE.
Diathesis-Stress in quantG×E Modeling
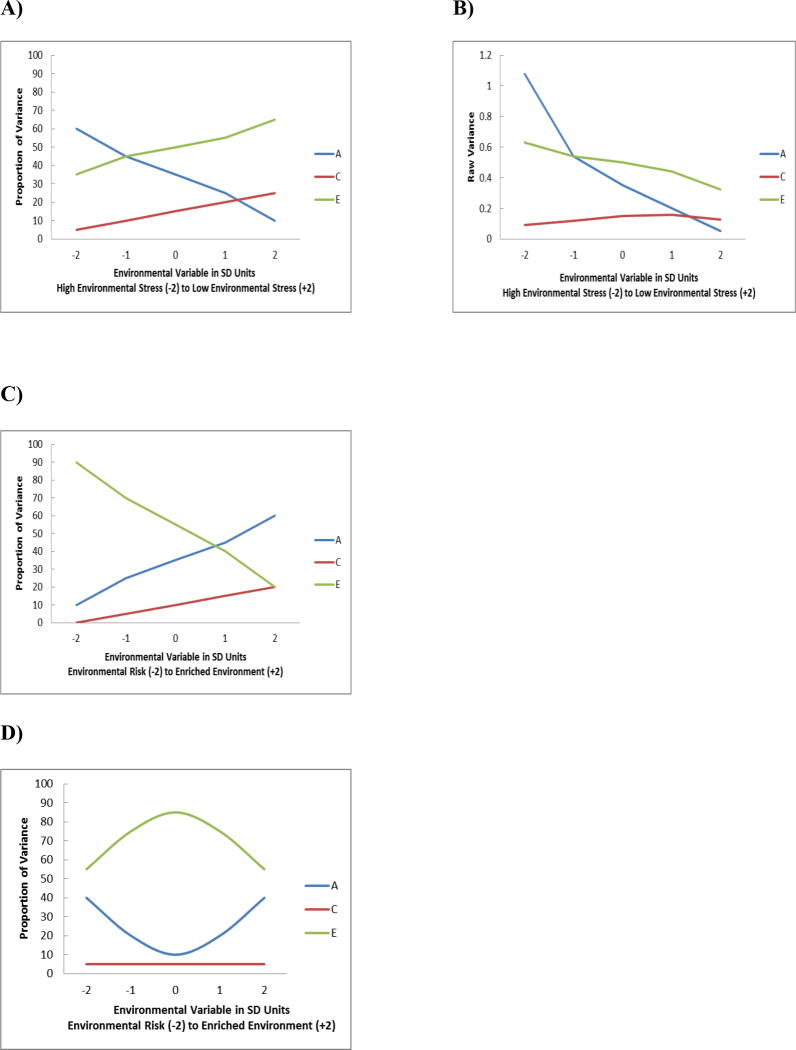
The diathesis-stress model would be supported if biometric modeling demonstrated that genetic influences were greater in the riskier environment. A possible hypothetical example is presented in Figure 2 (Panel A). As shown, our theoretical environmental moderator is on the X axis in standard deviation units, from a very risky environment marked by high levels of stress (-2 SD) to very low risk environment with little or no stressors (+2 SD). The environmental moderator variable could range from a more global risk factor that sits more distally in each individual's ecosystem (e.g., socioeconomic status) to a more proximal variable that affects the person's microsystem (e.g., relationships with romantic partners, friends, parents). Following from the diathesis-stress model, additive genetic influences are greatest in the environment marked by the most stress (at -2 SD from the mean) and decrease from high to low levels of stress. Conversely, the nonshared environment (E) and the shared environment (C) increase from high to low levels of stress. Of note, these are standardized proportions of variance, such that the total variance in the phenotype has to total to one at every level of the moderator. Purcell (2002) recommended also plotting and presenting the results of the unstandardized variance components. Shown in Figure 2 (Panel B) is one possible example of the raw ACE variance components that lead to the standardized proportions of our imaginary example in panel A. The raw genetic variance still peaks at the riskiest end of the moderator, and decreases until it is lowest at the “least stressful” levels of the moderator. Note, however, that the total amount of variance in the phenotype is greatest in the most stressful end of the population. This is found rather commonly in quantG×E models that use a moderator specifically designed to assess maladaptive contexts, like conflict with parents or stressful life events (e.g., Hicks, South, DiRago, Iacono, & McGue, 2009). It makes intuitive sense that there would simply be more variation in the outcome variable when the environment is marked by greater stress. Because there is more variation at the highest levels of environmental stress, even though the raw nonshared environmental variance decreases from high to low levels of stress and the shared environmental variance is essentially flat (Panel B), the standardized proportions of variance for both increase (Panel A). Again, this shows the importance of plotting and presenting both raw and standardized variance components.
Figure 2.
Example of patterns of quant G×E for different theoretical models. Genetic (A), shared environmental (C) and nonshared environmental (E) components of variance for an outcome are plotted as a function of an environmental moderator variable, shown on the X axis at five different levels: -2, -1, 0, 1, and 2 Standard Deviations from the Mean.
Panel (A): Diathesis-stress model plotted with standardized variance components.
Panel (B): Diathesis-stress model plotted with raw variance components.
Panel (C). Bioecological model.
Panel (D). Differential susceptibility model.
Illustrative Example: Body Mass Index
Much of the support for the diathesis-stress model in quantG×E comes from work on an important indicator of physical health—body mass index (BMI). In early work in this area, Johnson and Krueger (2005a, 2005b) used the bivariate Purcell (2002) biometric moderation model to examine whether body mass index and number of chronic illnesses were affected differentially by genetic and environmental influences dependent on income and perceived life control. Analyses were conducted using the twin sample from the Midlife in the United States Study (MIDUS; Kessler, Gilman, Thornton, & Kendler, 2004), notable for being one of the few nationally representative adult twin samples in the US. They found greater genetic variance in BMI at lower levels of income (used as a proxy for socioeconomic status), even after controlling for education level and presence vs. absence of insurance coverage. When perceived control was the moderator, genetic variance in BMI decreased from low to high levels of perceived control and shared environment increased slightly at higher levels of control. The authors concluded that the risky environment (one marked by low income and low control) is the stressor that ultimately changes a physiological mechanism, leading to the expression of a genetic predisposition to poor health. Of course, finding that genetic variance is greater in “riskier” environments does not necessarily mean that a stressful environmental context (e.g., low income) is triggering genetic influences. Indeed, it is possible that the less risky environment (e.g., high income) is acting to compress genetic variance (Johnson & Krueger, 2005a; see also Social Control and Social Compensation, below).
