Abstract
We report the structure of MoeN5, a unique prenyl transferase involved in moenomycin biosynthesis. MoeN5 catalyzes the reaction of geranyl diphosphate (GPP) with the cis-farnesyl group in phosphoglycolipid 5 to form the (C25) moenocinyl sidechain-containing lipid 7. GPP binds to an allylic site (S1) and aligns well with known S1 site inhibitors. Alkyl glycosides, glycolipids, can bind to both S1 as well as a second site, S2. Long sidechains in S2 are “bent” and co-locate with the homoallylic substrate isopentenyl diphosphate in other prenyl transferases. These observations support a MoeN5 mechanism in which 5 binds to S2 with its C6–C11 group poised to attack C1 in GPP to form the moenocinyl sidechain, the more distal regions of 5 aligning with the distal glucose in decyl maltoside. The results are of general interest because they provide the first structures of MoeN5 and a structural basis for its mechanism of action, results that will facilitate the design of new antibiotics.
Keywords: isoprenoid biosynthesis, protein structure, antibiotics, enzyme mechanisms, drug discovery
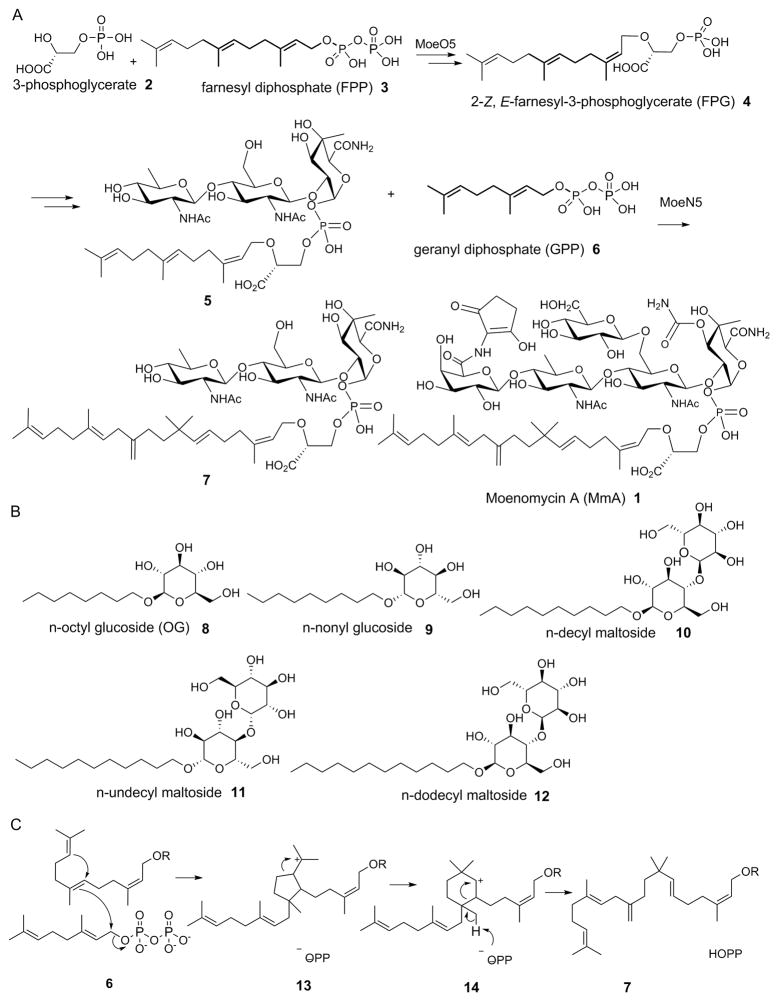
The moenomycins are a potentially important class of antibiotics, discovered ~60 years ago [1], that function by inhibiting bacterial cell wall biosynthesis [2]. Moenomycin A (MmA 1, Scheme 1) is one member of this class and in vitro is a more potent antibiotic than is vancomycin. It has, however, poor pharmacokinetics, but given the global increase in antibiotic resistance[3] there is renewed interest in the development of MmA analogs[4], as well as the possibility of using MmA to treat gastro-intestinal tract infections, such as that caused by Helicobacter pylori[5]. MmA biosynthesis is quite complex, with 17 enzymes involved [4a]. The first committed step involves condensation of 3-phosphoglycerate (2) with farnesyl diphosphate (3) to form 2-(Z,E)-farnesyl-3-phosphoglycerate [FPG, 4] in a reaction catalyzed by MoeO5 [6] and in recent work we reported the X-ray structure of MoeO5 and proposed a mechanism of action [6b]. 