Abstract
Background
The revolutionary concept of “jumping genes” was conceived by McClintock in the late 1940s while studying the Activator/Dissociation (Ac/Ds) system in maize. Transposable elements (TEs) represent the most abundant component of many eukaryotic genomes. Mobile elements are a driving force of eukaryotic genome evolution. McClintock’s Ac, the autonomous element of the Ac/Ds system, together with hobo from Drosophila and Tam3 from snapdragon define an ancient and diverse DNA transposon superfamily named hAT. Other members of the hAT superfamily include the insect element Hermes and Tol2 from medaka. In recent years, genetic tools derived from the ‘cut’ and ‘paste’ Tol2 DNA transposon have been widely used for genomic manipulation in zebrafish, mammals and in cells in vitro.
Results
We report the purification of a functional recombinant Tol2 protein from E.coli. We demonstrate here that following microinjection using a zebrafish embryo test system, purified Tol2 transposase protein readily catalyzes gene transfer in both somatic and germline tissues in vivo. We show that purified Tol2 transposase can promote both in vitro cutting and pasting in a defined system lacking other cellular factors. Notably, our analysis of Tol2 transposition in vitro reveals that the target site preference observed for Tol2 in complex host genomes is maintained using a simpler target plasmid test system, indicating that the primary sequence might encode intrinsic cues for transposon integration.
Conclusions
This active Tol2 protein is an important new tool for diverse applications including gene discovery and molecular medicine, as well as for the biochemical analysis of transposition and regulation of hAT transposon/genome interactions. The measurable but comparatively modest insertion site selection bias noted for Tol2 is largely determined by the primary sequence encoded in the target sequence as assessed through studying Tol2 protein-mediated transposition in a cell-free system.
Electronic supplementary material
The online version of this article (doi:10.1186/s13100-016-0062-z) contains supplementary material, which is available to authorized users.
Keywords: Tol2 transposase, hAT superfamily, Recombinant transposase protein, Zebrafish, Transposition site preference
Background
Our understanding of transposable elements (TEs) begins with McClintock’s revolutionary work with the Activator/Dissociation (Ac/Ds) system in maize [1]. Ac, the autonomous element of the Ac/Ds system, together with hobo from Drosophila and Tam3 from snapdragon define an ancient and diverse DNA transposon superfamily named hAT [2–4]. Widespread in plants and animals, hAT transposons are the most abundant DNA transposons in humans [5]. However, none of the human hAT elements have been active during the past 50 million years [5].
The first active DNA transposon discovered in vertebrates was the medaka fish (Oryzias latipes)-derived hAT element Tol2 [6]. Tol2 shares a number of features with other hAT members including transposases with a DDE (aspartate-aspartate-glutamate) catalytic motif, short terminal inverted repeats (TIRs) and formation of 8-bp host duplications upon transposition [2, 7]. Because derivatives from Tol2 have high cargo-capacity and low susceptibility to over-production inhibition, they have become popular gene transfer agents in a variety of animal systems, including zebrafish, African frog, chicken, mouse and human cell cultures including primary T cells [8], and for various genome biology applications (for reviews, see [9, 10].
TEs generally display very diverse patterns of target site selectivity. Studying the mechanisms for such selection is useful to gain insights on the biology of transposition, shedding light on the genome structure and designing better transposon tools for specific applications. Previous research has suggested that the mechanisms of target site selection are very complex and varied from mobile element to mobile element. In many cases, it involves the direct interaction between the transposase/recombinase and the target DNA or their indirect communication through accessory proteins [11]. However, specific factors that contribute to hAT element integration preference are largely unknown.
Our knowledge of transposition is largely based on bacterial TEs. Less is known about eukaryotic transposase proteins since they have been historically more difficult to express and reconstitute in vitro. In the present work, we establish a recombinant Tol2 protein-based system to serve as a tool for genome engineering and to probe the transposition mechanism of this vertebrate hAT transposon, focusing on the integration steps. We directly compare known Tol2 isoforms for activity in both human cells and zebrafish in vivo. We demonstrate that the highest activity variant can be epitope-tagged and retain full activity, and we purify epitope-tagged Tol2 protein (His-Tol2) from E. coli. The functionality of this recombinant Tol2 transposase is demonstrated in vivo in zebrafish using both somatic and germline transposition assays. Thus His-Tol2 protein is a viable new source of transposase for molecular medicine and genome engineering applications.
We further show that purified His-Tol2 can carry out both the excision and integration steps of transposition in vitro in the absence of any cellular co-factors. Tol2 displays a modest preference for AT-rich DNA in vivo [12]. Such insertion bias is also noted in a cell-free and defined assay when the insertion distribution of miniTol2 into a target plasmid was measured. miniTol2 contains the transposon end sequences necessary and sufficient in vivo for excision and integration [13, 14]. miniTol2 insertion is accompanied by 8-bp target site duplications as occurs in vivo and displays an insertion site preference in vitro, with a higher likelihood of insertion into AT-rich sequences similar to that noted for in vivo integrations [12]. These results suggest the target selection mechanism is at least in part maintained in this much simpler system.
Results
The 649 amino acid Tol2 isoform is the most active transposase in vivo and in vitro
Different coding sequences and activities have been described in previous works for the hAT transposase Tol2. Two distinct endogenous Tol2 mRNAs were identified in the original medaka fish isolate [15]. The shorter mRNA (Tol2-S, GenBank accession number AB031080) is encoded by exons 2–4 and results in a predicted protein of 576 amino acids [15] (Fig. 1a). The longer Tol2 mRNA (Tol2-L, GenBank accession number AB031079) is encoded by exons 1–4, and a protein of 685 amino acids is predicted using the first in-frame start codon [15] (Fig. 1a). Tol2-S inhibits excision by the Tol2-L protein, this inhibition was probably not through competition of DNA-binding but some other unknown mechanism, as Tol2-S protein lacks the majority of the BED zinc finger motif [16] (Fig. 1a). This also suggests that the sequences encoded by exon 1 are critical to Tol2 transposase function. When a copy of medaka Tol2 genomic sequence was introduced into zebrafish cells that do not harbor any endogenous Tol2 elements, a third Tol2 isoform of different length was identified (Tol2-M) [17] (Fig. 1a). This mRNA encodes a predicted protein of 649 amino acids that is shorter at the N-terminus than Tol2-L (Fig. 1a). This latter, heterologous mRNA (Tol2-M) is used in nearly all currently available Tol2-based genetic tools [13, 18–20]. The activity of the naturally occurring Tol2-L has not been explored in detail or compared to Tol2-M under similar conditions.