Subsequent work has largely acted to solidify these earlier findings on BMI across a variety of moderators. For instance, Mustelin and colleagues (2009), using the FinnTwinn16 sample (age range 22-27), found that the genetic variance in BMI decreased with increasing physical activity. In a study of adult twins from the University of Washington Twin Registry, researchers examined quantG×E for BMI as a function of sleep duration (Watson et al., 2012). They found significant moderation of genetic and shared environmental parameters, such that the proportion of variation due to genetics was greatest at the lowest levels of sleep duration and decreased as sleep duration increased, while shared environmental influences increased from low to high levels of sleep. Not all findings have replicated perfectly, however; using the Danish Twin Registry, Johnson and colleagues (Johnson, Kyvik, Skytthe, Deary, & Sorensen, 2011) examined education as a moderator of BMI. In partial replication of previous findings, they found that genetic variance was greater for women, but not men, with lower levels of education; in both genders, shared environmental variance decreased from low to high levels of education, resulting in a heritability of BMI that was greater at higher levels of education for both men and women (a finding in contrast to what would be expected for the diathesis stress model). The authors suggested that partial replication of previous work (particularly Johnson & Krueger, 2005a, 2005b, using the MIDUS study) could be due to several factors: lack of power resulting from the smaller US sample, cultural differences between the US and Denmark, or differences in the two moderators (income vs. education). An important take-home message for researchers from this early work may be the importance of combining data across samples, in order to increase sample size and to take into account possible cultural differences in the phenotypes (both moderator and outcome) of interest. For instance, the IGEMS (Interplay of Genes and Environment across Multiple Studies; Pedersen et al., 2013) consortium was recently formed to combine data across eight longitudinal twin studies. We look forward to the progress they make in examining a multitude of outcomes as a function of various factors related to social inequality, including early life adversity.
Bioecological Model
The bioecological model predicts that genetic influences are maximized in stable and adaptive environments (Bronfenbrenner & Ceci, 1994). Specifically, the model assumes that stable environments permit positive and enduring interactions—termed proximal processes—between individuals and their immediate surroundings, which enable them to actualize their genetic potentials. Two things should be noted about this theory. First, the authors were most interested in the development of effective physiological functioning, and what environments would allow individuals to flourish. Indeed, Shanahan and Hofer (2005) specifically refer to a G×E interaction in which the environment leads to an adaptive or beneficial outcome as “social context as enhancement.” Not surprisingly, many quantG×E studies that operate from this paradigm are interested in adaptive functioning (e.g., cognitive ability, see below), but many other studies have appeared in the literature that examine how genetic potential for maladaptive processes may be actualized under certain environmental contexts (e.g. genetic influences on psychopathology based on SES; South & Krueger, 2011; Tuvblad, Grann, & Lichtenstein, 2006). In this case, what researchers are examining is the “genetic potential not for expressing dysfunctional outcomes but for buffering against and thus reducing them” (Bronfenbrenner & Ceci, 1994, p. 582). Second, the authors approached this theory with a heavy emphasis on heritability, and as such had a very specific hypothesis about how genetic influences would be affected by the environment. In a low-risk, enriched environment, that is, one marked by lower levels of social inequality (poverty, poor access to health care and education) we would expect less variability in the phenotype and greater genetic influences. This idea is also consistent with the conceptually related “social push” model (Raine, 2002), which posits that genetic influences on maladaptive behavior are more evident in the absence of environmental risk factors that push individuals towards such behavior.
Bioecological Model in quantG×E Modeling
From Bronfenbrenner and Ceci's (1994) writings on the bioecological model, we can derive explicit hypotheses about the pattern of findings from quantG×E. As they hypothesized, when proximal processes (e.g., parent-child relationship) are strong, heritability will be greater. Figure 2 (Panel C) presents a hypothetical example of findings from a quantG×E model supporting the bioecological theory. As shown, the genetic influences on the outcome increase from an environment marked by risk to an environment that is relatively more enriched, while nonshared environmental influences are greatest at the riskiest levels of the environmental moderator. For illustrative purposes we have plotted the proportion of shared environmental variance such that it increases somewhat from high to low levels of risk; it is possible that shared environmental influences may not change as a function of the moderator (particularly in adult twins). One advantage of biometric moderation models is that they allow for an empirical test of whether moderation is significant only for the A and E variance components but not C. What we would expect for the bioecological model is a crossing of A and E influences, with E showing greater expression at the risky end of the moderator and A showing greater expression in the enriched end of the moderator.
Illustrative Example: Cognition
There is a long and extensive history of research examining the genetic and environmental influences on cognitive ability, particularly intelligence. Indeed, intelligence is one of the most consistently heritable individual difference phenotypes that has ever been studied, routinely demonstrating heritability coefficients ranging from 60-80% (Plomin, DeFries, Knopik, & Neiderhiser, 2012). More than a decade ago, however, researchers made headlines for a study that suggested the genetic influences on intelligence may differ as a function of socioeconomic status (SES). Using a large sample that was notable for including children from families at the extreme low end of SES, the researchers (Turkheimer et al., 2003) showed that the genetic influences on intelligence were greatest among individuals in the highest SES (a linear combination of occupational status, parental education, and income), while for individuals at the low end of SES, most of the variance in intelligence could be explained by nonshared environmental factors. The authors interpreted this finding as evidence of Bronfenbrenner and Ceci's (1994) bioecological sensitivity to context model—that genetic influences on a phenotype will be most expressed in an environment that allows for flourishing. There had been previous studies that examined whether genetic influences on cognitive ability varied as a function of aspects of SES (as reviewed in Hanscombe et al., 2012), but the Turkheimer et al. study was the first to use Purcell's (2002) moderation model.
