Abstract
Morphogenetic proteins are responsible for patterning the embryonic nervous system by enabling cell proliferation that will populate all the neural structures and by specifying neural progenitors that imprint different identities in differentiating neurons. The adoption of specific neurotransmitter phenotypes is crucial for the progression of neuronal differentiation, enabling neurons to connect with each other and with target tissues. Preliminary neurotransmitter specification originates from morphogen-driven neural progenitor specification through the combinatorial expression of transcription factors according to morphogen concentration gradients, which progressively restrict the identity that born neurons adopt. However, neurotransmitter phenotype is not immutable, instead trophic factors released from target tissues and environmental stimuli change expression of neurotransmitter-synthesizing enzymes and specific vesicular transporters modifying neuronal neurotransmitter identity. Here we review studies identifying the mechanisms of catecholaminergic, GABAergic, glutamatergic, cholinergic and serotonergic early specification and of the plasticity of these neurotransmitter phenotypes during development and in the adult nervous system. The emergence of spontaneous electrical activity in developing neurons recruits morphogenetic proteins in the process of neurotransmitter phenotype plasticity, which ultimately equips the nervous system and the whole organism with adaptability for optimal performance in a changing environment.
Introduction
The genesis of a neuron starts with the neural progenitor exiting the cell cycle followed by the first phases of neuronal differentiation and the specialization of the newborn neuron. For a long time it was believed that the neurotransmitter phenotype was predetermined with the specification of the neural progenitor and that this fate was sealed and unique, meaning that the neuron born from the specified progenitor will permanently express a specific and single neurotransmitter phenotype. However, many studies spanning through the last decades have challenged these dogmas, demonstrating that neurotransmitter phenotypes may be multiple for a single neuron and that the identity of these phenotypes may change developmentally and upon changes of the intrinsic and extrinsic environments through adulthood (Spitzer, 2012, 2015).
The specialization of neural progenitors consists in the combinatorial expression of a specific set of transcription factors which will control expression of target genes related to the identity of the developing neuron, including genes associated with neurotransmitter phenotype. The expression of a specific neurotransmitter identity in the differentiating neuron depends on the transcriptional regulation of the biosynthetic and release machinery necessary for implementing the specific transmission in the chemical synapse. However, progenitor cells and developing neurons are sensitive to a myriad of signaling mechanisms that are spatiotemporally dynamic and may add to the genetic program triggered in progenitors, intercept it or even switch it.
Here we review studies in diverse species ranging from zebrafish and Xenopus to mice and rats that identify the mechanisms of neurotransmitter specification through neural progenitor specialization and neuronal differentiation with particular emphasis on the findings that demonstrate that acquisition of neurotransmitter identity is plastic and subjected to dynamic changes. We focused on classical neurotransmitters: acetylcholine, biogenic amines and the amino acid transmitters. The review is centered on the role of morphogenetic proteins and trophic factors in the specification of neurotransmitter identity and their interaction with electrical activity when mediating the changes in neurotransmitter phenotype.
Catecholaminergic phenotype
Preliminary specification
Expression of the noradrenergic and dopaminergic phenotypes starts with the recruitment of specialized progenitors. The sympathetic noradrenergic neurons originate from neural crest-derived progenitors that become fate-restricted mostly by bone morphogenetic protein (BMP) signal (Howard, 2005). Transcription factors necessary for the expression of dopaminergic and noradrenergic phenotypes include Mash1, Phox2a, Phox2b, Hand2 and Gata2/Gata3 (Stanke et al., 1999; Goridis and Rohrer, 2002). Regulatory regions in genes encoding the biosynthetic enzymes for catecholamines, tyrosine hydroxylase (TH) and dopamine β-hydroxylase, contain binding sites for these transcription factors. Alternatively, some of them are upstream of those transcription factors that bind to the neurotransmitter identity target genes like neurotransmitter biosynthetic enzymes and vesicular transporters, becoming necessary for the expression of the catecholaminergic phenotype. For instance, BMP2 supports the persistent expression of Mash1 in neural progenitors from the fetal rat gut (Lo et al., 1997), and Mash1, in turn, promotes the expression of proneuronal genes, but Mash1 expression terminates after neuronal differentiation when other transcription factors take over to promote expression of noradrenergic and dopaminergic phenotypes (Lo et al., 1999; Goridis and Rohrer, 2002; Lo et al., 2002).
In the central nervous system, another morphogenetic protein, Wnt, specifically regulates the number of progenitors specified for the dopaminergic phenotype of diencephalic neurons, early during neural ectoderm patterning in zebrafish (Russek-Blum et al., 2008). Wnt activity restricts the number of dopaminergic neurons in the developing diencephalon by negatively regulating expression of the transcription factor Fezl (Russek-Blum et al., 2008), which in turn regulates the development of monoaminergic neurons (Levkowitz et al., 2003). In zebrafish forebrain and anterior hindbrain catecholaminergic neuron specification depends strongly on Nodal signaling and to a lesser extent on Sonic hedgehog (Shh) and fibroblast growth factor (FGF) 8 (Guo et al., 1999; Holzschuh et al., 2003). Specification of midbrain dopaminergic neurons is strongly regulated by Shh signaling through the transcriptional regulation of target genes at different stages of these neurons’ development (Abeliovich and Hammond, 2007). The recruited transcription factors include the canonical Shh pathway effectors Gli, which are required for neural progenitor proliferation (Zervas et al., 2004) and the expression of Phox2a (Blaess et al., 2006) as demonstrated in the developing mouse midbrain. Lmx1a is necessary for chick and mouse midbrain dopaminergic neuron specification and its expression is also dependent on Shh signaling (Andersson et al., 2006). On the other developmental end, expression of Nurr1 also mediated by Shh in postmitotic midbrain precursors induces TH expression allowing for the maturation of the midbrain dopaminergic phenotype (Wallen and Perlmann, 2003). Many other transcription factors contribute to the specification of the midbrain dopaminergic phenotype and it is through the complex interaction among these factors that the mature phenotype is established (Abeliovich and Hammond, 2007; Panman et al., 2011).
