Abstract
Gene-specific activation by enhancers involves their communication with the basal RNA polymerase II transcription machinery at the core promoter. Core promoters are diverse and may contain a variety of sequence elements such as the TATA box, the Initiator (INR), and the downstream promoter element (DPE) recognized, respectively, by the TATA-binding protein (TBP) and TBP-associated factors of the TFIID complex. Core promoter elements contribute to the gene selectivity of enhancers, and INR/DPE-specific enhancers and activators have been identified. Here, we identify a TATA box-selective activating sequence upstream of the human β-actin (ACTB) gene that mediates serum response factor (SRF)-induced transcription from TATA-dependent but not INR-dependent promoters and requires the TATA-binding/bending activity of TBP, which is otherwise dispensable for transcription from a TATA-less promoter. The SRF-dependent ACTB sequence is stereospecific on TATA promoters but activates in an orientation-independent manner a composite TATA/INR-containing promoter. More generally, we show that SRF-regulated genes of the actin/cytoskeleton/contractile family tend to have a TATA box. These results suggest distinct TATA-dependent and INR-dependent mechanisms of TFIID-mediated transcription in mammalian cells that are compatible with only certain stereospecific combinations of activators, and that a TBP-TATA binding mechanism is important for SRF activation of the actin/cytoskeleton-related gene family.
1. Introduction
Proper development and homeostasis of eukaryotic organisms rely on the accurate spatio-temporal regulation of specific gene transcription in response to diverse physiological and environmental signals. Specific gene transcription by RNA polymerase II (RNAPII) is controlled by the combinatorial assortment of a variety of regulatory DNA sequences located at promoter-proximal and more distal (i.e., enhancer) regions. These regulatory DNA elements are bound by sequence-specific regulators (activators or repressors) that in turn recruit a variety of coregulators (coactivators or corepressors) to influence transcription initiation by the RNAPII basal transcription machinery at the core promoter. Hence, the core promoter and the basal transcription machinery are the ultimate targets of signal-dependent regulator/coregulator complexes [1, 2].
The core promoter is the minimal DNA sequence surrounding the transcription start site (TSS) that is sufficient to direct a low (i.e., basal) level of accurate transcription initiation by the basal RNAPII transcription machinery [1]. The canonical basal transcription machinery for RNAPII is composed of six basal (or “general”) transcription initiation factors: TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH, which interact with core promoter DNA and RNAPII to form the preinitiation complex (PIC). Core promoters are structurally diverse and may contain a number of DNA sequence elements that are bound by the basal transcription factors, and primarily by TFIID and TFIIB, during the assembly of the canonical PIC. Core promoter elements determine both the position of the TSS and the basal activity of the core promoter [1, 3, 4]. Core promoter elements have been most extensively characterized in metazoans and include the TATA box, the Initiator (INR), the TFIIB recognition element (BRE) “upstream” and “downstream” (BREu and BREd), the DPE, the MTE, the DCE, the TCT, and others [4–6]. Except for the TATA box and INR-like sequences, similar elements have not been found in yeast promoters [7]. In metazoans, the TATA box is located at −30 bp upstream of the TSS [8] and is bound by the TATA-binding protein (TBP) subunit of the TFIID complex. TFIID is composed of TBP and up to fourteen TBP-associated factors (TAFs) that form stable complexes in metazoans [9–11]; in contrast, TBP in yeast can be recruited to certain promoters independently of TAFs [12, 13]. TBP binding to the TATA box is the first step in core promoter recognition and leads to a sharp DNA bend that is important for TFIIB recruitment and, hence, for the assembly and stability of the PIC [1, 3, 9, 14]. The BRE elements that flank the TATA box in certain promoters are recognized by TFIIB; this further stabilizes the bent TBP-DNA complex via both TFIIB-TBP and TFIIB-DNA interactions [14, 15]. The INR is located at the TSS (+1) and defines the transcription initiation site independently of, or in synergy with, the TATA box [4]. In contrast, the DCE, MTE and DPE elements are located downstream of the TSS and increase the overall activity of the promoter in synergy with either the TATA box or the INR element [5]. The INR and these downstream core promoter elements are all recognized by different TAFs of the TFIID complex: the INR is bound by TAF1 and TAF2; the DCE is recognized by TAF1; and the MTE and DPE were shown to crosslink to TAF6 and TAF9 [4–6]. Importantly, these elements are not present in all core promoters, and core promoters can be classified according the presence or absence of these elements (e.g., TATA, TATA/INR, INR, INR/DPE, etc.). For instance, the large group of housekeeping gene promoters that are often located within CpG islands do not have recognizable core promoter elements and direct transcription initiation from multiple dispersed TSSs [6]. The TCT element is found at the TSS of a select group of TATA-less genes such as the ribosomal protein genes, and is recognized by the gene-specific TBP-related factor 2 (TRF2) rather than by the canonical TBP/TFIID [16]. TRF2 also binds and stimulates certain DPE-dependent promoters in Drosophila [17]. Often, the members of the metazoan-specific family of TBP-related factors (TRFs) control core promoters of highly-specialized developmental and/or cell type-specific genes that do not depend on the TBP/TFIID basal transcription factor [18–20]. For the vast majority of canonical TBP/TFIID-dependent genes, the core promoter sequence also influences the mechanisms of transcription initiation by RNAPII. This was indicated, for instance, by (i) the differential requirements and functions of TBP and TAFs at TATA-dependent and TATA-less promoters [12, 13, 21–27]; and (ii) the requirement of accessory core promoter-specific cofactors for the TAF-dependent basal activity of mammalian INR-dependent and DPE-dependent core promoters [28–30] and for the TFIID-dependent synergy of TATA and INR elements [28, 29]. Hence, there is no universal mechanism (or general machinery) for specific core promoter recognition and transcription initiation by eukaryotic RNAPII, and this is also true for the canonical TFIID-dependent basal machinery in metazoans, which often relies on auxiliary core promoter-specific cofactors [31].
Besides influencing the basal transcription initiation process and the overall transcription output, increasing evidence also indicates an important regulatory role of the core promoter in the preferential response of specific genes to the regulatory signals of certain activators and enhancers [4, 32]. Many Drosophila enhancers were shown recently to preferentially activate a TCT/TRF2-dependent promoter rather than a TFIID-dependent core promoter [33]. However, enhancers and activators can also differentially stimulate canonical TFIID-dependent transcription in a core promoter-dependent manner. Early observations showed that different TATA box sequences have different abilities to convey the activating signals of certain enhancers and activators in mammalian cells [34, 35] and in yeast [36–38]. The activation domains of VP16 and SP1 fused to the yeast GAL4 DNA-binding domain were also reported to have different core promoter preferences in mammalian cells, although no strict core promoter selectivity was observed since both VP16 and SP1 could significantly activate all core promoter types tested [39]. In contrast, the upstream D′/Elf-1 sequence element of the mouse TdT gene promoter (a TATA-less, INR-containing promoter) was shown to activate selectively INR-containing core promoters (with or without a TATA box) but not a TATA-only core promoter [40, 41]. In Drosophila, the promoter preference of certain enhancers has also long been known [42–44]. Promoter competition experiments demonstrated that the Drosophila AE1 and IAB5 enhancers preferentially activated transcription from the TATA-containing even-skipped core promoter relative to the TATA-less white core promoter. However, there was no strict core promoter selectivity since in the absence of a competing TATA-containing promoter both enhancers could activate the TATA-less white promoter [45]. An enhancer-trapping screen in Drosophila identified three enhancers that selectively activated an INR/DPE core promoter and one enhancer that preferentially activated a TATA/INR core promoter [46]. However, whether the core promoter preference of the TATA/INR-specific enhancer was solely due to the intrinsic function of the TATA box (rather than the INR or the combined/cooperative activities of TATA and INR elements) remained unclear. More recently, investigation of TATA-less DPE-dependent promoters of Drosophila developmental genes identified the enhancer factors Caudal, Relish and Dorsal as activators that preferentially stimulate transcription through the DPE [47–49]. Although each of these activators could also weakly stimulate transcription from a TATA-dependent promoter (TATA/INR), Caudal could not activate a TATA promoter containing an upstream BRE (BREu), indicating that core promoter context and cognate basal transcription mechanisms are important for the extent and selectivity of activation by a specific activator [32, 47]. Natural TATA-selective activating DNA sequences and cognate TATA-specific activators that do not act through an INR-directed mechanism remained to be identified and characterized.