Since Turkheimer and colleagues (2003) published their findings, many research groups, including some of the original authors, have attempted to replicate these findings in different samples that vary in age, assessment of “intelligence” or “cognition”, and measure of SES. For instance, successful replications of the moderation of genetic variance have been found for cognitive ability in children as young as age 2 (Tucker-Drob, Rhemtulla, Harden, Turkheimer, & Fask, 2011), for math ability in 2- and 4-year old children (Rhemtulla & Tucker-Drob, 2012), and for cognitive outcomes (from the National Merit Scholastic Qualifying Test) in middle- to upper-class 17-year old twins as a function of parental income (but not necessarily parental education; Harden, Turkheimer, & Loehlin, 2007). In a test of the moderation of ACE influences in an adult sample (ranging in age from 24-84), researchers found that childhood SES moderated total and genetic variance in intelligence measured in adulthood, with the greatest phenotypic and genetic variance in intelligence found at the highest levels of childhood SES (Bates, Lewis, & Weiss, 2013).
There have also been failures to replicate Turkheimer and colleagues' original (2003) findings. Using a large sample of twins from the Twins Early Development Study based in the United Kingdom (Hanscombe et al., 2012), researchers examined the moderation of ACE influences on cognitive ability at eight different ages (2, 3, 4, 7, 9, 10, 12, 14) as a function of different indices of SES (a variation of parent education, occupation, and income). Out of 17 possible models (three SES indices at different ages), only one showed evidence of genetic moderation (age 10), and it failed to replicate when alternative SES indices were used. Instead, the greatest support across ages and indices of SES was for moderation of shared environmental variance, with less shared environmental variance found at the highest levels of SES. Similarly, another study in an adult sample drawn from the Netherlands Twin Registry also failed to find evidence of moderation of genetic influences on IQ, using both more distal (parents education level) and proximal (partner's education level, urbanization level, mean real estate price of residential area) indices of SES (van der Sluis, Willemsen, de Geus, Boomsma, & Posthuma, 2008); there was some evidence, however, that shared environmental influences were greater at higher levels of SES for older male twins. In a study using an all-male sample from the Vietnam Era Twin Registry, there was no evidence of moderation of genetic or environmental influences on an index of general cognitive ability (the Armed Forces Qualification Test) as a function of parental education (Grant et al., 2010).
In the most recent replication attempt, Kirkpatrick and colleagues (2014) used a combined sample of twins and non-twin sibling pairs from the United States. They used rearing-parent data from these offspring to determine family-level SES variables (parents' occupational status, educational attainment, annual household income) and examined all possible combinations of moderation on the A, C, and E paths to IQ. They concluded that there was moderation on genetic influences, no moderation of the shared environment, and moderation on the nonshared environment was equivocal at best; genetic moderation was such that genetic influences were greatest among those with highest family-of-origin SES. Moderation effects were not age dependent, meaning there was no evidence that moderation on certain parameters might be present in children but disappear by adulthood. They also suggested that differences in findings across the different studies to date could mean that the moderation of genetic influences on IQ is a result specific to a certain nationality (US) and SES variable (income). We point to the similarity in mixed findings for SES and BMI (see above), in which significant genetic moderation found when income and a US sample were used (Johnson & Krueger, 2005a) was not fully replicated when education and a European sample were used (Johnson et al., 2011). This again cements the importance of synthesizing both the outcome and the environmental measure in order to accurately compare and contrast results across studies.
Differential Susceptibility
What if the diathesis-stress and bioecological models are both right, for the same phenotype and environmental moderator? It is possible that the risky end of an environment (e.g., very low SES) allows for the expression of genetic vulnerability to poor outcomes, and that the enriched end of the environment (e.g., very high SES) also allows for the genes for good outcomes to “will out.” Ellis and Boyce (2008) refer to this model as the biological sensitivity to context model, or the Orchid hypothesis; like that very particular flower, some individuals may need just the right combination of variables in the environment to flourish, while others, like a dandelion, will do well in any environment. Belsky and Pluess (2009) have described the differential susceptibility model as one in which the same individual who may be genetically predisposed to suffer the most from risky environments may also benefit the most from environments without adversity. In other words, human beings differ in their plasticity to environments, with some being far more susceptible to the effect of both positive and negative environments. As an example, they point to the findings from the Caspi et al. (2003) study on depression, life stress, and 5-HTTLPR. While the focus of the findings was on greater depression in those with a combination of the risk allele and life stress, the results also demonstrated that individuals at lowest risk were those with the risk allele and no history of life stress. Belsky and Pluess suggest that these cross-over interactions, in which those who demonstrate the greatest likelihood of the outcome when they have the presence of genetic risk and environmental stressor also have the lowest likelihood of the outcome in the absence of either risk or stressor, are demonstrative of the differential susceptibility model. More recently, other researchers have laid out more explicit criteria for distinguishing diathesis-stress from differential susceptibility, with the concern that some findings may be incorrectly interpreted as diathesis-stress because they fail to evaluate for the cross-over inflection point (Roisman et al., 2012).
Differential Susceptibility in quantG×E Modeling
To our knowledge, there are no known guidelines for establishing differential susceptibility in quantG×E models. Since genes influence individuals' plasticity, individuals with certain genetic variants can have the worst outcomes in negative environments yet enjoy the best outcomes in positive environments, compared to individuals with other variants of the same gene. Extending the plasticity model to predictions about the cumulative effect of all genes, the model would predict that aggregate genetic influences are greatest at both ends of the environmental risk continuum. As shown in Figure 2 (Panel D), we would expect that genetic influences on the phenotype would be highest at the most extreme ends of the moderator variable, forming a U-shaped curve. In our example, we have artificially constrained C to be essentially flat across all levels of the moderator; because the total proportion of variance must add up to 100, nonshared environmental influences would parallel genetic influences in the opposite direction, increasing at the average levels of the moderator but decreasing at the extreme ends.
Illustrative Example: Marital Satisfaction and Physical Health
To date, there has been only one empirical example in quantG×E that supports the differential susceptibility hypothesis, along the guidelines we laid out above. Using the MIDUS adult twin sample, South and Krueger (2013) demonstrated that the etiological components of physical health differ depending on a person's marital relationship quality. The heritability of physical health, as measured by subjective perceptions of health, was greatest among those with very distressed marriages (h2=.38) and with very satisfying marriages (h2=.30). Nonshared environmental influences (as a proportion of total variance) were most elevated at average levels of marriage quality, and shared environmental influences actually increased from low to high levels of marital quality. Increases in the shared environment at the extremes of the moderator have also been found for parent-child conflict and adolescent personality (i.e., positive emotionality; Krueger, South, Johnson, & Iacono, 2008), and marital quality moderating internalizing psychopathology (South & Krueger, 2008) again suggesting that a notable benefit of these quantG×E models is the ability to identify when aspects of the rearing environment have the greatest impact on development.
