Abstract
Immediate early genes (IEGs) are rapidly activated after sensory and behavioral experience and are believed to be critical for converting experience into long-term memory. Neuronal PAS domain protein 4 (Npas4), a recently discovered IEG, has several characteristics that make it likely to be a particularly important molecular link between neuronal activity and memory: it is among the most rapidly induced IEGs, is expressed only in neurons, and is selectively induced by neuronal activity. By orchestrating distinct activity-dependent gene programs in different neuronal populations, Npas4 affects synaptic connections in excitatory and inhibitory neurons, neural circuit plasticity and memory formation. It may also be involved in circuit homeostasis through negative feedback and psychiatric disorders. We summarize these findings and discusses their implications.
Introduction to Npas4
A fleeting moment of experience can leave a memory lasting a lifetime. Conversion of a momentary event into permanently encoded information in the brain starts with groups of neurons that are activated by the transient sensory and behavioral stimulation. It is believed that the changes that subsequently occur in these activated neuronal ensembles, which require de novo protein synthesis, create long-term memory. Thus, acutely triggered activity-dependent gene transcription in specific neuronal ensembles is critical for recording a fleeting moment of experience.
Neuronal activity-triggered gene transcription, mediated by intracellular free calcium (Ca2+) as the second messenger, occurs within minutes and is spearheaded by a group of immediate early genes (IEGs, Box I) whose expression does not require de novo protein synthesis [1]. Beginning with the IEG-dominated first wave of gene transcription, a global gene transcription cascade is unleashed, resulting in transcription of a second wave of genes that usually requires new protein synthesis and carries out the specific cellular functions of the response, such as synapse modification and memory formation. Notably, many IEGs are transcription factors and directly regulate transcription of the second wave of effector genes. Not surprisingly, there has been an increasing interest in IEGs in the neuroscience field in the past few decades, not only because of the key molecular link they provide between experience and subsequent modification of the brain, but also because the robust and near-instantaneous activation of many IEGs provides a means of identifying the neuronal ensembles that presumably encode specific memories.
Box I.
The term immediate-early gene (IEG) was first used to describe the viral genes that are transcribed in host cells within 2 minutes after viral infection [49]. Since then, the term IEG has been more broadly used to describe genes that are activated rapidly and transiently in response to extracellular stimuli and independently of de novo protein synthesis [11], even if the transcription happens after the 2 minute mark.
In mammalian neurons, c-fos is arguably the best known IEG. It was first studied in non-neuronal cells in response to growth factors. Applying PDGF (platelet-derived growth factor) to cultured 3T3 cells induced c-fos transcription within minutes, and it became undetectable after 30 minutes [2]. The initial characterization of IEG expression in neuronal cells was done in the pheochromocytoma cell line PC12, in which c-fos was induced by growth factors or depolarizing stimuli [50]. This last finding suggested for the first time a potential role for IEGs in linking neuronal activity to long-term changes in gene expression. Since then some 30–40 different IEGs have been identified that are known to be induced in vitro by extracellular stimuli and/or activated in vivo under physiological conditions [51, 52]. The table below lists some well-known IEGs expressed in the nervous system. Most IEGs found in mammals are conserved across many species, from invertebrates such as C. elegans [53], Aplysia [54] and Drosophila [55] to vertebrates, but only a subset have been tested to see whether they are rapidly induced by neuronal activity.
While IEGs with rapid induction similar to c-fos, such as Arc and Zif268 (also called Egr1), usually come to mind when neuronal IEGs are mentioned, the induction of many neuronal IEGs, such as BDNF and Narp, is much slower. Npas4 belongs to the rapidly induced group. Since in most cases the point when transcription begins has not been determined, the distinction between quickly and slowly induced IEGs is usually based on when their mRNA levels peak after stimulation, with faster IEGs usually peaking within 30 minutes and slower IEGs around 3–6 hours in vivo. In neurons, the definition of an IEG may not be clear cut, because some slower IEGs may actually be activated by faster IEGs already expressed due to on-going basal activity, which strictly speaking disqualifies them from being true IEGs (since their expression requires de novo protein synthesis). Definitions aside, it is important to characterize the activity-dependent transcription cascades in order to understand how activity-dependent events unfold in neurons after neuronal stimulation.
Functionally, IEGs can be categorized into two major classes: transcription factors that regulate downstream gene expression (e.g. c-fos, Zif268, Npas4) and effector proteins that directly modulate cellular functions (e.g. Arc, BDNF, Homer1a, Narp). The majority of the transcription factor IEGs are induced rapidly, while effector IEGs, with the exception of Arc, are generally induced more slowly.
IEGs, particularly the rapidly induced ones, have been used as markers of neuronal activity in the brain, although the precise quantitative relationship between IEG expression and neuronal firing is unclear. As different IEGs are activated by distinct signaling pathways in some cases, the ensembles of neurons defined by expression of different IEGs are not necessarily identical [6]. A major problem is to determine how the transcription cascades led by individual IEGs function either independently or in concert to modify neural circuits. Recently, a picture of complicated crosstalk among them has started to emerge. For example, Npas4 and c-fos not only bind enhancer elements on each other’s promoter, they also both bind, often in the same region, to the promoters of a large number of other activity-regulated genes (including other IEGs), potentially directly regulating their expression [56, 57]. Furthermore, Npas4 has been shown to directly regulate the expression of BDNF and Narp, and BDNF in turn strongly induces the expression of c-fos and Arc, but not Npas4. Such interconnections between IEGs may contribute to feed-forward and feedback regulation within activity-dependent gene transcription cascades.
Long-term plasticity and memory generally require transcription during and immediately after stimulation. This period coincides with IEG induction, suggesting that IEGs may play important roles in experience-dependent modulation of synapses in order to encode memory. It is therefore unsurprising that mutant animals missing IEGs, including c-fos, Zif268, Arc and Npas4, are found to be deficient in learning and memory. However, the steps between IEG induction and memory formation are largely unknown and are likely to remain an area of intensive investigation for some time.
Many IEGs are rapidly and robustly induced by neuronal activity, including c-fos, arg3.1 (Arc) and zif268 (egr1), and have been studied for more than two decades [2–4]. This review focuses on a more recently identified IEG, Neuronal PAS domain protein 4 (Npas4, see Box II). Npas4 possesses several important features, some of which are unique among known IEGs (Figure 1). Firstly, unlike most IEGs, Npas4 is only expressed in neurons. Secondly, Npas4 is unique in that it is activated selectively by neuronal activity, not by extracellular stimuli such as growth factors and neurotrophins [5, 6]. Thirdly, Npas4 appears to directly control the expression of a very large number of activity-dependent genes [5–7]. Finally, Npas4 has been shown to be important for both glutamatergic and GABAergic synapse development in both glutamatergic and GABAergic neurons [5, 7–9], a set of functions that has not been reported for other IEGs. Npas4 appears to have substantial functional importance: it is involved in neural circuit plasticity, may be involved in maintaining circuit homeostasis, and is required for long-term memory formation [6, 10]. Thus, studying Npas4 provides an opportunity to greatly increase our understanding of activity-dependent mechanisms underlying learning and memory.