Here, we found that the natural serum-responsive upstream activating sequence (UAS) of the human β-actin gene (ACTB) selectively activates TATA box-dependent but not INR-dependent transcription via a mechanism that involves the serum response factor (SRF) activator and the TATA-binding/bending activity of TBP in live human cells. In contrast, the TATA-binding/bending activity of TBP is not essential for TFIID-dependent transcription of the endogenous TATA-less CDKN1A gene. We further show that activation by the ACTB UAS is unidirectional on TATA but bi-directional on TATA/INR core promoters, revealing a stereospecific INR dependency for activation by a natural SRF-responsive UAS. Consistent with these results, computational analyses further indicate that SRF-regulated promoters of the actin cytoskeleton and contractile gene family often contain a TATA box. Our results underscore the regulatory role of the core promoter and support the idea that specific core promoter elements facilitate the coordinated regulation of specific families of genes with related biological functions, and demonstrate the existence of distinct TBP/TFIID-mediated mechanisms of transcription initiation at TATA-dependent and TATA-independent promoters in mammalian cells.
2. Materials and methods
2.1 Plasmid DNAs
The ACTB promoter-luciferase construct was previously described [29]. The ACTB[+INR] luciferase construct was constructed using the QC-PCR kit (Stratagene) to mutate the transcription start site (TSS) region of ACTB construct into a consensus mouse TdT initiator using primer: 5′-GGCGGCGCGACGCGCCctcattctCGAGACCGCGTCCGCCCC-3′(mutated sequence in bold) according to manufacturer’s instructions. The HSV TK luciferase construct has was described previously [50]. The HSV TK[+INR] was constructed, using the QC-PCR kit (Stratagene) as above, to mutate the TSS region of HSV TK into a consensus mouse TdT Initiator with the primer: 5′-GTGACGCGTGTGGCCTCCctcattctGCGACCCTGCAGCGACC-3′. The G5-TATA, G5-TATA/INR and G5-INR luciferase constructs were generated by inserting a HindIII DNA fragment containing the five Gal4-binding sites and the core promoter from pG5TdT(-41TATA/+33), pG5TdT(-41TATA/Inr+33) and pG5TdT(-41Inr/+33) [28] into the HindIII site of pGL3-basic luciferase expression vector (Promega). The mammalian expression vectors for Gal4(1–147), Gal4-SP1(132–243), Gal4-CTF1(399–499) and Gal4-VP16(411–490) were provided by Dr. Nicolas Mermod [51]. The upstream activating sequence of human ACTB promoter (−120 to −40 bp), UASACTB, was amplified by PCR (primers: 5′-TCTAGTggtaccGCGAAGCCGGTGAGTGAGCG-3′ and 5′-TACATAggtaccGCGGCCGCTCGAGCCATAAAAGGC-3′) and inserted into the KpnI site of TATA, TATA/INR or INR core promoter-luciferase plasmids [29] in the forward or reverse direction to generate UASACTB-TATA, UASACTB-TATA/INR, UASACTB-INR and their reverse constructs. The mut.CCAAT, mut.CArG, mut.Both, 5xCCAT, 5xCArG and 5xGC constructs were generated by inserting synthetic DNA oligonuleotides into the KpnI site of TATA, TATA/INR and INR core promoter-luciferase plasmids (see all the synthetic oligonucleotide sequences in Supplementary Materials and Methods). The CDKN1A-Luciferase reporter was the p21P plasmid described previously [52]. All constructs were confirmed by DNA sequencing.
2.2 Cell culture, transient transfection, luciferase assay, and RNAi
HEK293 cells, HeLa S3 cells, and the derivative HeLa S3+T210K cell line described previously [22] were cultured in Dulbecco’s modified Eagles medium supplied with 10% fetal bovine serum and 5% CO2 at 37°C. Transient transfections of HEK293 cells and HeLa S3 cells were performed with Lipofectamine 2000 (Invitrogen), and luciferase assays were performed as described previously [53]. Cells in a 6-well plate were transfected with luciferase reporter plasmids (0.8 μg to 2 μg) and 0.2 μg of pCMV-β-Galactosidase internal control plasmid. Luciferase assays were performed 20 to 48 hours after transfection. Luciferase activities were analyzed in triplicates from three or more independent transfections and were normalized to β-Gal activity and to the luciferase activity of cells transfected with the promoter-less pGL3-basic vector (arbitrarily set to 1.0). Control siRNAs were purchased from Ambion (AM4635#1 or AM4637#2). SRF and NFYB siRNAs were previously described: siSRF797 [54] and siNF-YB2 in [55], respectively. The siRNA sequences for siNFYA were 5′-CGUCUAUCAACCAGUUAAUdTdT-3′ and 5′-AUUAACUGGUUGAUAGACGdTdT-3′ for sense and anti-sense strands. The sequences of TBP 3′-UTR siRNA were 5′-GUGACUGUGAGUUGCUCAUdTdT-3′ and 5′-AUGAGCAACUCACAGUCACdTdT-3′ for sense and anti-sense strand. All siRNAs were synthesized by Ambion, except the SP1 siRNA, which was from Cell Signaling (Cat#12104). For TBP knockdown, cells were seeded over night to reach 70% confluency at the time of transfection. 200 nM TBP siRNA was transfected into Hela S3 or Hela S3+T210K cells at time 0 hour. Twelve hours later, 200 nM TBP siRNA was transfected again together with the indicated luciferase constructs. Twenty hours after the second transfection the cells were harvested and half of the cells were used for luciferase assay and the other half for Western blot. Alternatively, to analyze expression of specific endogenous genes total cellular protein and RNA were isolated from cells transfected with control or TBP siRNAs and analyzed by Western blot and RT-qPCR (see below). For the knockdown of all other factors, 200nM siRNA was transfected only once into HEK293 cells together with 0.8 μg of ACTB-Luc. Forty-eight hours after transfection, half of the cells were used for luciferase assay and the other half for Western blotting. Luciferase activities in of Fig. 6 were normalized to total cellular protein.
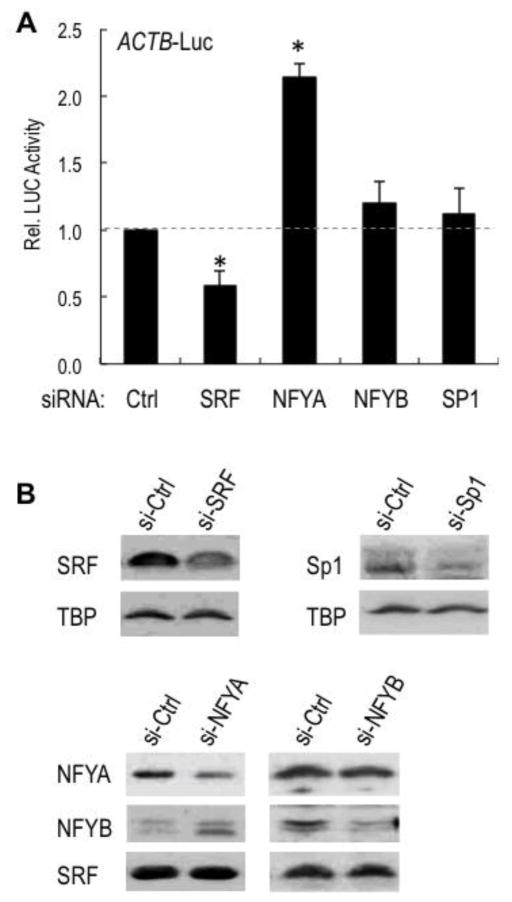
Figure 6. ACTB promoter activity requires SRF but not NFY or SP1.
A. Relative luciferase activity from the natural ACTB promoter-luciferase construct (see Fig. 1A) in cells transfected with control (Ctrl), SRF, NFYA, NFYB or SP1 siRNAs. Luciferase activity was normalized to total protein. Luciferase activity in cells transfected with the siRNA Ctrl was arbitrarily set to 1. Results are the means from three independent experiments. Asterisks indicate significant differences (P<0.05) relative to Ctrl.
B. Western blot analysis of SRF, NFYA, NFYB and SP1 proteins in extracts of cells transfected with the control siRNA (si-Ctrl) and specific siRNAs as indicated.
2.3 Antibodies and Western blots
The antibodies SRF(sc-335), NFYB(sc-13045x), NFYA(sc-10779x), SP1(sc-59x), CRSP70(sc-48776) and TAF9/TAFII31(sc-1248) were purchased from Santa Cruz Biotechnology. Vinculin (V4505) monoclonal antibody was purchased from Sigma. Rabbit polyclonal antibodies against TBP, TAF4/TAFII135 and TAF5/TAFII100 were gifts from Robert G. Roeder. Western blots were performed essentially as previously described [53].
2.4 In Vitro Transcription and Primer Extension
Transcription reactions with HeLa nuclear extracts and primer extension were described previously [28]. The extension primer for luciferase transcripts was described previously [29]. Total RNA was purified from HEK293 cells using Qiagen RNeasy kit (Cat# 74104). Ten μg of total cellular RNA was used in primer extension analyses as previously described [29]. The X-ray films were scanned with HP Precisionscan Pro 3.1 scanner and signals were quantitated with NIH ImageJ.