Social Control and Social Compensation
So far, we have reviewed a model that focuses on genetic expression in a risky environmental context (diathesis-stress), genetic expression in an enriched environment (bioecological), and genetic expression at the extremes of a moderator that ranges from very bad to very good. Our final model focuses on dampening of genetic expression in the presence of the right environmental contexts. Here we group together two types of interaction posited by Shanahan and Hofer (2005) as they are conceptually overlapping and result in similar quantG×E results. Both involve the presence of a genetic diathesis and a context that prevents the expression of that diathesis. In the case of social control, the environment is one where constraints are imposed by structural processes or social norms. For social compensation, the environment is one that is notable either for the absence of stress or the presence of enriching properties. In essence, the control/compensation models result in the same pattern as the diathesis-stress model, but focusing on the opposite end of the interaction. Whereas the diathesis-stress model emphasizes the combination of genetic predisposition and presence of stress, control/compensation focuses on the circumstances that inhibit or lower genetic influences on an undesirable outcome. Thus, a quantG×E model supporting control/compensation would look much like Panel A in Figure 2. But instead of an environmental risk factor where the environment is a stressful trigger (e.g., delinquent peers) we could substitute an environmental context that constrains the possibility of the outcome for any individual (e.g., parental monitoring). In the case of an outcome like adolescent smoking, we would posit that at high levels of parental monitoring (+2 SD of the X axis) fewer individuals in general would smoke and any genetic influences on smoking would be dampened by the constraints imposed by the environment. In fact, this is exactly what happens; genetic influences on adolescent smoking decrease from low to high levels of parental monitoring (Dick et al., 2007). Again, there are many similarities between the diathesis-stress and control/compensation models, and determining whether the findings from a quantG×E study support one versus the other may be dependent on the environmental measure, and whether it assesses a putatively stressful or maladaptive risk factor or a protective factor that either constraints or possibly enriches individuals in that context.
Implications for Theory and Research
In this review, we have argued for the continued relevance of biometric modeling techniques, even in this age of increasingly common molecular genetic studies, particularly as applied to phenotypes related to social inequality. Specifically, we contend that biometric moderation modeling of latent genetic and environmental influences as a function of measured environmental contexts has the potential not only to inform the search for measured genes for things like personality, psychopathology, well-being and other indicators of social inequality, but also to add to our knowledge base of developmental phenotypes related to social inequality through theory testing. In this section, we lay out our final thoughts on how the study of social inequality can incorporate quantG×E to test theory and ultimately develop interventions that can be applied at multiple levels.
We have reviewed how early failures and mixed findings from candidate gene studies led to the search for cG×E. There are many practical advantages of quantG×E over cG×E, and given the turn in the field toward much stricter scrutiny of cG×E, quantG×E might fit well as a first step for identifying the presence of genetic moderation and the environmental context of that moderation. We readily acknowledge that more recent work in molecular genetics is faring much better than early linkage, candidate, and association studies (Sullivan, Daly, & O'Donovan, 2012). Unlike these earlier techniques, GWAS search across the genome for differences in single nucleotide polymorphisms (SNPs) and are unbound from a priori hypotheses about candidate genes. GWAS studies have improved upon previous methods and have produced replicable results for physical health (e.g., the FTO gene link to body mass index, Frayling et al., 2007) and psychiatric disorders (Sullivan et al., 2012). For instance, one of the most recent studies of schizophrenia used genome-wide genotype data from 36, 989 cases and 113,075 controls and identified 128 significant associations across 108 loci; of note, most (75%) were in protein-coding regions of the genome and many had strong expression in the brain or in tissues with important roles in immunity (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014).
Finding individual genetic variants that contribute to the outcome of interest remains challenging, but studies suggest the importance of sets of polymorphisms that collectively contribute to variance in the phenotype (e.g., Purcell et al., 2014). Earlier, we briefly mentioned the use of polygenic risk scores—these composite scores are a sum across a number of genetic markers (i.e., risk alleles) that may not achieve significance on an individual basis in reasonably sized samples, but as a group are significantly related to a trait or outcome of interest (for a more complete review of this method, see Wray et al., 2014). The genetic markers for the polygenic score are often chosen based on having the strongest p values in a GWAS discovery sample (and are often weighted according to effect size), so determining the initial composite still requires the time and resources of large discovery samples. Further, results will be limited by the size of the sample (some may be underpowered to detect effects) and the percentage of variance in the trait explained in that sample (Dudbridge, 2013). The advantage of polygenic scores is that once the composite is determined (possibly from markers identified from previous GWAS) that composite score can be tested in a new target sample of individuals; further, the alleles chosen for inclusion in the polygenic risk composite do not have to meet the stringent criteria often required for GWAS significance (and in fact, risk scores at times include all SNP scores, some of which may simply be noise). To date, polygenic scores have been used to examine the variance explained in personality, psychopathology, and cancer (Dudbridge, 2013; Wray et al., 2014). Only very recently, researchers have begun to test whether the effect of these “gene sets” on a phenotype are moderated by a measured environmental variable (Li et al., 2010; Salvatore et al., online ahead of print). We see this as directly analogous to quantG×E; both examine if environmental contexts change the relative importance of a genetic composite, whether it is all genes (quantG×E) or a subset of genes that may or may not contribute to the phenotype under study (polygenic G×E). Indeed, quantG×E may serve as an important first step in a program of research that aims to determine when and where to direct molecular genetic efforts.
