Abstract
Apolipoprotein E (APOE) gene has been the most replicated longevity-associated gene in humans. Two common APOE alleles are either significantly depleted (ε4 allele) or enriched (ε2 allele) in long-lived individuals as compared to controls. We performed high-throughput sequencing analysis of exons and 2 kb proximal promoter of APOE in 450 centenarians and 500 controls of Ashkenazi Jewish decent. We found two common regulatory variants, rs405509 (p=0.006) and rs769449 (p=0.036), that were significantly depleted in centenarians. Genotyping analysis of rs7412 and rs429358 showed significant enrichment of ε2 allele (p=0.003) and ε2/ε3 genotype (p= 0.005), and significant depletion of ε3/ε4 genotype (p=0.005) in centenarians. Our findings support the hypothesis that variants in both coding and regulatory regions of APOE may contribute to longevity in humans.
Keywords: APOE, Centenarian, Genetic variant, Longevity, Pooled target capture sequencing
Description
Genome-wide association studies (GWAS) have achieved great success in identifying common genetic variants robustly associated with increased risk of complex traits and diseases (http://www.genome.gov/gwastudies). Thus far, GWAS involving long-lived individuals have identified the chromosome 19q13.2 harboring APOE gene as the single most replicated longevity-associated locus, confirming the previous single gene association studies. (Beekman et al., 2013; Deelen et al., 2011; Nebel et al., 2011; Sebastiani et al., 2012). APOE has two common missense variants, rs429358 (Cys130Arg) and rs7412 (Arg176Cys), and a combination of the two determines functional alleles of APOE: ε2 (Cys130, Cys176), ε3 (Cys130, Arg176) and ε4 (Arg130, Arg176). While depletion of APOE ε4 allele and/or enrichment of APOE ε2 allele has been found to be associated with longevity, APOE ε4 allele has been associated with risk of multiple age-related disease such as Alzheimer’s disease and cardiovascular disease (Christensen et al., 2006), and supporting the role of the common functional APOE variants in healthspan and lifespan.
In this study, to further discover all possible variants in coding and regulatory region of APOE gene, we took advantage of an efficient pooled target capture next-generation sequencing approach (Pool-seq) (Bansal et al., 2011). Our study population consist of 450 Ashkenazi Jewish centenarians (mean age: 98) older than 95 years old and 500 controls (mean age: 73) without family history of longevity. Using this population, we have successfully identified longevity associated-phenotypes, -genotypes, and their association with health outcomes (Atzmon et al., 2006; Barzilai et al., 2006; Barzilai et al., 2003). In particular, we identified two rare missense variants in the IGF-1 receptor gene (IGF1R) that were enriched in Ashkenazi Jewish centenarians as compared to younger elderly and we further showed that they were true reduced-function variants (Suh et al., 2008; Tazearslan et al., 2011). These results highlight the value of resequencing candidate genes in our Ashkenazi Jewish centenarian cohort to identify clinically relevant targets in humans. As a result of this study, we discovered 33 common and rare variants in both coding and regulatory region of APOE gene, 17 of them were novel variants not identified previously (Figure 1 and Supplementary Table 1). Among them, longevity-associated common variants were rs405509 (p=0.006) and rs769449 (p=0.036) in upstream and intronic region respectively, that were depleted in centenarians and may be functional based on the prediction by RegulomeDB score (http://regulomedb.org/) (Supplementary Table 1).
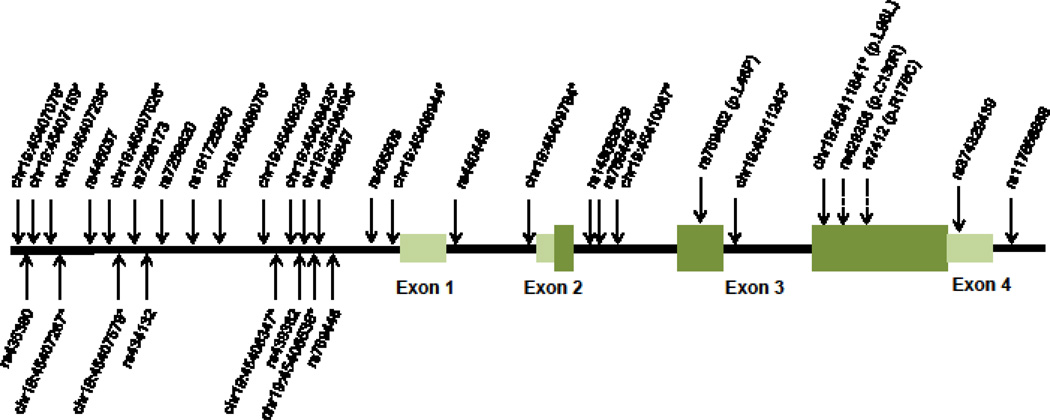
Figure 1. Discovered variants in APOE gene from Pool-seq and genotyping in centenarians and controls.
