Abstract
Purpose
To test second-line personalized medicine combination therapies, based on genomic and proteomic data, in patient-derived xenograft (PDX) models.
Methods
We established 12 PDX from BRAF inhibitor progressed melanoma patients. Following expansion, PDX were analyzed using targeted sequencing and reverse phase protein arrays (RPPA). By using multi-arm pre-clinical trial designs, we identified efficacious precision medicine approaches.
Results
We identified alterations previously described as drivers of resistance: NRAS mutations in 3 PDX, MAP2K1 (MEK1) mutations in 2, BRAF amplification in 4, and aberrant PTEN in 7. At the protein level, re-activation of phospho MAPK predominated, with parallel activation of PI3K in a subset. Second line efficacy of the pan-PI3K inhibitor BKM120 with either BRAF (encorafenib) /MEK (binimetinib) inhibitor combination or the ERK inhibitor VX-11e was confirmed in vivo. Amplification of MET was observed in 3 PDX models, a higher frequency than expected and a possible novel mechanism of resistance. Importantly, MET amplification alone did not predict sensitivity to the MET inhibitor capmatinib. In contrast, capmatinib as single agent resulted in significant but transient tumor regression in a PDX with resistance to BRAF/MEK combination therapy and high pMET. The triple combination capmatinib/encorafenib/binimetinib resulted incomplete and sustained tumor regression in all animals.
Conclusions
Genomic and proteomic data integration identifies dual core pathway inhibition as well as MET as combinatorial targets. These studies provide evidence for biomarker development to appropriately select patients' personalized therapies and avoid treatment failures.
Keywords: melanoma, patient-derived xenografts, targeted therapy, BRAF, MET
Introduction
The treatment of advanced melanoma has been significantly improved in recent years, enabled by BRAF and MEK inhibitors as new standard therapies in melanomas with BRAF-V600E/K mutations (1, 2) and immune checkpoint inhibitors showing remarkably durable responses in a subset of patients (3-5). Although the majority of patients treated with BRAF or BRAF/MEK inhibitors experience a robust initial response, the excitement about the therapeutic success is dampened by the relapse of most patients. This is due to the development of acquired (secondary) resistance mediated by multiple mechanisms (6-10). Therefore, rational second line combination therapies are urgently needed and we expect that these therapies require individualization to the spectrum of each patient's resistance mechanism (11). There is a lack of translational models to study precision medicine approaches to resistance mechanisms found in patients, although a range of preclinical mouse melanoma models, including patient-derived xenografts (PDX), are in use (12). PDX have been successfully established for solid tumors including melanoma by implanting fresh tumor material from patients directly into immune deficient mice (13). Success rates vary significantly between tumor types, yet melanoma is highly suited to this experimental approach possibly due to the fact that even a few melanoma cells are sufficient to establish a tumor in NSG (NOD. Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice (14). Tumor grafts generated in this way and used as “avatars”, can predict therapeutic responses in cancer patients (15). Melanoma PDX recapitulate the tumor architecture and genotype of the patient tumor (16), and metastatic behavior of these PDX correlates with clinical outcome in patients (17). In this study, we developed PDX from a cohort of patients who became resistant to and progressed on BRAF inhibitors. Using genomic and proteomic analysis we were able to identify targets and test combinations of compounds in clinical development. However, we had an added advantage in that we were able to test multiple combinations in parallel due to an in vivo expansion strategy. These “pre-clinical trials” allowed us to define effective double and triple combination therapies, leading to complete tumor regression in all tumors of one PDX model treated. This translational approach towards improving personalized medicine in melanoma highlights the potential use of MET inhibitor combination therapy in a defined subset of melanoma patients.