Box II. Npas4 and bHLH-PAS family transcription factors.
Npas4 belongs to the basic-helix-loop-helix (bHLH)-PER-ARNT-SIM (PAS) family of transcription factors, which consist of a bHLH domain for DNA binding and tandem PAS domains for dimerization and protein-protein interaction. bHLH-PAS transcription factors are generally engaged in acute responses to extracelluar stimuli and environmental changes. Well-studied members of the bHLH-PAS family include those involved in response to xenobiotic stress (AhR), hypoxia (HIF1α), and circadian regulation (Clock and BMAL). The mechanisms by which these proteins initiate an adaptive response are different. AhR resides in the cytoplasm and translocates into the nucleus upon ligand binding to activate downstream gene expression. The activity of HIF1α is tightly controlled at the protein level: it is constitutively degraded under normoxia, but stabilized under hypoxic conditions. Clock and BMAL form an autoregulatory transcriptional feedback loop in response to day-night light cycles that results in the circadian rhythm [78].
To activate transcription, bHLH-PAS proteins must form a dimer with one of the bHLH-PAS binding partners ARNT, ARNT2, BMAL1 or BMAL2. In experiments conducted in vitro, Npas4 appears to form dimers with ARNT and ARNT2, but not with BMAL1 or BMAL2 [79, 80]. Since ARNT and ARNT2 have different expression patterns in the brain, different heterodimers may be responsible for downstream gene expression in different brain regions and possibly during different behaviors, and may induce expression of different downstream genes.
Npas4 was named as the fourth member of the Neural PAS domain protein (NPAS) group. Note that proteins in the NPAS group, although they are all bHLH-PAS family proteins, are not closely related homologs and appear to have distinct biological functions.
Figure 1. Schematic drawing showing the activity-dependent induction and synaptic functions of Npas4.
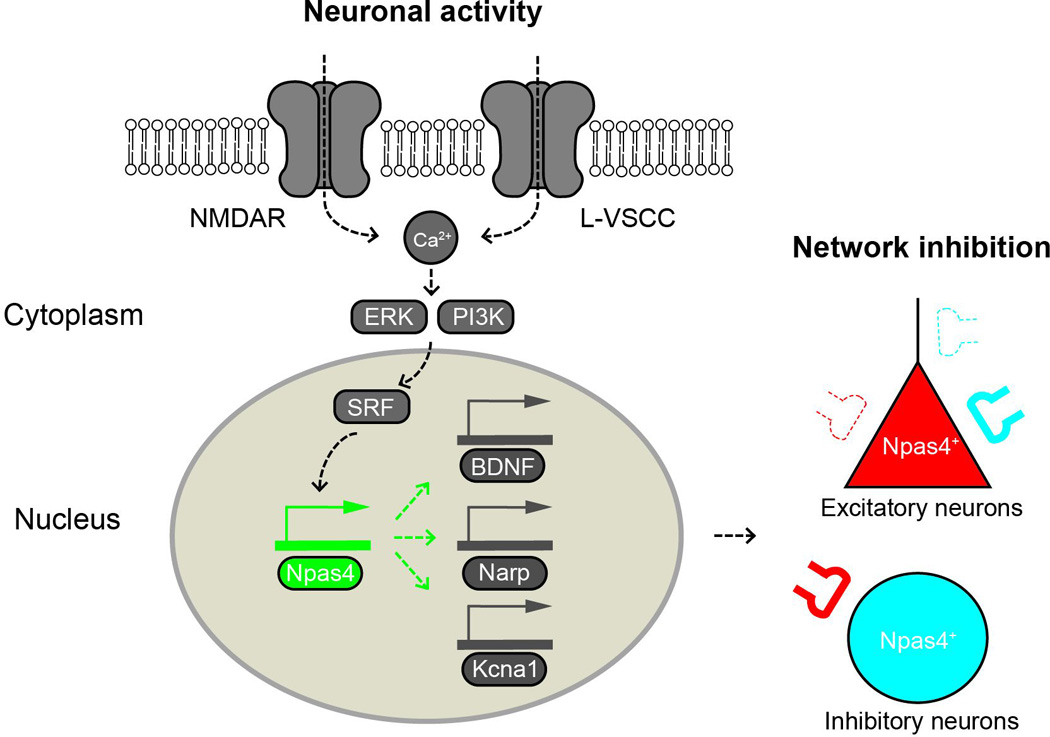
Npas4 is induced by calcium influx triggered by neuronal activity. As a transcription factor, Npas4 regulates the expression of a large number of downstream genes (e.g. BDNF, Narp, Kcna1), which mediate the diverse effects of Npas4 on synapses. In excitatory neurons that have been activated to express Npas4 (Npas4+), Npas4 upregulates perisomatic inhibitory synapses, diminishes distal dendritic inhibitory synapses and downregulates excitatory synapses. In Npas4+ inhibitory neurons, Npas4 may upregulate excitatory synapses in response to activity, as shown here in somatostatin-expressing neurons. In general, Npas4 appears to scale down the level of network activity in response to neuronal excitation.
Activity-dependent induction of Npas4
When IEGs were first identified 3 decades ago, it was postulated that different sets of IEGs would be found to carry out immediate responses to different stimuli in different cell types [11]. It was therefore surprising that in subsequent decades the core set of IEGs was found to be induced in almost all cell types by a wide range of stimuli. Npas4 is one of the few exceptions.
Npas4 is expressed exclusively in neurons [5, 12]. Its expression in other cell types is actively suppressed by the binding of RE-1 silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF) to Npas4’s promoter and intron regions [13]. When Npas4 was selected in our screen for neuronal activity-regulated genes in cultured neurons, we found it to be responsive to depolarization only, not to growth factors, neurotrophins or several kinase pathways that are known to induce other IEGs such as c-fos, zif268 and Arc [5, 6]. Calcium influx and nuclear calcium signaling are required for the activation of Npas4 expression [5, 14].
We currently know only a little about the upstream transcriptional pathways that induce Npas4 expression. The mitogen-activated protein kinase (MAPK) and phosphatidylinositol-3 kinase (PI3K) pathways have been shown to be required for the induction of Npas4 by pharmacologically-induced LTP (long term potentiation) and LTD (long term depression), respectively, in mouse hippocampal slices [15]. A recent study showed that serum response factor (SRF) is a possible upstream regulator of Npas4, as its deletion led to a significant decrease in Npas4 expression [16]. Npas4’s promoter region contains two SRF binding sites, suggesting that SRF directly regulates Npas4 expression. However, since many signaling pathways, both activity-dependent and -independent, lead to the activation of SRF-dependent transcription, SRF alone does not seem sufficient to explain the activity-dependence of Npas4 induction. It is quite possible that induction of Npas4 expression requires the interaction of multiple upstream pathways.