2.5 Reverse transcription and real-time quantitative PCR (RT-qPCR)
Total cell RNA from three biological replicates was purified as above. Reverse transcription and real time PCR were performed with the BioRad iScript™ Reverse Transcription Supermix (#170–8840) and iQ™ SYBR® Green Supermix (#170–8882) per manufacturer’s instructions. Real time PCR was performed in triplicates on a BioRad MiniOpticon system. PCR primers (forward and reverse) were ACTB mRNA: 5′-TGACGGGGTCACCCACACTGTGCCCA-3′ and 5′-CTAGAAGCATTTGCGGTGGACGATGGAGGG-3′; ACTB pre-mRNA: 5′-GGCACCACACCTTCTACAATG-3′ (exon) and 5′-CCACCAGAAGAGGTAGCGGG-3′ (intron); MYC mRNA: 5′-CGACGCGGGGAGGCTATTCTGC-3′ and 5′-CCCGCCACCGCCGTCGTTGTCT-3′; and CDKN1A mRNA: 5′-GGCAGACCAGCATGACAGATT-3′ and 5′-GCGGATTAGGGCTTCTCTTT-3′. Relative expression of specific mRNAs in siRNA-treated cells was normalized to cells transfected with the control siRNA and was obtained using the Pfaffl method. The normalized means (±S.D.) are presented for three independent experiments.
2.6 Statistics and computational analyses of core promoters
Significant differences (P<0.05) in gene expression analyses were analyzed by two-tailed Student’s t-test of at least three independent experiments. Global frequencies of 9010 human core promoters and 7995 conserved orthologous mouse core promoters with experimentally validated transcription start sites were reported previously [56] and used to classify the core promoters of SRF target genes (below) into TATA-containing and TATA-less groups. All SRF target genes were obtained from previous ChIP-seq analyses in mouse fibroblasts [Table 5 in [57]]. The list of SRF target genes of the cytoskeleton-contractile family was obtained from a previous report [Table 1 in [58]]. Significant deviations from the global TATA-containing/TATA-less core promoter frequencies for all SRF target genes and for the specific group of SRF-associated cytoskeleton-contractile genes were analyzed by Chi-square test.
3. Results
3.1 The ACTB gene upstream activating sequence (UAS) stimulates TATA box-dependent but not Initiator-dependent transcription
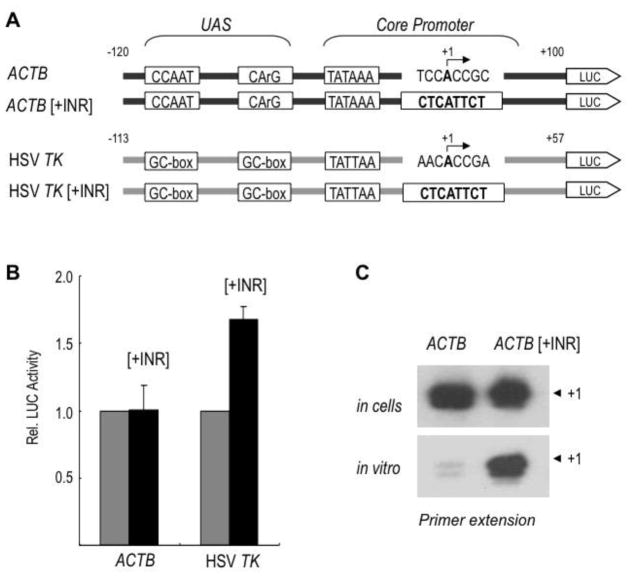
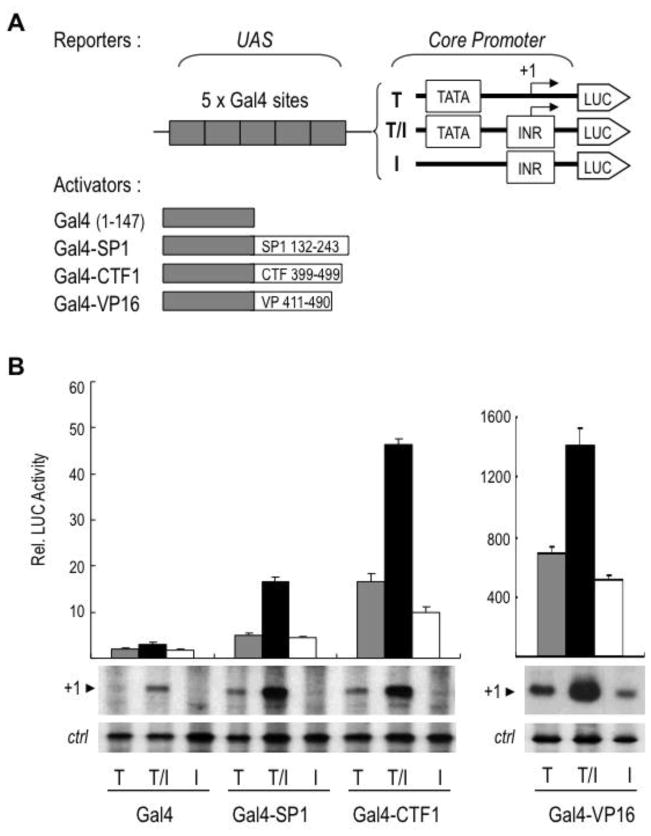
In an attempt to generate a highly active ACTB promoter for reporter gene assays, we introduced by site-directed mutagenesis a perfect Initiator (INR) element at the transcription start site downstream of the TATA box of the ACTB promoter in the context of a luciferase reporter plasmid (Fig. 1A). To our surprise the activity of the INR-containing ACTB promoter (ACTB[+INR]) in transfected HEK293 cells was not stimulated by the INR, whereas the activity of the TATA-containing HSV TK promoter was, as expected, increased by the INR (Fig. 1B). Notably, the INR sequence was functional and stimulated transcription in the context of the ACTB promoter (ACTB[+INR]) in transcription experiments in vitro using HeLa cell nuclear extracts (Fig. 1C). We reasoned that the different behavior of the ACTB[+INR] promoter in transfected cells and in vitro could be due to the differential activity of the ACTB upstream activating sequence (UAS) and associated activators/coactivators in cells vs. in nuclear extracts (see further below). Hence, these results suggested the possibility that the UAS of ACTB and associated sequence-specific activators could be unable to stimulate INR-dependent transcription under more physiological conditions in live cells. This was highly intriguing since, to our knowledge, all activators tested so far in mammalian cells support INR-dependent transcription to some extent. We further investigated this in our cell system by analyzing the ability of distinct activation domains fused to the yeast Gal4 DNA-binding domain to activate three core promoters that differed by only the presence or absence of a TATA and/or INR element: i.e., TATA-only (T), INR-only (I), and combined TATA/INR (T/I) core promoters (Fig. 2A). Consistent with previous observations, the glutamine-rich activation domain of SP1, the proline-rich activation domain of CTF1, and the acidic activation domain of the viral VP16 activator were all able to activate these three types of core promoters, and the activity of the composite T/I core promoter was always the highest, as expected (Fig. 2B; and Supplemental Fig. S1A). We then substituted the Gal4 UAS sequences with the UAS of ACTB (−120/−40), which include a CCAAT box and a CArG box, upstream of the three heterologous core promoters (T, T/I and I; Fig. 3, top) and analyzed their activities similarly. As shown in Fig. 3 (bottom, WT lanes 1–3), the UAS of ACTB did not significantly activate the TATA-less INR-only (I) core promoter and activated TATA-only (T) and composite TATA/INR (T/I) core promoters to a similar extent, as measured by both the relative luciferase activities and the levels of correctly initiated transcripts (+1) by primer extension. Similar results were obtained in HeLa cells indicating that this core promoter selectivity is not cell type-specific (Supplemental Fig. S1B). These results demonstrated the requirement of a TATA box for activation by the UAS of ACTB and the inability of these activating sequences and cognate activator/coactivators to stimulate an INR-dependent transcription pathway.
Figure 1. An INR cannot stimulate transcription from the human ACTB promoter in vivo.
A. Schematic diagrams of luciferase reporters containing the natural ACTB promoter (−120/+100 bp) or the natural HSV TK promoter (−113/+57bp) and derivatives [+INR] containing a consensus INR sequence (CTCATTCT) indicated in bold at the transcription start site (bent arrow; +1). The TATA box sequence of ACTB is TATAAA and that of HSV TK is TATTAA. The upstream activating sequence (UAS) of ACTB contains two essential elements: a CCAAT box and a CArG box. The UAS of HSV TK promoter contains two essential GC-box elements.