Ultimately, any gene finding efforts directed to variables related to social inequality are interested in identifying the biological pathways and mechanisms that lead to things like poverty, poor physical and mental health, and subjective well-being, among other outcomes. Studies of quantG×E can help with this, not only as a step in identifying genetic variants, but also by empirically testing theoretical models of gene-environment interplay. In this article, we have reviewed four developmental paradigms that lead to specific predictions about the pattern of quantG×E that would be found for each model. Three models—diathesis-stress, bioecological, and differential susceptibility—each posit the expression of genetic influences (i.e., higher levels of heritability) as a function of the right environmental contexts. The fourth, social control/compensation, shares the same shape as the diathesis-stress model, but focuses on how genetic influences might be dampened or diminished as a function of an environmental moderator. Thus, the pattern of moderation found when examining a phenotype of interest and an environmental context of interest can be matched to one of these models as a direct test of how genetic influences exert an effect on the outcome. We readily acknowledge two important caveats to these theoretical models. First, most have explicit predictions about how genetic influences would change as a function of an environmental moderator, but are silent as to how or why environmental influences would change depending on context. For instance, we have found support for the diathesis-stress model such that genetic influences on internalizing psychopathology are diminished among those in high-quality marriages; instead, the variance in internalizing among those with satisfying marriages is mainly explained by individual differences in the family and nonshared environment (South & Krueger, 2008). What those particular environmental influences are has been a point of contention among behavior geneticists for decades (Turkheimer & Waldron, 2000), and we can only speculate as to what environmental conditions might lead to depression and anxiety among individuals in a happy marriage (e.g., work strain, physical health problems, caring for elderly parents). The second caveat is that future work may find patterns of moderation that are more complex than a simple linear increase or decrease of genetic or environmental influences across the range of the moderator. Briley and colleagues (in press) recently introduced a new nonparameteric approach to testing for nonlinear G×E. As an example, they demonstrated that genetic variance in kindergarten reading achievement peaked between 0 and 1 standard deviations above the mean of SES (on a z-score scale), before dropping dramatically by +2 SD. Their local structural equation modeling (LOSEM) application of nonparametric techniques is currently only available when the moderator is shared between family members, but they recommend extension to situations where the moderator differs between twins. To address these two important caveats, future researchers in this field should test hypotheses about how genetic and environmental influences are changing in these models of quantG×E. In doing so, it is worth being open minded about the possibility of complex (e.g., non-linear) relations that may not be optimally captured by all G×E models.
As with any method, replication across different samples that include different developmental periods will be paramount. To date, most quantG×E studies have been cross-sectional or have examined aspects of the childhood environment on now-adult twins. An important next step will be to examine the pattern of quantG×E over time, ideally using the same sample measured for the same constructs over time. This would serve several purposes: 1) finding the same pattern of quantG×E in constructs measured over time would be strong evidence that the effect is not due to Type I error; and 2) determining whether a pattern of quantG×E persists throughout development or is only apparent at certain stages would speak to the importance of sensitive developmental periods; and 3) testing G×E longitudinally will identify if something that looks like a diathesis-stress effect at one point might be differential susceptibility, for instance, at another (Roisman et al., 2012). Key to determining the distinction between the models will be measuring the entire spectrum of the environment. Too many studies focus on evaluating the risk end of the spectrum; a commensurate focus on positive aspects of these relationships is necessary to capture all possible forms of G×E. Going forward, researchers should also attempt to utilize the same phenotype and environmental moderator across different studies of quantG×E. It is important to determine, when there are mixed findings across studies, if the effect is not replicating because it truly is not there or because it is specific to one type of moderator (i.e., income instead of education as a proxy for SES).
The four models that we have outlined provide researchers with theory with which to ground future predictions about specific combinations of phenotypes and outcomes. The difficulty, however, is that the predictions of competing models may seem equally plausible a priori. For instance, even though most of the studies examining biometric moderation of BMI support a diathesis stress model, one could easily imagine predicting a priori that genetic predispositions for good health would be most expressed in an enriched environment (i.e., a bioecological model). To guide thinking on how environments may impact the etiology of an outcome, it may be necessary to think about the functionality of that trait, particularly from an evolutionary standpoint. Johnson and Krueger (2005b) suggested that the direction of effect found for a particular combination of moderator and trait might be related to the relative adaptiveness of a trait. As they posited, if the trait in question is relatively adaptive, like high IQ, then favorable environments will allow for genetic expression of that trait and unfavorable environments will suppress genetic influences (our addition in italics). If the trait is not adaptive, like mental illness or physical disability/disease, favorable environments will suppress genetic expression and unfavorable environments will allow genetic influences to will out. To their original interpretation we would add the corollary that there may be times when a genetic predisposition has the potential for being both adaptive and maladaptive (see the arguments that mental illness may have persisted evolutionarily because it brings certain advantages; (Keller & Miller, 2006), leading to genetic influences being expressed in favorable and unfavorable environments.
Ultimately, we would hope that testing quantG×E would lead to applied work that will reduce social inequality. The past ten years have seen a surge in the use of these biometric moderation models since they were first introduced, particularly for a handful of phenotypes (e.g., cognition, BMI, behavioral and substance use disorders). The findings from some of this work have direct practical applications; for instance, findings from quantG×E for cognition and related phenotypes suggest that environmental interventions are best aimed at those from the lowest SES groups. We see the potential for an expansion of this work into more phenotypes that have relevance to social inequality. Researchers could examine, for instance, what the pattern of quantG×E is for education, income or career attainment, and overall well-being. Many existing twin databases can provide a wealth of resources for examining quantG×E for these phenotypes, and more importantly, have excellent measures of the environment from greatest risk to most positive enrichment. Further, an exciting aspect of quantG×E models is the potential to inform interventions at the individual level. Again, we emphasize that quantG×E does not tell us how important genes are for any one specific person, but it does get us closer and closer to identifying, for specific subsets of individuals, the relative importance of genetics and environment. The administration of the current President of the United States has recently announced an investment in “precision” or “personalized” medicine, in which interventions are tailored to a patient based on individual differences in lifestyles, genetics, and environment. In this vein, we see results from work using quantG×E leading to the identification of certain “sensitive periods” during development (e.g., Roisman et al., 2012), when genetic influences are most susceptible to lifestyle and environmental contexts, and when interventions for the most at-risk members of the population can do the most good. We are still not at the point of being able to estimate an “individual heritability” but these models of G×E interplay are getting us closer and closer to a form of personalized medicine for social science phenotypes.
