Abstract
Sigmatropic rearrangements, while rare in biology, offer opportunities for the efficient and selective synthesis of complex chemical motifs. A “P411” serine-ligated variant of cytochrome P450BM3 has been engineered to initiate a sulfimidation/[2,3]-sigmatropic rearrangement sequence in whole E. coli cells, a non-natural function for any enzyme, providing access to enantioenriched, protected allylic amines. Five mutations in the enzyme substantially enhance its activity toward this new function, demonstrating the evolvability of the catalyst toward challenging nitrene transfer reactions. The evolved catalyst additionally performs the highly enantioselective imidation of non-allylic sulfides.
Keywords: biocatalysis, amination, nitrene transfer, cytochrome P450, directed evolution
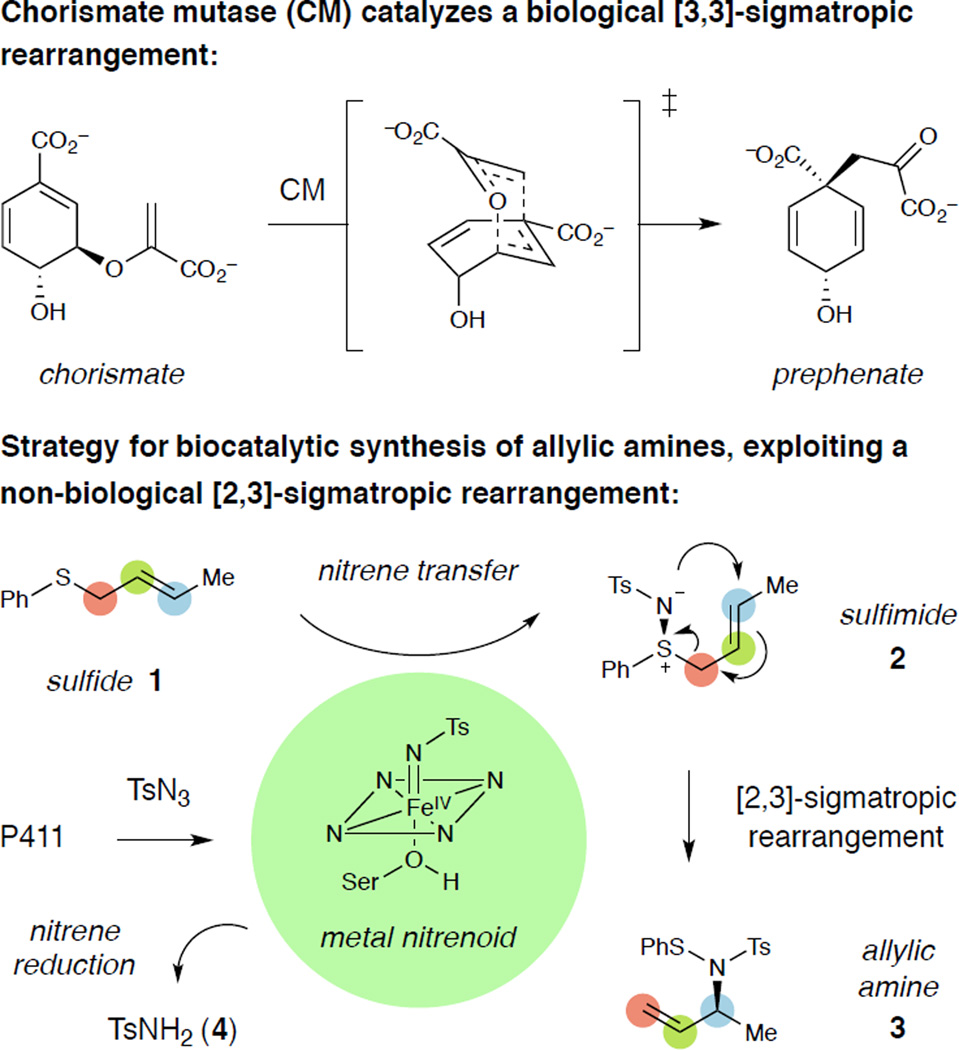
Sigmatropic rearrangements, a class of pericyclic reactions in which one σ bond is exchanged for another, are highly valuable reactions in synthetic chemistry due to their ability to forge challenging chiral centers with high stereospecificity in complex structural motifs.1 While useful in chemical synthesis, sigmatropic rearrangements are remarkably rare in biology.2 Only a handful of enzymes that exploit such rearrangements have been identified,3 a notable example being chorismate mutase, which catalyzes the Claisen rearrangement of chorismate to prephenate in the biosynthesis of tyrosine and phenylalanine (Scheme 1).4 Alternatively, certain biological pathways involve sigmatropic rearrangements that are spontaneous once a given precursor has been assembled; for instance, Claisen5 and Cope rearrangements6 have been implicated in the prenylation of aromatic amino acid side chains.