4 then undergoes a series of glycosylations and other modifications to form a phosphoglycolipid, the FPG-trisaccharide 5 which then reacts with geranyl diphosphate (GPP, 6) to produce the (C25) moenocinyl trisaccharide 7 [7] which after further transformations [4a] results in formation of 1. The 5 → 7 reaction is catalyzed by MoeN5 [4a] and has been proposed [4a] to involve two carbocations. However, the structure of MoeN5 has not been reported and a BLAST (Basic Local Alignment Search Tool) search reveals no homologs with known structure. We thus sought to determine the structure of MoeN5—in the absence and presence of GPP, FPG and a series of model glycolipids— in order to help clarify the mechanism of action of this very unusual “head-to-middle” prenyl transferase.
Scheme 1.
Structures of molecules of interest, and the MoeN5 mechanism. A, Selected steps in the biosynthesis of moenomycin A showing the reaction catalyzed by MoeN5. B, Structures of the 5 β-n-alkyl glycosides investigated. C, The MoeN5 mechanism proposed by Walker et al. [ref 4a].
Recombinant MoeN5 from Streptomyces ghanaensis was first expressed with a 35-residue N-terminal tag, hereinafter denoted as MoeN5-NT, and its structure was solved by using single-wavelength anomalous diffraction (SAD) with a monoclinic C2 Se-Met data set (3.3 Å; Supporting Information Table S1). The structure was highly α-helical. However, several regions were disordered and the resolution was low. We thus next sought to find better crystal forms by adding one of four protein fusion tags: thioredoxin from Escherichia coli; small ubiquitin-like modifier (SUMO, from Saccharomyces cerevisiae), and chromosomal proteins 7d from the hyperthermophiles Sulfolobus acidocaldarius (Sac7d), and Sulfolobus solfataricus (Sso7d). The construct containing Sso7d linked by 5 residues to the MoeN5 C-terminus together with a 12 residue N-terminal His-tag, hereinafter referred to as MoeN5-CS, was successfully expressed and purified, and crystallized in two different orthorhombic space groups: C222 and I222. Both crystals yielded improved diffraction data sets: 2.29 Å for the C222 form, and 2.80 Å for the I222 form. The two structures were solved by molecular replacement, and the improved electron density maps allowed full elucidation of the MoeN5 structure. Data acquisition and refinement details are given in Table S2, and full experimental details are given in the Supporting Information.