Fig. 1.
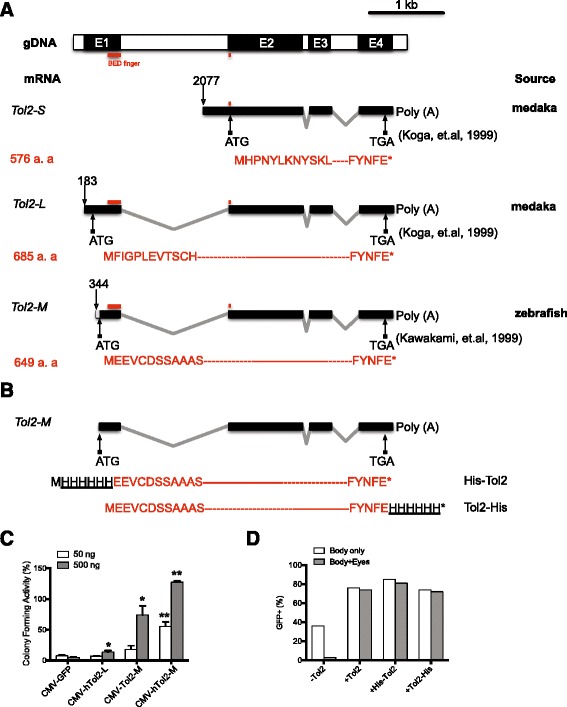
The highest activity Tol2 isoform and its modification by 6X His tag. a Tol2 genomic DNA (GenBank: D84375.2) structure and expressed mRNA from various sources (Tol2-S GenBank: AB031080; Tol2-L GenBank: AB031079; Tol2-M GenBank: AB032244). The first nucleotide location is numbered in relation to genomic DNA sequence in each mRNA and indicated by an open-ended arrow. The start and stop codons of the longest open reading frame in each mRNA are also indicated. Tol2-L spans all 4 exons (black bar, E1-4) while Tol2-S covers only the last three (E2-4). Tol2-M starts further into exon 1 than L. The predicted Tol2-M protein is slightly shorter at the amino-terminus than Tol2-L. Corresponding sequences encoding the BED zinc finger DNA binding motif are indicated on genomic DNA and mRNA. b In this paper we used a modified Tol2-M mRNA [13] encoding the same protein as Tol2-M, but all the nucleotides 5′ to the first start codon in Tol2-M (Fig. 1a, Tol2-M gray box) were deliberately removed to eliminate any possible effects from an out-of-frame ATG within those sequences. 6X His tags were added to either 5′- or 3′ of the modified mRNA, and the deduced N-terminus or C-terminus modified Tol2 protein sequences were outlined. c Human codon-optimized Tol2-M was most effective for gene transfer in HeLa cells. Tol2-M was tested for activity in human cell culture by colony forming assay. A Tol2 transposon vector carrying a Zeocin resistance gene cassette was co-transfected with different version of Tol2 or GFP control driven by CMV promoter at 50 ng or 500 ng. The numbers of Zeocin-resistant colonies formed in each treatment were compared to the treatment with the highest colony numbers (500 ng CMV-hTo2-M) as percentages. Bars represent mean values ± SEM of three independent experiments. * or ** indicate data is significantly different from respective CMV-GFP controls (* t-test P < 0.05; ** t-test P < 0.01). d Tol2 transposase could be modified by 6XHis epitope without loss of activity. A GFP-reporter [13] was co-injected with synthesized mRNA corresponding to untagged Tol2 or Tol2 tagged with 6X-His at either N-terminus (His-Tol2) or C-terminus (Tol2-His). Somatic GFP expression was scored at 3 dpf and the percentages of injected fish demonstrating GFP only in the body (body only) or demonstrating strong GFP in eyes and anywhere in the body (body + eyes) were recorded
To determine whether the isoforms of Tol2 might encode different functional outcomes, we compared Tol2 isoforms in human HeLa cell transposition assays upon co-transfection of transposase helper and transposon donor plasmids [13]. For Tol2-M, the gene-transfer activity is positively related to the amount of Tol2 transposase provided as plasmids (Fig. 1c). In addition, through codon optimization [21], we increased gene transfer in HeLa cells by ‘humanizing’ the Tol2-M codon usage (hTol2-M). The same strategy was applied to Tol2-L. However, even this ‘humanized’ Tol2-L (hTol2-L) showed reduced activity compared to Tol2-M in human cells (Fig. 1c). Tol2-M also showed higher activity than Tol2-L in zebrafish transposition assays [13] (data not shown). Thus the 649 amino acid Tol2-M isoform shows the highest activity of the three described protein forms (Fig. 1a), and will be referred to hereafter as Tol2 unless otherwise stated.
Tol2 can be modified on either the N- or C- terminus by 6XHis and retain full activity
The ability to identify and readily purify a protein is critical for the development of a system for biochemical analysis. Targeted modification of vertebrate transposase enzymes has been a challenge in the field. The addition of new sequences normally results in either reduced or limited enzymatic activity, as was the case for Sleeping Beauty, the most studied vertebrate transposable element [22, 23] or with Tol2 in prior studies [24, 25]. Here we chose a small 6X-His protein tag for use with Tol2 (Fig. 1b).
To test if such modification will compromise transposase activity, zebrafish embryos were injected with synthetic Tol2 transposase mRNA encoding Tol2, 5′-His (6X) tagged Tol2, or 3′-His (6X) tagged Tol2 as well as with a transposon reporter vector [13]. Embryos were scored for GFP-positive cells or eyes with green fluorescence at 3 days post-fertilization (dpf) as described [13]. Visualization of net gene transfer into the eyes of injected zebrafish embryos is a convenient and robust assay system for determining Tol2-mediated transposition in somatic tissues [13]. Importantly, the 6X-His tagged versions of Tol2 showed the same activity as the untagged Tol2 proteins in this in vivo transposition assay (Fig. 1d).