Scheme 1.
Design of an enzymatic sulfimidation/sigmatropic rearrangement sequence
Biocatalysis requires new enzymes for the green and economical synthesis of chemical products that are often not accessible using natural enzymatic reactions.7 The design of enzymes capable of catalyzing or initiating sigmatropic rearrangements would introduce valuable, complexity-building reactions into biocatalysis, but work in this area has largely been limited to catalytic antibodies.8,9 Recently, our laboratory10 and Fasan’s group11 demonstrated that heme-containing proteins can perform nitrene transfer, a class of reactions for which there is no biological precedent. In these reactions, the ferrous state of the heme cofactor first reacts with a nitrene precursor (such as a sulfonyl azide) to generate an iron nitrenoid; this reactive intermediate is then intercepted by a nucleophile to yield an aminated product. Variants of cytochrome P450BM3, a soluble P450 from Bacillus megaterium, in which the axial ligand to iron is mutated from cysteine to serine are particularly active catalysts for nitrene transfer.10 As these variants display a ferrous-CO Soret peak at 411 nm, we term them “cytochrome P411s.”12 Here, we describe a strategy that merges cytochrome P411-catalyzed amination with a sigmatropic rearrangement for the synthesis of chiral allylic amines.
In particular, we aimed to exploit the spontaneous sigmatropic rearrangement of allylic sulfimides; these species undergo a [2,3]-rearrangement in which an N–S bond is exchanged for an N–C bond, delivering protected allylic amines.13 This type of sigmatropic rearrangement is not a feature of any known biological pathway. Previously, we demonstrated that cytochrome P411s catalyze nitrene transfer to simple sulfides.10b We thus envisioned using a P411 to perform the enantioselective imidation of prochiral allylic sulfide 1 to generate allylic sulfimide 2 (Scheme 1). This species would then spontaneously undergo the [2,3]-rearrangement to deliver the protected allylic amine 3, in which a new chiral center is forged. Since sigmatropic rearrangements often proceed with high stereospecificity, the stereochemistry set in the P411-catalyzed sulfimidation step would be transferred to the allylic amine product.14 This strategy would establish a biocatalytic route to chiral allylic amines, compounds that are biologically active15 as well as valuable synthetic intermediates.16,17 Furthermore, expanding the breadth of enzymatic amination reactions will facilitate the construction of non-natural in vivo pathways for chiral amine production.
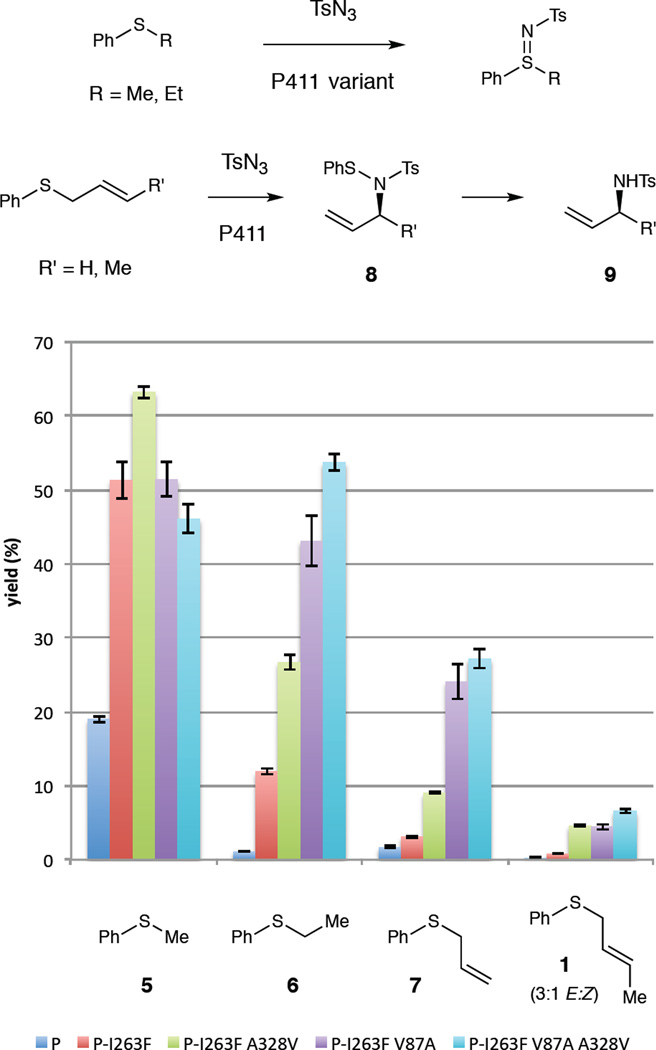