Typical features of IEG activation are that it is both rapid (occurring within minutes) and transient (lasting only a few hours). In depolarized cultured neurons, Npas4 is one of the most rapidly induced genes [5]. The Npas4 mRNA level in cultured neurons reaches its maximum between 30 minutes and 1 hour after stimulation, and the protein level peaks between 1 and 2 hours [5]. Although its temporal induction profile in vitro is very similar to that of c-fos, in vivo in the hippocampus Npas4 appears to be have a much steeper induction profile than other IEGs [6]: after contextual fear conditioning (CFC), the level of Npas4 mRNA in the hippocampus reaches its peak within 5 minutes, compared to 30 minutes for c-fos and Arc. Protein levels follow the same trend: Npas4 protein reaches its maximum by 30 minutes (the earliest time point we examined), compared to 1.5 hours for c-fos. We do not know why the Npas4 transcript reaches its peak much earlier in vivo than in cultured neurons or the significance of its extremely steep induction dynamics for activity-dependent transcription programs. The rapid induction of Npas4 could be achieved by the release of RNA polymerase II molecules that were staging at the transcription start site, ready to transcribe the gene [17]. Interestingly, it was shown recently that formation of a double strand break in the promoter region can facilitate the induction of a select few IEGs, including c-fos, zif268 and Npas4 [18].
Behaviorally-induced Npas4 expression
In vivo, Npas4 is induced in many regions of the brain by physiological [5, 6, 10, 19–22] or pathological stimuli [23–26]. Under CFC, in which mice learn to associate a novel context with aversive stimuli (foot shocks) during a brief training session, Npas4’s expression in the mouse hippocampus was different from that of other well-known IEGs. Npas4 was induced only when mice were engaged in contextual learning. Treatments that didn’t lead to long-term contextual memory, such as foot shocks alone, did not significantly induce Npas4, although they activated expression of c-fos and Arc [6]. Furthermore, CFC-induced Npas4 expression was largely limited to the CA3 subregion, with very low expression in the dentate gyrus and almost none in CA1 [6]. As the CFC paradigm is known to require the CA3 subregion [27–29], the very rapid and regionally specific expression of Npas4 in CA3 seems likely to be important in the formation of contextual memories (this is discussed further below).
Consistent with the idea that Npas4 expression may often be induced selectively by associative learning, during tone-fear conditioning Npas4 expression in the amygdala is induced only when a tone stimulus is paired with an aversive foot shock [10]. In the barrel cortex, Npas4 is induced by sensory conditioning in which whisker stimulation (CS: conditional stimulus) is paired with an aversive stimulus (US: unconditioned stimulus), but not by unpaired CS and US or by US alone [30]. Similarly, in the striatum Npas4 expression was induced in the nucleus accumbens only by repeated, and not by acute, amphetamine (AMPH) treatments, which are believed to be associated with long-term circuit change and drug-seeking behavior. In contrast, c-fos was induced by both acute and repeated AMPH treatment [31].
Several recent studies have examined the expression of Npas4 under stressful conditions. Treatment with corticosterone or physical stress exposure down-regulates Npas4 expression in both prefrontal cortex and hippocampus in vivo [21, 32–34]. This may result from the activation of glucocorticoid pathways that suppress Npas4 expression [35]. However, it is unclear whether down-regulation of Npas4 is involved in the maladaptive changes that lead to stress-induced disorders or the stress adaption response that reinstates homeostasis. Results from studies using Npas4 knockout animals [32, 36], although not entirely consistent with each other, generally appear to support the latter idea, although mice deficient in Npas4 exhibited impaired adult neurogenesis in the hippocampal subventricular zone, consistent with the former possibility [32]. Further studies are needed to investigate Npas4’s function in stress in a region-specific manner, as it might play different roles in different brain regions.
Synaptic functions of Npas4
Npas4 was initially thought to preferentially regulate GABAergic synapses on excitatory neurons [5]. However, it has now become clear that it plays a broader role in modulating activity-dependent synaptic connections. Depending on the circumstances and the neuronal cell type, Npas4 may modulate either GABAergic, glutamatergic, or both types of synapses. Npas4 does appear to be preferentially involved in negatively regulating synaptic drive in response to activity, suggesting that it is part of negative feedback mechanisms and may be involved in synaptic homeostasis to maintain neural circuit balance [5, 7].
In cultured hippocampal pyramidal neurons, reducing Npas4 expression by RNAi leads to a decrease in the number of inhibitory synapses formed on these neurons, while over-expression of Npas4 increases their number [5]. These results suggest that the level of Npas4 expressed in an excitatory neuron dictates the amount of inhibitory drive it receives. Since Npas4 expression was manipulated before or during the major wave of synaptogenesis that occurs in these cultures, these data suggest a role for Npas4 in inhibitory synaptogenesis during early development. However, in organotypic hippocampal slices, in addition to modulating the number and strength of GABAergic synapses on CA1 pyramidal neurons, manipulation of the Npas4 expression level also leads to a change in the number and strength of excitatory synapses on those neurons, although to a much lesser extent [5]; a reduced Npas4 level leads to less inhibition and more excitation, while a higher Npas4 level results in more inhibition and less excitation. It remains to be seen whether Npas4’s effect on excitatory synapses is a secondary effect resulting from its effect on inhibitory synapses. However, it is clear that Npas4’s effects on both inhibitory and excitatory synapses result in reduced excitability of pyramidal neurons.
Npas4’s role in modulating GABAergic synapses on excitatory neurons was later confirmed in vivo. Based on mIPSCs recorded from CA1 pyramidal neurons in acute hippocampal slices, in response to heightened activity following epileptic seizure or enriched environment exposure, Npas4 appeared to be required for activity-dependent upregulation of GABAergic synapses onto these neurons [37]. Npas4’s effect on glutamatergic synapses was not examined in that study. Further investigation showed that Npas4 expression in CA1 pyramidal neurons actually increased the number of perisomatic GABAergic synapses while decreasing the number on distal apical dendrites. This finding implies that Npas4 is involved in modulating the neuron’s subsequent response to inputs received in different subcellular compartments. We do not yet know how Npas4 manages to differentially regulate GABAergic synapses in different subcellular domains.
Npas4 is also induced by neuronal activity in GABAergic neurons. For example, in the primary visual cortex Npas4 is robustly induced after light exposure in both excitatory neurons and major subtypes of GABAergic neurons [5, 7]. Consistent with a general role for Npas4 in negatively modulating the overall synaptic drive of neural circuits in response to activity, deletion of Npas4 from somatostatin (SST)-expressing GABAergic neurons, both in culture and in vivo, resulted in a reduction in the number of glutamatergic synapses on these neurons, without affecting the number of GABAergic synapses [7]. This finding, together with the results described above, suggests that Npas4 might be involved in homeostatic plasticity, maintaining the excitatory/inhibitory balance of neural circuits by recruiting inhibitory synapses onto excitatory neurons and excitatory synapses onto inhibitory neurons in response to increased activity.