Purified His-Tol2 from E.coli
We examined the expression of both N- and C-terminally tagged Tol2 variants in E. coli. The C-terminally tagged version gave predominantly the full-length form with multiple, smaller protein products. In contrast, the amino-terminally tagged Tol2 (Fig. 2a) yielded a single protein product at the predicted size (~74 kD) when purified from E. coli (Fig. 2b, c). This His-Tol2 protein was used in all subsequent studies unless noted otherwise.
Fig. 2.

Expression and detection of recombinant Tol2 transposase protein. a Tol2 transpoase sequence was cloned into an N-terminal 6XHis expression vector pET-21a (Novagen). The expression cassette driven by T7 promoter is shown, and a fusion protein of ~74 kD was expected. rbs: ribosome binding site. b Expression of His-Tol2 in E.coli BL21-AI strain. unind: cell lysate from uninduced cells harboring Tol2-expression vector; t: total induced crude cell lysate; i: insoluble protein fraction from induced cell lysate; s: soluble protein fraction from induced cell lysate. Equal volume of cell culture was loaded in each lane. p: ~ 350 ng purified His-Tol2 protein (arrow head). c Immunoblot analysis of His-Tol2 expression with anti-His antibody. The same protein samples as in (b) were loaded except that only ~ 6 ng purified His-Tol2 was used for detection by western blot
Recombinant Tol2 protein is a fully functional hAT transposase in vivo
Transposon tools are often used as a two-component system in gene discovery and gene delivery applications (Fig. 3a). One component is donor DNA containing a genetic cargo of interest flanked by transposon terminal sequences, and the other component is a transposase source. In previously described vertebrate applications, transposase was provided by either a DNA-encoding plasmid or in vitro transcribed transposase-encoding mRNA. To determine if purified recombinant His-Tol2 protein is fully functional and robust enough to be an alternative transposase source for practical genetic applications, we tested His-Tol2 function in vivo using this two-component system in both somatic and germline gene transfer assays.
Fig. 3.
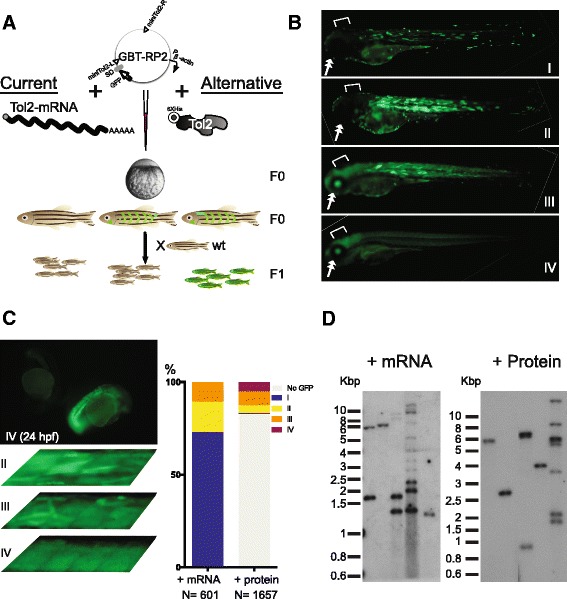
His-Tol2 is a fully functional transposase in vivo. a Diagram showing microinjection of zebrafish embryo at one-cell stage to generate transgenic animals. GBT-RP2 plasmid containing GFP-reporter gene trap was co-injected with either Tol2 mRNA or His-Tol2 protein. Mosaic somatic GFP signals were seen in F0 injected fish. F0 embryos were raised to adult and outcrossed to obtain F1 generation. Ubiquitous GFP would be detected in F1 fish if the integration was due to a gene trap event and was passed through the germline. Molecular analysis of transposon insertion numbers and genomic locations was carried out on tissue from F1 generation animals. Pβ-actin: β-actin promoter; SD: splice donor. b Representative images of F0 embryos demonstrating a catalog of GFP-positive somatic patterns that could be observed at 3 dpf from microinjections of different methods. Notice the signal difference in eyes (double arrow) and brain regions (open end arrow) for each category. With vector-only injections, notice only Cat. I and II patterns were observed. c A small number of His-Tol2 protein injected embryos showing ubiquitous whole body green fluorescence as early as 24 hpf. Those fish generally displayed uniform GFP expression throughout the body, with few uneven myotome GFP stripes at 3 dpf (Cat. IV). Also shown are 3 dpf injected fish somites at higher magnification, demonstrating the difference in uniformity of GFP signals in tissues from various injection categories. Injection GFP pattern distribution from typical RNA or protein mediated injection was shown. d GFP-positive F1 fish from either founder family generated by mRNA injection or His-Tol2 injection were subjected to fin-clip and Southern blot analysis with a GFP probe. Each lane represents one individual F1 adult fish. Southern blots from 5 adult fish selected from random founder family are shown for each method. See also Additional file 1
For gene transfer testing in somatic cells, we used a high stringency transposition test system in which insertions of a specialized miniTol2 element, GBT-RP2 [26], in transcriptionally competent genomic loci result in enhanced Green Fluorescent Protein (GFP) expression in the resulting animals. The GBT-RP2 element was based on the 3′ gene trap derived from previously described gene-breaking transposon (GBT) vectors [27, 28]. Transposon DNA was co-injected with either Tol2 mRNA or His-Tol2 protein into one-cell zebrafish embryos (Fig. 3a). The F0 somatic GFP patterns of injected embryos generally can be characterized into different categories based on signal uniformity at 3 days post-fertilization (dpf): a category I pattern is characterized by embryos showing minimal GFP signals, usually only a couple of bright stripes in the myotomes; a category II pattern is characterized by large areas of the fish body with GFP signals, but with little or no GFP-positive signals in the eyes or brain region; and a category III pattern is characterized by whole body green fluorescence, including the brain and eyes (Fig. 3b).
