Abstract
We cultured enterococci from urinary tract infections in Iranian hospitals. Seven different Enterococcus species (E. raffinosus, E. durans, E. hirae, E. avium, E. mundtii, E. faecium and E. faecalis) were found. Seven strains were vancomycin resistant, leading to an overall vancomycin resistance rate of 3.9%. The enterococcal infection rate was high and vancomycin-resistant enterococci incidence low. We report the first vanA-positive E. mundtii urinary tract infections.
Keywords: Enterococcal infection, E. mundtii, urinary tract infection, vanA-positive, Vancomycin-resistant enterococci
Enterococci are members of the gastrointestinal flora and emerged in the 1970s as leading causes of multidrug-resistant hospital acquired infections [1]. Enterococcal colonization may result in asymptomatic bacteriuria but also in overt urinary tract infection (UTI) [2]. Vancomycin-resistant enterococci (VRE), the most important class of drug-resistant enterococci, mainly belong to the species Enterococcus faecium and contain vanA and/or vanB genes [3]. There are four additional vancomycin resistance genes: vanC, vanD, vanE and vanG [4], [5]. Detection of VRE is possible with culture or PCR [6]. We investigated UTI-related VRE and their vanA and vanB content from samples of East Iranian origin.
We obtained 177 isolates from 358 urine specimens of patients with UTI from two hospitals (Amir Al-Momenin and Imam Khomeini) in Zabol, Iran (2013–2014). Diagnostics included Gram staining, catalase production, pyrrolidonyl arylamidase detection and growth on bile-esculin/6.5% NaCl media. Vancomycin susceptibility testing included screening on brain–heart infusion (BHI) agar (Difco, USA) containing 6 μg/mL vancomycin (Sigma, Germany). Teicoplanin susceptibility testing was performed using 30 μg teicoplanin disks on Mueller-Hinton agar (Difco; Mast, UK). Minimum inhibitory concentrations (MICs) were determined by agar dilution on BHI agar [7]. The MIC breakpoint for both vancomycin and teicoplanin resistance was ≥32 μg/mL. Susceptibility to vancomycin (MIC ≤4 μg/mL) and to teicoplanin (MIC ≤8 μg/mL) was according to the Clinical and Laboratory Standards Institute.
DNA was extracted according to Perez-Hernandez et al. [8] and used for vanA and vanB PCR. All enterococci were PCR assayed for vanA and vanB genes using previously described primers and protocols [9]. Resistant Enterococcus spp. strains E. faecalis E206 (vanA positive) and E. faecium E2781 (vanB positive) were used as positive controls; Enterococcus faecalis ATCC 29212 was used as negative control.
We identified seven different species of Enterococcus: E. raffinosus 3.3% (n = 6), E. durans 2.8% (n = 5), E. hirae 3.9% (n = 7), E. avium 4.5% (n = 8), E. mundtii 10% (n = 18), E. faecium 27% (n = 48), and E. faecalis 48% (n = 85) (n = Table 1). E. faecalis and E. faecium accounted for 75% of the isolates. Out of 94 isolates from Amir Al-Momenin hospital, 4 (4.2%) were resistant to vancomycin: E. raffinosus (n = 1), E. faecalis (n = 2) and E. mundtii (n = 1) (see Fig. 1). Imam Khomeini Hospital showed an even lower resistance rate of 3.6% (n = 3), including two E. raffinosus and one E. faecium strains. The overall vancomycin resistance rate in the two hospitals was 3.9% (Table 2).
Table 1.
Frequency of enterococcal species in two hospitals in Zabol, Iran
| Hospital | No. of Enterococcus spp. |
|||||||
|---|---|---|---|---|---|---|---|---|
| E. raffinosus | E. durans | E. hirae | E. avium | E. mundtii | E. faecium | E. faecalis | Total | |
| Amir Al-Momenin | 6 | 4 | 1 | 6 | 12 | 26 | 46 | 94 |
| Imam Khomeini | 0 | 1 | 6 | 2 | 6 | 24 | 39 | 83 |
| Total | 6 | 5 | 7 | 8 | 18 | 48 | 85 | 177 |
Fig. 1.