Acknowledgments
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
Footnotes
Susan C. South, Department of Psychological Sciences, Purdue University; Nayla R. Hamdi, Department of Psychology, University of Minnesota; Robert F. Krueger, Department of Psychology, University of Minnesota. The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Contributor Information
Susan C. South, Purdue University
Nayla Hamdi, University of Minnesota
Robert F. Krueger, University of Minnesota
References
- Bates TC, Lewis GJ, Weiss A. Childhood socioeconomic status amplifies genetic effects on adult intelligence. Psychological Science. 2013;24:2111–2116. doi: 10.1177/0956797613488394. [DOI] [PubMed] [Google Scholar]
- Belsky J, Pluess M. Beyond diathesis stress: Differential susceptibility to environmental influences. Psychological Bulletin. 2009;135:885–908. doi: 10.1037/a0017376. [DOI] [PubMed] [Google Scholar]
- Benjamin DJ, Cesarini D, van der Loos MJHM, Dawes CT, Koellinger PD, Magnusson PKE, et al. Visscher PM. The genetic architecture of economic and political preferences. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(21):8026–8031. doi: 10.1073/pnas.1120666109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briley DA, Harden KP, Bates TC, Tucker-Drob EM. Nonparametric estimates of gene x environment interaction using local structural equation modeling. Behavior Genetics. doi: 10.1007/s10519-015-9732-8. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronfenbrenner U, Ceci SJ. Nature-nurture reconceptualized in developmental perspective: A bioecological model. Psychological Review. 1994;101:568–586. doi: 10.1037/0033-295x.101.4.568. [DOI] [PubMed] [Google Scholar]
- Burns RA, Machin MA. Psychological wellbeing and the diathesis-stress hypothesis model: The role of psychological functioning and quality of relations in promoting subjective well-being in a life events study. Personality and Individual Differences. 2013;54(3):321–326. [Google Scholar]
- Burt SA. A mechanistic explanation of popularity: Genes, rule-breaking, and evocative gene-environment correlations. Journal of Personality & Social Psychology. 2009;96:783–794. doi: 10.1037/a0013702. [DOI] [PubMed] [Google Scholar]
- Chabris CF, Lee JJ, Benjamin DJ, Beauchamp JP, Glaeser EL, Borst G, et al. Laibson DI. Why it is hard to find genes associated with social science traits: Theoretical and empirical considerations. American Journal of Public Health. 2013;103(Suppl 1):S152–S166. doi: 10.2105/ajph.2013.301327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland HH. Disadvantaged neighborhoods and adolescent aggression: Behavioral genetic evidence of contextual effects. Journal of Research on Adolescence. 2003;13(2):211–238. [Google Scholar]
- Dick DM, Viken R, Purcell S, Kaprio J, Pulkkinen L, Rose RJ. Parental monitoring moderates the importance of genetic and environmental influences on adolescent smoking. Journal of Abnormal Psychology. 2007;116(1):213–218. doi: 10.1037/0021-843X.116.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genetics. 2013;9(3):e1003348. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Keller MC. A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. The American Journal of Psychiatry. 2011;168(10):1041–1049. doi: 10.1176/appi.ajp.2011.11020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Pollastri AR, Smoller JW. Mind the gap: Why many geneticis and psychological scientists have discrepant views about gene-environment interaction (G×E) research. American Psychologist. 2014;69:249–268. doi: 10.1037/a0036320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis BJ, Boyce WT. Biological sensitivity to context. Current Directions in Psychological Science. 2008;17:183–187. [Google Scholar]
- Falconer DS. The inheritance of liability to certain diseases, estimated from the incidence among relatives. Annals of Human Genetics. 1965;29:51–76. [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. McCarthy MI. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant MD, Kremen WS, Jacobson KC, Franz C, Xian H, Eisen SA, et al. Lyons MJ. Does parental education have a moderating effect on the genetic and environmental influences of general cognitive ability in early adulthood? Behavior Genetics. 2010;40(4):438–446. doi: 10.1007/s10519-010-9351-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdi NR, Krueger RF, South SC. Socioeconomic status moderates genetic and environmental effects on the amount of alcohol use. Alcoholism: Clinical and Experimental Research. doi: 10.1111/acer.12673. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanscombe KB, Trzaskowski M, Haworth CMA, Davis OSP, Dale PS, Plomin R. Socioeconomic status (SES) and children's intelligence (IQ): In a UK-representative sample SES moderates the environmental, not genetic, effect on IQ. PLoS ONE. 2012;7(2) doi: 10.1371/journal.pone.0030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden KP, Turkheimer E, Loehlin JC. Geneotype by environment interaction in adolescents' cognitive aptitude. Behavior Genetics. 2007;37:273–283. doi: 10.1007/s10519-006-9113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Jardine R, Martin NG. Interactive effects of genotype and social environment on alcohol consumption in female twins. Journal of Studies on Alcohol. 1989;50(38-48) doi: 10.15288/jsa.1989.50.38. [DOI] [PubMed] [Google Scholar]
- Hewitt JK. Editorial policy on candidate gene association and candidate gene-by-enviornment interaction studies of complex traits. Behavior Genetics. 2012;42:1–2. doi: 10.1007/s10519-011-9504-z. [DOI] [PubMed] [Google Scholar]
- Hicks BM, South SC, DiRago AC, Iacono WG, McGue M. Environmental adversity and increasing genetic risk for externalizing disorders. Archives of General Psychiatry. 2009;66(6):640–648. doi: 10.1001/archgenpsychiatry.2008.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaekel J, Pluess M, Belsky J, Wolke D. Effects of maternal sensitivity on low birth weight children's academic achievement: a test of differential susceptibility versus diathesis stress. J Child Psychol Psychiatry. 2014 doi: 10.1111/jcpp.12331. [DOI] [PubMed] [Google Scholar]
- Jarnecke AM, South SC. Genetic and environmental influences on alcohol use: Moderation by some, but not all, forms of social support. Alcoholism: Clinical and Experimental Research. 2014;38:367–375. doi: 10.1111/acer.12263. [DOI] [PubMed] [Google Scholar]