In initial experiments, we evaluated the ability of a variant previously identified for the imidation of sulfides10b to promote the desired reaction (this variant, termed P411BM3-CIS T438S, contains 15 mutations relative to wild-type P450BM3 and will be abbreviated “P”). In whole-cell bioconversions using tosyl azide (TsN3) as the nitrene precursor, variant P only efficiently imidates aryl–methyl sulfides such as thioanisole (5, Figure 1). This variant provides essentially no product (<0.5% yield) when reacted with phenyl crotyl sulfide (1). In general, P411-catalyzed nitrene transfer reactions suffer from a competing reduction of tosyl azide to p-toluenesulfonamide (4), both via reduction of the azide by cellular agents and via electron transfer to the metal nitrenoid intermediate from the enzyme’s reductase.10 However, we reasoned that the enzyme could be engineered to accommodate allylic sulfide substrates, orient them in a manner suitable for productive bond formation with the iron nitrenoid, and in such a way outcompete undesired azide reduction pathways.
Figure 1.
Evolution of a P411 catalyst for nitrene transfer to progressively larger aryl-alkyl sulfides. Experiments were performed using whole cells overexpressing the P411 variant, resuspended to OD600 = 30, with 5 mM sulfide and 5 mM tosyl azide. Results are the average of experiments performed with duplicate cell cultures, each used to perform duplicate chemical reactions. P = P411BM3-CIS T438S.
To evolve catalysts for the desired reaction we performed a substrate walk, in which P411 variants were first evaluated for their ability to imidate phenyl ethyl sulfide (6) and the most active variants subsequently assayed for activity toward the larger sulfides 7 and 1 (Figure 1). We first found that introducing the active site mutation I263F into variant P is activating toward sulfimidation; this mutation was identified during studies on P411-catalyzed regioselective C–H amination.10c Importantly, P-I263F displays a sufficient level of activity on phenyl ethyl sulfide (6, 12% yield) for screening of enzyme variants. Taking P-I263F as the parent, we evaluated site-saturation libraries at residues V87, L181, A328, and S438 (all located near the active site on the distal face of the heme) for the ability to imidate sulfide 6, from which we identified the beneficial mutations V87A and A328V (Figure 1). Gratifyingly, in addition to displaying improved activity on sulfide 6, the variants incorporating these mutations also display appreciable activity for transformation of the desired substrate 1. Combining these mutations yielded an enzyme (P-I263F V87A A328V, or P-3) having further improved activity toward sulfide 1, delivering the rearrangement product in 7% yield with 220 turnovers, while also validating our reaction design and directed evolution strategy.