B. Relative luciferase activities of the constructs in panel A transfected into HEK293 cells. Activities of ACTB-Luc and HSV-TK-Luc lacking an INR (gray bars) were arbitrarily set to 1. Results are the means ±S.D. of three independent experiments.
C. Primer extension analyses of RNAs transcribed from the ACTB-Luc or ACTB[+INR]-Luc reporters transfected in HEK293 cells (top panel) or after in vitro transcription in a HeLa nuclear extract (bottom). Correctly initiated transcripts are indicated by “+1”.
Figure 2. The INR stimulates transcription activation by different types of activation domains in vivo.
A. Schematic diagram of promoter-luciferase reporters and Gal4-fusion activators. Reporters contain five Gal4 binding sites upstream of TATA (T), TATA/INR (T/I) or INR (I) core promoters. Gal4(1–147) is the yeast Gal4 DNA binding domain (amino acids 1–147). The activation domains of SP1 (132–243), CTF1 (399–499) and VP16 (411–490) were fused to Gal4(1–147) to generate Gal4-SP1, Gal4-CTF1 and Gal4-VP16 fusion activators, respectively.
B. The reporters above were co-transfected with Gal4(1–147), Gal4-SP1, Gal4-CTF1 or Gal4-VP16 in HEK293 cells. Luciferase activities are relative to the activity of the promoter-less pGL3 luciferase vector, which was arbitrarily set to 1, and are the means ±S.D. of three independent experiments. The basal activities without any activator are shown in supplemental Fig. S1A. Bottom panels show autoradiograms of representative primer extension analyses indicating correctly initiated transcripts (+1) in transfected cells, including an endogenous cellular transcript used as an internal control (ctrl). The autoradiogram for Gal4-VP16 resulted from a shorter X-ray film exposure.
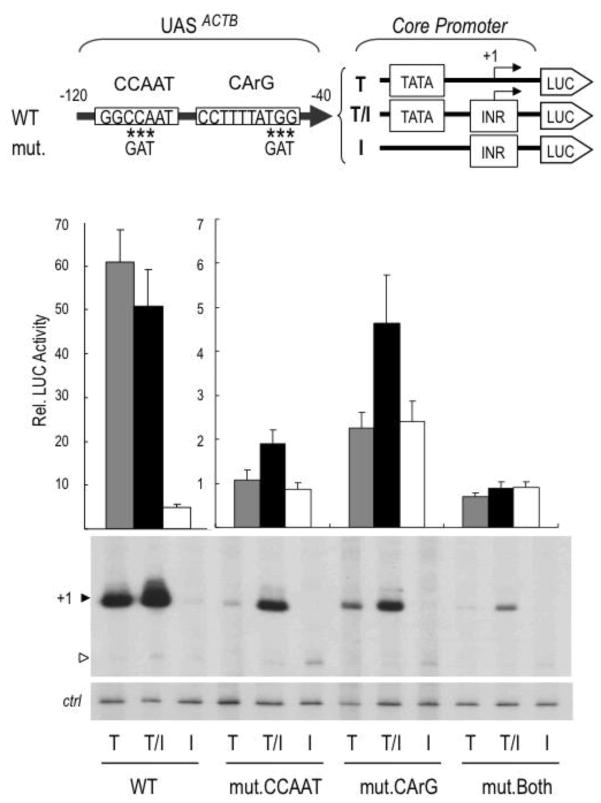
Figure 3. The UAS of ACTB is composed of a CCAAT box and a CArG box that synergistically activate TATA-dependent but not INR-dependent transcription.
Top, are diagrams of the reporters containing the wild type (WT) UAS of ACTB (−120 bp to −40 bp) or mutated (mut.) derivatives upstream of heterologous TATA (T), TATA/INR (T/I) or INR (I) core promoters. The specific nucleotides substituted in the CCAAT-box (mut.CCAAT), CArG-box (mut.CArG) or both boxes (mut.Both) are indicated with asterisks. Bottom, are the relative luciferase activities of wild type and mutant reporters transfected into HEK293 cells (n ≥ 3 independent experiments). The Luc activities are relative to that of the promoter-less pGL3 luciferase vector, which was arbitrarily set to 1. Note the two different scales for the Rel. Luc Activities of WT and mutated ACTB UAS reporters. A representative primer extension analysis is also presented showing the correctly initiated transcripts (closed arrowhead; +1). A low level of incorrectly initiated transcription is seen mostly in the INR (I) lanes (open arrowhead). An endogenous cellular transcript served as internal control (ctrl), as in Fig. 3.
The UAS of ACTB contains a CCAAT box and a CArG box, which were shown previously to be both important for the activity of the promoter [59–61]. To determine which of these elements is responsible for the TATA-selectivity and INR-independence of the UAS, we introduced mutations in each of these elements that impair binding of cognate transcription factors NFY and SRF [61] and analyzed the activity and core promoter selectivity of the mutated UAS, as above. As expected, mutation of either the CCAAT box (mut.CCAAT) or the CArG box (mut.CArG) strongly decreased the activity of the UAS from both TATA-only (T) and composite TATA/INR core promoters (T/I), while the TATA-less INR-only (I) core promoter remained inactive (Fig. 3 bottom). Notably, there was a residual activity of the mut.CCAAT and of the mut.CArG UAS above the basal activity of the promoters containing both elements mutated (mut.Both). Interestingly, although the residual activation by the mut.CCAAT and the mut.CArG UAS was still TATA-dependent (i.e. the TATA-less INR promoter was not stimulated), the INR was now required in addition to the TATA-box for efficient transcription (Fig. 3, bottom panel). To further analyze the intrinsic activity of the CCAAT box and CArG box separately and independently of natural flanking UAS sequences, 5 copies of each of these elements were cloned upstream of the different core promoters (Supplemental Fig. S2). We found that 5 copies of the CArG box by itself were not sufficient for any significant activation, while 5 copies of the CCAAT box activated transcription but were not sufficient for high levels of activation from the TATA-only promoter and required the INR element for increased activation; this was also observed for 5 copies of an SP1/GC-box and for activation by all Gal4 fusion activators, which activated transcription from all promoters as shown above (Fig. 2). This result is also consistent with previous observations that CCAAT boxes bound by NFY can activate both TATA-dependent and TATA-less promoters [62, 63]. In vitro transcription experiments with HeLa nuclear extracts and the above mutant UAS constructs confirmed that the CCAAT box (but not the CArG box) is sufficient for a low level of activated transcription and that HeLa nuclear extracts lack a cofactor for TATA-specific activation by the UAS of ACTB (Supplemental Fig. S3). Thus, we conclude that it is not the individual activities of each element/box but rather their concerted functions (perhaps also involving additional immediately adjacent sequences) that are responsible for the TATA-specific and INR-independent activity of this UAS.
3.2 Orientation-independent transcription activation by the UAS of ACTB is core promoter-specific and requires an INR element
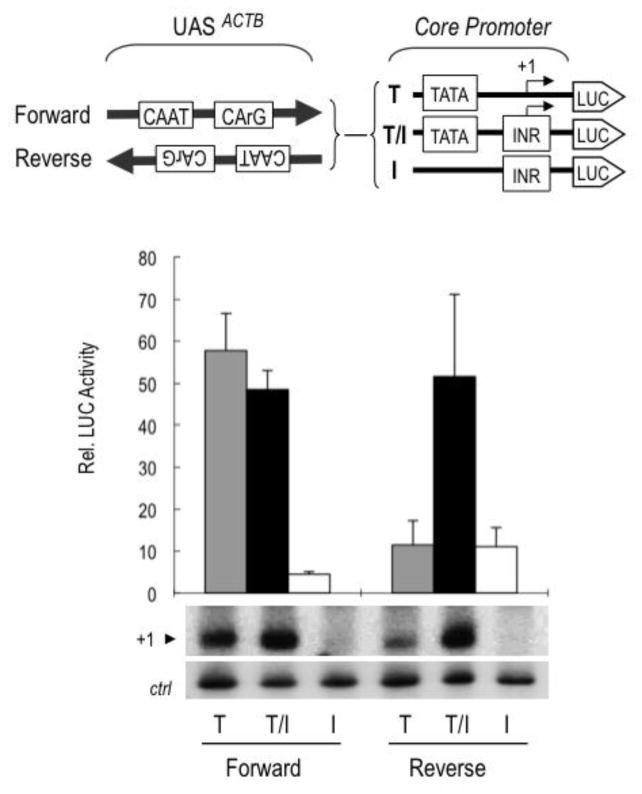
Our data above indicated that the CCAAT box and CArG box in the UAS of ACTB function cooperatively in core promoter selection, possibly as an enhancer-like unit. Indeed it was shown that the CCAAT box and the CArG box need to be on the same face of the DNA helix for UAS function, whereas the activity of the UAS was unaffected by increasing its distance form the TATA box by full or half helical turns [61]. To further address whether the UAS of ACTB has enhancer-like properties, we tested its ability to activate transcription when cloned in both orientations upstream of the different heterologous core promoters. As described above, in its natural “Forward” orientation the UAS activated the TATA and composite TATA/INR core promoters to a similar extent (Fig. 4; Forward, lanes T and T/I respectively) but not the TATA-less INR promoter (Forward, lane I). In contrast, in the “Reverse” orientation, activation of the TATA-only promoter was drastically reduced to ~20%, while efficient activation of the composite TATA/INR core promoter was retained (Fig. 4; Reverse, lanes T and T/I, respectively). We conclude that the UAS of ACTB does not have the orientation-independent properties of enhancers for TATA-only core promoters, but that it does have such bi-directional activity on selected core promoters having both TATA and INR elements. Thus, core promoter-selective activation is not only dictated by the identity and cooperativity of the DNA regulatory elements and cognate sequence-specific activators, but also by their stereo-specific arrangement relative to the core promoter. These observations further suggest the possibility that composite core promoters, containing both TATA and INR elements, could be targets for activation by a broader spectrum of regulatory and enhancer sequences.