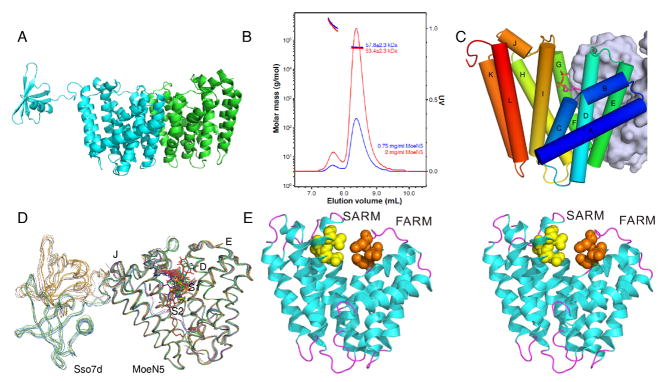
We show in Figure 1A the structure of MoeN5-CS (i.e. containing the Sso7d C-terminus tag; PDB ID code 5B02) where it can be seen that each molecule forms a dimer in the crystal. It is also clear from SEC-MALS (Size Exclusion Chomatography Multi-Angle Laser Light Scattering) results (Figure 1B) that MoeN5 without the fusion tag also forms a dimer, in solution. Each MoeN5 domain contains 12 α-helices, A–L (Figure 1C). Dimer formation mainly involves helices E, F, and G and, together with the C-terminal regions of helices A and D, the dimer interface buries 1830 Å2 of surface area on each monomer. The Sso7d fusion tag showed higher B-values than did MoeN5 and was visible for only one monomer in each MoeN5-CS dimer, Figures 1A,D, where it occupied one of two sites, Figure 1D. Using the PDBeFold server [8] we find that the MoeN5 structure has some similarity to a range of other prenyl synthases including a (C10) geranyl transferase from Thermotoga maritima (PDB ID code 2ftz [9], a 3.05 Cα root mean square deviation, rmsd, over 231 residues); (C20) geranylgeranyl diphosphate synthase from Sinapis alba (PDB ID code 2jlo [10], 2.78 Å/214 residues); as well as the catalytic domain in a polyprenyl diphosphate synthase from Shigella flexneri (PDB ID code 2for [11], 2.85 Å/221 residues). These (and many other) proteins catalyze well-known [12] “head-to-tail” trans-prenyl synthase reactions and contain two “Asp-rich” (DDXXD or DXXXD) domains: the first aspartate-rich domain (FARM) and the second aspartate-rich domain (SARM), both of which bind to Mg2+, and are involved in the ionization of allylic substrates. MoeN5 likewise contains two such Asp-rich domains, illustrated in the stereo-view in Figure 1, the orange (FARM; DDLMD) and yellow (SARM; DDLTD) spheres in Figure 1E. The active site is expected to be located near these Asp-rich domains.
Figure 1.
Streptomyces ghanaesis MoeN5 structures. A, MoeN5-CS dimer structure (PDB ID code 5BO2). The Sso7d tag is only ordered in one molecule in the dimer. B, SEC-MALS molecular weight determination for MoeN5-NT showing dimer (56 kD) formation in solution. Protein concentrations were 0.75 mg/mL and 2 mg/mL. C, The 12 α-helices (A–L) in MoeN5. D, Superposition of all MoeN5-CS structures showing increased B-factors for the Ssod7 domains. E, Stereo-view of MoeN5 (PBD ID code 5B00) showing the first (orange) and second (yellow) Asp-rich domains.
We next sought to see if we could determine the spatial locations of the two substrate binding sites which, by analogy to work on other (head-to-head and head-to-tail) prenyl synthases [12b, 13] we shall call S1, the allylic binding site (into which GPP should bind), and S2, into which the FPG-trisaccharide 5 should bind. As can be seen in Figure 2A, GPP binds to MoeN5-CS close to the Asp-rich domains and the ligand electron density is well resolved, Figure 2A. Similar results with GPP are found for all sites in two structures, PDB ID codes 5B00 and PDB ID code 5B03, Figures S1 and S2. There are actually slight differences in the site occupancies (in the unit cell) with all ligands investigated, and site occupancies (a total of 41) for all 7 systems are given in Table S3; ligand electron densities (there are 37 more and less well defined densities) are given in Figures S1–7; LLDF (local ligand density fit) values are also in shown in Table S3.
Figure 2.