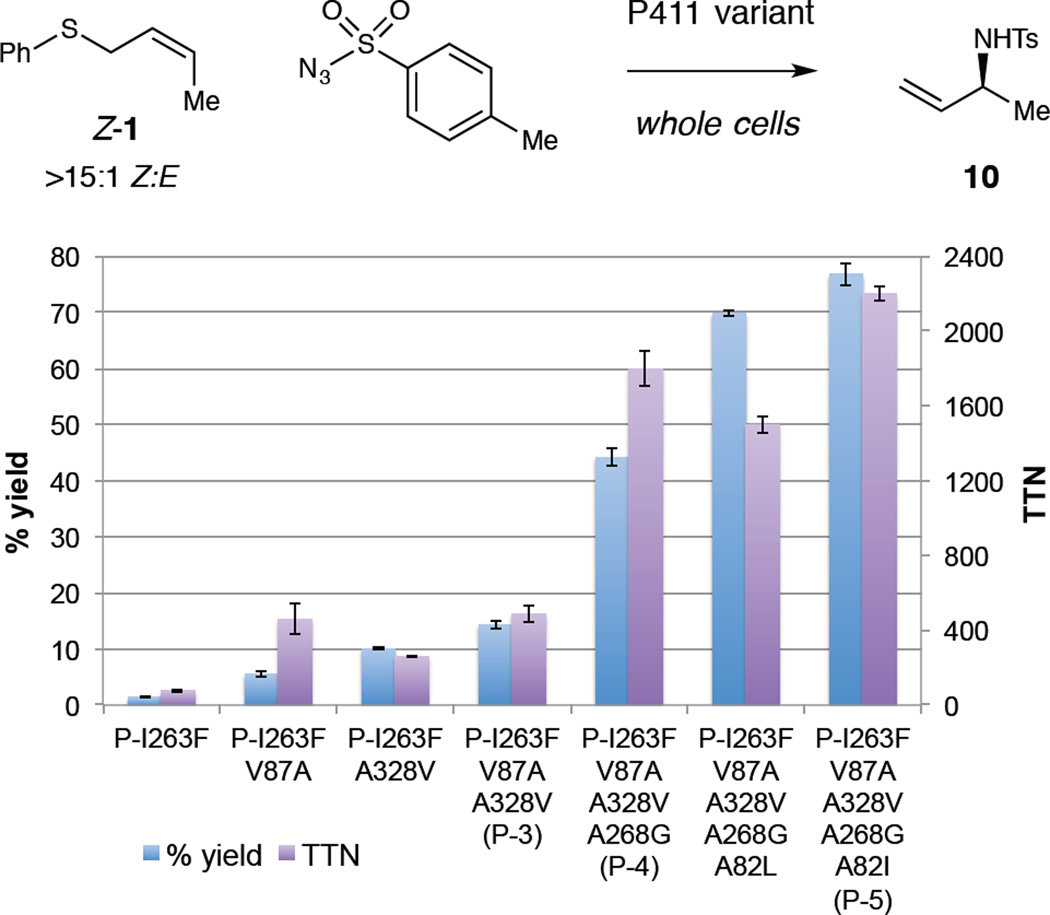
The observed product of these reactions, however, is not the phenylthiosulfonamide 8 (the initial product of the rearrangement) but rather the sulfonamide 9, in which the S–N bond of 8 has undergone reductive cleavage, presumably by cellular reductants such as NAD(P)H or glutathione. Furthermore, for the experiments described above, we employed sulfide 1 as a 3:1 E:Z mixture of alkene isomers.18 Considering that the P411 variants likely display differing activities toward the two alkene isomers, we prepared a batch of sulfide 1 enriched in the Z olefin (Z-1, >15:1 Z:E). Employing Z-1 in the amination reaction with P-3 improves the yield two-fold to 14% (490 turnovers), demonstrating that the enzyme prefers the Z-olefin over the E-olefin (Figure 2).
Figure 2.
Evolution of a catalyst for sulfimidation/[2,3]-rearrangement of Z-1. Reaction conditions as given in Figure 1, with a DTT work-up employed for reactions giving >30% yield.21
Having identified a catalyst active on the desired substrate for the rearrangement, we then performed a second round of evolution using P-3 as the parent and screening for activity on sulfide 1 (see SI for details). Upon evaluating site-saturation libraries at positions A268, L437, and S438, the mutation A268G was found to provide a 3-fold improvement in yield (Figure 2). The wild-type threonine residue at this position plays a critical role in the natural monooxygenation reaction,19 and the mutation T268A was previously shown to be highly activating toward nitrene as well as carbene transfer by P450BM3 variants.10,20 We then performed a further round of evolution by saturating active-site residues A78, A82, T260, and P329 in the variant P-I263F V87A A328V A268G (P-4). Mutagenesis at A82 yielded two variants that improve the reaction yield more than 1.5-fold: the mutations A82L and A82I both improve protein expression in E. coli compared to P-4, with the variant having the A82I mutation showing higher specific activity (Figure 2). This variant, P-5, promotes the sulfimidation/[2,3]-rearrangement of sulfide Z-1 in 77% yield with greater than 2,000 turnovers.21 The product 10 is obtained in moderate enantiomeric excess (68% ee, Table 1, entry 1). Notably, the mutations identified in this study both introduce (A328V, A82L/I) and remove (V87A, A268G) steric bulk in the active site. It thus seems likely that the new mutations not only enlarge the active site to accommodate substrates, but also constrict substrates and intermediates in a manner that facilitates bond formation.