- Johnson W, Krueger RF. Genetic effects on physical health: Lower at higher income levels. Behavior Genetics. 2005a;35(5):579–590. doi: 10.1007/s10519-005-3598-0. [DOI] [PubMed] [Google Scholar]
- Johnson W, Krueger RF. Higher perceived life control decreases genetic variance in physical health: Evidence from a national twin study. Journal of Personality and Social Psychology. 2005b;88(1):165–173. doi: 10.1037/0022-3514.88.1.165. [DOI] [PubMed] [Google Scholar]
- Johnson W, Kyvik KO, Skytthe A, Deary IJ, Sorensen TI. Education modifies genetic and environmental influences on BMI. PLoS ONE. 2011;6(1):e16290. doi: 10.1371/journal.pone.0016290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karg K, Burmeister M, Shedden K, Sen S. The serotonin transporter promoter variant (5-HTTLPR), stress, and depression metaanalysis revisited: Evidence of genetic moderation. Archives of General Psychiatry. 2011;68:444–454. doi: 10.1001/archgenpsychiatry.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MC, Miller G. Resolving the paradox of common, harmful, heritable mental disorders: Which evolutionary genetic models work best? Behavioral and Brain Sciences. 2006;29:385–452. doi: 10.1017/S0140525X06009095. [DOI] [PubMed] [Google Scholar]
- Kendler K, Baker JH. Genetic influences on measures of the environment: A systematic review. Psychological Medicine. 2007;37:615–626. doi: 10.1017/S0033291706009524. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Gilman SE, Thornton LM, Kendler KS. Health, wellbeing, and social responsibility in the MIDUS twin and sibling subsamples. In: Brim OG, Ryff CD, Kessler RC, editors. How healthy are we? A national study of wellbeing at midlife. Chicago: University of Chicago Press; 2004. pp. 124–152. [Google Scholar]
- Kirkpatrick RM, McGue M, Iacono WG. Replication of a gene-environment interaction via multimodal inference: Additive-genetic variance in adolescents' general cognitive ability increases with family-of-origin socioeconomic status. Behavior Genetics, Online first posting. 2014 doi: 10.1007/s10519-014-9698-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenen KC, Caspi A, Moffitt TE, Rijsdijk F, Taylor A. Genetic influences on the overlap between low IQ and antisocial behavior in young children. Journal of Abnormal Psychology. 2006;115(4):787–797. doi: 10.1037/0021-843X.115.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger RF, South SC, Johnson W, Iacono W. The heritability of personality is not always 50%: Gene-environment interactions and correlations between personality and parenting. Journal of Personality. 2008;76:1485–1522. doi: 10.1111/j.1467-6494.2008.00529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhao JH, Luan Ja, Ekelund U, Luben RN, Khaw KT, et al. Loos RJF. Physical Activity Attenuates the Genetic Predisposition to Obesity in 20,000 Men and Women from EPIC-Norfolk Prospective Population Study. PLoS Medicine. 2010;7(8):e1000332. doi: 10.1371/journal.pmed.1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein B, Hindroff LA, Hunter DJ, et al. Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuck SB, McCaffery JM. Gene-environment interaction. Annual Review of Psychology. 2014;65:41–70. doi: 10.1146/annurev-psych-010213-115100. [DOI] [PubMed] [Google Scholar]
- Martin NW, Medland SE, Verweij KJH, Lee SH, Nyholt DR, Madden PA, et al. Martin NG. Educational Attainment: A Genome Wide Association Study in 9538 Australians. PLoS ONE. 2011;6(6):e20128. doi: 10.1371/journal.pone.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt TE, Caspi A, Rutter M. Measured gene-environment interactions in psychopathology: Concepts, research strategies, and implications for research, intervention, and public understanding of genetics. Perspectives on Psychological science. 2006;1:5–27. doi: 10.1111/j.1745-6916.2006.00002.x. [DOI] [PubMed] [Google Scholar]
- Monroe SM, Simons AD. Diathesis-stress theories in the context of life stress research: Implications for the depressive disorders. Psychological Bulletin. 1991;110:406–425. doi: 10.1037/0033-2909.110.3.406. [DOI] [PubMed] [Google Scholar]
- Munafò MR, Durrant C, Lewis G, Flint J. Gene × Environment interactions at the serotonin transporter locus. Biological Psychiatry. 2009;65:211–219. doi: 10.1016/j.biopsych.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Mustelin L, Silventoinen K, Pietilainen K, Rissanen A, Kaprio J. Physical activity reduces the influence of genetic effects on BMI and waist circumference: A study in young adult twins. International Journal of Obesity. 2009;33:29–36. doi: 10.1038/ijo.2008.258. [DOI] [PubMed] [Google Scholar]
- Pedersen NL, Christensen K, Dahl AK, Finkel D, Franz CE, Gatz M, et al. Reynolds CA. IGEMS: the consortium on Interplay of Genes and Environment across Multiple Studies. Twin Res Hum Genet. 2013;16(1):481–489. doi: 10.1017/thg.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomin R, DeFries JC, Knopik V, Neiderhiser JM. Behavioral genetics. 6th. New York: Worth Publishers; 2012. [Google Scholar]
- Purcell S. Variance components models for gene-environment interaction in twin analysis. Twin Research. 2002;5:554–571. doi: 10.1375/136905202762342026. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. Sklar P. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506(7487):185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raine A. Biosocial studies of antisocial and violent behavior in children and adults: A review. Journal of Abnormal Child Psychology. 2002;30:311–326. doi: 10.1023/a:1015754122318. [DOI] [PubMed] [Google Scholar]
- Rathouz PJ, Van Hulle CA, Rodgers JL, Waldman ID, Lahey BB. Specification, testing, and interpretation of gene-by-measured-environment interaction models in the presence of gene-environment correlation. Behavior Genetics. 2008;38(3):301–315. doi: 10.1007/s10519-008-9193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhemtulla M, Tucker-Drob EM. Gene-by-socioeconomic status interaction on school readiness. Behavior Genetics. 2012;42(4):549–558. doi: 10.1007/s10519-012-9527-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld CA, Cesarini D, Benjamin DJ, Koellinger PD, De Neve JE, Tiemeier H, et al. Bartels M. Molecular genetics and subjective well-being. PNAS. 2013a;110:9692–9697. doi: 10.1073/pnas.1222171110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld CA, Medland SE, Derringer J, Yang J, Esko Tn, Martin NW, et al. Koellinger PD. GWAS of 126,559 Individuals Identifies Genetic Variants Associated with Educational Attainment. Science (New York, N Y) 2013b;340(6139):1467–1471. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Herrell R, Lehner T, Liang KY, Eaves L, Hoh J, et al. Merikangas KR. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: A meta-analysis. JAMA. 2009;301(23):2462–2471. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roisman GI, Newman DA, Fraley RC, Haltigan JD, Groh AM, Haydon KC. Distinguishing differential susceptibility from diathesis–stress: Recommendations for evaluating interaction effects. Development and Psychopathology. 2012;24(2):389–409. doi: 10.1017/S0954579412000065. [DOI] [PubMed] [Google Scholar]