Plasticity
The catecholaminergic phenotype has become a classic paradigm for the switch in neurotransmitter identity. Noradrenergic sympathetic axons innervating the rat sweat glands experience a switch to the cholinergic phenotype when they reach their target (Landis and Keefe, 1983; Schotzinger and Landis, 1990; Francis and Landis, 1999). This target-dependent switch in neurotransmitter phenotype is dependent on the expression of gp130 receptor in mouse sympathetic neurons and cytokine release from the sweat glands (Stanke et al., 2006). The transcriptional mechanism of this switch involves the upregulation of the expression of Satb2, chromatin architecture protein, when noradrenergic nerves contact the rat sweat glands, which in turn binds to responsive elements in the choline acetyltransferase (ChAT) locus becoming necessary and sufficient for the switch from noradrenergic to cholinergic phenotype (Apostolova et al., 2010). Moreover, Satb2 expression is regulated by the mitogen-activated protein kinase p38α/β activity, which is necessary for the upregulation of the cholinergic phenotype in noradrenergic sympathetic rat neurons grown in vitro and in mice in vivo (Loy et al., 2011). The participation of p38 both in neurotransmitter phenotype switching (Loy et al., 2011) and activity-dependent synaptic plasticity (Thomas and Huganir, 2004) poses it as a key signaling factor in the integration of short-term transcription-independent and long-term transcription-dependent plastic responses to the changing environment, challenging the presumed rigidity of the early neurotransmitter phenotype specification.
Indeed, the role of electrical activity in the specification of the catecholaminergic phenotype manifests in the central nervous system. A change in environmental stimuli changes the number of neurons expressing the dopaminergic phenotype of the ventral suprachiasmatic nucleus in Xenopus larva (Dulcis and Spitzer, 2008) and in the adult rat brain (Dulcis et al., 2013), suggesting a universal mechanism. In the amphibian the switch to the dopaminergic trait results in the expression of a dual neurotransmitter phenotype, NPY- and TH-expressing neurons (Dulcis and Spitzer, 2008). In the rat the increase in number of dopaminergic neurons happens at the expense of the decrease in the somatostatin phenotype (Dulcis et al., 2013).
The role of morphogens in contributing to the neurotransmitter phenotype specification continues beyond morphogenesis and early patterning. The transcription factors Engrailed 1 and 2 necessary for the midbrain dopaminergic phenotype are first expressed during mouse brain patterning by the action of FGF8, and then they induce expression of this morphogen to maintain the dopaminergic phenotype in already differentiated neurons (Alberi et al., 2004; Simon et al., 2004). Also, BMP4 induces the dopaminergic phenotype in cultured GABAergic neurons derived from the mouse cortical striatum during a sensitive period in vitro (Stull et al., 2001). Considering that BMPs (Swapna and Borodinsky, 2012), Shh (Belgacem and Borodinsky, 2011), Wnts (Varela-Nallar et al., 2010) and FGF modulate Ca2+ dynamics and kinase activity in developing neurons together with the electrical activity-dependent plasticity of neurotransmitter phenotype, the potential role of morphogenetic proteins in postmitotic neurons participating in neurotransmitters respecification through Ca2+-mediated signaling becomes apparent.
GABAergic/Glutamatergic phenotypes
Preliminary specification
GABAergic interneurons represent a diverse population in the central nervous system. Differentiation of GABAergic phenotypes is thought to be a default fate of differentiating neuronal precursors (Furmanski et al., 2009), which depends on the expression of particular transcription factors. For instance, the Dlx transcription factors promote differentiation of olfactory GABAergic interneurons in mice by regulating the expression of Wnt5a (Paina et al., 2011). Interestingly, the Wnt signaling switches from canonical, β-catenin-mediated, to non-canonical, Ca2+/PKC-mediated, signaling in the transition from proliferating neural precursors to differentiating neurons by the action of different Wnt ligands (Paina et al., 2011). In the mouse cerebellum the transcription factor Ptf1a is necessary and sufficient for driving GABAergic neuron differentiation of the cerebellar ventricular zone precursors (Hoshino et al., 2005), presumably through Shh action (Huang et al., 2010; Fleming et al., 2013). In the forebrain, the Nkx2.1-expressing medial ganglionic eminence progenitors, specified by the Shh signaling, generate GABAergic or cholinergic neurons but the expression of Lhx6, which feeds forward to promote neuronal production of Shh (Flandin et al., 2011), shifts them towards the GABAergic phenotype (Zhao et al., 2003; Manabe et al., 2005; Flandin et al., 2011). In the chick embryonic diencephalon, the thalamus develops into a rostral population of GABAergic neurons and a caudal region of glutamatergic thalamic neurons. These are patterned by different Shh levels through the regulatory control of Pax6 and Irx3 expression; Pax6 is necessary for the expression of the glutamatergic phenotype and inhibits the GABAergic phenotype (Robertshaw et al., 2013). In the dorsal spinal cord, Tlx1 and Tlx3, transcription factors upregulated by Wnt signaling (Kondo et al., 2011), act as selectors of the glutamatergic over the GABAergic phenotype in chick and mouse embryos (Cheng et al., 2004), while Lbx1 inverts the selection inducing the specification of the GABAergic phenotype in these spinal neurons (Cheng et al., 2005). BMPs are also involved in the differentiation of the GABAergic phenotype in mice. BMP2 enhances the expression of the GABA transporter gat1 by recruiting transcription factors Smad4 and YY1, which directly bind to the transporter regulatory region (Yao et al., 2010).
Plasticity
Many of the genetic programs summarized in the previous section are sensitive to electrical activity, thus enabling the process of glutamatergic and GABAergic phenotype specification to be activity-dependent. Ca2+ spikes in developing spinal neurons of Xenopus tropicalis phosphorylate the transcripton factor cJun that represses transcription of the glutamatergic/GABAergic transcription factor selector Tlx3, promoting the specification of the GABAergic phenotype over the glutamatergic one (Marek et al., 2010). In the rat hippocampus, the expression of the GABAergic phenotype is induced by depolarizing stimuli in glutamatergic granule cells (Gomez-Lira et al., 2005). Interestingly, during development the GABAergic and glutamatergic phenotypes overlap in the frog spinal cord (Root et al., 2008) and in the rat dentate gyrus (Gutierrez et al., 2003) and they become restricted to segregated neuronal populations as development progresses and the nervous system matures (Gutierrez, 2003; Root et al., 2008). Similarly, in the auditory system the GABAergic/glycinergic neurons of the medial nucleus of the trapezoid body that synapse in the rat lateral superior olive also express the glutamatergic phenotype, most prominently during the developmental peak of synapse elimination (Gillespie et al., 2005) that is important for acquiring precision of tonotopy in the inhibitory auditory pathway (Noh et al., 2010).