Methods
Patient samples and generation of PDX
Biopsies from patients with a BRAF-V600E mutation who had progressed by RECIST on either vemurafenib or dabrafenib were included in this study. Tissue collection was approved by Wistar IRB. Sterile tumor samples were placed in transport media (DMEM, Fungizone 0.1%, and 2mL Gentamicin 0.2%) on wet ice and processed within 24 hours under sterile conditions. Tumor tissue was finely minced using the cross blade technique, digested in collagenase IV for 20min at 37 degrees with repeated trituration, followed by a 2 minute incubation in trypsin. The tumor slurry was implanted with matrigel (Corning Life Sciences) s.c. in NSG mice. When tumors reached a volume of 1000 mm3 (determined by weekly caliper measurements using the formula WXWXL/2) animals were sacrificed and tumors harvested. Tumor grafts were digested as above and either re-implanted within 24 hours or banked. All animal experiments were approved by Wistar IACUC.
Targeted next generation sequencing
PDX tumors were massively parallel DNA sequenced by Foundation Medicine (http://foundationone.com) for 315 cancer gene exons and 28 cancer gene introns for base pair change, insertions, deletions, copy number changes, and select fusions by next gen sequencing (18). Copy number changes in genes known to be recurrently amplified in cancer were called as high-level (CN>8) and focal (CN>5) amplifications, non-focal lower-level (CN<=8) amplifications, and homozygous deletions of genes known to be recurrently deleted in cancer.
Reverse phase protein arrays (RPPA)
Frozen tumor tissue was ground in a mixer mill (Retsch, Haan, Germany) and lysed with 200ul ice-cold lysis buffer (1% Triton X-100, 50mM HEPES, pH 7.4, 150mM NaCl, 1.5mM MgCl2, 1mM EGTA, 100mM NaF, 10mM Na pyrophosphate, 1mM Na3VO4, 10% glycerol, freshly added protease and phosphatase cocktail tablets (Roche Applied Science Cat. # 05056489001 and # 04906837001)). After 2 flash freeze cycles, samples were centrifuged at 13000rpm for 15 minutes at 4°C and supernatants were collected. Protein concentration was determined by Bro-Rad protein assay (#500-0006). About 40μl cell lysate (protein adjusted to 1-1.5μg/μl) were mixed with 4X SDS sample buffer (40% Glycerol, 8% SDS, 0.25M Tris_HCL, pH 6.8; beta- mercaptoethanol at 1/10 of volume without bromophenol added before use). The samples were then heated for 10 minutes at 100°C in a heat block and submitted for RPPA processing. RPPA was performed by the MD Anderson Center RPPA core facility as previously described (19) and data reported as Normalized Log2. Several RPPA data sets were successfully merged using replicates based normalization (RBN)(20). Unsupervised hierarchical clustering was performed on RBN Log2 Median Centered protein values using Cluster 3.0 software (http://rana.lbl.gov/EisenSoftware.htm). Results were visualized using Treeview software (http://rana.lbl.gov/EisenSoftware.htm).
Compounds
PLX4720 200ppm chemical additive diet was irradiated and heat sealed (Research Diets, New Brunswick, NJ) and fed to mice once tumors were established. PLX4720 was provided by Plexxikon, Berkeley, CA. Encorafenib, binimetinib, VX-11e, capmatinib, and BKM120 were provided by Novartis. For in vivo oral gavage, compounds were suspended in 0.5% carboxymethylcellulose sodium (MC), 0.5% Tween 80 (encorafenib); 1% MC, 0.5% Tween 80 (binimetinib); 5% ethanol, 20% propylene glycol, 7.4% Tween80 (VX-11e); 0.5% MC, 0.1% Tween80 (capmatinib); 1% MC, 1% Tween80 (BKM120) in water and dosed using feeding tubes (Instech Laboratories, Inc. Plymouth Meeting, PA).