Background, non-transposase mediated gene transfer was measured by the injection of the RP2 vector without transposase, resulting in sporadic GFP signals seen in myotomal tissues of some F0 fish by 3 dpf. Usually, the majority of the background GFP-positive fish displayed category I patterns, and very few displayed category II patterns (Fig. 3b).
When transposase was provided by Tol2 mRNA, the overall somatic GFP signals were notably increased, especially with a higher number of embryos exhibiting the category III pattern, which was a very rare observation without the co-addition of transposase (Fig. 3b, c).
With His-Tol2 protein, the majority of the injected fish did not display any GFP signal. Among the GFP-positive F0 embryos, GFP patterns representing all three categories were noted. However, the number of category I animals, which were less likely to carry transposon-mediated germ-line integration, was much lower than the GFP-positive category II or category III (Fig. 3b, c). We also conducted multi-generational testing for germline gene transfer rate testing. F0 fish displaying category III somatic patterning from either mRNA or His-Tol2 transposase resulted in a similar gene trapping rate and low mosaicism (Table 1).
Table 1.
His-Tol2 and Tol2 mRNA as sources of transposase in vivo
| Somatic F0 pattern | Tol2 form | Germline trapping and expression frequency | Average F0 germline mosaicism |
|---|---|---|---|
| Cat II. | mRNA | 65 % (13/20) | 27 % |
| Cat III. | mRNA | 88 % (35/40) | 39 % |
| Cat III. | Protein | 76 % (13/17) | 39 % |
| Cat IV. | Protein | 90 % (9/10) | 51 % |
Analyses of somatic and germline transmission of reporter genes are shown. Fish injected with either mRNA or His-Tol2 were characterized by different F0 somatic patterns and assessed for corresponding germline trapping frequency, estimated as the percentage of F0 fish producing GFP-positive offspring. The number of fish screened is shown in parentheses. The F0 germline mosaicism rate was determined by the percentage of GFP-positive F1 offspring from a founder outcross. The average mosaicism rate for founder fish is listed from each different category
Interestingly, an additional pattern found only in protein-injected embryos was observed. A small portion of these injected animals (~5–10 %) exhibited ubiquitous GFP expression as early as 24 h after injection (Fig. 3c), and the signals were very uniform throughout the body at 3 dpf (Fig. 3c). Such F0 embryos were designated category IV (Table 1). Zebrafish embryos undergo rapid cell divisions every 20 min during the early development and reach ~ 1000 cells by 3 h post fertilization [29]. Thus the timing of transposition potentially plays a critical role in determining the mosaicism of resulting animals. We hypothesized that the uniform GFP signals of the category IV fish could be the result of very early integrations of the GFP reporter gene into the genome. Indeed, a higher percentage of germline transmission was obtained from such F0 fish (Table 1). More importantly, the average F0 germline mosaicism was much reduced for category IV fish (Table 1). Our observations of His-Tol2 mediated injection suggest that even though transposition events were rarer under current condition than when compared to mRNA as a source of transposase, protein injection can “jump start” integration inside the early zebrafish embryo.
An important additional measurement of the germline gene transfer rate is copy number. Transposon copy number was estimated by Southern blot analysis on individual fish (Fig. 3d). An average of ~ 4 unique transposon insertions per haploid F0 founder germline were observed (Additional file 1). The fact that category IV (Additional file 1, family 1,2,3,5,6 and 8) did not contain more transposon insertions than in other groups suggests that the uniform fluorescence is most likely due to earlier incorporation of the reporter gene into the embryo genome.
Transposase-mediated insertion can be distinguished from other insertion mechanisms by the target site duplications flanking the transposon sequences. We sequenced the insertion sites of the His-Tol2 protein-mediated RP2 transposon insertions, and the hAT signature 8-bp genomic duplications were present in all checked cases (Table 2). We conclude that His-Tol2 protein is a fully functional hAT transposase in both somatic and germline lineages in vivo.
Table 2.
Transposase-mediated germline integration
| Site | Sequence | Chr. | Refseq |
|---|---|---|---|
| 1 | aaatatttaccaagcaac-5′-Tol2-3′-caagcaacacgttcagtg | 8 | Intron of gfra2 |
| 2 | ataatttcctcttatttg-5′-Tol2-3′-cttatttgcatgtcagat | 13 | Intergenic |
| 3 | cgcatgctaacttataga-5′-Tol2-3′-cttatagaggaggtgccc | 8 | Exon of wu: fb79a07 |
| 4 | aaacgttcctcctaacac-5′-Tol2-3′-cctaacacagttagatgg | 3 | Intergenic |
| 5 | caacacatgactcgttgg-5′-Tol2-3′-ctcgttggccatatgcta | 15 | Intergenic |
| 6 | gggaatatgtgttattaa-5′-Tol2-3′-gttattaactgcgtccca | 4 | Repetitive sequences |
| 7 | agctgtctcttctgtgtc-5′-Tol2-3′-tctgtgtcattcagtctc | 3 | Intron of LOC557901 |
| 8 | tgtcagagatctaggtca-5′-Tol2-3′-ctaggtcagatggaggaa | 25 | Intergenic |
Tol2 insertion sites generated by His-Tol2 protein co-injection were cloned from F1 transgenic fish. Examples of 8-bp genomic sequence duplication (in bold) were shown with insertion loci information
Recombinant Tol2 protein can mediate concerted target joining in vitro
We continued to explore whether this recombinant Tol2 protein is functional in vitro, focusing on the target joining step by directly assaying the ability of His-Tol2 to join both ends of a DNA segment flanked by Tol2 end sequences to a target plasmid (Fig. 4a). Tol2 terminal sequences consisting of 261 bp from the left arm and 192 bp from the right arm (miniTol2) are sufficient transposon end sequences for efficient Tol2 transposition in vivo [13, 14]. We used a PCR-generated KanR gene flanked by these Tol2 ends (Additional file 2), miniTol2-KanR together with His-Tol2 protein to assess target joining in vitro. After incubation of blunt end miniTol2-KanR with an ampicillin resistant target plasmid (pGL) in the presence of His-Tol2 protein and Mn2+ ions, DNA was precipitated, transformed into E. coli, and plated on LB-Amp+ plates to determine the total number of target plasmids and on LB-Kan+ plates to identify integration products (Fig. 4a, b). The number of Tol2 integrants increased as the number of transposons in the reaction increased (Fig. 4c, Additional file 3), and we detected up to 0.8 % of integration ratio among the conditions tested. We sequenced large numbers of integrants obtained in multiple experiments and found that the 8-bp host duplications at the insertion site characteristic of hAT transposition occurred in 86 % of insertions (Total N = 333) (Fig. 4d). Thus His-Tol2 integration in vitro recapitulates Tol2 integration in vivo. The distribution of recovered insertions in the target plasmid was non-random with miniTol2 element insertion biased towards the AT-rich (AT content 64 %) SV40 polyA segment of the target vector (Fig. 5a; see also below). We also noticed that Tol2 protein was able to excise a miniTol2-flanking fragment out of a plasmid donor (data not shown), indicating His-Tol2 is fully competent gene transfer vector in vitro as well as in vivo.
