Abstract
Macrophages are important mediators of tumor progression and their function is broadly influenced by different microenvironmental stimuli. To understand the molecular basis of the tumor-supporting role of macrophages in aggressive breast cancer we co-cultured human peripheral monocytes with two breast cancer cell lines representing distinct aggressive cellular phenotype and transcriptionally profiled the changes occurring in both cells during in vitro activated crosstalk. Here we provide a detailed description of the experimental design, sample identity and analysis of the Illumina RNA-Seq data, which have been deposited into Gene Expression Omnibus (GEO): GSE75130.
Keywords: Breast cancer, Co-culture, RNA-Seq, Illumina
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo sapiens/breast cancer cell line/MDA-MB-231 and T47D Homo sapiens/peripheral blood monocyte/3 healthy donors |
| Sex | Female |
| Sequencer or array type | Illumina HiSeqTM 2000 |
| Data format | Raw data: fastq Processed data: xls |
| Experimental factors | Monocyte cancer cell co-culture vs. single culture |
| Experimental features | Transcriptome profiling of genes that modulate monocyte/cancer cell activation in aggressive breast cancer |
| Consent | Data are publicly available |
| Sample source location | Zurich, Switzerland |
1. Direct link to deposited data
2. Experimental design, materials and methods
2.1. Cells
Peripheral blood monocytes were isolated from three buffy coats (Blutspende Zürich, Zurich, Switzerland) by density gradient centrifugation (Ficoll-Paque PLUS (GE Healthcare)) and enriched for co-culture assays using Human Monocyte Isolation Kit II (MACS, Miltenyi Biotech). The enriched cells further specified as monocytes “donor1, donor2 and donor3” were used immediately after enrichment.
The breast cancer cell lines MDA-MB-231 and T47D cells (kindly provided by Dr. Nancy E. Hynes, FMI, Basel, Switzerland) were maintained in DMEM (Gibco) supplemented with 10% fetal bovine serum (Sigma) and penicillin/streptomycin.
2.2. Transwell co-culture assay
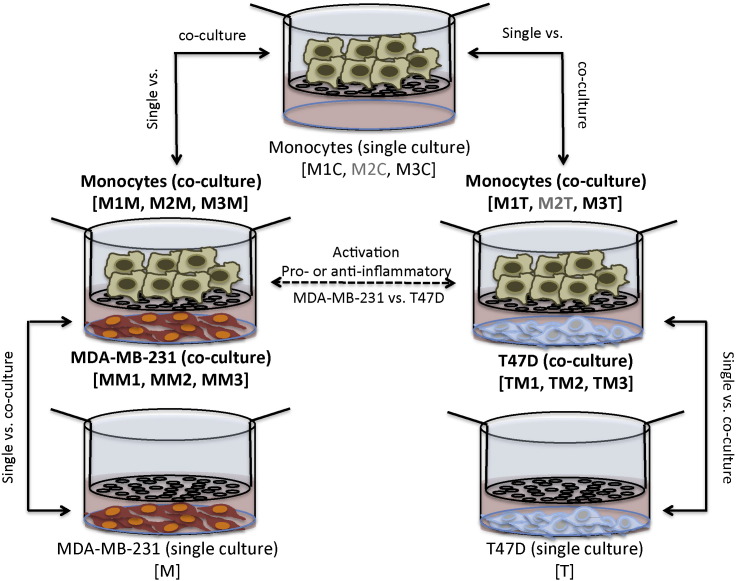
The breast cancer cell lines were plated 1 × 106 cells in the lower compartment of 75 mm polycarbonate transwell inserts pore size 0.4 μm (Corning) in maintenance medium one day prior monocyte isolation. The next day the medium was replaced by RPMI (Gibco) on hour before the addition of 4 × 106 monocytes into the upper compartment of the transwell inserts. The cells were co-cultured for five days in a humidified chamber at 37 °C. Control wells contained either cancer cells only in the lower compartment with RPMI in the upper compartment or RPMI in the lower compartment with only monocytes in the upper compartment. See Fig. 1 for experimental design.
Fig. 1.
Experimental design. Monocytes and breast cancer cell lines (MDA-MB-231 or T47D) were co-cultured for five days in a transwell system and the gene expression profile of co-cultured cells was compared to the gene expression of the same cells cultured alone. Sample names are indicated in the brackets. The samples marked in gray were not sequenced because of low RNA concentration. The middle dashed line demonstrates the experimental feature where the difference of macrophage activation in the presence of either MDA-MB-231 or T47D cells was analyzed.