Neurotrophins BDNF and NGF promote the acquisition of the GABAergic and cholinergic phenotypes of mouse basal forebrain neurons in vitro and in vivo. The neurotrophin-induced GABAergic phenotype is non-cell autonomous, mediated by factors released from p75 receptor-expressing cholinergic neurons (Lin et al., 2007). The promotion of the GABAergic phenotype by neurotrophins is not dependent on proliferation since the total number of neurons remains the same. Instead it seems to recruit cells that are non-cholinergic and non-GABAergic to change their transmitter specification (Lin et al., 2007).
These changes in neurotransmitter specification do not appear to be dissociated from the earlier developmental processes and cues that regulate specification of progenitors. Morphogens also participate in the respecification of neurotransmitter identity by recruiting Ca2+-mediated activity in their signaling. For instance, Shh switches from the canonical to non-canonical signaling during Xenopus laevis spinal cord development, transitioning from the proliferative Gli-dependent pathway to the Ca2+ spike activity-mediated neuronal differentiation (Belgacem and Borodinsky, 2011, 2015). This change in pathways results in an increase in number of GABAergic neurons in the developing spinal cord mediated by the Shh-Ca2+ spikes signaling axis (Belgacem and Borodinsky, 2011). Remarkably, the intercalation of Ca2+ activity in Shh signaling represses Gli activity, thus inverting Shh action on its own canonical pathway (Figure 1) (Belgacem and Borodinsky, 2015).
Figure 1. Interplay between Shh and Ca2+ spike activity regulates neurotransmitter specification in developing spinal neurons.
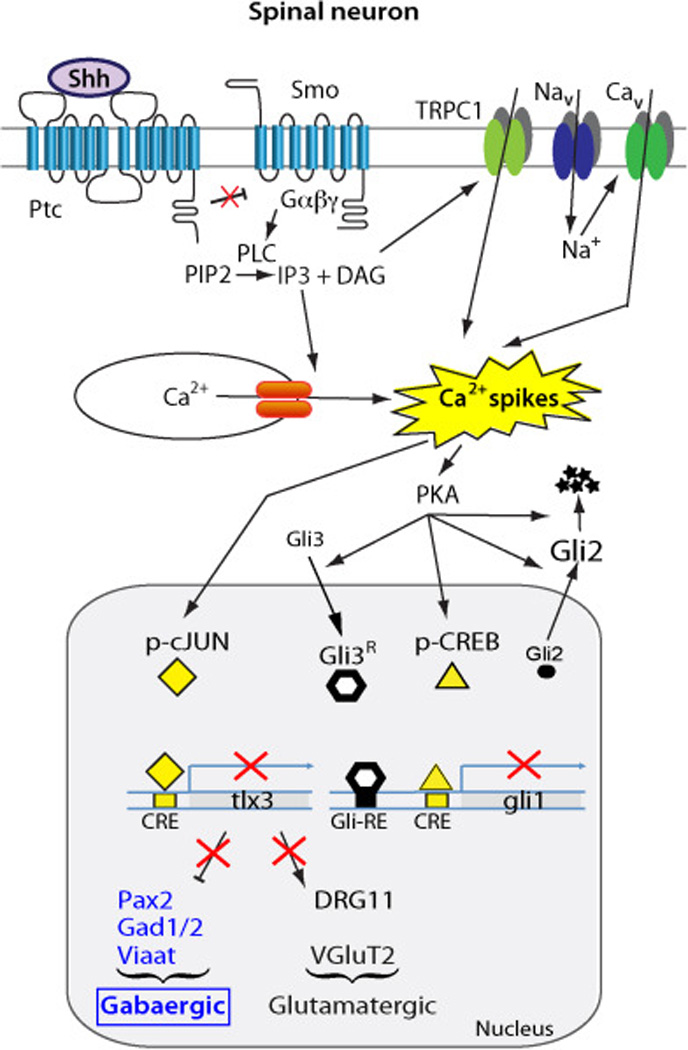
Shh binds to Patched (Ptc) releasing the constitutive inhibition on the coreceptor Smoothened (Smo) which activates phopholipase C (PLC) that increases inositol triphosphate (IP3) and diacylglycerol (DAG) levels thus enhancing Ca2+ spike activity through the activation of transient receptor potential channel 1 (TRPC1), voltage-gated Na+ and Ca2+ channels (Nav, Cav) and Ca2+ release from stores. Enhanced Ca2+ spike activity intercalates in Shh canonical pathway, inverting Shh action and leading to Gli2 cytosolic localization, Gli2 and Gli3 processing into repressor forms, repression of Gli1 transcription and an overall downregulation of Gli activity. In contrast, Ca2+ spikes activate transcription factors cAMP-responsive element binding protein (CREB) and cJun, which promote the expression of the GABAergic over glutamatergic phenotype by regulating expression of the transcription factor selector tlx3. Based on Cheng et al., 2004, Marek et al., 2010, Belgacem and Borodinsky, 2011, 2015.
Cholinergic phenotype
Preliminary specification
The molecular mechanisms of the specification of the cholinergic phenotype are still unclear. Nevertheless, some key players have been identified from studies focused on the specification of vertebrate motor neurons, which are cholinergic. MNR2 is a homeobox gene upregulated by Shh signaling, expressed by chick motor neuron progenitors and transiently by postmitotic neurons that is necessary and sufficient for the expression of ChAT, acetylcholine synthetic enzyme (Tanabe et al., 1998). Also, the cholinergic/motor neuron phenotype can be elicited in mouse embryonic stem cells in vitro by recapitulating the morphogenetic environment of motor neuron progenitors in vivo, particularly retinoic acid and Shh (Wichterle et al., 2002). In the mouse basal forebrain, the specification of cholinergic neurons is dependent on the expression of the transcription factor Lhx8 (Mori et al., 2004).