Fig. 4.
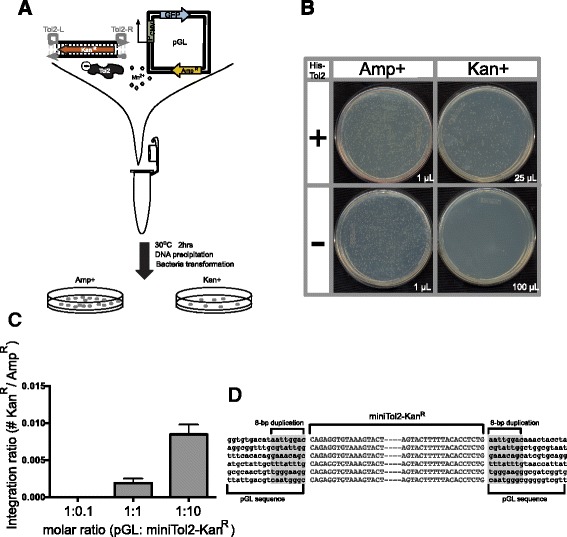
Tol2 in vitro integration assay. a Scheme of in vitro integration assay. b One example miniTol2-Kan R integration, revealing different kanamycin-resistant colony recovery from assays with versus without His-Tol2 protein. The amount of transformed bacteria spread on each plate is indicated. c Tol2-mediated miniTol2-Kan R integration. The amount of target plasmid (pGL) was kept at 0.5 pMol, while PCR fragments were mixed at three different ratios. The ratio of colony numbers (Additional file 3) recovered from LB-Kan+ to LB-Amp+ was used as an indicator of integration activity. Bars represent mean values ± SEM from three independent experiments. d Plasmids isolated from colonies on LB-Kan+ plate were sequenced at both junctions of miniTol2-Kan R insertion. 8-bp target plasmid duplications were indicated. Six examples of the integration junction are shown here
Fig. 5.
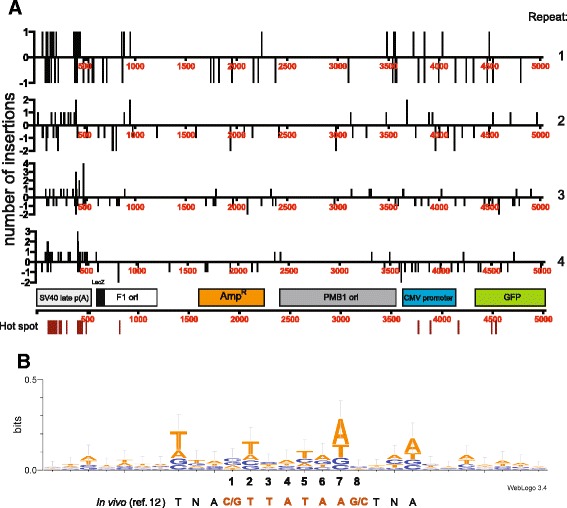
miniTol2 insertion sites and consensus sequence. a Tol2-mediated insertions from four independent experiments were mapped to target plasmid (pGL) for miniTol2-Kan R (n = 266). Y-axis indicates the number of insertion events and orientation (positive: sense orientation; negative: anti-sense orientation) Major features of the pGL vector were also indicated. Red lines below the sequences indicate insertion “hot spots”, defined as the same locations that are discovered in more than one experiment, regardless of the insertion orientation or locations in one experiment that have transposons inserted in both directions (also see Additional file 5). b Tol2 integration site motif analyzed by WebLog (version 3.0) (n = 266) and aligned to a previous indentified weak AT-rich consensus in vivo [12]
Target site preference of Tol2 is conserved using a simplified target plasmid system
As described above, the miniTol2-KanR-insertions generated in vitro were not randomly distributed on the target plasmid, and they clustered at the SV40 poly (A) rich region. Analysis of these patterns by Monte Carlo simulation demonstrated that the bias was highly significant (p-value by simulation < 0.0001, see Methods) in each of four independent experiments (Fig. 5a; Additional file 4 and Additional file 5). This observed preference in vitro is in agreement with prior work showing that Tol2 has a preference for AT-rich regions in vivo. More importantly, the in vivo observed integration signature, TNA(C/G)TTATAA(G/C)TNA (bold: insertion site) [12], was similar to the findings reported here using the plasmid insertion system (Fig. 5b). This observation suggests a primary sequence of the target site is likely to play a critical role in Tol2/hAT integration site selection.
Discussion
In the past decade, the Tol2-based transposition system has become a versatile tool for gene delivery in commonly used model organisms, especially in zebrafish, which has seen many cases of successful genetic screening with various highly efficient Tol2 systems [30, 31]. Recently, Tol2 transposon tools have been applied to pioneer gene transfer in novel biological systems, such as haplochromine cichlid [32, 33] and African killifish [33]; or as an alternative to viral vectors showing promise in engineered T-cell therapy [34]. Here we report the purification of recombinant Tol2 hAT transposase from E. coli and demonstrate that it is fully functional in vivo and in vitro. A functional recombinant Tol2 protein will be a useful addition to the Tol2-based genome engineering kit. Researchers will have more choices over what form of transposase will work best for the organisms/cells of their interest. Transposase provided as readily made protein, in some systems, may be advantageous over an mRNA-based delivery method. For example, when the translation of the Tol2 mRNA is not efficient or optimized for the intended heterologous system. And even in zebrafish where mRNA is an effective method for Tol2 delivery, we noted advantages of using the functional Tol2 transposase protein.