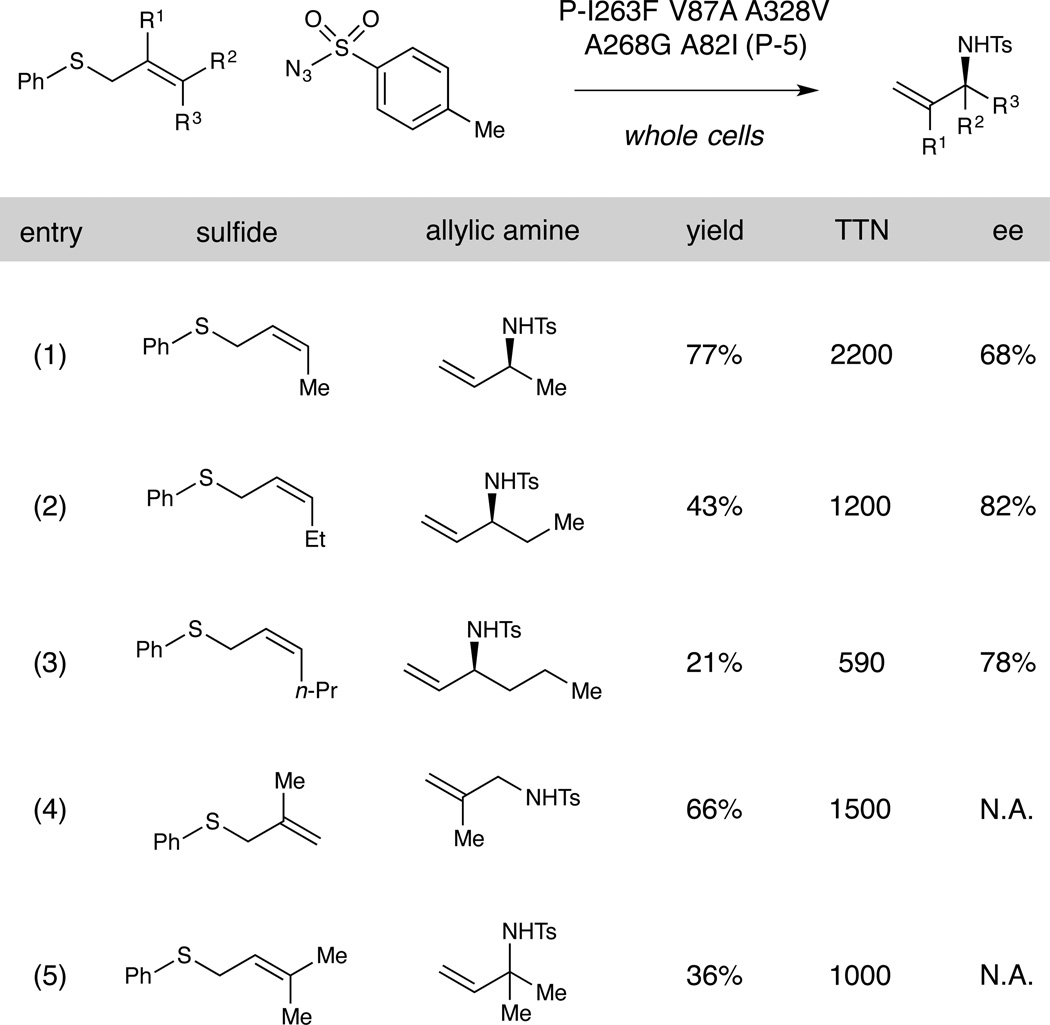
Table 1.
Substrate scope of the P411-catalyzed sulfimidation/[2,3]-rearrangement.a
We then evaluated the ability of the best evolved variant (P-5) to perform the sulfimidation/rearrangement of other allylic sulfides. While the enzyme shows a strong preference for amination of the substrate for which it was evolved (Z-1), it also catalyzes the amination of a range of other sulfides to form both chiral (entries 1–3, Table 1) and achiral (entries 4 and 5) tosyl-protected allylic amines. Interestingly, given that the parent variant (P) is a poor catalyst for the amination of sulfides as small as phenyl ethyl sulfide (6), the evolved variant P-5 is capable of nitrene transfer to substrates having alkyl chains as long as six carbons (Table 1, entry 3). The reaction may be carried out on semi-preparative scale with reduced catalyst loadings (whole cells at OD600 = 10, ~0.6 µM enzyme) to afford amine 10 in 71% isolated yield and 6,100 turnovers (see SI for details). Finally, the nitrene transfer activity of the variant P and other cytochrome P411s is generally inhibited in the presence of oxygen, leading us to employ anaerobic conditions for the above biotransformations. In contrast, the evolved variant P-5 performs nitrene transfer under aerobic conditions with no loss of activity, delivering allylic amine 10 in 79% yield.