Npas4’s role in activity-dependent synapse formation during development has also been examined in neurons born during adult neurogenesis, when neuronal activity is important for their incorporation into the surrounding established neural circuit. In adult-born dentate gyrus granule neurons, Npas4 has been found to be a major mediator of the effect of neural activity on both the GABAergic and glutamatergic synaptic inputs the neurons receive: increasing the excitability of individual newborn neurons significantly altered their GABAergic synaptic inputs and dendritic spines (the sites of the majority of glutamatergic synapses), and these effects were completely abolished by deleting the Npas4 gene [8]. In adult-born olfactory bulb granule cells (OBGCs), which are GABAergic, Npas4 is induced by odorant exposure and is required for dendritic spine formation in these cells following the sensory experience [9]. Since dendritic spines on OBGCs are the sites of synapses between OBGCs and mitral/tuft cells that relay olfactory sensory information to the cortex, by regulating the experience-dependent formation of these dendritic spines Npas4 could play an important role in the processing of olfactory information.
Transcriptional targets of Npas4: mechanisms underlying its synaptic functions
Being a transcription factor, Npas4 exerts its biological function by controlling the expression of its downstream transcriptional targets. To uncover genes that are involved in activity-dependent development of GABAergic synapses, RNAi was used to acutely knock down Npas4 expression in cultured hippocampal neurons prior to transcription profiling using DNA microarrays. About 300 genes were found to be significantly altered when Npas4 was knocked down, more than half of which were also acutely regulated by neuronal activity [5].
Among these possible Npas4 targets, brain-derived neurotrophic factor (BDNF) stood out because of its established role in GABAergic synapse development [38]. Npas4 directly regulates the activity-dependent expression of BDNF by binding to its activity-dependent promoters [5]. BDNF appears to be a major downstream effector of Npas4 in modulation of GABAergic synapses, because when BDNF is knocked down by RNAi the ability of Npas4 overexpression to elevate GABAergic synapses is greatly diminished, although not completely abolished [5]. BDNF was subsequently shown to be required only for the activity-dependent up-regulation of perisomatic inhibitory synapses on CA1 pyramidal neurons that is mediated by Npas4, but not for its down-regulation of dendritic inhibitory synapses [37]. Surprisingly, BDNF was not involved in the activity-dependent modulation of either GABAergic or glutamatergic synapses on adult-born dentate gyrus granule neurons, which requires Npas4 [8]. In adult-born OBGCs, Npas4 regulates dendritic spine density through the E3-ubiquitin ligase murine double minute 2 (Mdm2) [9]. These various findings suggest that Npas4 engages different downstream transcriptional targets in different cell types to modify synaptic connections.
A thorough investigation of Npas4-dependent genetic programs in excitatory and inhibitory neurons revealed that Npas4 regulates different sets of downstream targets in those different cell types [7]. BDNF, which is normally expressed in excitatory but not in inhibitory neurons, could not be induced in inhibitory cortical neurons even when Npas4 was over-expressed. Therefore, other Npas4 targets are responsible for its role in modulating synapses in GABAergic neurons. It would be very enlightening to understand how Npas4 orchestrates different transcriptional programs in each cell type to help maintain the overall homeostasis of the network.
Npas4 appears to be important for the activity-dependent regulation of a very large number of genes. Acute removal of the Npas4 gene from cultured Npas4 conditional knockout neurons by Cre recombinase greatly reduced the activity-dependent induction of a large number of activity-regulated genes, including most of the well-known IEGs [6]. This is supported by ChIP-seq data showing that Npas4 co-localizes with RNA polymerase II (Pol II) on promoters and enhancers of thousands of activity-regulated genes [39]. In the absence of Npas4, the recruitment of Pol II to promoter and enhancer regions of both c-fos and BDNF was abolished [6]. It is unclear whether Npas4 is directly involved in or plays a permissive role at the level of chromatin structure to facilitate Pol II recruitment. Because Npas4 appears to have such a broad impact on activity-regulated gene expression, it would be interesting to investigate if it mediates many types of adaptive responses of neurons to activity.
Npas4’s role in plasticity and memory
Activity-dependent transcription mechanisms are believed to underlie experience-induced synaptic changes both during development and in the mature brain. It is possible that Npas4 plays a role in both processes. While Npas4 is known to directly regulate genes such as BDNF and Narp that have been shown to be important for plasticity [40–42], Npas4’s role in neural circuit plasticity has not yet been completely elucidated.
Npas4 has recently been implicated in the restoration of ocular dominance (OD) plasticity to the juvenile level in adult mice after fluoxetine treatment [43], presumably through a reduction in intracortical inhibition [44, 45]. Npas4 expression was found to be elevated by fluoxetine treatment and Npas4 knockdown prevented the fluoxetine-induced reactivation of plasticity in adult mice [43]. Furthermore, overexpression of Npas4 alone mimicked the fluoxetine effect. Interestingly, expression of the Npas4 target BDNF was transiently increased by fluoxetine treatment and blocking BDNF-TrkB signaling also abolished the fluoxetine effect [46]. These findings strongly suggest that the Npas4-dependent transcription program in the relevant cells includes components that promote neural circuit plasticity. However, these experiments do not tell us whether Npas4 promotes adult plasticity by modulating inhibitory synapses. It would be interesting to determine whether Npas4 is also important for critical period OD plasticity, which has been shown to be dependent on the development of GABAergic synapses.
Npas4 has been shown to be important for long-term memory formation in two regions of the brain: hippocampus and amygdala. CFC was used to test the involvement of Npas4 in long-term memory formation in the hippocampus. Npas4 knockout mice fail to form contextual fear memories and this result does not seem to be due to impairment of their locomotive activity, ability to sense pain (from the footshocks) or an abnormal anxiety level [6]. As mentioned earlier, Npas4 induction in the hippocampus following CFC is largely restricted to the CA3 subregion, suggesting that Npas4 might be required specifically in CA3 for contextual memory formation. Both loss-of-function and gain-of-function experiments showed that this is indeed the case. Deletion of Npas4 from CA3 in Npas4 conditional knockout mice led to impairment of long-term contextual memory formation, and re-expression of Npas4 in CA3 in Npas4 total knockout animals restored memory formation capacity [6]. Moreover, Npas4 is not required in CA1 for contextual memory formation. This selective induction of Npas4 in CA3 is unusual, because other activity-dependent genes such as c-fos and Arc are induced throughout the hippocampus during CFC. This is consistent with the fact that the CA3 region is known to be required for the rapid encoding of contextual information in the CFC paradigm, but until now the molecular pathways involved had not been elucidated [27–29].
While it is clear that Npas4 is required in CA3 but not in CA1 for contextual memory formation, we do not yet know what distinct signaling pathway in CA3 leads to the induction of Npas4 here but not in CA1, or exactly why Npas4 is required only in CA3 for contextual memory formation. It would be interesting to find out whether Npas4 is important in other hippocampus-dependent learning and memory tasks that don’t require CA3.
Auditory fear memory, which depends on the amygdala but not the hippocampus, is normal in Npas4 total knockout mice, suggesting that the amygdala circuit is intact [10]. This may be due to compensatory pathways that produce normal amygdala function in the absence of germline Npas4, since when Npas4 is acutely knocked down in the lateral amygdala (LA) by adeno-associated virus (AAV) expressing a short hairpin RNA (shRNA) that specifically targets Npas4, auditory fear memory formation is impaired [10].