We observed robust somatic and germline transposition in zebrafish using E.coli -purified Tol2 transposase. Our experiments suggest using His-Tol2 may be advantageous in generating low mosaic transgenic animals with higher germline transmission rate. Transposition using a protein source of transposase may “jumpstart” transposition as it avoids possible delays from transcription, translation and assembly of a functional transpososome containing transposase multimers when transposase is supplied in a ready form. Transposase multimers are the active form of transposase in other studied systems [7, 35, 36]. Static light scattering analysis has shown that purified Tol2 is a dimer (A. Voth and F. Dyda, personal communication). This recombinant Tol2 protein is an excellent tool for future experimental methods for gene delivery. As documented using this initial system, the percentage of GFP-positive F0 embryos was lower using protein as a transposase source when compared to the well-established mRNA co-injection methods,. However, the current protein co-injection method has yet to be fully optimized, such as experimentally establishing the optimal protein to donor plasmid ratio.
Functional Tol2 transposase protein is also of use for studying transposon biology science, enabling experimental testing of this hAT element both in vivo and in vitro. Such work will shed more light on our understanding of how this and related transposons work in the future. In this paper, we demonstrated that Tol2 protein could be used in an assay to probe the integration site preference for the transposon and identified a weak consensus that are largely conserved between in vivo chromosomes and a cell-free target plasmid. Two other commonly used transposases for genome engineering, Sleeping Beauty and piggyBac use obligate TA and TTAA core consensus targeting sequence respectively [37, 38]. In contrast, Tol2 target site selection is still considered promiscuous on the level of primary DNA sequence [39]. The preference noticed towards AT-rich DNA context could also reflect a preference towards integration into regions of higher DNA flexiblity as suggested previously [40–42].
Conclusions
We presented here a fully functional recombinant transposase protein, both in vitro and in vivo, for the popular vertebrate gene transfer transposon Tol2. Our initial work in zebrafish indicated that His-Tol2 has a high potential for genome engineering applications. His-Tol2 can also serve as a tool for probing transposon biology in vitro and help confirm an integration site consensus shared between in vivo and cell-free systems.
Methods
Ethics statement
Zebrafish larvae were raised within the Mayo Clinic’s Zebrafish Facility with adherence to the NIH Guide for the Care and Use of Laboratory Animals and approval by Mayo Clinic’s Institutional Animal Care and Use Committee.
Important constructs
Details of how important constructs were made are included in Additional file 6: Supplemental Experimental Procedures.
HeLa cell transfection assays
Transfection of HeLa cells with a Zeocin resistant vector (pTol2miniZeo) with either control vector pCMV-GFP or vectors providing Tol2 transposase (pCMV-Tol2-M, pCMV-hTol2-M or pCMV-hTol2-L) was following the same protocol as described in [21]. Zeocin-resistant colony forming assay was conducted as in [21] as well.
Zebrafish somatic transposition assays
To compare transposition of a GFP-reporter mediated by tagged vs. untagged Tol2 mRNA (Fig. 1d), zebrafish embryos were injected at one-cell stage with a mixture of 25 pg pTol2-S2EF1α-GM2 (pDB591, [13]) plasmid DNA with 25 pg or 100 pg T3TS-Tol2-M (Tol2), T3TS-His-Tol2, or T3TS-His-Tol2 RNA (Fig. 1d) respectively. Surviving normal-looking embryos were scored for GFP fluorescence at 3 dpf.
Protein expression and immunoblotting
Small scale detection of protein expression was conducted according to Qiagen recommended protocol. Detail procedures are included in Additional file 6: Supplemental Experimental Procedures. His-Tol2 protein expression was detected by penta-His antibody (Qiagen) following the recommended western protocol. An adapted protocol [43] was used to generate large-scale nuclease-free protein fractions.
Zebrafish one-cell GBT microinjection
One-cell stage zebrafish microinjection was performed as described [44]. The injected amount of reagents were 25 pg GBT-RP2 plasmid co-injected with 25 pg T3TS-Tol2 mRNA [13]; or co-injected with ~ 0.2 ng His-Tol2 protein.
Southern blot
Genomic DNA isolation and Southern blot hybridization was conducted as previously described [28]. Tail fins from individual GFP-positive F1 fish generated by co-injection of Tol2 mRNA or His-Tol2 protein at F0 were clipped and genomic DNA was isolated followed by digestion with BamHI and BglII for Southern blotting.
Molecular analysis of genomic integration sites
Cloning of insertion site in F1 individual fish was conducted through adapted inverse and linker-mediated PCR (LM-PCR) [44]. Information of primer sequences and PCR conditions are included in Additional file 6: Supplemental Experimental Procedures.
In vitro integration assay
Blunt end miniTol2-KanR fragments were generated by PCR with platinum Pfx DNA polymerase (Invitrogen) with forward primer Tol2-mini5′ (5′- CAGAGGTGTAAAGTACTTGA-3′), reverse primer Tol2-mini3′ (5′- CAGAGGTGTAAAAAGTACTC-3′) and a template (pTol2miniKan(-)amp). PCR product was treated with DpnI to get rid of the template before purified by QIAquick PCR purification kit (Qiagen). Freshly prepared plasmids and PCR products were used in the assay. In a 20 μL reaction, 0.5 pMol plasmid pGL, corresponding PCR fragments and 27 pMol His-Tol2 (~2 μg) were mixed in MOPS buffer (25 mM MOPS, pH 7.0; 1 mM MnCl2; 50 mM NaCl; 5 % glycerol; 2 mM DTT; 100 ng/ul BSA). The reaction was incubated for 2 h at 30 °C. DNA was phenol-chloroform extracted using Maxtract High Density columns (Qiagen) and resuspended in 5 μL TE buffer. 1 μL or 5 μL DNA was used to transform Top10 competent cells (Invitrogen). The number of colonies grew on Amp-resistant plates was used to calculate the total number recovered plasmids, and the number of colonies that grew on Kan-resistant plates was used to calculate the number of plasmid with KanR integration. MOPS buffer with MgCl2 or no added cations were tested as well, and the integration activity was modestly higher with MnCl2.