Figure 4. Transcription activation by the ACTB UAS is unidirectional on TATA but bidirectional on a composite TATA/INR core promoter.
The top part shows the luciferase reporters containing the UAS of ACTB (−120 to −40 bp) inserted upstream of heterologous TATA, TATA/INR or INR core promoters in forward or reverse orientation. The lower part shows the luciferase activities of the above reporters in transfected HEK293 cells. Luciferase activities are the means of three independent experiments. Primer extension analyses of correctly initiated Luc transcripts (+1), including an internal control (ctrl) transcript (as in Figs. 2 and 3), are shown at the bottom.
3.3 The TATA-binding/bending activity of TBP is important in vivo for the TATA-dependent promoters of ACTB and MYC but not for the TATA-independent CDKN1A promoter
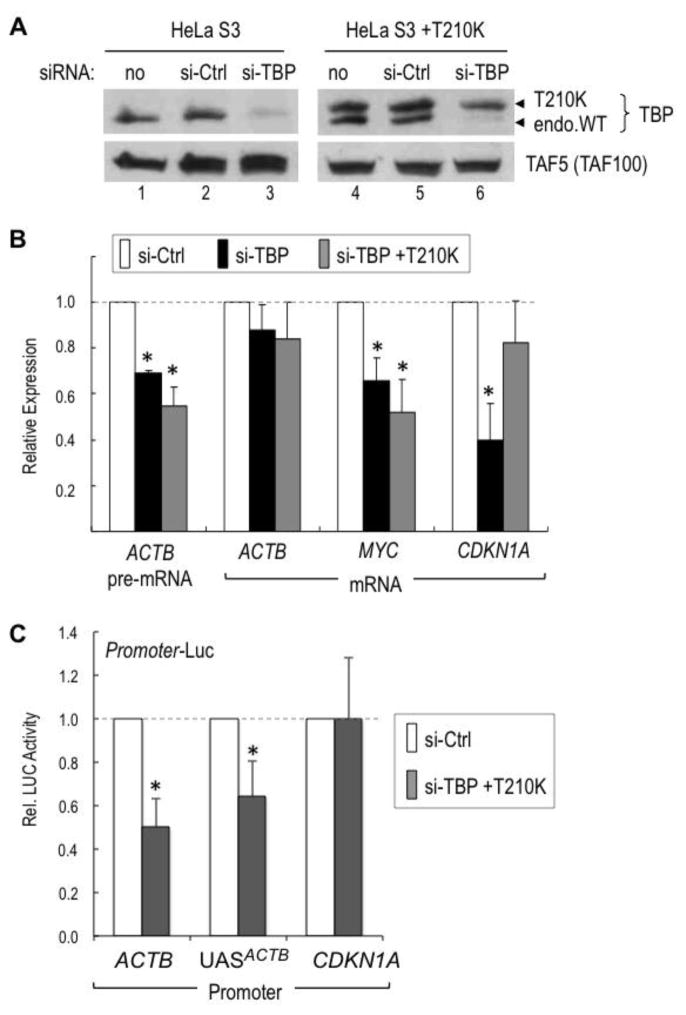
Having established the requirement of the TATA box for the activity of the UAS of ACTB in transfection/reporter gene assays, we then investigated the importance of such TATA box-dependent pathway for transcription of the endogenous ACTB gene in human cells. We used the HeLa S3 cell line and a derivative (HeLa S3+T210K) that expresses similar physiologic levels of both the endogenous wild type TBP and an ectopic mutant TBP (TBP-T210K) carrying the T210K amino acid substitution within its TATA-binding domain [22]. The T210K substitution (equivalent to the T112K mutation in yeast TBP) is the most radical TBP mutation reported in terms of abolishing the TATA-specific DNA-binding activity of TBP [27, 64–66], and also prevents TBP-mediated DNA bending [67], but does not affect TBP interaction with TAFs and with other components of the basal transcription machinery [22, 66]. To test the requirement of TBP and of the TBP-TATA interaction for endogenous ACTB gene transcription in HeLa cells, TBP expression was knocked down by transient transfection with a specific siRNA that targets the 3′-UTR of human TBP mRNA. This siRNA efficiently and selectively reduced endogenous TBP protein levels in wild type HeLa S3 cells and in HeLa S3+T210K cells (Fig. 5A, lanes 3 and 6, endo. WT; see also supplemental Fig. S4) but did not affect the expression of the ectopic mutant TBP-T210K (Fig. 5A, lane 6, T210K). This provided us with an assay to determine whether replacement of endogenous TBP with TBP-T210K affects transcription of ACTB and other genes. After knocking down TBP in wild type cells we analyzed both steady-state ACTB mRNA and pre-mRNA levels by reverse transcription and quantitative real-time PCR (RT-qPCR). ACTB steady state mRNA levels were not affected by the transient knockdown of TBP, probably due to the high abundance and/or stability of ACTB mRNA; in contrast, ACTB pre-mRNA levels, which better reflect de novo transcription, were reduced significantly and to similar extents in both wild type HeLa S3 cells and in HeLa S3+T210K cells (Fig 5B, ACTB black and gray bars, respectively). Thus, the TBP-T210K mutant protein could not rescue transcription of ACTB in cells depleted of endogenous TBP, confirming the requirement of the TBP-TATA interaction in vivo for endogenous ACTB gene transcription. Similar results were also obtained for the TATA-containing MYC gene. In contrast, TBP-dependent transcription of the TATA-less CDKN1A gene was rescued by the mutant TBP-T210K (Fig. 5B, MYC vs. CDKN1A). Consistent with these results, promoter-luciferase assays in TBP knockdown cells indicated that the activities of the wild type ACTB promoter and of its UAS cloned upstream of a heterologous TATA-containing core promoter could not be rescued by the TBP-T210K mutant, while the activity of the CDKN1A promoter was rescued by TBP-T210K (Fig. 5C). These results demonstrate that the in vivo TBP-TATA interaction is important for activation of the ACTB and other TATA-dependent promoters but is dispensable for TBP-dependent transcription from at least some TATA-less (or TATA-independent) promoters in mammalian cells.
Figure 5. The TATA-binding activity of TBP is required in vivo for the TATA-containing ACTB and MYC promoters but not for the TATA-less CDKN1A promoter.
A. Western blot analyses of endogenous TBP knockdown in Hela S3 and HeLa S3+T210K cell lines. The top stripe was probed with a TBP antibody. Endogenous TBP wild type (endo.WT) and HA-tagged TBP-T210K mutant (T210K) proteins are indicated with arrowheads. The bottom stripe was probed with a TAF5/TAF100 antibody. No siRNA (lanes 1 and 4), a control siRNA (lanes 2 and 5) or the TBP 3′-UTR siRNA were transfected into both cell lines, as indicated. TBP knockdown under the conditions used (see Materials and Methods) did not affect expression of TAFs or other proteins (see also Supplemental Fig. S4).
B. Expression levels of ACTB pre-mRNA, and ACTB, MYC and CDKN1A mRNAs were quantitated by RT-qPCR in HeLa S3 (black bars) and HeLa S3+T210K (gray bars) cells after transfection with the TBP siRNA (si-TBP) and are relative to the levels of expression of each gene transcript in cells transfected with the control siRNA (si-Ctrl; white bars), arbitrarily set to 1. The asterisks indicate significant differences relative to si-Ctrl transfected cells (P<0.05).