It is not known for which step of long-term memory formation and storage Npas4 is required. Acute deletion of Npas4 does not impair short-term CFC memory, suggesting that the initial learning is intact [2]. In the amygdala Npas4 is not required for memory retrieval, because infusing shRNA-expressing AAV 3 days after auditory fear conditioning to knock down Npas4 in LA had no effect on fear memory reactivation 21 days later. Npas4 might therefore be involved in post-learning consolidation and reconsolidation. Consistent with this idea, Npas4 knockdown impaired retention of the auditory fear memory after reactivation [10]. Npas4’s effect on synaptic plasticity in the hippocampus or amygdala under these paradigms remain to be examined in order for us to understand how Npas4 is involved in memory formation at the cellular level.
Note that there is no reason to think that Npas4’s role in memory is limited to the paradigms we have discussed involving the hippocampus and amygdala; it may very well be involved in the formation of other type of memories in other parts of the brain.
Future directions: active ensembles and linking neuronal activity to the memory trace
As neuroscience pursues an understanding of the mechanisms that underlie long-term memory, a looming challenge is to identify the sites in the brain where particular memories are encoded and stored (the memory trace). A recently emerging view is that IEG-expressing neural ensembles might represent key components of the memory trace. In recent studies optogenetic reactivation or inactivation of IEG-defined ensembles led to retrieval or suppression of memories, respectively [47, 48].
We have seen that Npas4 expression is regulated selectively by neural activity [5, 6], that its expression leads to modification of synaptic connections, that it is induced in specific neuronal populations by various learning experiences [6, 30, 31], and its deletion results in impairment of memory formation [6, 10]. It is therefore reasonable to speculate that experience-dependent changes in neural circuits that are important for memory formation are likely to take place in neurons in which Npas4 is induced. The development of tools to identify and manipulate Npas4-expressing neural ensembles will allow us to test this hypothesis and investigate the mechanism of memory. Note that ensembles defined by expression of Npas4 may not be identical to those defined by c-fos or Arc. Ensembles defined by different IEGs may play distinct roles in memory. Given the unique features of Npas4 that distinguish it from other IEGs, neuronal ensembles defined by Npas4 expression may play a unique role in memory formation and provide particularly enlightening information about the process.
Having identified the active neuronal population associated with a specific experience, plasticity occurring in the active neurons and their synapses and the gene expression programs required for long-lasting cellular changes in these neurons can be isolated and investigated as being specifically relevant to the experience being encoded. This information is key to our understanding of how a memory is encoded by the ensemble neurons acting collectively as a neural circuit. From the various roles of Npas4 in activity-dependent synapse modification in several different types of neurons, a picture of how an experience may shape the memory-encoding circuit by modulating the interactions between ensemble neurons is starting to emerge (Figure 2). However, as illustrated in Figure 2, many missing links need to be found before we can move closer to fully understanding how a transient experience is recorded permanently in the brain.
Figure 2. Schematic drawing showing the potential roles of Npas4 in regulating neural circuits.
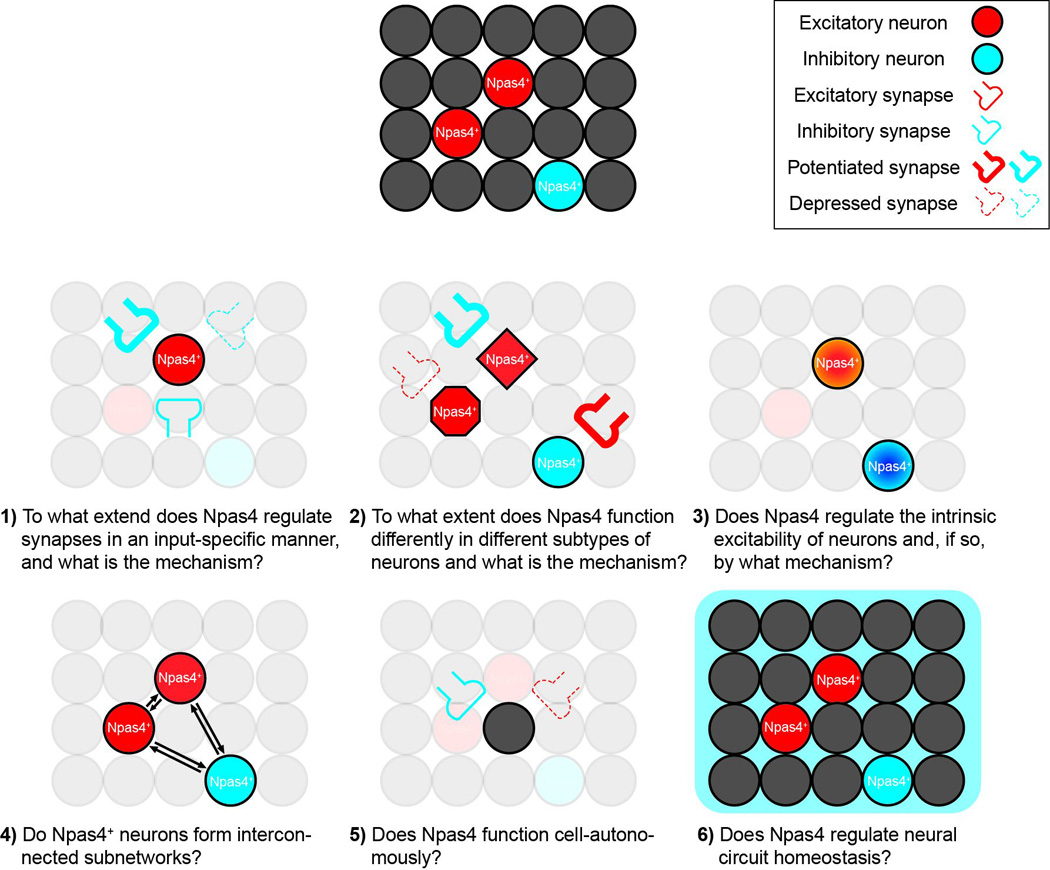
Npas4 has been shown to regulate both excitatory and inhibitory synapses, but how this synaptic modulation contributes to neural circuit plasticity is largely unknown. Isn’t changing synapses in this way plasticity? Six questions regarding the potential roles of Npas4 in regulating neural circuits are listed, and are discussed further in the Outstanding Questions Box.
Table I.