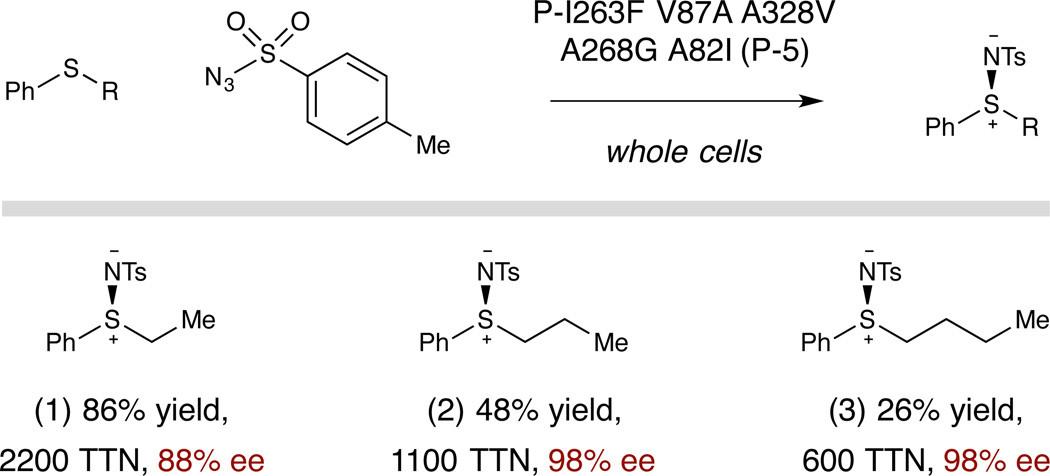
We rationalized that two possibilities exist for the moderate enantioselectivities obtained in the amination of 1: either the sulfimidation event is only moderately selective and the rearrangement proceeds with high stereospecificity, or the sulfimidation is more highly selective and erosion of enantiopurity occurs in the rearrangement step. In examining the imidation of non-allylic sulfides by the variant P-5, we found that these substrates react with very high levels of enantioinduction. For example, phenyl n-butyl sulfide, which differs from sulfide 1 only by saturation of the alkene, is converted to its corresponding sulfimide in 98% ee (Table 2, entry 3). It is therefore likely that in the sulfimidation/rearrangement reactions, the imidation event also proceeds with high enantioselectivity, while the rearrangement proceeds with imperfect stereofidelity, via competing endo and exo rearrangement transition states.14 Notably, however, these results also represent the first highly enantioselective enzymatic imidation of sulfides.
Table 2.
Enantioselective imidation of sulfides catalyzed by an evolved P411 variant.a
Reaction conditions as given in Figure 1.
We have thus demonstrated that cytochrome P411BM3 variants can promote a sulfimidation/[2,3]-rearrangement sequence in whole cells, introducing a new sigmatropic rearrangement into the synthetic repertoire of biocatalysis. While the P411 does not catalyze the rearrangement itself, the enzyme enables the reaction by assembling the required allylic sulfimide intermediate; by performing this step with high enantioselectivity, the P411 ultimately delivers enantioenriched allylic amine products. It may be possible to improve the enantiospecificity of the rearrangement by evolving the enzyme to preferentially bind one of the competing rearrangement transition states.8h The activity of the catalyst was substantially enhanced by directed evolution: for the transformation of sulfide Z-1, the five mutations identified in this study improve the conversion more than 200-fold over the activity of the parent P as well as confer the ability to function under aerobic conditions. Starting from a variant (P) that performs azide reduction almost exclusively (>99%) in preference to nitrene transfer to sulfide 1, directed evolution delivered a variant (P-5) that efficiently promotes the desired nitrene transfer process (up to 77% yield for amination of Z-1). As both nitrene transfer and nitrene reduction are reaction pathways that are absent in nature, we have demonstrated that only a few mutations in P411BM3 can partition reactivity between two competing non-natural pathways. This study thus highlights the ability of enzymes to adapt, through directed evolution, to facilitate valuable reaction pathways for which no natural enzymes have evolved.