C. Relative activities of promoter-luciferase constructs containing the natural ACTB or CDKN1A promoters or UAS-ACTB-TATA. Luciferase reporters were transfected into Hela S3+T210K cells in the conditions of mock or knockdown of endogenous TBP. Luciferase activity was averaged from three independent experiments. Luciferase activity from si-Ctrl samples was arbitrarily set to 1.
3.4 Serum response factor (SRF)-dependent actin cytoskeleton and contractile genes tend to have TATA-containing core promoters
Transcription activators that stimulate selectively TATA-dependent but not INR-directed transcription have not yet been described. The identification of the UAS of ACTB as a TATA-specific activating unit suggested a role for the DNA-binding factors SRF and NFY, which were shown originally to bind, respectively, the CArG box and CCAAT box sequences of this UAS in vitro [61]. These sequences are also bound in vivo by SRF and NFY in a variety of human cell lines, including in HeLa S3 cells, as indicated by the ENCODE transcription factor ChIP-seq datasets at UCSC Genome Browser. In addition, a GC box partially overlapping with the CCAAT box is also bound by SP1 in a subset of cell lines (ENCODE data not shown). While the binding of SRF to the CArG box and SRF-mediated activation of mammalian ACTB genes in response to serum-induced signaling is now well established [57, 58], the role of NFY or SP1 is still unclear. As expected, siRNA-mediated knockdown of SRF reduced the activity of the ACTB promoter (Fig. 6A and B). However, knockdown of two NFY subunits (NFYA and NFYB) or SP1 did not inhibit the ACTB promoter (Fig. 6A and B), while a control 5xGC-box reporter was inhibited (data not shown). Surprisingly, knockdown of NFYA but not NFYB increased ACTB promoter activity, suggesting a possible repressive role of NFYA. Thus, SRF and a still elusive cooperating factor(s) that binds this GC/CAAT box (Fig. 3) and appears to be distinct from conventional NFY or SP1 is required for the TATA-selective activity of the UAS of ACTB.
To determine whether SRF is more generally involved in regulation of TATA box-dependent transcription, we analyzed the promoters of all SRF target genes identified by ChIP-seq in mouse fibroblasts [57]. We only considered genes with experimentally validated transcription start sites and well characterized core promoters classified previously as either TATA-containing or TATA-less [56]. As shown in Table 1, the global frequency of TATA-containing core promoters in mouse and human genes was found to be only ~16% [56]. In contrast, by the same criteria about 22% of all SRF target gene promoters in mouse fibroblasts contain a TATA box. Although this moderate enrichment was statistically significant (P=0.0024), it also suggested that only a fraction of SRF-regulated genes might be TATA-dependent, as expected from the fact that SRF cooperates with other factors to elicit the TATA selectivity at the UAS of ACTB. Hence, we then focused our analysis more specifically on the promoters of the actin-related group of SRF-regulated genes having cytoskeleton or contractile functions since SRF is a well established master regulator of the actin cytoskeleton [58]. Interestingly, a larger fraction, i.e., about 40% (38% in human and 43% in mouse) of the core promoters for these cytoskeleton-contractile genes have been classified as TATA-containing (Table 1). This is a substantial and statistically very significant enrichment compared to the low global frequency of TATA-containing promoters in the genome (P<10−7). Altogether these results show that SRF has a regulatory bias for TATA-containing promoters genome-wide and, more specifically, for its targeted group of genes encoding the actin cytoskeleton and contractile apparatus.
Table 1.
Frequencies of core promoter types for mammalian SRF target genes
| Core promoter type | All | TATA-containing | TATA-less |
|---|---|---|---|
| Global frequencies : All Characterizeda | |||
| Human | 9010 (100%) | 1483 (16.5%) | 7527 (83.5%) |
| Mouse | 7995 (100%) | 1304 (16.3%) | 6691 (83.7%) |
| SRF target genes (Mouse): Allb | 476 (100%) | 103 (21.7%)* | 373 (78.3%)* |
| SRF target genes: Cytoskeleton-contractilec | |||
| Human | 97 (100%) | 37 (38.1%)** | 60 (61.9%)** |
| Mouse | 82 (100%) | 35 (42.7%)*** | 47 (57.3%)*** |
Asterisks indicate significant deviation from global genomic frequencies analyzed by Chi-square test:
P=2.4 E-3;
P =2.7 E-8;
P =4.4 E-10 .
Global frequencies of core promoter types for human and mouse orthologous genes with experimentally validated transcription start sites (Jin et al., 2006).
All SRF target genes identified by ChIP-seq in mouse fibroblasts (Esnault et al., 2014) with characterized core promoters in Jin et al., 2006.
Cytoskeleton-contractile SRF target genes (from Miano et al., 2006) with characterized core promoters in Jin et al., 2006.
4. Discussion
The importance of core promoter DNA elements in regulation of transcription by upstream promoter-bound activators and distal enhancers has long been proposed. However, only few natural cis-acting DNA sequences and trans-acting factors with core promoter selectivity have been reported and core promoter-specific mechanisms for transcription initiation by the canonical TBP/TFIID-dependent basal transcription machinery have remained poorly documented in vivo (see introduction). Here, we have demonstrated the importance of a TATA box, and of the TATA-binding/bending activity of TBP, for in vivo activation by the SRF-dependent UAS of the human ACTB gene and the inability of these activating sequences to stimulate transcription through an INR-dependent mechanism. To our knowledge, this is the first report of a TATA box-selective activating sequence and cognate activator. Our results further indicate that it is not simply the individual activities of the SRF-bound CArG box or the adjacent essential GC/CAAT box but rather their concerted functions, as a unit, that are responsible for the TATA-specific and INR-independent activity of this UAS. However, the GC/CAAT box-associated factors that cooperate with SRF remain to be identified. Indeed, although SP1 and NFY factors bind this GC/CAAT box in various cell types (from data of the ENCODE project), our RNAi analyses suggest that neither SP1 nor the conventional NFY trimeric complex (composed of the NFYA, B and C subunits) is required for activation by this UAS. Instead, knockdown of NFYA stimulated the activity of the UAS, while knockdown of NFYB had no apparent effect, suggesting a repressive function of NFYA and dispensability of NFYB at the ACTB UAS. This would be consistent with the reported dual positive and negative transcription activities of NFY [68], and with previous observations indicating that the activities of NFYA and NFYB do not always overlap since these two NFY subunits are required for regulation of not only common but also distinct target genes, and have different cellular functions [55, 69].
Although activation by the ACTB UAS is not influenced by the presence or absence of an INR element in the core promoter, we found that an INR is required for efficient activation when the UAS is in the non-native reverse orientation relative to the core promoter. Hence, it is not only the nature of the activating sequences and cognate DNA-binding factors but also their stereospecific alignment relative to the core promoter that influences the requirement of specific core promoter elements for efficient activation. These observations have implications for the functional definitions of enhancer sequences, which are characterized by their ability to activate transcription of heterologous promoters in a position- and orientation-independent manner. The ACTB UAS was shown previously to activate transcription of heterologous TATA-containing promoters at varying and long distances from the promoter [61]. We now show that if the core promoter also contains an INR, the UAS of ACTB can function in an orientation-independent manner as well. Thus, UAS sequences such as the one in ACTB may function as enhancers for only certain core promoters, and composite core promoters that contain both TATA and INR elements could be targets for activation by a broader spectrum of regulatory and enhancer sequences. It is also interesting to note that more than half of mammalian promoters are bidirectional and drive divergent transcription from sense and upstream anti-sense core promoters that are separated and controlled by a common UAS sequence [70, 71]. We propose that bidirectional transcription at those promoters is dependent on the compatibility of the shared UAS and cognate activators/coactivators with both the sense and upstream antisense core promoter elements.
SRF binds thousands of sites in the mammalian genome and activates a variety of genes in response to serum-induced intracellular Rho-actin or Ras signaling pathways, and functions via concerted interactions with signal-dependent cofactors (MRTFs or TCFs) and a variety of other cooperating factors that interact with adjacent DNA motifs. Hence, the mechanisms of SRF-dependent activation are gene-/context-specific [57, 58]. Accordingly, we found that the core promoter structure of mammalian SRF target genes is also diverse, albeit with a moderate bias for TATA-containing promoters when SRF target genes are analyzed globally. However, SRF is a well established master regulator of the specific family of genes encoding the actin cytoskeleton and contractile apparatus [58], and we found that ~40% of the core promoters for these genes contain a TATA box, which is a significant enrichment compared to the low overall frequency of TATA-containing promoters in human and mouse genomes (Table 1). These results suggest that a TATA box-dependent transcription mechanism is essential for SRF regulation of the actin cytoskeleton/contractile-related gene family and support the idea that core promoter elements play a role in the coordinated regulation of specific families of functionally related genes or genes networks [6, 32].