In vivo experiments
Human melanoma RPDX tumors were expanded in vivo using NSG mice prior to the therapy experiments. Pooled tumor chunks banked from early (3-5) mouse passages (MP) were implanted into 50 NSG mice (1:10 expansion). These tumors were harvested when reaching the max volume allowed on the protocol (1000mm3), digested and banked as live cells. The larger part of this stock was retained as a master bank and the other part was implanted at a 1:5 ratio into NSG mice to use in the therapy experiments. The expansion phase was under continuous drug pressure with PLX4720 200ppm chemical additive diet at approximately clinical plasma levels. The plasma levels of PLX4720 (103.7ug/ml ±3.2 after 7 days) were similar to steady state levels in patients treated with vemurafenib 960mg BID (130.6ug/ml ± 71.78) (21). When tumors have reached 200mm3 per caliper measurement, animals were randomized into treatment groups followed by a 3-day washout phase. Tumor size was assessed twice weekly per caliper measurement. Mice were sacrificed after two weeks of treatment or when necessary for animal welfare. Dosing was prolonged when tumor control was achieved as indicated. Tumor tissue was conserved in formalin (for FFPE) and snap frozen in liquid N2 for protein extraction. Treatment groups were sacrificed 4 hours post last dose.
IHC
Tumor tissue was fixed in 10% formalin, dehydrated and embedded in paraffin. The immune histochemical staining procedure followed the manufacturer's protocol (Vector R.T.U Vectastain Kit, Universal Elite ABC kit #PK-7200). Primary antibody (Ki67: Vector #VP-RM04, 1:500; cleaved caspase 3: Cell signaling #9664s, 1:300) was added to each section and incubated overnight at 4°C in a humidity chamber. The color visualization was Vector Impact DAB kit (SK-4105), followed by counterstaining with hematoxylin.
Western blot
Protein extraction was performed as described for RPPA. 15ug of protein extracts were subjected to electrophoresis on 10% SDS-Page gels and transferred on nitrocellulose membranes in the Bio-Rad Trans-Blot Turbo transfer system. The membranes were blocked with ODYSSEY Blocking Buffer (#:927-40000, 1:1 diluted in TBS; LI-COR, Lincoln, NE) for 1hr at room temperature and incubated at 4°C overnight with the following primary antibodies: pMet #3129, pAKT (s473) #4060s, pAKT (Thr308) #13038P, pERK #4370s, pMEK #9121S, MEK #2352, RSK #8408, pRSK #9344 (all Cell Signaling Technology, Danvers, MA), and B-actin (Sigma #A5441). All primary antibodies were diluted 1:1000 in 5% BSA TBS-0.1%Tween20 buffer except B-actin which was diluted 1:10000. After washing and incubating with secondary antibodies (Thermo Scientific # PI35571, Thermo Scientific #PI35518, diluted 1:10000 in 1:4 ODYSSEY Blocking Buffer), the bands were visualized by the LI-COR Odyssey infrared imaging system.
Statistics
Patient's PFS and OS were calculated using the Kaplan Meier method. For in vivo experiments, statistical significance was determined using the trends of mean tumor volume over time. Treatment groups were compared using linear mixed models and a likelihood ratio testing nested model was used to examine if trends were overall significantly different among groups. P<0.05 was considered to be significant.