Some well-known IEGs
| Gene | Full Name | Neuron- specific |
mRNA peak time |
Cellular function |
Roles in synaptic plasticity |
Learning and memory paradigm tested |
References |
|---|---|---|---|---|---|---|---|
| c-Fos | FBJ osteosarcoma oncogene | No | Early | Transcription factor | LTP | Morris water maze; Fear conditioning; | [2, 58] |
| Zif268 (Egr1) | Early growth response 1 | No | Early | Transcription factor | LTP | Morris water maze; conditioned taste aversion; objection recognition | [59, 60] |
| C/EBP | CCAAT/enhancer binding protein | No | Late | Transcription factor | LTF, LTP | Inhibitory avoidance; Morris water maze | [61–63] |
| Npas4 | Neuronal PAS domain protein 4 | Yes | Early | Transcription factor | Not characterized | Fear conditioning | [5, 6, 10] |
| NR4A1 (Nur77) | Nuclear receptor subfamily 4 group A member 1 | No | Early | Transcription factor | LTP | Fear conditioning | [64, 65] |
| Arc | Activity-regulated cytoskeleton- associated protein | No | Early | Cytoskeletal associated protein | LTP, LTD, Homeostatic scaling | Morris water maze; fear conditioning; objection recognition | [4, 66, 67] |
| BDNF | Brain-derived neurotrophic factor | No | Late | Neurotrophic factors | LTP | Morris water maze; fear conditioning | [68, 69] |
| Narp (Nptx2) | Neuronal pentraxin 2 | No | Late | Secreted proteins | Homeostatic scaling | Food devaluation; extinction of conditioned place preference | [40, 70, 71] |
| Homer1a | Homer scaffolding protein 1 | No | Late | Synaptic scaffolding proteins | Homeostatic scaling | Morris water maze | [72–74] |
| Cpg15 | Neuritin 1 | No | Late | Extracellular surface proteins | Not determined | Morris water maze; fear conditioning | [75–77] |
mRNA peak time: “Early” indicates mRNA peaks 5–60 minutes after stimulation; “Late” indicates mRNA peaks after 60 minutes.
Outstanding Questions Box.
- To what extent does Npas4 regulate synapses in an input-specific manner, and what is the mechanism?
- In hippocampal CA1 pyramidal neurons, Npas4 may differentially regulate perisomatic and distal inhibitory inputs onto the cells. To what extent does similar input-specific regulation occur for other types of synaptic inputs (such as excitatory synapses), in other cell types (such as inhibitory neurons) and in other regions of the brain? Also, by what mechanism is this input-specificity achieved?
- To what extent does Npas4 function differently in different subtypes of neurons, and what is the mechanism?
- Npas4 has been shown to upregulate inhibitory inputs in excitatory neurons, but increase excitatory inputs in SST+ GABAergic interneurons. Does Npas4 have the same function in all subtypes of excitatory neurons? How about in other subtypes of inhibitory neurons? By what mechanism are these cell type differences achieved?
- Does Npas4 regulate the intrinsic excitability of neurons and, if so, by what mechanism?
- Given that Npas4 is known to affect the expression of several ion channels, it seems quite likely that Npas4’s functions include regulation of intrinsic excitability. It would be interesting to determine whether Npas4 does in fact have this function and, if so, what mechanism is used.
- Do Npas4+ neurons form interconnected subnetworks?
- In Npas4+ excitatory neurons, do the inhibitory inputs that are upregulated in response to activity come directly from Npas4+ inhibitory neurons? Similarly, do activated Npas4+ excitatory neurons send stronger excitatory inputs onto Npas4+ inhibitory neurons? The formation of such subnetworks may facilitate memory encoding within larger neural ensembles.
- Does Npas4 function cell-autonomously?
- Npas4 is expressed only in a sparse population of neurons under physiological conditions. Do activity-dependent responses in Npas4+ neurons modulate the function of Npas4− neurons, for example through secreted factors or modification of the synaptic connections between them.
- Does Npas4 regulate neural circuit homeostasis?
- Npas4 has been implicated in maintaining homeostasis of activity levels in neural circuits, probably by increasing inhibitory inputs on excitatory neurons and elevating excitatory drive onto inhibitory neurons. Future studies are needed to test this idea directly using a homeostatic plasticity paradigm. Additionally, are these homeostatic responses restricted to Npas4+ neurons, with Npas4+ and Npas4− neurons perhaps displaying different homeostatic responses?
Trends Box.
Memory formation requires converting experience-induced neuronal activity into long lasting changes in the brain. This process generally requires activity-dependent gene transcription.
A neuron-specific immediate-early gene (IEG), Npas4 is induced only by neuronal activity.
In learning and memory paradigms, Npas4 induction is more tightly associated with manipulations that will result in long term memory than other well-known IEGs, making it a unique molecular link for memory formation.
Npas4 is involved in activity-dependent synaptic modulation in both excitatory and inhibitory neurons. It regulates the expression of a large number of activity-regulated genes that mediate diverse effects on synapses.
Npas4 function is required for memory formation in multiple brain regions. It has also been implicated in neural circuit plasticity, although here detailed studies are still lacking.
Acknowledgments
The authors wish to thank C. M. Fletcher for critical reading of the manuscript. Supported by the John Merck Scholar Program and NIH grant MH091220-01. Y.L. is a Fred and Carole Middleton Career Development Professor.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Morgan JI, Curran T. Calcium as a modulator of the immediate-early gene cascade in neurons. Cell Calcium. 1988;9(5–6):303–311. doi: 10.1016/0143-4160(88)90011-5. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg ME, Ziff EB. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311(5985):433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 3.Saffen DW, et al. Convulsant-induced increase in transcription factor messenger RNAs in rat brain. Proc Natl Acad Sci U S A. 1988;85(20):7795–7799. doi: 10.1073/pnas.85.20.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyford GL, et al. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14(2):433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- 5.Lin Y, et al. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455(7217):1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramamoorthi K, et al. Npas4 regulates a transcriptional program in CA3 required for contextual memory formation. Science. 2011;334(6063):1669–1675. doi: 10.1126/science.1208049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spiegel I, et al. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell. 2014;157(5):1216–1229. doi: 10.1016/j.cell.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sim S, et al. Increased cell-intrinsic excitability induces synaptic changes in new neurons in the adult dentate gyrus that require Npas4. J Neurosci. 2013;33(18):7928–7940. doi: 10.1523/JNEUROSCI.1571-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshihara S, et al. Npas4 regulates Mdm2 and thus Dcx in experience-dependent dendritic spine development of newborn olfactory bulb interneurons. Cell Rep. 2014;8(3):843–857. doi: 10.1016/j.celrep.2014.06.056. [DOI] [PubMed] [Google Scholar]