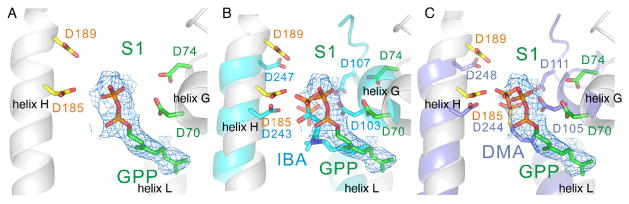
The S1 (GPP-binding) site in MoeN5. A, GPP electron density Fo−Fc omit maps shown in a mesh representation (cyan, contoured at 1 sigma), together with several close-by amino-acid residues. Maps contoured at 1 and 2.5 sigma are in SI Figure S2. B, Alignment of GPP in MoeN5 with the S1-site inhibitor ibandronate in human FPPS. C, Alignment of GPP in MoeN5 with the S1-site inhibitor S-thiolo-dimethylallyl diphosphate, in E. coli FPPS.
As can be seen in Figures 2B,C, there is good accord between the position of GPP in MoeN5 and the positions of known S1 site inhibitors (and substrate) found in other trans-prenyl synthases. For example, both the bisphosphonate ibandronate bound to farnesyl diphosphate synthase (PDB ID code 2F94 [14]; Cα rmsd 3.58 Å/236 residues)]), as well as S-thiolo-dimethylallyl diphosphate (DMA) bound to FPPS [15] (PDB ID code 1RQI; Cα rmsd 2.83 Å/218 residues), bind to the allylic site S1, as shown in the superposition (with GPP in MoeN5, PDB ID code 5B00) in Figures 2B,C. This similarity helps confirm the location (and function) of the allylic, GPP-binding site S1 in MoeN5. The locations of the conserved Asp residues are also shown in the Figure and are similar in both MoeN5 and the FPPSs.
We also anticipate a second, S2-like site in MoeN5 that would be analogous to that seen in other trans-prenyl transferases, although it would not formally be a “homoallylic” site. However, attempts to obtain structures of FPG (4), a model for 5 that might be expected to bind to an S2 site, as well as its mono and trimethyl esters, and MmA, were not successful. We thus adopted an alternate strategy which was to determine the binding sites of a series of glycolipid models, the alkyl glycosides (8–12; Scheme 1B). The idea was that it might be possible to map out the hydrophobic side-chain binding pocket (S2) into which the hydrophobic side-chain of 5 should bind and perhaps, it might also be possible to suggest where the sugar residues (in 5) would locate. Presumably, S2 should accomodate bent-sidechain species that would be close to the GPP C1 in S1, for catalysis, plus, the sugars might extend into water.
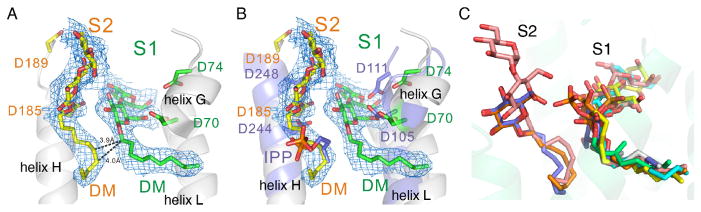
We determined 5 structures (MoeN5-CS bound to one of ligands 8–12) and as can be seen in Figures S3–S7, the electron densities in several cases are very well defined. Surprisingly, however, many of the glycosides bound to S1, and some bound to both S1 as well as a new site that we assign to S2. The two sites can be clearly seen in the electron density map of decyl maltoside (DM, 10) bound to MoeN5-CM (PDB ID code 5BOK), Figure 3A (and SI Figure 5B. This occupancy of both S1 and S2 sites is of interest since as can be seen in Figure 3A, the alkyl side-chains have close contacts—as low as ~4 Å—as would be required for reaction of S1 and S2 site substrates. What is even more interesting is the observation that the longer n-alkyl side-chains of the ligands (9,10) that bind in S2 are bent at their C-terminus, as proposed [4a] for 5 during catalysis and as illustrated in Scheme 1C. The bend is induced by a wall consisting of F149, L150 and I181, Figure S8. The S2 site in MoeN5 overlaps the isopentenyl diphosphate S2 site, in FPPS [PDB ID code 1RQI [15]], Figure 3B. However, MoeN5 does not contain the highly conserved basic residue motifs (containing RR and K) that bind to diphosphates since the phosphate in 5 is expected to be far removed from the typical IPP phosphate-binding site location. When the ligands from all seven structures are superimposed, Figure 3C, it is clear that there are indeed 2 distinct ligand binding sites in MoeN5. The S1 domain corresponds to the allylic substrate, bisphosphonate-inhibitor and allylic S-thiolo-diphosphate inhibitor binding sites seen in e.g. FPPS, while S2 can house alkyl glycosides with a bent C-terminus, and this bent C-terminus is close to the position occupied by the GPP C1 seen in the superpositions. These structural results immediately suggest that the phosphoglycolipid 5 binds to S2 in a “bent” conformation with its C6,7 (and C10,11) double bond poised to attack the geranyl carbocation formed on diphosphate release from 6.