Other studies have shown that BMPs, and particularly BMP9, enhance the expression of the cholinergic phenotype in mouse septum and spinal cord neurons, both in vitro and in vivo (Lopez-Coviella et al., 2000). The mechanisms of BMP-induced cholinergic specification in mice and rat basal forebrain may involve the upregulation of c-Fos expression (Lopez-Coviella et al., 2005), which in turn enhances expression of the cholinergic phenotype genes, ChAT, VAChT and AChE (Kaufer et al., 1998). Trophic factors like NGF also regulate the expression of the cholinergic phenotype in the rat pheochromocytoma cell line (PC12) and in primary neuronal cultures from the mouse embryonic septum, potentially by recruiting the Akt/PI3K pathway and by regulating GSK3 activity (Madziar et al., 2008), which in turn may control CREB activity that also regulates the expression of the cholinergic locus in mouse and rat neurons (Brock et al., 2007; Liu et al., 2008).
Plasticity
The cholinergic phenotype is probably the paradigm of neurotransmitter phenotype respecification. The switch from noradrenergic to cholinergic upon innervation of the footpad sweat glands reviewed above represents the first evidence of the plasticity of the cholinergic phenotype specification. This is not restricted to the peripheral nervous system, mouse and rat glutamatergic hypothalamic neurons induce expression of the cholinergic phenotype when glutamate signaling is inhibited through NMDAR-mediated and Ca2+/CREB/NF-κB-dependent mechanism (Belousov et al., 2001; Belousov et al., 2002; Liu et al., 2008). In contrast, excessive stimulation of brain excitability by stress or by inhibiting the acetylcholine esterase in vivo and in hippocampal slices respectively, downregulates expression of the cholinergic gene locus through a Ca2+ and c-Fos-mediated pathway (Kaufer et al., 1998).
Moreover, the cholinergic phenotype in the developing Xenopus laevis spinal cord is also sensitive to the levels of Ca2+-mediated electrical activity. Enhancement of Ca2+ spike frequency leads to lower number of cholinergic neurons while suppressing Ca2+ spike activity increases the number of cholinergic neurons (Borodinsky et al., 2004). These changes in neurotransmitter phenotype follow a homeostatic rule with excitatory neurotransmitter phenotype, glutamatergic and cholinergic, increasing when activity is suppressed and viceversa for the inhibitory neurotransmitter phenotypes, GABAergic and glycinergic (Borodinsky et al., 2004). The changes in the cholinergic phenotype of motor neurons upon changes in activity levels (Borodinsky et al., 2004) are accompanied by corresponding changes in the neurotransmitter receptor expression in the skeletal muscle resulting in non-cholinergic neuromuscular junctions in the Xenopus tadpole (Borodinsky and Spitzer, 2007).
Serotonergic
Preliminary Specification
The generation of serotonergic neurons starts with the specification of neural progenitors of the mouse ventral hindbrain that express the transcription factor Nkx2.2 induced by Shh (Briscoe et al., 1999). Gata-2 is induced by the Shh-Nkx2.2 pathway and is necessary for enhancing expression of Gata-3, Pte-1 and Lmx1b which all participate in the specification of the serotonergic phenotype in the mouse and chick hindbrain (van Doorninck et al., 1999; Cheng et al., 2003; Ding et al., 2003; Craven et al., 2004).
Plasticity
The number of serotonergic, tryptophan hydroxylase-expressing, neurons in vitro increases upon serotonin stimulation through the upregulation of BDNF expression and signal transduction through trk-C receptor in embryonic rat raphe-derived cultured cells (Eaton et al., 1995; Galter and Unsicker, 2000). In contrast, the trophic factors CNTF and LIF reduce the expression of the serotonergic phenotype at the expense of increasing expression of the cholinergic phenotype of embryonic raphe nuclei neuronal cultures (Rudge et al., 1996).
In the Xenopus laevis tadpole hindbrain the number of serotonergic neurons depends on the level of spontaneous Ca2+ spike activity. Enhanced activity downregulates expression of Lmx1b and thus decreases the number of cells expressing the serotonin synthesizing enzyme tryptophan hydroxylase (Demarque and Spitzer, 2010). These changes in serotonergic neuron number modify the swimming pattern of frog larva (Demarque and Spitzer, 2010).
Concluding remarks
Early specification of neural progenitors sets the organizing structural principles of the developing nervous system. However, many aspects of neuronal differentiation are not terminally fated by the specialization assumed by progenitor cells and the neurotransmitter phenotype is a paradigmatic example (Figure 2). The cues that implement nervous system embryonic patterning do not shut off or disappear but instead they participate from the plastic events driving the changes in neurotransmitter phenotype specification. This is achieved by interactions of morphogenetic proteins with the changing intrinsic and external environment as the nervous system develops and matures. The emergence during neural development of pathways mediated by trophic factors, electrical and calcium activity that in turn are sensitive to persisting morphogenetic protein signaling, orchestrates neuronal differentiation by modulating activity of critical transcription factors. Moreover, the intercalation of the emerging signaling pathways in the maturing neurons allows to switching gear on the morphogenetic protein action and repurposing them for implementing distinct responses to different stimuli.
Figure 2. Intrinsic and extrinsic stimuli change neurotransmitter specification in developing and mature neurons.