- 10.Ploski JE, et al. The neuronal PAS domain protein 4 (Npas4) is required for new and reactivated fear memories. PLoS One. 2011;6(8):e23760. doi: 10.1371/journal.pone.0023760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herschman HR. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- 12.Bersten DC, et al. Human variants in the neuronal basic helix-loop-helix/Per-Arnt-Sim (bHLH/PAS) transcription factor complex NPAS4/ARNT2 disrupt function. PLoS One. 2014;9(1):e85768. doi: 10.1371/journal.pone.0085768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bersten DC, et al. Regulation of the neuronal transcription factor NPAS4 by REST and microRNAs. Biochim Biophys Acta. 2014;1839(1):13–24. doi: 10.1016/j.bbagrm.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Zhang SJ, et al. Nuclear calcium signaling controls expression of a large gene pool: identification of a gene program for acquired neuroprotection induced by synaptic activity. PLoS Genet. 2009;5(8):e1000604. doi: 10.1371/journal.pgen.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coba MP, et al. Kinase networks integrate profiles of N-methyl-D-aspartate receptor-mediated gene expression in hippocampus. J Biol Chem. 2008;283(49):34101–34107. doi: 10.1074/jbc.M804951200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuzniewska B, et al. Adult Deletion of SRF Increases Epileptogenesis and Decreases Activity-Induced Gene Expression. Mol Neurobiol. 2015 doi: 10.1007/s12035-014-9089-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saha RN, et al. Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nat Neurosci. 2011;14(7):848–856. doi: 10.1038/nn.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madabhushi R, et al. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell. 2015;161(7):1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo ML, et al. Upregulation of Npas4 protein expression by chronic administration of amphetamine in rat nucleus accumbens in vivo. Neurosci Lett. 2012 doi: 10.1016/j.neulet.2012.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin TA, et al. Methamphetamine causes differential alterations in gene expression and patterns of histone acetylation/hypoacetylation in the rat nucleus accumbens. PLoS One. 2012;7(3):e34236. doi: 10.1371/journal.pone.0034236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yun J, et al. Chronic restraint stress impairs neurogenesis and hippocampus-dependent fear memory in mice: possible involvement of a brain-specific transcription factor Npas4. J Neurochem. 2010;114(6):1840–1851. doi: 10.1111/j.1471-4159.2010.06893.x. [DOI] [PubMed] [Google Scholar]
- 22.Ramamoorthi K, Lin Y. The contribution of GABAergic dysfunction to neurodevelopmental disorders. Trends Mol Med. 2011;17(8):452–462. doi: 10.1016/j.molmed.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flood WD, et al. Nxf and Fbxo33: novel seizure-responsive genes in mice. Eur J Neurosci. 2004;20(7):1819–1826. doi: 10.1111/j.1460-9568.2004.03646.x. [DOI] [PubMed] [Google Scholar]
- 24.Ooe N, et al. Functional characterization of basic helix-loop-helix-PAS type transcription factor NXF in vivo: putative involvement in an "on demand" neuroprotection system. J Biol Chem. 2009;284(2):1057–1063. doi: 10.1074/jbc.M805196200. [DOI] [PubMed] [Google Scholar]
- 25.Shamloo M, et al. Npas4, a novel helix-loop-helix PAS domain protein, is regulated in response to cerebral ischemia. Eur J Neurosci. 2006;24(10):2705–2720. doi: 10.1111/j.1460-9568.2006.05172.x. [DOI] [PubMed] [Google Scholar]
- 26.Leong WK, et al. Upregulation of the neuronal Per-Arnt-Sim domain protein 4 (Npas4) in the rat corticolimbic system following focal cerebral ischemia. Eur J Neurosci. 2013;37(11):1875–1884. doi: 10.1111/ejn.12163. [DOI] [PubMed] [Google Scholar]
- 27.Lee I, Kesner RP. Differential contributions of dorsal hippocampal subregions to memory acquisition and retrieval in contextual fear-conditioning. Hippocampus. 2004;14(3):301–310. doi: 10.1002/hipo.10177. [DOI] [PubMed] [Google Scholar]
- 28.Nakashiba T, et al. Hippocampal CA3 output is crucial for ripple-associated reactivation and consolidation of memory. Neuron. 2009;62(6):781–787. doi: 10.1016/j.neuron.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakashiba T, et al. Transgenic inhibition of synaptic transmission reveals role of CA3 output in hippocampal learning. Science. 2008;319(5867):1260–1264. doi: 10.1126/science.1151120. [DOI] [PubMed] [Google Scholar]
- 30.Kaliszewska A, Kossut M. Npas4 expression in two experimental models of the barrel cortex plasticity. Neural Plast. 2015;2015:175701. doi: 10.1155/2015/175701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo ML, et al. Upregulation of Npas4 protein expression by chronic administration of amphetamine in rat nucleus accumbens in vivo. Neurosci Lett. 2012;528(2):210–214. doi: 10.1016/j.neulet.2012.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coutellier L, Gilbert V, Shepard R. Npas4 deficiency increases vulnerability to juvenile stress in mice. Behav Brain Res. 2015 doi: 10.1016/j.bbr.2015.04.027. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, et al. Hippocampal expression of aryl hydrocarbon receptor nuclear translocator 2 and neuronal PAS domain protein 4 in a rat model of depression. Neurological Sciences. 2013;35(2):277–282. doi: 10.1007/s10072-013-1505-7. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z, et al. Decreased expression of neuronal Per-Arnt-Sim domain protein 4 gene in the hippocampus of a post-stroke depression rat model. Exp Ther Med. 2014;7(4):1045–1049. doi: 10.3892/etm.2014.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furukawa-Hibi Y, et al. Transcriptional suppression of the neuronal PAS domain 4 (Npas4) gene by stress via the binding of agonist-bound glucocorticoid receptor to its promoter. Journal of Neurochemistry. 2012;123(5):866–875. doi: 10.1111/jnc.12034. [DOI] [PubMed] [Google Scholar]
- 36.Jaehne EJ, et al. Effects of Npas4 deficiency on anxiety, depression-like, cognition and sociability behaviour. Behav Brain Res. 2015;281:276–282. doi: 10.1016/j.bbr.2014.12.044. [DOI] [PubMed] [Google Scholar]
- 37.Bloodgood BL, et al. The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature. 2013;503(7474):121–125. doi: 10.1038/nature12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang ZJ, et al. BDNF regulates the maturation of inhibition and the critical period of plasticity in mouse visual cortex. Cell. 1999;98(6):739–755. doi: 10.1016/s0092-8674(00)81509-3. [DOI] [PubMed] [Google Scholar]
- 39.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2012;465(7295):182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang MC, et al. Narp regulates homeostatic scaling of excitatory synapses on parvalbumin-expressing interneurons. Nat Neurosci. 2010;13(9):1090–1097. doi: 10.1038/nn.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greenberg ME, et al. New insights in the biology of BDNF synthesis and release: implications in CNS function. J Neurosci. 2009;29(41):12764–12767. doi: 10.1523/JNEUROSCI.3566-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shepherd JD, et al. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52(3):475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maya-Vetencourt JF, et al. Experience-dependent expression of NPAS4 regulates plasticity in adult visual cortex. J Physiol. 2012 doi: 10.1113/jphysiol.2012.234237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guidotti G, et al. Developmental influence of the serotonin transporter on the expression of npas4 and GABAergic markers: modulation by antidepressant treatment. Neuropsychopharmacology. 2012;37(3):746–758. doi: 10.1038/npp.2011.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maya Vetencourt JF, et al. The antidepressant fluoxetine restores plasticity in the adult visual cortex. Science. 2008;320(5874):385–388. doi: 10.1126/science.1150516. [DOI] [PubMed] [Google Scholar]