The variants in APOE gene discovered from Pool-seq were indicated on gene structure. Discovered variants were named by NCBI rsSNP ID for known variants and chromosomal coordinates of human genome (hg19) for novel variants not reported on database. Novel variants were indicated by asterisk and variants for genotyping were indicated as dotted arrow.
Our target capture sequencing performed poorly in exon 4 region of APOE gene harboring previously reported longevity-associated alleles (rs429358 (c.T388C:p.C130R) and rs7412 (c.C526T:p.R176C)) because of the high GC content (Supplementary Figure 1). Therefore, to validate previously reported longevity-associated APOE alleles, we performed genotyping of rs7412 and rs427580 using an independent Taqman (Life Technologies) SNP genotyping assay. As a result of a successful genotyping in 438 centenarians and 515 controls, rs7412 was significantly enriched (p-value: 0.001), while rs429358 was significantly depleted (p-value: 0.038) in centenarians (Table 1). Based on the genotyped variants, APOE ε2 (rs429328: T / rs7412: C>T), ε3 (rs429328: T / rs7412: C), and ε4 (rs429328: T>C / rs7412: C) alleles were determined. We examined the association of APOE alleles with longevity using allelic test of association and found that ε2 allele was significantly enriched in centenarians (9.82%) as compared to controls (6.02%) (Odd Ratio (OR): 1.70, p-value: 0.003), whereas ε4 allele was depleted in centenarians (6.62%) as compared to controls (9.03%) (OR: 0.71, p-value: 0.061) (Table 1). In addition, the genotypic test of association revealed that ε2/ε3 genotype was enriched in centenarians (16.4%) as compared to controls (10.1%) (OR: 1.75, p-value: 0.005), conversely, ε3/ε4 genotype was significantly depleted in centenarians (9.59%) compared to controls (15.7%) (OR: 0.57, p-value: 0.005) (Table 1). These results are in concordance with previous reports that APOE ε2 is enriched and APOE ε4 is depleted in centenarians (Christensen et al., 2006). Here we validated longevity-associated APOE ε2 allele in our Ashkenazi Jewish population.
Table 1.
Association analysis of APOE genotypes and alleles with longevity and lipid profile.
| Type | Frequency (centenarians n=438, controls n=515) | LDL level | |||||
|---|---|---|---|---|---|---|---|
| Centenarians (%) |
Controls (%) |
OR (95% CI) | p-value | Type | The rest | p-value | |
| ε2 | 86 (9.82) | 62 (6.02) | 1.70 (1.21–2.39) | 0.003* | 91.7+/−5.26 | 109+/−1.51 | 0.002* |
| ε3 | 732 (83.6) | 875 (85) | 0.90 (0.70–1.15) | 0.412 | 108+/−1.57 | 102+/−3.67 | 0.11 |
| ε4 | 58 (6.62) | 93 (9.03) | 0.71 (0.51–1.00) | 0.061 | 112+/−5.09 | 107+/−1.52 | 0.41 |
| ε2/ε2 | 4 (0.91) | 2 (0.39) | 2.36 (0.43–13.0) | 0.422 | 74.5+/−21.6 | 108+/−2.07 | 0.13 |
| ε2/ε3 | 72 (16.4) | 52 (10.1) | 1.75 (1.20–2.57) | 0.005* | 93.9+/−5.75 | 109+/−2.19 | 0.01* |
| ε2/ε4 | 6 (1.37) | 6 (1.17) | 1.18 (0.38–3.68) | 0.780 | 94+/−27.8 | 108+/−2.08 | 0.63 |
| ε3/ε3 | 309 (70.5) | 371 (72) | 0.93 (0.70–1.23) | 0.616 | 109+/−2.4 | 103+/−3.86 | 0.15 |
| ε3/ε4 | 42 (9.59) | 81 (15.7) | 0.57 (0.38–0.85) | 0.005* | 113+/−5.63 | 107+/−2.2 | 0.30 |
| ε4/ε4 | 5 (1.14) | 3 (0.58) | 1.97 (0.47–8.29) | 0.481 | 109+/−19.7 | 107+/−2.09 | 0.92 |
Bold numerals with asterisk indicate p-value < 0.05.