Figure 3.
Ligands bind to both the S1 and S2 sites in MoeN5. A, β-n-decyl maltoside (DM, 10) binds to both S1 and S2. Electron density Fo−Fc omit map (cyan, contoured at 1 sigma), together with several close-by amino-acid residues. The chains approach to within ~4 Å. For additional density results see Figure S5B. B, Alignment of DM (10) in MoeN5 with the canonical trans-prenyl transferase S2-site ligand IPP in human FPPS. C, Superposition of all 6 ligands investigated (GPPS and the 5 glycosides), bound to MoeN5. The ligands shown are those with the most complete electron densities in Figures S1–S7.
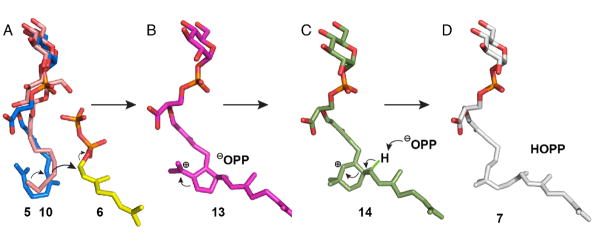
The S1 and S2 site structures we have obtained support the structure-based model for the interaction of the phosphoglycolipid 5 with GPP to form the moenocinyl phosphoglycolipid 7, shown in Figure 4. Figure 4A shows a manual alignment of 5 (containing the proposed “bent” C6–C11 sidechain geometry [7], in blue) to the alkyl side-chain in 10 (pink) in S2 in which the 1st sugar in 5 is aligned to the distal glucose in the maltoside 10. Also shown is GPP (in yellow), in S1. On diphosphate ionization, C6, 7 [7] attacks the GPP C1 and the 5 and 6 membered ring carbocations (13 and 14), form, Figures 4B, C. The diphosphate group released from 6 is then poised to remove the methyl proton in 14 (Scheme 1C; Figure 4C) to form the product 7, reminiscent of the general-base role of the diphosphate group (in dimethylallyl diphosphate) in isoprene synthase [16]. We find no bad clashes with any protein residues, strongly supporting this structural model and, consequently, the mechanism of action first proposed by Arigoni et al. [7].
Figure 4.
Structural models for MoeN5 mechanism. A, The phosphoglycolipid 5 (blue, shown for simplicity with a single sugar) binds to S2 with its bent (C6–11) sidechain occupying the same region as does 10 (purple); the proximal sugar in 5 overlaps the distal glucose on the maltoside. Other sugars will be in water. Also shown is GPP 6 (yellow). B, Diphosphate release from GPP 6 and attack by C6,7/C10,11 in 5 yields the cyclopentyl carbocation 13. C, 13 rearranges to the cyclohexyl carbocation 14. D, Removal of a methyl proton in 14 by PPi yields the product 7.