- 46.Maya Vetencourt JF, et al. Serotonin triggers a transient epigenetic mechanism that reinstates adult visual cortex plasticity in rats. Eur J Neurosci. 2011;33(1):49–57. doi: 10.1111/j.1460-9568.2010.07488.x. [DOI] [PubMed] [Google Scholar]
- 47.Liu X, et al. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484(7394):381–385. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denny CA, et al. Hippocampal memory traces are differentially modulated by experience, time, and adult neurogenesis. Neuron. 2014;83(1):189–201. doi: 10.1016/j.neuron.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jayaraman R. Transcription of bacteriophage T4 DNA by Escherichia coli RNA polymerase in vitro: identification of some immediate-early and delayed-early genes. J Mol Biol. 1972;70(2):253–263. doi: 10.1016/0022-2836(72)90537-2. [DOI] [PubMed] [Google Scholar]
- 50.Curran T, Morgan JI. Superinduction of c-fos by nerve growth factor in the presence of peripherally active benzodiazepines. Science. 1985;229(4719):1265–1268. doi: 10.1126/science.4035354. [DOI] [PubMed] [Google Scholar]
- 51.Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loebrich S, Nedivi E. The function of activity-regulated genes in the nervous system. Physiol Rev. 2009;89(4):1079–1103. doi: 10.1152/physrev.00013.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hiatt SM, et al. Caenorhabditis elegans FOS-1 and JUN-1 regulate plc-1 expression in the spermatheca to control ovulation. Mol Biol Cell. 2009;20(17):3888–3895. doi: 10.1091/mbc.E08-08-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cyriac A, et al. An Aplysia Egr homolog is rapidly and persistently regulated by long-term sensitization training. Neurobiol Learn Mem. 2013;102:43–51. doi: 10.1016/j.nlm.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guan Z, et al. Genome-wide transcriptional changes associated with enhanced activity in the Drosophila nervous system. Neuron. 2005;48(1):91–107. doi: 10.1016/j.neuron.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 56.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malik AN, et al. Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nat Neurosci. 2014;17(10):1330–1339. doi: 10.1038/nn.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fleischmann A, et al. Impaired long-term memory and NR2A-type NMDA receptor-dependent synaptic plasticity in mice lacking c-Fos in the CNS. J Neurosci. 2003;23(27):9116–9122. doi: 10.1523/JNEUROSCI.23-27-09116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richardson CL, et al. Correlation between the induction of an immediate early gene, zif/268, and long-term potentiation in the dentate gyrus. Brain Res. 1992;580(1–2):147–154. doi: 10.1016/0006-8993(92)90938-6. [DOI] [PubMed] [Google Scholar]
- 60.Jones MW, et al. A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat Neurosci. 2001;4(3):289–296. doi: 10.1038/85138. [DOI] [PubMed] [Google Scholar]
- 61.Alberini CM, et al. C/EBP is an immediate-early gene required for the consolidation of long-term facilitation in Aplysia. Cell. 1994;76(6):1099–1114. doi: 10.1016/0092-8674(94)90386-7. [DOI] [PubMed] [Google Scholar]
- 62.Taubenfeld SM, et al. The consolidation of new but not reactivated memory requires hippocampal C/EBPbeta. Nat Neurosci. 2001;4(8):813–818. doi: 10.1038/90520. [DOI] [PubMed] [Google Scholar]
- 63.Chen A, et al. Inducible enhancement of memory storage and synaptic plasticity in transgenic mice expressing an inhibitor of ATF4 (CREB-2) and C/EBP proteins. Neuron. 2003;39(4):655–669. doi: 10.1016/s0896-6273(03)00501-4. [DOI] [PubMed] [Google Scholar]
- 64.Hawk JD, et al. NR4A nuclear receptors support memory enhancement by histone deacetylase inhibitors. J Clin Invest. 2012;122(10):3593–3602. doi: 10.1172/JCI64145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bridi MS, Abel T. The NR4A orphan nuclear receptors mediate transcription-dependent hippocampal synaptic plasticity. Neurobiol Learn Mem. 2013;105:151–158. doi: 10.1016/j.nlm.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Plath N, et al. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52(3):437–444. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 67.Shepherd JD, Bear MF. New views of Arc, a master regulator of synaptic plasticity. Nat Neurosci. 2011;14(3):279–284. doi: 10.1038/nn.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Korte M, et al. Hippocampal long-term potentiation is impaired in mice lacking brain-derived neurotrophic factor. Proc Natl Acad Sci U S A. 1995;92(19):8856–8860. doi: 10.1073/pnas.92.19.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu Y, Christian K, Lu B. BDNF: a key regulator for protein synthesis-dependent LTP and long-term memory? Neurobiol Learn Mem. 2008;89(3):312–323. doi: 10.1016/j.nlm.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsui CC, et al. Narp, a novel member of the pentraxin family, promotes neurite outgrowth and is dynamically regulated by neuronal activity. J Neurosci. 1996;16(8):2463–2478. doi: 10.1523/JNEUROSCI.16-08-02463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson AW, et al. A selective role for neuronal activity regulated pentraxin in the processing of sensory-specific incentive value. J Neurosci. 2007;27(49):13430–13435. doi: 10.1523/JNEUROSCI.4320-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brakeman PR, et al. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386(6622):284–288. doi: 10.1038/386284a0. [DOI] [PubMed] [Google Scholar]
- 73.Jaubert PJ, et al. Complex, multimodal behavioral profile of the Homer1 knockout mouse. Genes Brain Behav. 2007;6(2):141–154. doi: 10.1111/j.1601-183X.2006.00240.x. [DOI] [PubMed] [Google Scholar]
- 74.Hu JH, et al. Homeostatic scaling requires group I mGluR activation mediated by Homer1a. Neuron. 2010;68(6):1128–1142. doi: 10.1016/j.neuron.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nedivi E, et al. A set of genes expressed in response to light in the adult cerebral cortex and regulated during development. Proc Natl Acad Sci U S A. 1996;93(5):2048–2053. doi: 10.1073/pnas.93.5.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fujino T, Lee WC, Nedivi E. Regulation of cpg15 by signaling pathways that mediate synaptic plasticity. Mol Cell Neurosci. 2003;24(3):538–554. doi: 10.1016/s1044-7431(03)00230-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fujino T, et al. CPG15 regulates synapse stability in the developing and adult brain. Genes Dev. 2011;25(24):2674–2685. doi: 10.1101/gad.176172.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Crews ST, Fan CM. Remembrance of things PAS: regulation of development by bHLH-PAS proteins. Curr Opin Genet Dev. 1999;9(5):580–587. doi: 10.1016/s0959-437x(99)00003-9. [DOI] [PubMed] [Google Scholar]
- 79.Moser M, et al. LE-PAS, a novel Arnt-dependent HLH-PAS protein, is expressed in limbic tissues and transactivates the CNS midline enhancer element. Brain Res Mol Brain Res. 2004;128(2):141–149. doi: 10.1016/j.molbrainres.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 80.Ooe N, Saito K, Kaneko H. Characterization of functional heterodimer partners in brain for a bHLH-PAS factor NXF. Biochim Biophys Acta. 2009;1789(3):192–197. doi: 10.1016/j.bbagrm.2009.01.003. [DOI] [PubMed] [Google Scholar]