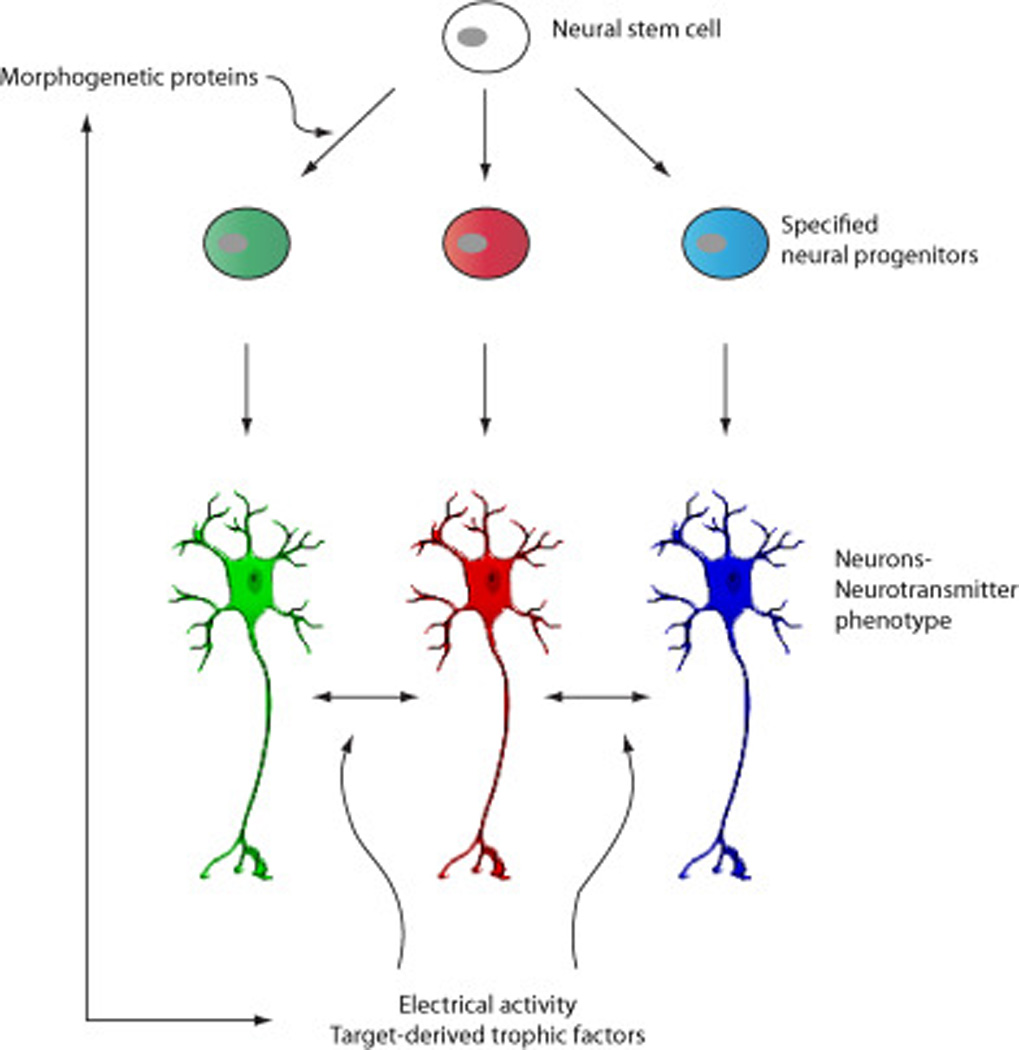
Preliminary specification of neural progenitors during the embryonic development is instrumental to morphogenesis of different nervous system structures and functional nuclei. However, the identity of developing neurons is not sealed. Instead features like neurotransmitter phenotype are sensitive to the changing internal and external environment; the emerging stimuli and developmental cues crosstalk with morphogenetic proteins to implement plasticity of neurotransmitter phenotype expression necessary for adapting to the ever-changing environment.
Highlights.
Neuronal activity recruits morphogens for neurotransmitter phenotype plasticity.
Trophic factors and calcium signaling modify neurotransmitter identity.
Emerging signaling cascades change morphogen action during neural development.
Acknowledgments
Research in the lab has been supported by the Basil O’Connor Starter Scholar Research Award Grant 5-FY09-131 from the March of Dimes Foundation, Klingenstein Foundation Award in Neuroscience, NSF 1120796, NIH-NINDS R01NS073055 and SHC 86500-NCA and 85220-NCA grants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abeliovich A, Hammond R. Midbrain dopamine neuron differentiation: factors and fates. Dev Biol. 2007;304:447–454. doi: 10.1016/j.ydbio.2007.01.032. [DOI] [PubMed] [Google Scholar]
- Alberi L, Sgado P, Simon HH. Engrailed genes are cell-autonomously required to prevent apoptosis in mesencephalic dopaminergic neurons. Development. 2004;131:3229–3236. doi: 10.1242/dev.01128. [DOI] [PubMed] [Google Scholar]
- Andersson E, Tryggvason U, Deng Q, Friling S, Alekseenko Z, Robert B, Perlmann T, Ericson J. Identification of intrinsic determinants of midbrain dopamine neurons. Cell. 2006;124:393–405. doi: 10.1016/j.cell.2005.10.037. [DOI] [PubMed] [Google Scholar]
- Apostolova G, Loy B, Dorn R, Dechant G. The sympathetic neurotransmitter switch depends on the nuclear matrix protein Satb2. J Neurosci. 2010;30:16356–16364. doi: 10.1523/JNEUROSCI.3502-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgacem YH, Borodinsky LN. Sonic hedgehog signaling is decoded by calcium spike activity in the developing spinal cord. Proc Natl Acad Sci U S A. 2011;108:4482–4487. doi: 10.1073/pnas.1018217108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgacem YH, Borodinsky LN. Inversion of Sonic hedgehog action on its canonical pathway by electrical activity. Proc Natl Acad Sci U S A. 2015;112:4140–4145. doi: 10.1073/pnas.1419690112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belousov AB, O'Hara BF, Denisova JV. Acetylcholine becomes the major excitatory neurotransmitter in the hypothalamus in vitro in the absence of glutamate excitation. J Neurosci. 2001;21:2015–2027. doi: 10.1523/JNEUROSCI.21-06-02015.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belousov AB, Hunt ND, Raju RP, Denisova JV. Calcium-dependent regulation of cholinergic cell phenotype in the hypothalamus in vitro. J Neurophysiol. 2002;88:1352–1362. doi: 10.1152/jn.2002.88.3.1352. [DOI] [PubMed] [Google Scholar]
- Blaess S, Corrales JD, Joyner AL. Sonic hedgehog regulates Gli activator and repressor functions with spatial and temporal precision in the mid/hindbrain region. Development. 2006;133:1799–1809. doi: 10.1242/dev.02339. [DOI] [PubMed] [Google Scholar]
- Borodinsky LN, Spitzer NC. Activity-dependent neurotransmitter-receptor matching at the neuromuscular junction. Proc Natl Acad Sci U S A. 2007;104:335–340. doi: 10.1073/pnas.0607450104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodinsky LN, Root CM, Cronin JA, Sann SB, Gu X, Spitzer NC. Activity-dependent homeostatic specification of transmitter expression in embryonic neurons. Nature. 2004;429:523–530. doi: 10.1038/nature02518. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Sussel L, Serup P, Hartigan-O'Connor D, Jessell TM, Rubenstein JL, Ericson J. Homeobox gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature. 1999;398:622–627. doi: 10.1038/19315. [DOI] [PubMed] [Google Scholar]
- Brock M, Nickel AC, Madziar B, Blusztajn JK, Berse B. Differential regulation of the high affinity choline transporter and the cholinergic locus by cAMP signaling pathways. Brain Res. 2007;1145:1–10. doi: 10.1016/j.brainres.2007.01.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Chen CL, Luo P, Tan M, Qiu M, Johnson R, Ma Q. Lmx1b, Pet-1, and Nkx2.2 coordinately specify serotonergic neurotransmitter phenotype. J Neurosci. 2003;23:9961–9967. doi: 10.1523/JNEUROSCI.23-31-09961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Samad OA, Xu Y, Mizuguchi R, Luo P, Shirasawa S, Goulding M, Ma Q. Lbx1 and Tlx3 are opposing switches in determining GABAergic versus glutamatergic transmitter phenotypes. Nat Neurosci. 2005;8:1510–1515. doi: 10.1038/nn1569. [DOI] [PubMed] [Google Scholar]
- Cheng L, Arata A, Mizuguchi R, Qian Y, Karunaratne A, Gray PA, Arata S, Shirasawa S, Bouchard M, Luo P, Chen CL, Busslinger M, Goulding M, Onimaru H, Ma Q. Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates. Nat Neurosci. 2004;7:510–517. doi: 10.1038/nn1221. [DOI] [PubMed] [Google Scholar]