In summary: we report the first x-ray structures of S. ghanaensis MoeN5, a prenyl transferase involved in biosynthesis of the antibiotic moenomycin A. Structure determination was made possible by the use of chromosomal protein 7d from the hyperthermophile Sulfolobus solfataricus as a fusion tag—an approach that may be applicable to other systems. The MoeN5 substrate GPP binds to the allylic site S1 seen in other prenyl transferases; amphiphilic glycosides can bind to both S1 and S2 with 9 and 10 binding to S2 in a “bent” C-terminus conformation. This conformation serves as a template for binding of the second MoeN5 substrate, 5, enabling attack of C6, 7 in 5 at the C1 of GPP, forming 7. The first glycoside group in 5 locates to the distal glucose in 10, with additional sugars (in 5 and 7) extending into water. Overall, the results are of broad general interest since they can be expected to facilitate the future development of MmA analogs as new anti-infective drug leads, plus, the use of small fusion tags such as Sso7d can be expected to facilitate structure determination of other proteins.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 31200053, 31300615, 31400678 and 31470240), the Chinese Academy of Sciences (grant KSZD-EW-Z-015-2), the United States Public Health Service (NIH grants CA158191 and GM065307), a Harriet A. Harlin Professorship and the University of Illinois Foundation/Oldfield Research Fund. We thank the National Synchrotron Radiation Research Center of Taiwan for beam-time allocation and data-collection assistance.
Footnotes
Supporting information for this article is given via a link at the end of the document.
Contributor Information
Lilan Zhang, Industrial Enzymes National Engineering Laboratory, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin 300308, China.
Prof. Dr. Chun-Chi Chen, Industrial Enzymes National Engineering Laboratory, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin 300308, China.
Dr. Tzu-Ping Ko, Institute of Biological Chemistry, Academia Sinica, Taipei 11529, Taiwan.
Jian-Wen Huang, AsiaPac Biotechnology Co., Ltd., Dongguan, 523808, China.
Prof. Dr. Yingying Zheng, Industrial Enzymes National Engineering Laboratory, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin 300308, China
Prof. Dr. Weidong Liu, Industrial Enzymes National Engineering Laboratory, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin 300308, China
Dr. Iren Wang, Institute of Biological Chemistry, Academia Sinica, Taipei 11529, Taiwan
Dr. Satish R. Malwal, Department of Chemistry, University of Illinois, Urbana, IL 61801, USA
Dr. Xinxin Feng, Department of Chemistry, University of Illinois, Urbana, IL 61801, USA
Dr. Ke Wang, Department of Chemistry, University of Illinois, Urbana, IL 61801, USA
Prof. Dr. Chun-Hsiang Huang, Industrial Enzymes National Engineering Laboratory, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin 300308, China
Prof. Dr. Shang-Te Danny Hsu, Institute of Biological Chemistry, Academia Sinica, Taipei 11529, Taiwan
Prof. Dr. Andrew H.-J. Wang, Institute of Biological Chemistry, Academia Sinica, Taipei 11529, Taiwan
Prof. Dr. Eric Oldfield, Email: eo@chad.scs.uiuc.edu, Department of Chemistry, University of Illinois, Urbana, IL 61801, USA
Prof. Dr. Rey-Ting Guo, Email: guo_rt@tib.cas.cn, Industrial Enzymes National Engineering Laboratory, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin 300308, China
References