To investigate the effect of the APOE gene alleles and genotypes on phenotypes such as lipid metabolism, we tested its association with lipid profiles including serum level of cholesterol, HDL, LDL, and triglyceride. Carriers of the APOE ε2 allele showed significantly lower LDL levels compared to the ε3 and ε4 carriers (91.7 +/− 5.26 vs.109 +/− 1.51 mg/dL, p=0.002) (Table 1). Serum cholesterol levels were lower in ε2 allele carriers with marginal significance (p-value: 0.052) (Supplementary Table 2). On the other hand, the ε3 and ε4 alleles were not associated with lipid profile even though ε3 and ε4 allele carriers showed a trend of increased level compared to ε2 allele carriers (Table 1). In addition, LDL level was significantly lower among ε2/ε3 genotype carriers compared with the other genotype carriers (93.9 +/− 5.75 vs.109 +/− 2.19 mg/dL, p=0.01) that is concordant with the effect of ε2 allele (Table 1).
In conclusion, we discovered all possible variants in coding and regulatory region of APOE gene in centenarians and controls by an efficient Pool-seq in large population. Interestingly, Poolseq with target region including regulatory region such as proximal upstream and intronic region identified longevity-associated common variants, rs405509 and rs769449 that were previously associated with longevity and other aging-related disease phenotypes (Lu et al., 2014; Soerensen et al., 2013). They are also predicted (Supplementary Figure 2) and experimentally proved (Supplementary Table 3) to be functional, implicating the potential role of regulatory variants in a longevity-association. Moreover, by independent genotyping, we replicated the significant enrichment of longevity-associated APOE ε2 allele and ε2/ε3 genotype among our Ashkenazi Jewish cohort for the first time, and we also validated the association of APOE ε2 allele with beneficial low LDL level as reported previously (Ferreira et al., 2010; Volcik et al., 2006), suggesting a functional effect of the APOE allele and genotype that may lead to longevity. Therefore, it would be interesting to further understand a protective role of longevity-associated, predicted functional variants in APOE gene.
Supplementary Material
Highlights.
-
►
Discovery of variants in APOE gene region by pooled target capture sequencing
-
►
Identification of longevity-associated variants in regulatory region of APOE gene
-
►
Validation of longevity association of APOE ε2 allele in Ashkenazi population
-
►
Association of longevity-associated allele with beneficial lipid profiles
Acknowledgments
We would like to thank AT for critical reading of the manuscript. This work was supported by NIH grants AG017242, GM104459, and CA180126, Einstein Glenn Center and the Paul Glenn foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors declare that they have no competing interests.
References
- Atzmon G, Rincon M, Schechter CB, Shuldiner AR, Lipton RB, Bergman A, Barzilai N. Lipoprotein genotype and conserved pathway for exceptional longevity in humans. PLoS biology. 2006;4:e113. doi: 10.1371/journal.pbio.0040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal V, Tewhey R, Leproust EM, Schork NJ. Efficient and cost effective population resequencing by pooling and in-solution hybridization. PLoS One. 2011;6:e18353. doi: 10.1371/journal.pone.0018353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzilai N, Atzmon G, Derby CA, Bauman JM, Lipton RB. A genotype of exceptional longevity is associated with preservation of cognitive function. Neurology. 2006;67:2170–2175. doi: 10.1212/01.wnl.0000249116.50854.65. [DOI] [PubMed] [Google Scholar]
- Barzilai N, Atzmon G, Schechter C, Schaefer EJ, Cupples AL, Lipton R, Cheng S, Shuldiner AR. Unique lipoprotein phenotype and genotype associated with exceptional longevity. JAMA. 2003;290:2030–2040. doi: 10.1001/jama.290.15.2030. [DOI] [PubMed] [Google Scholar]