We demonstrated that the native ACTB gene and a heterologous TATA-containing promoter controlled by the SRF-dependent UAS of ACTB require TBP and its TATA-binding/bending activity in vivo. During these analyses we found that the TATA-containing MYC gene behaved similarly but that the TATA-less CDKN1A gene functioned differently. Native CDKN1A and an artificial CDKN1A promoter-luciferase construct were transcribed in a TBP-dependent manner in human cells but did not rely on the TATA-binding/bending activity of TBP (which was inactivated by the T210K substitution in TBP). Similarly, it was shown recently in yeast that several TBP mutants defective in TATA binding were unable to sustain expression of the TATA-containing HIS4 and SNZ1 genes but could direct transcription of the ribosomal protein genes RPS5 and RPL5 that do not have a consensus TATA element [27]. These findings together with previous evidence indicating that TBP can be recruited to promoters via alternative TAF-dependent mechanisms [21–23, 25, 26, 37, 38] suggest that the in vivo TBP-TATA interaction (including TBP-induced DNA bending) is critical for activation of TATA-dependent promoters but is dispensable for TBP-dependent transcription from at least some TATA-less (or TATA-independent) promoters in all eukaryotes. Hence, the canonical TBP-TATA binding/bending mechanism for PIC assembly characterized on selected TATA-containing promoters in vitro is not universal in vivo. It will be important to determine what fraction of yeast and metazoan genomes do in fact depend on the TATA-binding/bending activity of TBP, especially since most metazoan promoters lack a TATA-like element.
Supplementary Material
BBA Highlights.
The ACTB upstream activating sequence (UAS) only activates TATA-containing promoters
Serum response factor (SRF) and the TATA box-binding activity of TBP are important
An Initiator (INR) core element confers bi-directional activity to the UAS of ACTB
SRF regulates the cytoskeleton/contractile gene family via TATA-containing promoters
Gene-specific regulation involves matching regulatory and core promoter elements
Acknowledgments
We would like to thank Dr. Joseph Dhahbi and Dr. Haimao Zhan for their assistance with the computational and statistical analyses; Drs. Nicolas Mermod, Robert G. Roeder, and Xiao-Fan Wang for generous gifts of reagents; and E.M. lab members for their encouragement. This work was supported by a grant from the National Science Foundation [MCB-1021696]. E.G.H. was supported by a MARCU-STAR training grant from the National Institutes of Health [T34GM062756 to E.M.].
Footnotes
Supplementary data to this article can be found online at…
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 2.Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat Struct Mol Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 5.Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter - the gateway to transcription. Curr Opin Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kadonaga JT. Perspectives on the RNA polymerase II core promoter. Wiley Interdiscip Rev Dev Biol. 2012;1:40–51. doi: 10.1002/wdev.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang C, Bolotin E, Jiang T, Sladek FM, Martinez E. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene. 2007;389:52–65. doi: 10.1016/j.gene.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldberg ML. Sequence analysis of Drosophila histone genes. Stanford University; California: 1979. [Google Scholar]
- 9.Martinez E. Multi-protein complexes in eukaryotic gene transcription. Plant Mol Biol. 2002;50:925–947. doi: 10.1023/a:1021258713850. [DOI] [PubMed] [Google Scholar]
- 10.Papai G, Weil PA, Schultz P. New insights into the function of transcription factor TFIID from recent structural studies. Curr Opin Genet Dev. 2011;21:219–224. doi: 10.1016/j.gde.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tora L. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 2002;16:673–675. doi: 10.1101/gad.976402. [DOI] [PubMed] [Google Scholar]
- 12.Li XY, Bhaumik SR, Green MR. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science. 2000;288:1242–1244. doi: 10.1126/science.288.5469.1242. [DOI] [PubMed] [Google Scholar]
- 13.Kuras L, Kosa P, Mencia M, Struhl K. TAF-Containing and TAF-independent forms of transcriptionally active TBP in vivo. Science. 2000;288:1244–1248. doi: 10.1126/science.288.5469.1244. [DOI] [PubMed] [Google Scholar]
- 14.Lagrange T, Kapanidis AN, Tang H, Reinberg D, Ebright RH. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng W, Roberts SG. A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev. 2005;19:2418–2423. doi: 10.1101/gad.342405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang YL, Duttke SH, Chen K, Johnston J, Kassavetis GA, Zeitlinger J, Kadonaga JT. TRF2, but not TBP, mediates the transcription of ribosomal protein genes. Genes Dev. 2014;28:1550–1555. doi: 10.1101/gad.245662.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kedmi A, Zehavi Y, Glick Y, Orenstein Y, Ideses D, Wachtel C, Doniger T, Waldman Ben-Asher H, Muster N, Thompson J, Anderson S, Avrahami D, Yates JR, 3rd, Shamir R, Gerber D, Juven-Gershon T. Drosophila TRF2 is a preferential core promoter regulator. Genes Dev. 2014;28:2163–2174. doi: 10.1101/gad.245670.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao L, Kim M, DeJong J. Developmental and cell type-specific regulation of core promoter transcription factors in germ cells of frogs and mice. Gene Expr Patterns. 2006;6:409–419. doi: 10.1016/j.modgep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 19.DeJong J. Basic mechanisms for the control of germ cell gene expression. Gene. 2006;366:39–50. doi: 10.1016/j.gene.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 20.Goodrich JA, Tjian R. Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat Rev Genet. 2010;11:549–558. doi: 10.1038/nrg2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez E, Chiang CM, Ge H, Roeder RG. TATA-binding protein-associated factor(s) in TFIID function through the initiator to direct basal transcription from a TATA-less class II promoter. EMBO J. 1994;13:3115–3126. doi: 10.1002/j.1460-2075.1994.tb06610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez E, Zhou Q, L’Etoile ND, Oelgeschlager T, Berk AJ, Roeder RG. Core promoter-specific function of a mutant transcription factor TFIID defective in TATA-box binding. Proc Natl Acad Sci U S A. 1995;92:11864–11868. doi: 10.1073/pnas.92.25.11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verrijzer CP, Chen JL, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 24.Hansen SK, Tjian R. TAFs and TFIIA mediate differential utilization of the tandem Adh promoters. Cell. 1995;82:565–575. doi: 10.1016/0092-8674(95)90029-2. [DOI] [PubMed] [Google Scholar]
- 25.Kaufmann J, Smale ST. Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes Dev. 1994;8:821–829. doi: 10.1101/gad.8.7.821. [DOI] [PubMed] [Google Scholar]
- 26.Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, Roeder RG. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell. 2013;152:1021–1036. doi: 10.1016/j.cell.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamenova I, Warfield L, Hahn S. Mutations on the DNA binding surface of TBP discriminate between yeast TATA and TATA-less gene transcription. Mol Cell Biol. 2014;34:2929–2943. doi: 10.1128/MCB.01685-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez E, Ge H, Tao Y, Yuan CX, Palhan V, Roeder RG. Novel cofactors and TFIIA mediate functional core promoter selectivity by the human TAFII150-containing TFIID complex. Mol Cell Biol. 1998;18:6571–6583. doi: 10.1128/mcb.18.11.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu M, Sharma P, Pan S, Malik S, Roeder RG, Martinez E. Core promoter-selective function of HMGA1 and Mediator in Initiator-dependent transcription. Genes Dev. 2011;25:2513–2524. doi: 10.1101/gad.177360.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis BA, Sims RJ, 3rd, Lane WS, Reinberg D. Functional characterization of core promoter elements: DPE-specific transcription requires the protein kinase CK2 and the PC4 coactivator. Mol Cell. 2005;18:471–481. doi: 10.1016/j.molcel.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 31.Martinez E. Core promoter-selective coregulators of transcription by RNA polymerase II. Transcription. 2012;3:295–299. doi: 10.4161/trns.21846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol. 2010;339:225–229. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zabidi MA, Arnold CD, Schernhuber K, Pagani M, Rath M, Frank O, Stark A. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature. 2015;518:556–559. doi: 10.1038/nature13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon MC, Fisch TM, Benecke BJ, Nevins JR, Heintz N. Definition of multiple, functionally distinct TATA elements, one of which is a target in the hsp70 promoter for E1A regulation. Cell. 1988;52:723–729. doi: 10.1016/0092-8674(88)90410-2. [DOI] [PubMed] [Google Scholar]