Insertion site distribution statistics
The simulation distributions of the number of insertions over the target features was done in R. We assume that every feature gets a number of distributions proportional to its size in bp. We simulated 75 counts with the probability of a count falling into a feature being the length of that feature divided by the length of all features types. We then ran this simulation 10,000 times. To check where the experimental data lie with respect to the simulated data, we determined how often the simulated counts are bigger or smaller than the experimental data.
Acknowledgments
We thank Gary Moulder and Dr. Karl Clark for help with zebrafish injections and Southern blotting. We thank Drs. Liqin Zhou, Xianghong Li and Nancy L. Craig for active scientific discussions on this project. We thank Ekker lab members, especially Tammy Greenwood and Dr. Henning Schneider for critical evaluation of the manuscript. The work was supported by the Mayo Foundation and the U.S. National Institutes of Health grants DA14546, HG006431 and GM63904 to Stephen C. Ekker.
Abbreviations
- Ac/Ds
Activator/Dissociation
- TEs
transposable elements
- TIRs
terminal inverted repeats
- GBT
gene-breaking transposon
- dpf
days post-fertilization
- KanR
kanamycin resistance
Additional files
Estimation of transposon insertion numbers by Southern analysis. Individual GFP-positive F1 fish from random selected F0 founder families were fin-clipped for gDNA isolation. Southern blotting with a GFP probe was conducted for each fish gDNA and the number of clear-labeled Southern bands was recorded as estimation of transposon insertions. Total number of unique Southern bands was used to estimate total transposon insertions for each F0 founder family. An average insertion number over all the families analyzed was calculated for both injection methods. (DOCX 91 kb)
miniTol2 sequences of the in vitro assays. DNA sequences of the left arm and right arm of miniTol2 flanking the KanR used in the in vitro integration assays. (DOCX 55 kb)
Colony counting numbers for in vitro integration assays. Colony numbers counted for the three independent experiments in Fig. 4c of different target plasmid and PCR insert ratios. (DOCX 49 kb)
P-value by simulation. For each independent repeat of miniTol2 mediated KanR insertion distribution, p-value was generated by simulation (see Methods) for different features on the target plasmid. (DOCX 61 kb)
Insertion “hot spots” for miniTol2. “Hot spots” were arranged according to the insertion loci along the target plasmid. 8-bp target duplication and flanking sequnces were shown. Forward (F) or reverse (R) insertion copies were counted. Repeats coverage indicate the “hot spot” indentified by how many independent experiments. (XLSX 45 kb)
Supplemental Experimental Procedures. (DOCX 131 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JN, KJW, DN, DB, KJS and MU carried out the experiments, and JN, KJW, DN, DB and SCE analyzed the data. KJW, DN and DB tested Tol2 isoforms and modifications, and KJW and DN conducted the Tol2 protein preparation. KJS and MU participated in the in vitro insertion site sequencing work and tests to determine transposition efficiency in vivo. JN and SCE drafted the manuscript. All authors read and approved the final manuscript.
Contributor Information
Jun Ni, Email: junni@stanford.edu.
Kirk J. Wangensteen, Email: Kirk.Wangensteen@uphs.upenn.edu
David Nelsen, Email: nels0433@umn.edu.
Darius Balciunas, Email: darius@temple.edu.
Kimberly J. Skuster, Email: Skuster.kimberly@mayo.edu
Mark D. Urban, Email: Urban.Mark@mayo.edu
Stephen C. Ekker, Phone: 507-284-5530, Email: ekker.stephen@mayo.edu
References
- 1.McClintock B. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci U S A. 1950;36(6):344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calvi BR, Hong TJ, Findley SD, Gelbart WM. Evidence for a common evolutionary origin of inverted repeat transposons in Drosophila and plants: hobo, Activator, and Tam3. Cell. 1991;66(3):465–471. doi: 10.1016/0092-8674(81)90010-6. [DOI] [PubMed] [Google Scholar]
- 3.Rubin E, Lithwick G, Levy AA. Structure and evolution of the hAT transposon superfamily. Genetics. 2001;158(3):949–957. doi: 10.1093/genetics/158.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warren WD, Atkinson PW, O’Brochta DA. The Hermes transposable element from the house fly, Musca domestica, is a short inverted repeat-type element of the hobo, Ac, and Tam3 (hAT) element family. Genet Res. 1994;64(2):87–97. doi: 10.1017/S0016672300032699. [DOI] [PubMed] [Google Scholar]
- 5.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 6.Koga A, Suzuki M, Inagaki H, Bessho Y, Hori H. Transposable element in fish. Nature. 1996;383(6595):30. doi: 10.1038/383030a0. [DOI] [PubMed] [Google Scholar]
- 7.Hickman AB, et al. Molecular architecture of a eukaryotic DNA transposase. Nat Struct Mol Biol. 2005;12(8):715–721. doi: 10.1038/nsmb970. [DOI] [PubMed] [Google Scholar]
- 8.Huang X, et al. Gene transfer efficiency and genome-wide integration profiling of Sleeping Beauty, Tol2, and piggyBac transposons in human primary T cells. Mol Ther. 2010;18(10):1803–1813. doi: 10.1038/mt.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawakami K. Transposon tools and methods in zebrafish. Dev Dyn. 2005;234(2):244–254. doi: 10.1002/dvdy.20516. [DOI] [PubMed] [Google Scholar]
- 10.Kawakami K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 2007;8 Suppl 1:S7. doi: 10.1186/gb-2007-8-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craig NL. Target site selection in transposition. Annu Rev Biochem. 1997;66:437–474. doi: 10.1146/annurev.biochem.66.1.437. [DOI] [PubMed] [Google Scholar]
- 12.Kondrychyn I, Garcia-Lecea M, Emelyanov A, Parinov S, Korzh V. Genome-wide analysis of Tol2 transposon reintegration in zebrafish. BMC Genomics. 2009;10:418. doi: 10.1186/1471-2164-10-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balciunas D, et al. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2006;2(11):e169. doi: 10.1371/journal.pgen.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174(2):639–649. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koga A, Suzuki M, Maruyama Y, Tsutsumi M, Hori H. Amino acid sequence of a putative transposase protein of the medaka fish transposable element Tol2 deduced from mRNA nucleotide sequences. FEBS Lett. 1999;461(3):295–298. doi: 10.1016/S0014-5793(99)01479-9. [DOI] [PubMed] [Google Scholar]