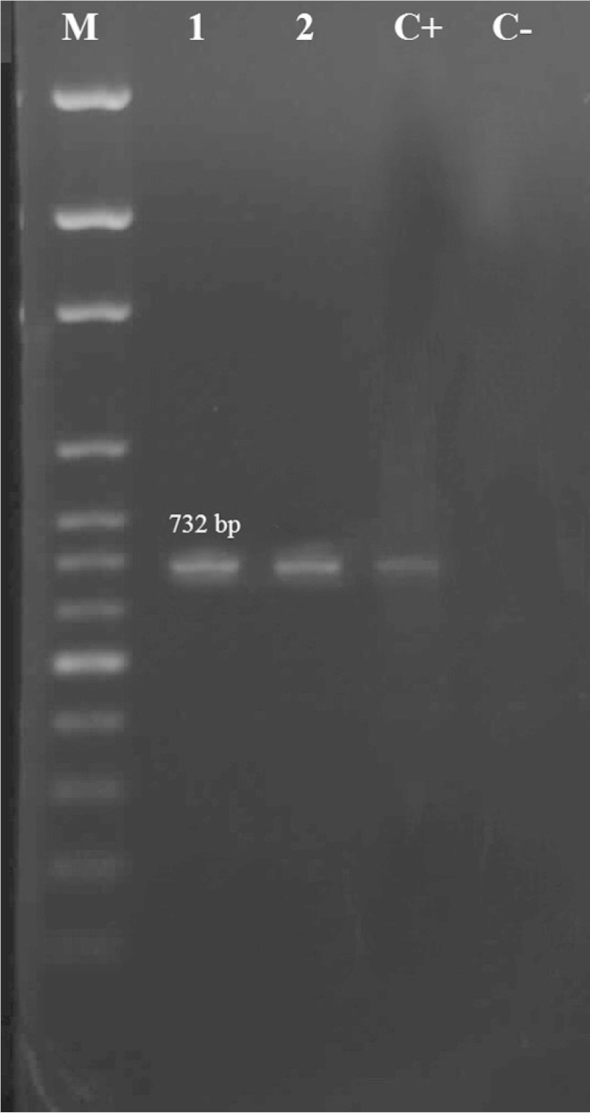
Patterns of agarose gel electrophoresis showing PCR amplification products for vancomycin resistant enterococci (VRE) lanes 1: E. faecalis; lane 2: E. mundtii. Lane M, DNA 100 basepair molecular size marker; C+: positive control; C−: negative control.
Table 2.
Distribution and VRE species diversity in two hospitals in Zabol, Iran
| Hospital | No. of isolates | VRE species, n (%) | No. of Enterococcus spp. |
|||
|---|---|---|---|---|---|---|
| E. raffinosus | E. faecium | E. faecalis | E. mundtii | |||
| Amir Al-Momenin | 94 | 4 (4.2) | 1 | 0 | 2 | 1 |
| Imam Khomeini | 83 | 3 (3.6) | 2 | 1 | 0 | 0 |
| Total | 177 | 7 (3.9) | 3 | 1 | 2 | 1 |
VRE, vancomycin-resistant enterococci.
All strains resistant to vancomycin showed an MIC of >512 μg/mL, while MICs obtained for teicoplanin resistant strains varied. E. faecium showed the highest teicoplanin MIC (98 μg/mL). E. faecalis (69 μg/mL), E. mundtii (60 μg/mL) and E. raffinosus (18 μg/mL) showed lower MICs.
Hospital-based VRE endemicity differs between countries, within a country and between different cities or hospitals. Here we show that the enterococcal UTI rate in two Iranian medical centers was relatively high. The VRE incidence, however, was not excessive. We show further evidence for the existence of infectious vanA-positive E. mundtii in humans beyond the single case previously reported from India [10].
Conflict of Interest
None declared.
References
- 1.Van Tyne D., Gilmore M.S. Friend turned foe: evolution of enterococcal virulence and antibiotic resistance. Annu Rev Microbiol. 2014;68:337–356. doi: 10.1146/annurev-micro-091213-113003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tristan O.D., Crank C.W. Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infect Drug Resist. 2015;8:217. doi: 10.2147/IDR.S54125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fram D., Castrucci F.M., Taminato M., Godoy-Martinez P., Freitas M.C.S., Belasco A. Cross-transmission of vancomycin-resistant Enterococcus in patients undergoing dialysis and kidney transplant. Braz J Med Biol Res. 2010;43:115–119. doi: 10.1590/s0100-879x2009007500023. [DOI] [PubMed] [Google Scholar]
- 4.Naas T., Fortineau N., Snanoudj R., Spicq C., Durrbach A., Nordmann P. First nosocomial outbreak of vancomycin-resistant Enterococcus faecium expressing a vanD-like phenotype associated with a vanA genotype. J Clin Microbiol. 2005;43:3642–3649. doi: 10.1128/JCM.43.8.3642-3649.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zirakzadeh A., Patel R. Vancomycin-resistant enterococci: colonization, infection, detection, and treatment. Mayo Clin Proc. 2006;81:529–536. doi: 10.4065/81.4.529. [DOI] [PubMed] [Google Scholar]
- 6.Seo J.Y., Kim P.W., Lee J.H., Song J.H., Peck K.R., Chung D.R. Evaluation of PCR-based screening for vancomycin-resistant enterococci compared with a chromogenic agar–based culture method. J Med Microbiol. 2011;60:945–949. doi: 10.1099/jmm.0.029777-0. [DOI] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute (CLSI) CLSI; Wayne, PA: 2010. Performance standards for antimicrobial susceptibility testing: 20th informational supplement. CLSI document M100–S20. [Google Scholar]
- 8.Perez-Hernandez X., Mendez-Alvarez S., Claverie-Martin F. A PCR assay for rapid detection of vancomycin-resistant enterococci. Diagn Microbiol Infect Dis. 2002;42:273–277. doi: 10.1016/s0732-8893(01)00360-1. [DOI] [PubMed] [Google Scholar]
- 9.Dutka-Malen S., Evers S., Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Praharaj I., Sujatha S., Parija S.C. Phenotypic and genotypic characterization of vancomycin resistant Enterococcus isolates from clinical specimens. Indian J Med Res. 2013;138:549–556. [PMC free article] [PubMed] [Google Scholar]