- Craven SE, Lim KC, Ye W, Engel JD, de Sauvage F, Rosenthal A. Gata2 specifies serotonergic neurons downstream of sonic hedgehog. Development. 2004;131:1165–1173. doi: 10.1242/dev.01024. [DOI] [PubMed] [Google Scholar]
- Demarque M, Spitzer NC. Activity-dependent expression of Lmx1b regulates specification of serotonergic neurons modulating swimming behavior. Neuron. 2010;67:321–334. doi: 10.1016/j.neuron.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding YQ, Marklund U, Yuan W, Yin J, Wegman L, Ericson J, Deneris E, Johnson RL, Chen ZF. Lmx1b is essential for the development of serotonergic neurons. Nat Neurosci. 2003;6:933–938. doi: 10.1038/nn1104. [DOI] [PubMed] [Google Scholar]
- Dulcis D, Spitzer NC. Illumination controls differentiation of dopamine neurons regulating behaviour. Nature. 2008;456:195–201. doi: 10.1038/nature07569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulcis D, Jamshidi P, Leutgeb S, Spitzer NC. Neurotransmitter switching in the adult brain regulates behavior. Science. 2013;340:449–453. doi: 10.1126/science.1234152. [DOI] [PubMed] [Google Scholar]
- Eaton MJ, Staley JK, Globus MY, Whittemore SR. Developmental regulation of early serotonergic neuronal differentiation: the role of brain-derived neurotrophic factor and membrane depolarization. Dev Biol. 1995;170:169–182. doi: 10.1006/dbio.1995.1205. [DOI] [PubMed] [Google Scholar]
- Flandin P, Zhao Y, Vogt D, Jeong J, Long J, Potter G, Westphal H, Rubenstein JL. Lhx6 and Lhx8 coordinately induce neuronal expression of Shh that controls the generation of interneuron progenitors. Neuron. 2011;70:939–950. doi: 10.1016/j.neuron.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming JT, He W, Hao C, Ketova T, Pan FC, Wright CC, Litingtung Y, Chiang C. The Purkinje neuron acts as a central regulator of spatially and functionally distinct cerebellar precursors. Dev Cell. 2013;27:278–292. doi: 10.1016/j.devcel.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Landis SC. Cellular and molecular determinants of sympathetic neuron development. Annual review of neuroscience. 1999;22:541–566. doi: 10.1146/annurev.neuro.22.1.541. [DOI] [PubMed] [Google Scholar]
- Furmanski O, Gajavelli S, Lee JW, Collado ME, Jergova S, Sagen J. Combined extrinsic and intrinsic manipulations exert complementary neuronal enrichment in embryonic rat neural precursor cultures: an in vitro and in vivo analysis. J Comp Neurol. 2009;515:56–71. doi: 10.1002/cne.22027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galter D, Unsicker K. Sequential activation of the 5-HT1(A) serotonin receptor and TrkB induces the serotonergic neuronal phenotype. Mol Cell Neurosci. 2000;15:446–455. doi: 10.1006/mcne.2000.0841. [DOI] [PubMed] [Google Scholar]
- Gillespie DC, Kim G, Kandler K. Inhibitory synapses in the developing auditory system are glutamatergic. Nat Neurosci. 2005;8:332–338. doi: 10.1038/nn1397. [DOI] [PubMed] [Google Scholar]
- Gomez-Lira G, Lamas M, Romo-Parra H, Gutierrez R. Programmed and induced phenotype of the hippocampal granule cells. J Neurosci. 2005;25:6939–6946. doi: 10.1523/JNEUROSCI.1674-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goridis C, Rohrer H. Specification of catecholaminergic and serotonergic neurons. Nat Rev Neurosci. 2002;3:531–541. doi: 10.1038/nrn871. [DOI] [PubMed] [Google Scholar]
- Guo S, Brush J, Teraoka H, Goddard A, Wilson SW, Mullins MC, Rosenthal A. Development of noradrenergic neurons in the zebrafish hindbrain requires BMP, FGF8, and the homeodomain protein soulless/Phox2a. Neuron. 1999;24:555–566. doi: 10.1016/s0896-6273(00)81112-5. [DOI] [PubMed] [Google Scholar]
- Gutierrez R. The GABAergic phenotype of the "glutamatergic" granule cells of the dentate gyrus. Prog Neurobiol. 2003;71:337–358. doi: 10.1016/j.pneurobio.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Gutierrez R, Romo-Parra H, Maqueda J, Vivar C, Ramirez M, Morales MA, Lamas M. Plasticity of the GABAergic phenotype of the "glutamatergic" granule cells of the rat dentate gyrus. J Neurosci. 2003;23:5594–5598. doi: 10.1523/JNEUROSCI.23-13-05594.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzschuh J, Hauptmann G, Driever W. Genetic analysis of the roles of Hh, FGF8, and nodal signaling during catecholaminergic system development in the zebrafish brain. J Neurosci. 2003;23:5507–5519. doi: 10.1523/JNEUROSCI.23-13-05507.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino M, Nakamura S, Mori K, Kawauchi T, Terao M, Nishimura YV, Fukuda A, Fuse T, Matsuo N, Sone M, Watanabe M, Bito H, Terashima T, Wright CV, Kawaguchi Y, Nakao K, Nabeshima Y. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron. 2005;47:201–213. doi: 10.1016/j.neuron.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Howard MJ. Mechanisms and perspectives on differentiation of autonomic neurons. Dev Biol. 2005;277:271–286. doi: 10.1016/j.ydbio.2004.09.034. [DOI] [PubMed] [Google Scholar]
- Huang X, Liu J, Ketova T, Fleming JT, Grover VK, Cooper MK, Litingtung Y, Chiang C. Transventricular delivery of Sonic hedgehog is essential to cerebellar ventricular zone development. Proc Natl Acad Sci U S A. 2010;107:8422–8427. doi: 10.1073/pnas.0911838107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufer D, Friedman A, Seidman S, Soreq H. Acute stress facilitates long-lasting changes in cholinergic gene expression. Nature. 1998;393:373–377. doi: 10.1038/30741. [DOI] [PubMed] [Google Scholar]