- 16.Tsutsumi M, Koga A, Hori H. Long and short mRnas transcribed from the medaka fish transposon Tol2 respectively exert positive and negative effects on excision. Genet Res. 2003;82(1):33–40. doi: 10.1017/S0016672303006335. [DOI] [PubMed] [Google Scholar]
- 17.Kawakami K, Shima A. Identification of the Tol2 transposase of the medaka fish Oryzias latipes that catalyzes excision of a nonautonomous Tol2 element in zebrafish Danio rerio. Gene. 1999;240(1):239–244. doi: 10.1016/S0378-1119(99)00444-8. [DOI] [PubMed] [Google Scholar]
- 18.Parinov S, Kondrichin I, Korzh V, Emelyanov A. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev Dyn. 2004;231(2):449–459. doi: 10.1002/dvdy.20157. [DOI] [PubMed] [Google Scholar]
- 19.Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci U S A. 2000;97(21):11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawakami K, et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7(1):133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Keng VW, et al. Efficient transposition of Tol2 in the mouse germline. Genetics. 2009;183(4):1565–1573. doi: 10.1534/genetics.109.100768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yant SR, Huang Y, Akache B, Kay MA. Site-directed transposon integration in human cells. Nucleic Acids Res. 2007;35(7):e50. doi: 10.1093/nar/gkm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu SC, et al. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc Natl Acad Sci U S A. 2006;103(41):15008–15013. doi: 10.1073/pnas.0606979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shibano T, et al. Recombinant Tol2 transposase with activity in Xenopus embryos. FEBS Lett. 2007;581(22):4333–4336. doi: 10.1016/j.febslet.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Meir YJ, et al. Genome-wide target profiling of piggyBac and Tol2 in HEK 293: pros and cons for gene discovery and gene therapy. BMC Biotechnol. 2011;11:28. doi: 10.1186/1472-6750-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark KJ, et al. In vivo protein trapping produces a functional expression codex of the vertebrate proteome. Nat Methods. 2011. In press. [DOI] [PMC free article] [PubMed]
- 27.Sivasubbu S, et al. Gene-breaking transposon mutagenesis reveals an essential role for histone H2afza in zebrafish larval development. Mech Dev. 2006;123(7):513–529. doi: 10.1016/j.mod.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Petzold AM, et al. Nicotine response genetics in the zebrafish. Proc Natl Acad Sci U S A. 2009;106(44):18662–18667. doi: 10.1073/pnas.0908247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 30.Asakawa K, Kawakami K. The Tol2-mediated Gal4-UAS method for gene and enhancer trapping in zebrafish. Methods. 2009;49(3):275–281. doi: 10.1016/j.ymeth.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark KJ, et al. In vivo protein trapping produces a functional expression codex of the vertebrate proteome. Nat Methods. 2011;8(6):506–515. doi: 10.1038/nmeth.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juntti SA, Hu CK, Fernald RD. Tol2-mediated generation of a transgenic haplochromine cichlid, Astatotilapia burtoni. PLoS One. 2013;8(10):e77647. doi: 10.1371/journal.pone.0077647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valenzano DR, Sharp S, Brunet A. Transposon-Mediated Transgenesis in the Short-Lived African Killifish Nothobranchius furzeri, a Vertebrate Model for Aging. G3 (Bethesda) 2011;1(7):531–538. doi: 10.1534/g3.111.001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsukahara T, et al. The Tol2 transposon system mediates the genetic engineering of T-cells with CD19-specific chimeric antigen receptors for B-cell malignancies. Gene Ther. 2015;22(2):209–215. doi: 10.1038/gt.2014.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizuuchi M, Baker TA, Mizuuchi K. Assembly of the active form of the transposase-Mu DNA complex: a critical control point in Mu transposition. Cell. 1992;70(2):303–311. doi: 10.1016/0092-8674(92)90104-K. [DOI] [PubMed] [Google Scholar]
- 36.Sakai J, Chalmers RM, Kleckner N. Identification and characterization of a pre-cleavage synaptic complex that is an early intermediate in Tn10 transposition. EMBO J. 1995;14(17):4374–4383. doi: 10.1002/j.1460-2075.1995.tb00112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson MH, Coates CJ, George AL., Jr PiggyBac transposon-mediated gene transfer in human cells. Mol Ther. 2007;15(1):139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 38.Vigdal TJ, Kaufman CD, Izsvak Z, Voytas DF, Ivics Z. Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J Mol Biol. 2002;323(3):441–452. doi: 10.1016/S0022-2836(02)00991-9. [DOI] [PubMed] [Google Scholar]
- 39.Grabundzija I, et al. Comparative analysis of transposable element vector systems in human cells. Mol Ther. 2010;18(6):1200–1209. doi: 10.1038/mt.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vrljicak P, et al. Genome-Wide Analysis of Transposon and Retroviral Insertions Reveals Preferential Integrations in Regions of DNA Flexibility. G3.115.026849; Early Online: 2016. [DOI] [PMC free article] [PubMed]
- 41.Yant SR, et al. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25(6):2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu G, et al. Target-site preferences of Sleeping Beauty transposons. J Mol Biol. 2005;346(1):161–173. doi: 10.1016/j.jmb.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 43.Zhou L, et al. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature. 2004;432(7020):995–1001. doi: 10.1038/nature03157. [DOI] [PubMed] [Google Scholar]
- 44.Davidson AE, et al. Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon. Dev Biol. 2003;263(2):191–202. doi: 10.1016/j.ydbio.2003.07.013. [DOI] [PubMed] [Google Scholar]