- 35.Wefald FC, Devlin BH, Williams RS. Functional heterogeneity of mammalian TATA-box sequences revealed by interaction with a cell-specific enhancer. Nature. 1990;344:260–262. doi: 10.1038/344260a0. [DOI] [PubMed] [Google Scholar]
- 36.Iyer V, Struhl K. Mechanism of differential utilization of the his3 TR and TC TATA elements. Mol Cell Biol. 1995;15:7059–7066. doi: 10.1128/mcb.15.12.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li XY, Bhaumik SR, Zhu X, Li L, Shen WC, Dixit BL, Green MR. Selective recruitment of TAFs by yeast upstream activating sequences. Implications for eukaryotic promoter structure. Curr Biol. 2002;12:1240–1244. doi: 10.1016/s0960-9822(02)00932-6. [DOI] [PubMed] [Google Scholar]
- 38.Mencia M, Moqtaderi Z, Geisberg JV, Kuras L, Struhl K. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol Cell. 2002;9:823–833. doi: 10.1016/s1097-2765(02)00490-2. [DOI] [PubMed] [Google Scholar]
- 39.Emami KH, Navarre WW, Smale ST. Core promoter specificities of the Sp1 and VP16 transcriptional activation domains. Mol Cell Biol. 1995;15:5906–5916. doi: 10.1128/mcb.15.11.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garraway IP, Semple K, Smale ST. Transcription of the lymphocyte-specific terminal deoxynucleotidyltransferase gene requires a specific core promoter structure. Proc Natl Acad Sci U S A. 1996;93:4336–4341. doi: 10.1073/pnas.93.9.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ernst P, Hahm K, Trinh L, Davis JN, Roussel MF, Turck CW, Smale ST. A potential role for Elf-1 in terminal transferase gene regulation. Mol Cell Biol. 1996;16:6121–6131. doi: 10.1128/mcb.16.11.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, Noll M. Compatibility between enhancers and promoters determines the transcriptional specificity of gooseberry and gooseberry neuro in the Drosophila embryo. EMBO J. 1994;13:400–406. doi: 10.1002/j.1460-2075.1994.tb06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merli C, Bergstrom DE, Cygan JA, Blackman RK. Promoter specificity mediates the independent regulation of neighboring genes. Genes Dev. 1996;10:1260–1270. doi: 10.1101/gad.10.10.1260. [DOI] [PubMed] [Google Scholar]
- 44.Corbin V, Maniatis T. The role of specific enhancer-promoter interactions in the Drosophila Adh promoter switch. Genes Dev. 1989;3:2191–2120. doi: 10.1101/gad.3.12b.2191. [DOI] [PubMed] [Google Scholar]
- 45.Ohtsuki S, Levine M, Cai HN. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 1998;12:547–556. doi: 10.1101/gad.12.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Butler JE, Kadonaga JT. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Juven-Gershon T, Hsu JY, Kadonaga JT. Caudal, a key developmental regulator, is a DPE-specific transcriptional factor. Genes Dev. 2008;22:2823–2830. doi: 10.1101/gad.1698108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zehavi Y, Kuznetsov O, Ovadia-Shochat A, Juven-Gershon T. Core promoter functions in the regulation of gene expression of Drosophila dorsal target genes. J Biol Chem. 2014;289:11993–12004. doi: 10.1074/jbc.M114.550251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shir-Shapira H, Sharabany J, Filderman M, Ideses D, Ovadia-Shochat A, Mannervik M, Juven-Gershon T. Structure-Function Analysis of the Drosophila melanogaster Caudal Transcription Factor Provides Insights into Core Promoter-preferential Activation. J Biol Chem. 2015;290:17293–17305. doi: 10.1074/jbc.M114.632109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang YL, Faiola F, Xu M, Pan S, Martinez E. Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J Biol Chem. 2008;283:33808–33815. doi: 10.1074/jbc.M806936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pankiewicz R, Karlen Y, Imhof MO, Mermod N. Reversal of the silencing of tetracycline-controlled genes requires the coordinate action of distinctly acting transcription factors. J Gene Med. 2005;7:117–132. doi: 10.1002/jgm.644. [DOI] [PubMed] [Google Scholar]
- 52.Datto MB, Yu Y, Wang XF. Functional analysis of the transforming growth factor beta responsive elements in the WAF1/Cip1/p21 promoter. J Biol Chem. 1995;270:28623–28628. doi: 10.1074/jbc.270.48.28623. [DOI] [PubMed] [Google Scholar]
- 53.Faiola F, Liu X, Lo S, Pan S, Zhang K, Lymar E, Farina A, Martinez E. Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription. Mol Cell Biol. 2005;25:10220–10234. doi: 10.1128/MCB.25.23.10220-10234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Werth D, Grassi G, Konjer N, Dapas B, Farra R, Giansante C, Kandolf R, Guarnieri G, Nordheim A, Heidenreich O. Proliferation of human primary vascular smooth muscle cells depends on serum response factor. Eur J Cell Biol. 2010;89:216–224. doi: 10.1016/j.ejcb.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 55.Benatti P, Basile V, Merico D, Fantoni LI, Tagliafico E, Imbriano C. A balance between NF-Y and p53 governs the pro- and anti-apoptotic transcriptional response. Nucleic Acids Res. 2008;36:1415–1428. doi: 10.1093/nar/gkm1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin VX, Singer GA, Agosto-Perez FJ, Liyanarachchi S, Davuluri RV. Genome-wide analysis of core promoter elements from conserved human and mouse orthologous pairs. BMC Bioinformatics. 2006;7:114. doi: 10.1186/1471-2105-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Esnault C, Stewart A, Gualdrini F, East P, Horswell S, Matthews N, Treisman R. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev. 2014;28:943–958. doi: 10.1101/gad.239327.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol Cell Physiol. 2007;292:C70–81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- 59.Ng SY, Gunning P, Liu SH, Leavitt J, Kedes L. Regulation of the human beta-actin promoter by upstream and intron domains. Nucleic Acids Res. 1989;17:601–615. doi: 10.1093/nar/17.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frederickson RM, Micheau MR, Iwamoto A, Miyamoto NG. 5′ flanking and first intron sequences of the human beta-actin gene required for efficient promoter activity. Nucleic Acids Res. 1989;17:253–270. doi: 10.1093/nar/17.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Danilition SL, Frederickson RM, Taylor CY, Miyamoto NG. Transcription factor binding and spacing constraints in the human beta-actin proximal promoter. Nucleic Acids Res. 1991;19:6913–6922. doi: 10.1093/nar/19.24.6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dolfini D, Zambelli F, Pavesi G, Mantovani R. A perspective of promoter architecture from the CCAAT box. Cell Cycle. 2009;8:4127–4137. doi: 10.4161/cc.8.24.10240. [DOI] [PubMed] [Google Scholar]
- 63.Martinez E. The NF-Y angle to the CCAAT’s tale. Cell Cycle. 2010;9:642–643. doi: 10.4161/cc.9.4.10828. [DOI] [PubMed] [Google Scholar]
- 64.Choukrallah MA, Kobi D, Martianov I, Pijnappel WW, Mischerikow N, Ye T, Heck AJ, Timmers HT, Davidson I. Interconversion between active and inactive TATA-binding protein transcription complexes in the mouse genome. Nucleic Acids Res. 2012;40:1446–1459. doi: 10.1093/nar/gkr802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reddy P, Hahn S. Dominant negative mutations in yeast TFIID define a bipartite DNA-binding region. Cell. 1991;65:349–357. doi: 10.1016/0092-8674(91)90168-x. [DOI] [PubMed] [Google Scholar]
- 66.Schultz MC, Reeder RH, Hahn S. Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II, and III promoters. Cell. 1992;69:697–702. doi: 10.1016/0092-8674(92)90233-3. [DOI] [PubMed] [Google Scholar]
- 67.Zhao X, Herr W. A regulated two-step mechanism of TBP binding to DNA: a solvent-exposed surface of TBP inhibits TATA box recognition. Cell. 2002;108:615–627. doi: 10.1016/s0092-8674(02)00648-7. [DOI] [PubMed] [Google Scholar]
- 68.Ceribelli M, Dolfini D, Merico D, Gatta R, Vigano AM, Pavesi G, Mantovani R. The histone-like NF-Y is a bifunctional transcription factor. Mol Cell Biol. 2008;28:2047–2058. doi: 10.1128/MCB.01861-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Benatti P, Dolfini D, Vigano A, Ravo M, Weisz A, Imbriano C. Specific inhibition of NF-Y subunits triggers different cell proliferation defects. Nucleic Acids Res. 2011;39:5356–5368. doi: 10.1093/nar/gkr128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scruggs BS, Gilchrist DA, Nechaev S, Muse GW, Burkholder A, Fargo DC, Adelman K. Bidirectional Transcription Arises from Two Distinct Hubs of Transcription Factor Binding and Active Chromatin. Mol Cell. 2015;58:1101–1112. doi: 10.1016/j.molcel.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Duttke SH, Lacadie SA, Ibrahim MM, Glass CK, Corcoran DL, Benner C, Heinz S, Kadonaga JT, Ohler U. Human promoters are intrinsically directional. Mol Cell. 2015;57:674–684. doi: 10.1016/j.molcel.2014.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.