- Rowe DC. Environmental and genetic influences on dimensions of perceived parenting: A twin study. Developmental Psychology. 1981;17:203–208. [Google Scholar]
- Salvatore JE, Aliev F, Bucholz K, Agrawal A, Hesselbrock V, Hesselbrock M, et al. Dick DM. Polygenic risk for externalizing disorders: Gene-by-development and gene-by-environment effects in adolescents and young adults. Clinical Psychological Science. doi: 10.1177/2167702614534211. online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schousboe K, Willemsen G, Kyvik KO, Mortensen J, Boomsma DI, Cornes BK, et al. Harris JR. Sex Differences in Heritability of BMI: A Comparative Study of Results from Twin Studies in Eight Countries. Twin Research and Human Genetics. 2003;6(05):409–421. doi: 10.1375/twin.6.5.409. [DOI] [PubMed] [Google Scholar]
- Shanahan MJ, Hofer SM. Social Context in Gene–Environment Interactions: Retrospect and Prospect. The Journals of Gerontology Series B: Psychological Sciences and Social Sciences. 2005;60(Special Issue 1):65–76. doi: 10.1093/geronb/60.Special_Issue_1.65. [DOI] [PubMed] [Google Scholar]
- South SC, Krueger RF. Marital quality moderates genetic and environmental influences on the internalizing spectrum. Journal of Abnormal Psychology. 2008;117:826–837. doi: 10.1037/a0013499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South SC, Krueger RF. Genetic and environmental influences on internalizing psychopathology vary as a function of economic status. Psychological Medicine. 2011;41:107–118. doi: 10.1017/S0033291710000279. [DOI] [PubMed] [Google Scholar]
- South SC, Krueger RF. Marital satisfaction and physical health: Evidence for an Orchid effect. Psychological Science. 2013 doi: 10.1177/0956797612453116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spotts EL, Pedersen NL, Neiderhiser JM, Reiss D, Lichtenstein P, Hansson K, Cederblad M. Genetic effects on women's positive mental health: Do marital relationships and social support matter? Journal of Family Psychology. 2005;19:339–349. doi: 10.1037/0893-3200.19.3.339. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: The emerging picture and its implications. Nature Reviews Genetics. 2012;13(8):537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Health and Retirement Study: A Longitudinal Study of Health, R., and Aging. Produced and distributed by the University of Michigan with funding from the National Institute on Aging (grant number NIA U01AG009740) Ann Arbor, MI. [WWW Document], n.d. URL http://hrsonline.isr.umich.edu/. Retrieved March 2, 2015.
- Tucker-Drob EM, Rhemtulla M, Harden KP, Turkheimer E, Fask D. Emergence of a Gene × Socioeconomic Status interaction on infant mental ability between 10 months and 2 years. Psychological Science. 2011;22:125–133. doi: 10.1177/0956797610392926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk DC. A diathesis-stress model of chronic pain and disability following traumatic injury. Pain Res Manag. 2002;7(1):9–19. doi: 10.1155/2002/252904. [DOI] [PubMed] [Google Scholar]
- Turkheimer E. Three laws of behavior genetics and what they mean. Current Directions in Psychological Science. 2000;9:160–164. [Google Scholar]
- Turkheimer E, Haley A, Waldron M, D'Onofrio B, Gottesman II. Socioeconomic status modifies heritability of IQ in young children. Psychological Science. 2003;14:623–628. doi: 10.1046/j.0956-7976.2003.psci_1475.x. [DOI] [PubMed] [Google Scholar]
- Turkheimer E, Waldron M. Nonshared environment: A theoretical, methodological, and quantitative review. Psychological Bulletin. 2000;126(1):78–108. doi: 10.1037/0033-2909.126.1.78. [DOI] [PubMed] [Google Scholar]
- Tuvblad C, Grann M, Lichtenstein P. Heritability for adolescent antisocial behavior differs with socioeconomic status: Gene-environment interaction. Journal of Child Psychology and Psychiatry. 2006;47:734–743. doi: 10.1111/j.1469-7610.2005.01552.x. [DOI] [PubMed] [Google Scholar]
- van der Loos MJHM, Rietveld CA, Eklund N, Koellinger PD, Rivadeneira F, Abecasis GaR, et al. Thurik AR. The Molecular Genetic Architecture of Self-Employment. PLoS ONE. 2013;8(4):e60542. doi: 10.1371/journal.pone.0060542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluis S, Posthuma D, Dolan CV. A note on false positives and power in G × E modelling of twin data. Behavior Genetics. 2012;42:170–186. doi: 10.1007/s10519-011-9480-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluis S, Willemsen G, de Geus EJC, Boomsma DI, Posthuma D. Gene-environment interaction in adults' IQ scores: Measures of past and present environment. Behavior Genetics. 2008;38(4):348–360. doi: 10.1007/s10519-008-9212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hulle CA, Lahey BB, Rathouz PJ. Operating characteristics of alternative statistical models for detecting gene-by-measured environment interaction in the presence of gene-environment correlatin in twin and sibling studies. Behavior Genetics. 2013;43:71–84. doi: 10.1007/s10519-012-9568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson NF, Harden KP, Buchwald D, Vitiello MV, Pack AI, Weigle DS, Goldberg J. Sleep Duration and Body Mass Index in Twins: A Gene-Environment Interaction. Sleep. 2012;35(5):597–603. doi: 10.5665/sleep.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A, Bates TC, Luciano M. Happiness is a personality thing: The genetics of personality and well-being in a representative sample. Psychological Science. 2008;19:205–210. doi: 10.1111/j.1467-9280.2008.02068.x. [DOI] [PubMed] [Google Scholar]
- Wray NR, Lee SH, Mehta D, Vinkhuyzen AAE, Dudbridge F, Middeldorp CM. Research Review: Polygenic methods and their application to psychiatric traits. Journal of Child Psychology and Psychiatry. 2014;55(10):1068–1087. doi: 10.1111/jcpp.12295. [DOI] [PubMed] [Google Scholar]