- Kondo T, Matsuoka AJ, Shimomura A, Koehler KR, Chan RJ, Miller JM, Srour EF, Hashino E. Wnt signaling promotes neuronal differentiation from mesenchymal stem cells through activation of Tlx3. Stem Cells. 2011;29:836–846. doi: 10.1002/stem.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis SC, Keefe D. Evidence for neurotransmitter plasticity in vivo: developmental changes in properties of cholinergic sympathetic neurons. Dev Biol. 1983;98:349–372. doi: 10.1016/0012-1606(83)90365-2. [DOI] [PubMed] [Google Scholar]
- Levkowitz G, Zeller J, Sirotkin HI, French D, Schilbach S, Hashimoto H, Hibi M, Talbot WS, Rosenthal A. Zinc finger protein too few controls the development of monoaminergic neurons. Nat Neurosci. 2003;6:28–33. doi: 10.1038/nn979. [DOI] [PubMed] [Google Scholar]
- Lin PY, Hinterneder JM, Rollor SR, Birren SJ. Non-cell-autonomous regulation of GABAergic neuron development by neurotrophins and the p75 receptor. J Neurosci. 2007;27:12787–12796. doi: 10.1523/JNEUROSCI.3302-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Popescu IR, Denisova JV, Neve RL, Corriveau RA, Belousov AB. Regulation of cholinergic phenotype in developing neurons. J Neurophysiol. 2008;99:2443–2455. doi: 10.1152/jn.00762.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo L, Sommer L, Anderson DJ. MASH1 maintains competence for BMP2-induced neuronal differentiation in post-migratory neural crest cells. Curr Biol. 1997;7:440–450. doi: 10.1016/s0960-9822(06)00191-6. [DOI] [PubMed] [Google Scholar]
- Lo L, Morin X, Brunet JF, Anderson DJ. Specification of neurotransmitter identity by Phox2 proteins in neural crest stem cells. Neuron. 1999;22:693–705. doi: 10.1016/s0896-6273(00)80729-1. [DOI] [PubMed] [Google Scholar]
- Lo L, Dormand E, Greenwood A, Anderson DJ. Comparison of the generic neuronal differentiation and neuron subtype specification functions of mammalian achaete-scute and atonal homologs in cultured neural progenitor cells. Development. 2002;129:1553–1567. doi: 10.1242/dev.129.7.1553. [DOI] [PubMed] [Google Scholar]
- Lopez-Coviella I, Berse B, Krauss R, Thies RS, Blusztajn JK. Induction and maintenance of the neuronal cholinergic phenotype in the central nervous system by BMP-9. Science. 2000;289:313–316. doi: 10.1126/science.289.5477.313. [DOI] [PubMed] [Google Scholar]
- Lopez-Coviella I, Follettie MT, Mellott TJ, Kovacheva VP, Slack BE, Diesl V, Berse B, Thies RS, Blusztajn JK. Bone morphogenetic protein 9 induces the transcriptome of basal forebrain cholinergic neurons. Proc Natl Acad Sci U S A. 2005;102:6984–6989. doi: 10.1073/pnas.0502097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy B, Apostolova G, Dorn R, McGuire VA, Arthur JS, Dechant G. p38alpha and p38beta mitogen-activated protein kinases determine cholinergic transdifferentiation of sympathetic neurons. J Neurosci. 2011;31:12059–12067. doi: 10.1523/JNEUROSCI.0448-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madziar B, Shah S, Brock M, Burke R, Lopez-Coviella I, Nickel AC, Cakal EB, Blusztajn JK, Berse B. Nerve growth factor regulates the expression of the cholinergic locus and the high-affinity choline transporter via the Akt/PKB signaling pathway. Journal of neurochemistry. 2008;107:1284–1293. doi: 10.1111/j.1471-4159.2008.05681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe T, Tatsumi K, Inoue M, Matsuyoshi H, Makinodan M, Yokoyama S, Wanaka A. L3/Lhx8 is involved in the determination of cholinergic or GABAergic cell fate. Journal of neurochemistry. 2005;94:723–730. doi: 10.1111/j.1471-4159.2005.03261.x. [DOI] [PubMed] [Google Scholar]
- Marek KW, Kurtz LM, Spitzer NC. cJun integrates calcium activity and tlx3 expression to regulate neurotransmitter specification. Nat Neurosci. 2010;13:944–950. doi: 10.1038/nn.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Yuxing Z, Takaki H, Takeuchi M, Iseki K, Hagino S, Kitanaka J, Takemura M, Misawa H, Ikawa M, Okabe M, Wanaka A. The LIM homeobox gene, L3/Lhx8, is necessary for proper development of basal forebrain cholinergic neurons. Eur J Neurosci. 2004;19:3129–3141. doi: 10.1111/j.0953-816X.2004.03415.x. [DOI] [PubMed] [Google Scholar]
- Noh J, Seal RP, Garver JA, Edwards RH, Kandler K. Glutamate co-release at GABA/glycinergic synapses is crucial for the refinement of an inhibitory map. Nat Neurosci. 2010;13:232–238. doi: 10.1038/nn.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paina S, Garzotto D, DeMarchis S, Marino M, Moiana A, Conti L, Cattaneo E, Perera M, Corte G, Calautti E, Merlo GR. Wnt5a is a transcriptional target of Dlx homeogenes and promotes differentiation of interneuron progenitors in vitro and in vivo. J Neurosci. 2011;31:2675–2687. doi: 10.1523/JNEUROSCI.3110-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panman L, Andersson E, Alekseenko Z, Hedlund E, Kee N, Mong J, Uhde CW, Deng Q, Sandberg R, Stanton LW, Ericson J, Perlmann T. Transcription factor-induced lineage selection of stem-cell-derived neural progenitor cells. Cell Stem Cell. 2011;8:663–675. doi: 10.1016/j.stem.