2.3. RNA isolation and RNA-Seq
The total RNA from 17 samples was isolated using the Trizol protocol (Invitrogen). See Table 1 for sample identity. The RNA quantity was determined by NanoDrop spectrophotometer and quality assessed on the Agilent 2100 BioAnalyzer (Agilent) using an RNA 6000 Nano Chip. Two samples, donor2 monocytes cultured as single culture (M2C) and with T47D co-culture (M2T) did not reach sufficient amount of RNA and were excluded from further processing. At this point the samples were sent to BGI China where they were processed according to BGI's standard RNA-Seq sample preparation. Shortly, magnetic beads with Oligo (dT) were used to isolate mRNA, which was mixed with the fragmentation buffer to fragment it into short fragments. The cleaved RNA fragments were synthesized into single-strand cDNA using superscript II reverse transcriptase and random hexamers followed by second strand synthesis with DNA polymerase I and Escherichia coli RNase H. After the second strand synthesis, with end repair and A-tailing, the synthesized double-stranded cDNA fragments were subjected to purification, ligated to Illumina adapters using Quick ligation TM kit (NEB) and DNA ligase. The resultant cDNA adapter-modified cDNA libraries were fractionated on agarose gel, 200-bp fragments were excised and amplified by 15 cycles of polymerase chain reaction. After purification the quality of cDNA libraries was checked by Bioanalyzer 2100 (Agilent). The concentration of the cDNA libraries was measured and diluted to 10 nM in Tris–HCl buffer prior to cluster generation. Cluster formation, primer hybridization and sequencing reactions were performed sequentially according to the manufacturer's recommended protocol. In the present study, we used pair-end sequencing by Illumina HiSeqTM 2000 (Illumina, San Diego, CA, USA).
Table 1.
Sample identity.
| Sample name | Source name | Cell type | Culture condition | Classification | RIN |
|---|---|---|---|---|---|
| M1C | Donor1 | Monocyte | Single | / | 8.20 |
| M1M | Donor1 | Monocyte | Co-culture | / | 8.60 |
| M1T | Donor1 | Monocyte | Co-culture | / | 7.60 |
| M2C | Donor2 | Monocyte | Single | / | 9.40 |
| M2M | Donor2 | Monocyte | Co-culture | / | 9.40 |
| M2T | Donor2 | Monocyte | Co-culture | / | N/A |
| M3C | Donor3 | Monocyte | Single | / | 9.50 |
| M3M | Donor3 | Monocyte | Co-culture | / | 8.50 |
| M3T | Donor3 | Monocyte | Co-culture | / | 9.10 |
| M | MDA-MB-231 | Breast cancer | Single | TNBC | 9.10 |
| MM1 | MDA-MB-231 | Breast cancer | Co-culture | TNBC | 8.60 |
| MM2 | MDA-MB-231 | Breast cancer | Co-culture | TNBC | 8.70 |
| MM3 | MDA-MB-231 | Breast cancer | Co-culture | TNBC | 8.50 |
| T | T47D | Breast cancer | Single | ER pos | 9.40 |
| TM1 | T47D | Breast cancer | Co-culture | ER pos | 9.10 |
| TM2 | T47D | Breast cancer | Co-culture | ER pos | 9.40 |
| TM3 | T47D | Breast cancer | Co-culture | ER pos | 9.00 |
TNBC, triple-negative breast cancer; ER pos, estrogen receptor positive; RIN, RNA integrity number; N/A, not applicable.
2.4. Data analysis
High-quality reads were aligned to the human reference genome with SOAPaligner/SOAP2 [1]. The matched reads were aligned to Human Refseq mRNA (NCBI). The sequences aligned with individual transcripts were counted digitally. The expression level for each gene was normalized to reads per kilobase per million mapped reads (RPKM) to facilitate the comparison of transcripts among samples. A mean log2 ratio [RPKM of monocytes co-cultured with MDA-MB-231 or T47D cells/monocytes cultured alone; RPKM of MDA-MB231 or T47D cells co-cultured with monocytes/MDA-MB231 or T47D cells cultured alone] of each gene was calculated. To identify genes differentially expressed between groups we used an algorithm based on Ref. [2] with a correction for false positive (type I) and false negative (type II) errors using the FDR method [3]. The genes were regarded as differentially expressed when their FDRs were less than 0.05. Further, genes were classified as up regulated when their mean log2 ratio was larger than 0.5 or down regulated when their log2 ratio was less than − 0.5.
3. Discussion
We describe here a dataset composed of RNA-Seq gene expression profiling of macrophage-cancer cell crosstalk using two different breast cancer cell lines with distinct aggressive phenotype. With this data we could show that macrophage response to different cancer cells does not necessarily promote a tumor-supporting inflammatory response since a pro-inflammatory gene signature was activated in macrophages co-cultured with estrogen positive breast cancer cells [4].
Conflict of interest
The authors declare no conflict of interests.
Acknowledgments
MH was supported by a Sigrid Jusélius fellowship and the Instrumentarium foundation.
References
- 1.Li R., Yu C., Li Y., Lam T.W., Yiu S.M., Kristiansen K. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25(15):1966–1967. doi: 10.1093/bioinformatics/btp336. Epub 2009/06/06. PubMed PMID: 19497933) [DOI] [PubMed] [Google Scholar]
- 2.Audic S., Claverie J.M. The significance of digital gene expression profiles. Genome Res. 1997;7(10):986–995. doi: 10.1101/gr.7.10.986. (Epub 1997/10/23. PubMed PMID: 9331369) [DOI] [PubMed] [Google Scholar]
- 3.Kim K.I., van de Wiel M.A. Effects of dependence in high-dimensional multiple testing problems. BMC Bioinforma. 2008;9:114. doi: 10.1186/1471-2105-9-114. (Epub 2008/02/27. PubMed PMID: 18298808; PubMed Central PMCID: PMC2375137) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hollmén M., Roudnicky F., Karaman S., Detmar M. Characterization of macrophage — cancer cell crosstalk in estrogen receptor positive and triple-negative breast cancer. Sci. Rep. 2015;5:9188. doi: 10.1038/srep09188. (PubMed PMID: 25776849) [DOI] [PMC free article] [PubMed] [Google Scholar]