- 1.a) Lindner F, Wallhausser KH. Archiv fur Mikrobiol. 1955;22:219–234. [PubMed] [Google Scholar]; b) Lindner F, Wallhausser K, Huber G. German Patent. 1960:1–113. [Google Scholar]; c) Wallhausser KH, Nesemann G, Prave P, Steigler A. Antimicrob Agents Chemother. 1965;5:734–736. [PubMed] [Google Scholar]
- 2.a) Huber G, Nesemann G. Biochem Biophys Res Commun. 1968;30:7. doi: 10.1016/0006-291x(68)90704-3. [DOI] [PubMed] [Google Scholar]; b) Jackson GED, Strominger JL. J Biol Chem. 1984;259:1483–1490. [PubMed] [Google Scholar]
- 3.a) CDC. 2014 http://www.cdc.gov/drugresistance/threat-report-2013/; b) Oldfield E, Feng X. Trends Pharmacol Sci. 2014;35:664–674. doi: 10.1016/j.tips.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Ostash B, Walker S. Nat Prod Rep. 2010;27:1594–1617. doi: 10.1039/c001461n. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gampe CM, Tsukamoto H, Doud EH, Walker S, Kahne D. J Am Chem Soc. 2013;135:3776–3779. doi: 10.1021/ja4000933. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zuegg J, Muldoon C, Adamson G, McKeveney D, Le Thanh G, Premraj R, Becker B, Cheng M, Elliott AG, Huang JX, Butler MS, Bajaj M, Seifert J, Singh L, Galley NF, Roper DI, Lloyd AJ, Dowson CG, Cheng TJ, Cheng WC, Demon D, Meyer E, Meutermans W, Cooper MA. Nat Commun. 2015;6 doi: 10.1038/ncomms8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Riess G, Seibert G, Hedtmann U. Aventis Pharma Deutschland GmbH. Frankfurt am Main; Germany: 2001. [Google Scholar]; b) Tseng YY, Liou JM, Hsu TL, Cheng WC, Wu MS, Wong CH. Bioorg Med Chem Lett. 2014;24:2412–2414. doi: 10.1016/j.bmcl.2014.04.041. [DOI] [PubMed] [Google Scholar]
- 6.a) Doud EH, Perlstein DL, Wolpert M, Cane DE, Walker S. J Am Chem Soc. 2011;133:1270–1273. doi: 10.1021/ja109578b. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ren F, Ko TP, Feng X, Huang CH, Chan HC, Hu Y, Wang K, Ma Y, Liang PH, Wang AHJ, Oldfield E, Guo RT. Angew Chem Int Ed. 2012;51:4157–4160. doi: 10.1002/anie.201108002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuricht U, Hennig L, Findeisen M, Welzel P, Arigoni D. Tetrahedron Lett. 2001;42:3835–3837. [Google Scholar]
- 8.Krissinel E, Henrick K. Acta Crystallogr D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 9.Joint Center for Structural Genomics. Worldwide Protein Data Bank. 2006 [Google Scholar]
- 10.Kloer DP, Welsch R, Beyer P, Schulz GE. Biochemistry. 2006;45:15197–15204. doi: 10.1021/bi061572k. [DOI] [PubMed] [Google Scholar]
- 11.Minasov G, Brunzelle JS, Shuvalova L, Collart FR, Joachimiak A, Anderson WF. Midwest Center for Structural Genomics. Worldwide Protein Data Bank. 2006 [Google Scholar]
- 12.a) Christianson DW. Chem Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]; b) Oldfield E, Lin FY. Angew Chem Int Ed. 2012;51:1124–1137. doi: 10.1002/anie.201103110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin F-Y, Liu C-I, Liu Y-L, Zhang Y, Wang K, Jeng W-Y, Ko T-P, Cao R, Wang AHJ, Oldfield E. Proc Natl Acad Sci. 2010;107:21337–21342. doi: 10.1073/pnas.1010907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rondeau J-M, Bitsch F, Bourgier E, Geiser M, Hemmig R, Kroemer M, Lehmann S, Ramage P, Rieffel S, Strauss A, Green JR, Jahnke W. Chemmedchem. 2006;1:267–273. doi: 10.1002/cmdc.200500059. [DOI] [PubMed] [Google Scholar]
- 15.Hosfield DJ, Zhang YM, Dougan DR, Broun A, Tari LW, Swanson RV, Finn J. J Biol Chem. 2004;279:8526–8529. doi: 10.1074/jbc.C300511200. [DOI] [PubMed] [Google Scholar]
- 16.Koeksal M, Zimmer I, Schnitzler JP, Christianson DW. J Mol Biol. 2010;402:363–373. doi: 10.1016/j.jmb.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.