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Robertshaw E, Matsumoto K, Lumsden A, Kiecker C. Irx3 and Pax6 establish differential competence for Shh-mediated induction of GABAergic and glutamatergic neurons of the thalamus. Proc Natl Acad Sci U S A. 2013;110:E3919–E3926. doi: 10.1073/pnas.1304311110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root CM, Velazquez-Ulloa NA, Monsalve GC, Minakova E, Spitzer NC. Embryonically expressed GABA and glutamate drive electrical activity regulating neurotransmitter specification. J Neurosci. 2008;28:4777–4784. doi: 10.1523/JNEUROSCI.4873-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge JS, Eaton MJ, Mather P, Lindsay RM, Whittemore SR. CNTF induces raphe neuronal precursors to switch from a serotonergic to a cholinergic phenotype in vitro. Mol Cell Neurosci. 1996;7:204–221. doi: 10.1006/mcne.1996.0016. [DOI] [PubMed] [Google Scholar]
- Russek-Blum N, Gutnick A, Nabel-Rosen H, Blechman J, Staudt N, Dorsky RI, Houart C, Levkowitz G. Dopaminergic neuronal cluster size is determined during early forebrain patterning. Development. 2008;135:3401–3413. doi: 10.1242/dev.024232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotzinger RJ, Landis SC. Acquisition of cholinergic and peptidergic properties by sympathetic innervation of rat sweat glands requires interaction with normal target. Neuron. 1990;5:91–100. doi: 10.1016/0896-6273(90)90037-g. [DOI] [PubMed] [Google Scholar]
- Simon HH, Thuret S, Alberi L. Midbrain dopaminergic neurons: control of their cell fate by the engrailed transcription factors. Cell Tissue Res. 2004;318:53–61. doi: 10.1007/s00441-004-0973-8. [DOI] [PubMed] [Google Scholar]
- Spitzer NC. Activity-dependent neurotransmitter respecification. Nat Rev Neurosci. 2012;13:94–106. doi: 10.1038/nrn3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer NC. Neurotransmitter Switching? No Surprise. Neuron. 2015;86:1131–1144. doi: 10.1016/j.neuron.2015.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Junghans D, Geissen M, Goridis C, Ernsberger U, Rohrer H. The Phox2 homeodomain proteins are sufficient to promote the development of sympathetic neurons. Development. 1999;126:4087–4094. doi: 10.1242/dev.126.18.4087. [DOI] [PubMed] [Google Scholar]
- Stanke M, Duong CV, Pape M, Geissen M, Burbach G, Deller T, Gascan H, Otto C, Parlato R, Schutz G, Rohrer H. Target-dependent specification of the neurotransmitter phenotype: cholinergic differentiation of sympathetic neurons is mediated in vivo by gp 130 signaling. Development. 2006;133:141–150. doi: 10.1242/dev.02189. [DOI] [PubMed] [Google Scholar]
- Stull ND, Jung JW, Iacovitti L. Induction of a dopaminergic phenotype in cultured striatal neurons by bone morphogenetic proteins. Brain Res Dev Brain Res. 2001;130:91–98. doi: 10.1016/s0165-3806(01)00216-4. [DOI] [PubMed] [Google Scholar]
- Swapna I, Borodinsky LN. Interplay between electrical activity and bone morphogenetic protein signaling regulates spinal neuron differentiation. Proc Natl Acad Sci U S A. 2012;109:16336–16341. doi: 10.1073/pnas.1202818109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe Y, William C, Jessell TM. Specification of motor neuron identity by the MNR2 homeodomain protein. Cell. 1998;95:67–80. doi: 10.1016/s0092-8674(00)81783-3. [DOI] [PubMed] [Google Scholar]
- Thomas GM, Huganir RL. MAPK cascade signalling and synaptic plasticity. Nat Rev Neurosci. 2004;5:173–183. doi: 10.1038/nrn1346. [DOI] [PubMed] [Google Scholar]
- van Doorninck JH, van Der Wees J, Karis A, Goedknegt E, Engel JD, Coesmans M, Rutteman M, Grosveld F, De Zeeuw CI. GATA-3 is involved in the development of serotonergic neurons in the caudal raphe nuclei. J Neurosci. 1999;19:RC12. doi: 10.1523/JNEUROSCI.19-12-j0002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela-Nallar L, Alfaro IE, Serrano FG, Parodi J, Inestrosa NC. Wingless-type family member 5A (Wnt-5a) stimulates synaptic differentiation and function of glutamatergic synapses. Proc Natl Acad Sci U S A. 2010;107:21164–21169. doi: 10.1073/pnas.1010011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallen A, Perlmann T. Transcriptional control of dopamine neuron development. Ann N Y Acad Sci. 2003;991:48–60. doi: 10.1111/j.1749-6632.2003.tb07462.x. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- Yao M, Niu G, Sheng Z, Wang Z, Fei J. Identification of a Smad4/YY1-recognized and BMP2-responsive transcriptional regulatory module in the promoter of mouse GABA transporter subtype I (Gat1) gene. J Neurosci. 2010;30:4062–4071. doi: 10.1523/JNEUROSCI.2964-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervas M, Millet S, Ahn S, Joyner AL. Cell behaviors and genetic lineages of the mesencephalon and rhombomere 1. Neuron. 2004;43:345–357. doi: 10.1016/j.neuron.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Marin O, Hermesz E, Powell A, Flames N, Palkovits M, Rubenstein JL, Westphal H. The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proc Natl Acad Sci U S A. 2003;100:9005–9010. doi: 10.1073/pnas.1537759100. [DOI] [PMC free article] [PubMed] [Google Scholar]