Supplementary Material
Acknowledgments
Our research is supported by the National Science Foundation, Division of Molecular and Cellular Biosciences (grant MCB-1513007). C.K.P. thanks the Resnick Sustainability Institute for a postdoctoral fellowship. T.H.K. and C.C.F. were supported by a Ruth L. Kirschstein National Research Service Award (F32GM108143) and an NSF Graduate Research Fellowship, respectively. We thank Dr. Scott Virgil and the Center for Catalysis and Chemical Synthesis at Caltech for assistance with chiral chromatography.
References
- 1.Carey FA, Sundberg RJ. Advanced Organic Chemistry, Part B. Reactions and Synthesis. 5th. New York, NY: Springer; 2007. pp. 473–617. [Google Scholar]
- 2.a) Silverman RB. The Organic Chemistry of Enzyme-Catalyzed Reactions. San Diego, CA: Academic Press; 2000. pp. 505–561. [Google Scholar]; b) Pindur U, Schneider GH. Chem. Soc. Rev. 1994;23:409. [Google Scholar]
- 3.a) Shipman LW, Li D, Roessner CA, Scott AI, Sacchettini JC. Structure. 2001;9:587. doi: 10.1016/s0969-2126(01)00618-9. [DOI] [PubMed] [Google Scholar]; b) DeClue MS, Baldridge KK, Künzler DE, Kast P, Hilvert D. J. Am. Chem. Soc. 2005;127:15002. doi: 10.1021/ja055871t. [DOI] [PubMed] [Google Scholar]; c) Li S, Lowell AN, Yu F, Raveh A, Newmister SA, Bair N, Schaub JM, Williams RM, Sherman DH. J. Am. Chem. Soc. 2015;137:15366. doi: 10.1021/jacs.5b10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee AY, Stewart JD, Clardy J, Ganem B. Chem. Biol. 1995;2:195. doi: 10.1016/1074-5521(95)90269-4. [DOI] [PubMed] [Google Scholar]
- 5.McIntosh JA, Donia MS, Nair SK, Schmidt EW. J. Am. Chem. Soc. 2011;133:13698. doi: 10.1021/ja205458h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Luk LYP, Qian Q, Tanner ME. J. Am. Chem. Soc. 2011;133:12342. doi: 10.1021/ja2034969. [DOI] [PubMed] [Google Scholar]; b) Tanner ME. Nat. Prod. Rep. 2015;32:88. doi: 10.1039/c4np00099d. [DOI] [PubMed] [Google Scholar]
- 7.Wohlgemuth R. Curr. Opin. Biotechnol. 2010;21:713. doi: 10.1016/j.copbio.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 8.a) Hilvert D, Carpenter SH, Nared KD, Auditor M-TM. Proc. Natl. Acad. Sci. U.S.A. 1988;85:4953. doi: 10.1073/pnas.85.14.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Jackson DY, Jacobs JW, Sugasawara R, Reich SH, Bartlett PA, Schultz PG. J. Am. Chem. Soc. 1988;110:4841. [Google Scholar]; c) Haynes MR, Stura EA, Hilvert D, Wilson IA. Science. 1994;263:646. doi: 10.1126/science.8303271. [DOI] [PubMed] [Google Scholar]; d) Braisted AC, Schultz PG. J. Am. Chem. Soc. 1994;116:2211. [Google Scholar]; e) Yoon SS, Oei Y, Sweet E, Schultz PG. J. Am. Chem. Soc. 1996;118:11686. [Google Scholar]; f) Zou ZS, Jiang N, Hilvert D. J. Am. Chem. Soc. 1997;119:3623. [Google Scholar]; g) Ulrich HD, Mundorff E, Santarsiero BD, Driggers EM, Stevens RC, Schultz PG. Nature. 1997;389:271. doi: 10.1038/38470. [DOI] [PubMed] [Google Scholar]; h) Zhou ZS, Flohr A, Hilvert D. J. Org. Chem. 1999;64:8334. doi: 10.1021/jo991299a. [DOI] [PubMed] [Google Scholar]
- 9.For other examples of enzyme-promoted, non-natural sigmatropic rearrangements, see: Latham JA, Jr, Branchaud BP, Chen Y-CJ, Walsh C. J. Chem. Soc. Chem. Commun. 1986:528. ; Tyagi V, Bonn RB, Fasan R. Chem. Sci. 2015;6:2488. doi: 10.1039/c5sc00080g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) McIntosh JA, Coelho PS, Farwell CC, Wang ZJ, Lewis JC, Brown TR, Arnold FH. Angew. Chem. Int. Ed. 2013;52:9309. doi: 10.1002/anie.201304401. [DOI] [PMC free article] [PubMed] [Google Scholar]; McIntosh JA, Coelho PS, Farwell CC, Wang ZJ, Lewis JC, Brown TR, Arnold FH. Angew. Chem. 2013;125:9479. doi: 10.1002/anie.201304401. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Farwell CC, McIntosh JA, Hyster TK, Wang ZJ, Arnold FH. J. Am. Chem. Soc. 2014;136:8766. doi: 10.1021/ja503593n. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Hyster TK, Farwell CC, Buller AR, McIntosh JA, Arnold FH. J. Am. Chem. Soc. 2014;136:15505. doi: 10.1021/ja509308v. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Farwell CC, Zhang RK, McIntosh JA, Hyster TK, Arnold FH. ACS Cent. Sci. 2015;1:89. doi: 10.1021/acscentsci.5b00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Singh R, Bordeaux M, Fasan R. ACS Catal. 2014;4:546. doi: 10.1021/cs400893n. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bordeaux M, Singh R, Fasan R. Bioorg. Med. Chem. 2014;22:5697. doi: 10.1016/j.bmc.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Singh R, Kolev JN, Sutera PA, Fasan R. ACS Catal. 2015;5:1685. doi: 10.1021/cs5018612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coelho PS, Wang ZJ, Ener ME, Baril SA, Kannan A, Arnold FH, Brustad EM. Nat. Chem. Biol. 2013;9:485. doi: 10.1038/nchembio.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilchrist TL, Moody CJ. Chem. Rev. 1977;77:409. [Google Scholar]
- 14.Hoffmann RW. Angew. Chem. Int. Ed. 1979;18:563. [Google Scholar]; Hoffmann RW. Angew. Chem. 1979;91:625. [Google Scholar]
- 15.Nanavati SM, Silverman RB. J. Am. Chem. Soc. 1991;113:9341. [Google Scholar]
- 16.Other biocatalytic approaches to amine synthesis have not yielded general routes to allylic amines: Kohls H, Steffen-Munsberg F, Höhne M. Curr. Opin. Chem. Biol. 2014;19:180. doi: 10.1016/j.cbpa.2014.02.021.
- 17.Small-molecule catalysts for this transformation have been reported: Takada H, Nishibayashi Y, Ohe K, Uemura S. Chem. Commun. 1996:931. ; b) Takada H, Nishibayashi Y, Ohe K, Uemura S, Baird CP, Sparey TJ, Taylor PC. J. Org. Chem. 1997;62:6512. [Google Scholar]; c) Murakami M, Katsuki T. Tetrahedron Lett. 2002;43:3947. [Google Scholar]; d) Murakami M, Uchida T, Saito B, Katsuki T. Chirality. 2003;15:116. doi: 10.1002/chir.10156. [DOI] [PubMed] [Google Scholar]; e) Wang J, Frings M, Bolm C. Angew. Chem. Int. Ed. 2013;52:8661. doi: 10.1002/anie.201304451. [DOI] [PubMed] [Google Scholar]; Wang J, Frings M, Bolm C. Angew. Chem. 2013;125:8823. doi: 10.1002/anie.201304451. [DOI] [PubMed] [Google Scholar]
- 18.The alkene isomers are inseparable using standard column chromatography methods.
- 19.a) Schlichting I, Berendzen J, Chu K, Stock AM, Maves SA, Benson DE, Sweet RM, Ringe D, Petsko GA, Sligar SG. Science. 2000;287:1615. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]; b) Nagano S, Poulos TL. J. Biol. Chem. 2005;280:31659. doi: 10.1074/jbc.M505261200. [DOI] [PubMed] [Google Scholar]
- 20.a) Coelho PS, Brustad EM, Kannan A, Arnold FH. Science. 2013;339:307. doi: 10.1126/science.1231434. [DOI] [PubMed] [Google Scholar]; b) Wang ZJ, Renata H, Peck NE, Farwell CC, Coelho PS, Arnold FH. Angew. Chem. Int. Ed. 2014;53:6810. doi: 10.1002/anie.201402809. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang ZJ, Renata H, Peck NE, Farwell CC, Coelho PS, Arnold FH. Angew. Chem. 2014;126:6928. doi: 10.1002/anie.201402809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.At the higher conversions (>30% yield) obtained with the evolved variants, the cell’s endogenous reductants are no longer sufficient to completely reduce intermediate 8, and therefore a work-up with dithiothreitol (DTT) was implemented to convert the product mixture entirely to the sulfonamide 9 prior to analysis (see SI for details).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.