- Beekman M, Blanche H, Perola M, Hervonen A, Bezrukov V, Sikora E, Flachsbart F, Christiansen L, De Craen AJ, Kirkwood TB, Rea IM, Poulain M, Robine JM, Valensin S, Stazi MA, Passarino G, Deiana L, Gonos ES, Paternoster L, Sorensen TI, Tan Q, Helmer Q, van den Akker EB, Deelen J, Martella F, Cordell HJ, Ayers KL, Vaupel JW, Tornwall O, Johnson TE, Schreiber S, Lathrop M, Skytthe A, Westendorp RG, Christensen K, Gampe J, Nebel A, Houwing-Duistermaat JJ, Slagboom PE, Franceschi C consortium, G. Genome-wide linkage analysis for human longevity: Genetics of Healthy Aging Study. Aging cell. 2013;12:184–193. doi: 10.1111/acel.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen K, Johnson TE, Vaupel JW. The quest for genetic determinants of human longevity: challenges and insights. Nat Rev Genet. 2006;7:436–448. doi: 10.1038/nrg1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelen J, Beekman M, Uh HW, Helmer Q, Kuningas M, Christiansen L, Kremer D, van der Breggen R, Suchiman HE, Lakenberg N, van den Akker EB, Passtoors WM, Tiemeier H, van Heemst D, de Craen AJ, Rivadeneira F, de Geus EJ, Perola M, van der Ouderaa FJ, Gunn DA, Boomsma DI, Uitterlinden AG, Christensen K, van Duijn CM, Heijmans BT, Houwing-Duistermaat JJ, Westendorp RG, Slagboom PE. Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging cell. 2011;10:686–698. doi: 10.1111/j.1474-9726.2011.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira CN, Carvalho MG, Fernandes AP, Lima LM, Loures-Valle AA, Dantas J, Janka Z, Palotas A, Sousa MO. Comparative study of apolipoprotein-E polymorphism and plasma lipid levels in dyslipidemic and asymptomatic subjects, and their implication in cardio/cerebro-vascular disorders. Neurochem Int. 2010;56:177–182. doi: 10.1016/j.neuint.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Lu F, Guan H, Gong B, Liu X, Zhu R, Wang Y, Qian J, Zhou T, Lan X, Wang P, Lin Y, Ma S, Lin H, Zhu X, Chen R, Zhu X, Shi Y, Yang Z. Genetic variants in PVRL2-TOMM40-APOE region are associated with human longevity in a Han Chinese population. PLoS One. 2014;9:e99580. doi: 10.1371/journal.pone.0099580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebel A, Kleindorp R, Caliebe A, Nothnagel M, Blanche H, Junge O, Wittig M, Ellinghaus D, Flachsbart F, Wichmann HE, Meitinger T, Nikolaus S, Franke A, Krawczak M, Lathrop M, Schreiber S. A genome-wide association study confirms APOE as the major gene influencing survival in long-lived individuals. Mechanisms of ageing and development. 2011;132:324–330. doi: 10.1016/j.mad.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Sebastiani P, Solovieff N, Dewan AT, Walsh KM, Puca A, Hartley SW, Melista E, Andersen S, Dworkis DA, Wilk JB, Myers RH, Steinberg MH, Montano M, Baldwin CT, Hoh J, Perls TT. Genetic signatures of exceptional longevity in humans. PLoS One. 2012;7:e29848. doi: 10.1371/journal.pone.0029848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soerensen M, Dato S, Tan Q, Thinggaard M, Kleindorp R, Beekman M, Suchiman HE, Jacobsen R, McGue M, Stevnsner T, Bohr VA, de Craen AJ, Westendorp RG, Schreiber S, Slagboom PE, Nebel A, Vaupel JW, Christensen K, Christiansen L. Evidence from case-control and longitudinal studies supports associations of genetic variation in APOE, CETP, and IL6 with human longevity. Age (Dordr) 2013;35:487–500. doi: 10.1007/s11357-011-9373-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y, Atzmon G, Cho MO, Hwang D, Liu B, Leahy DJ, Barzilai N, Cohen P. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3438–3442. doi: 10.1073/pnas.0705467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazearslan C, Huang J, Barzilai N, Suh Y. Impaired IGF1R signaling in cells expressing longevity-associated human IGF1R alleles. Aging cell. 2011;10:551–554. doi: 10.1111/j.1474-9726.2011.00697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volcik KA, Barkley RA, Hutchinson RG, Mosley TH, Heiss G, Sharrett AR, Ballantyne CM, Boerwinkle E. Apolipoprotein E polymorphisms predict low density lipoprotein cholesterol levels and carotid artery wall thickness but not incident coronary heart disease in 12,491 ARIC study participants. Am J Epidemiol. 2006;164:342–348. doi: 10.1093/aje/kwj202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.