Results
Establishment of PDX from BRAF inhibitor progressed patient samples
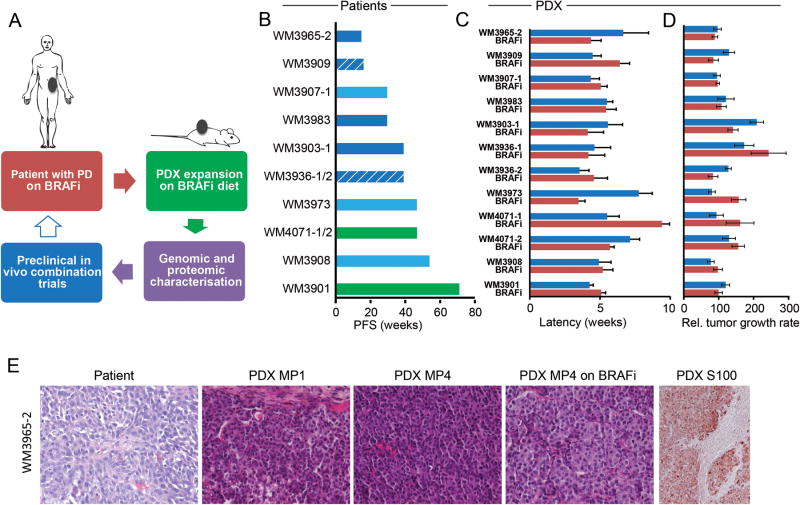
We collected 12tumorsamples from 10melanoma patients post-progression on a BRAF inhibitor (Fig 1A). In one patient, tissue from the same lesion was collected twice at different time points, and in another patient a bowel and a brain metastasis sample was collected. The distribution between male and female was 6:4 and the median age at biopsy was 64.5 years. All patients, except one with an unknown primary, had cutaneous primaries and all had distant metastatic disease, from which the PDX models were established, with 7 biopsies from subcutaneous metastases, one each from the parotid gland and bowel, and 3 from brain metastases (Supplementary Table S1). Two patients had surgical complete responses after excision of their progressing lesions, 5 had partial responses, and 3 patients had stable disease as best response to BRAF inhibitor therapy. The median progression free survival (PFS) of all patients in this set was 39 weeks with a wide standard deviation (SD) of 7.3 weeks (Fig 1B). Median overall survival (OS) was 97.57 weeks with a SD of 45.64 weeks.
Figure 1. establishment of 12 PDX from BRAF inhibitor progressed patients.
A Schematic of BRAF inhibitor resistant PDX generation, expansion, characterization, and in vivo testing. The arrow shown in blue outline denotes possible future clinical translation. B. Patient's progression free survival (PFS) is measured from first day on drug (vemurafenib/solid bar or dabrafenib/hatched bar) to progression by RECIST. Best response is denoted by the color of each bar: SD- light blue, PR- blue, surgical CR- green. Each bar represents one patient, 1/2 denote two samples collected from the same patient. C. PDX tumor grafts are followed from implantation until palpable, blue bars are untreated mice, red bars are mice continuously dosed with BRAF inhibitor (PLX4720) diet. Mean of mouse cohorts (n>5) is shown, error bars are SEM. D. the growth rate of the same tumor grafts from palpable until sacrifice is calculated by max tumor volume in mm3/time in weeks, thus higher values indicate a faster growth rate; blue bars are untreated mice, red bars are mice continuously dosed with BRAF inhibitor (PLX4720) diet. Mean of mouse cohorts (n>5) is shown, error bars are SEM. E. Histology of patient tumor tissue used to generate the PDX and subsequent PDX passages harvested;WM3965-2 is shown as a representative model, all others are shown in supplementary figure S2; MP1: first mouse passage; MP4: 4th serial transplantation in mice; on BRAFi: mice were continuously dosed with BRAF inhibitor diet; H&E staining except PDX S100, a melanoma marker.
Whereas the majority of samples were from patients progressed on vemurafenib, 2 samples were from patients who had progressed on dabrafenib, a drug with similar clinical efficacy 22). All samples were successfully established as tumor grafts with a median latency until palpable of 5.75 weeks (Fig 1C). The median growth rate was 120.3mm3/weeks to sacrifice, measured from palpability to last follow-up (Fig 1D). We did not observe any significant growth delay between untreated and chronically PLX4720 treated tumor grafts (Supplementary Figure S1). The histology of the original patient tumor and the tumor grafts grown in mice showed similarities with respect to morphology and histo-pathological criteria. Further, PDX serially transplanted up to 5 passages in mice still resembled the initial lesion, even when these were grown under continuous drug pressure (Fig 1E, Supplementary Figure S2).
Identification of targetable resistance mechanisms
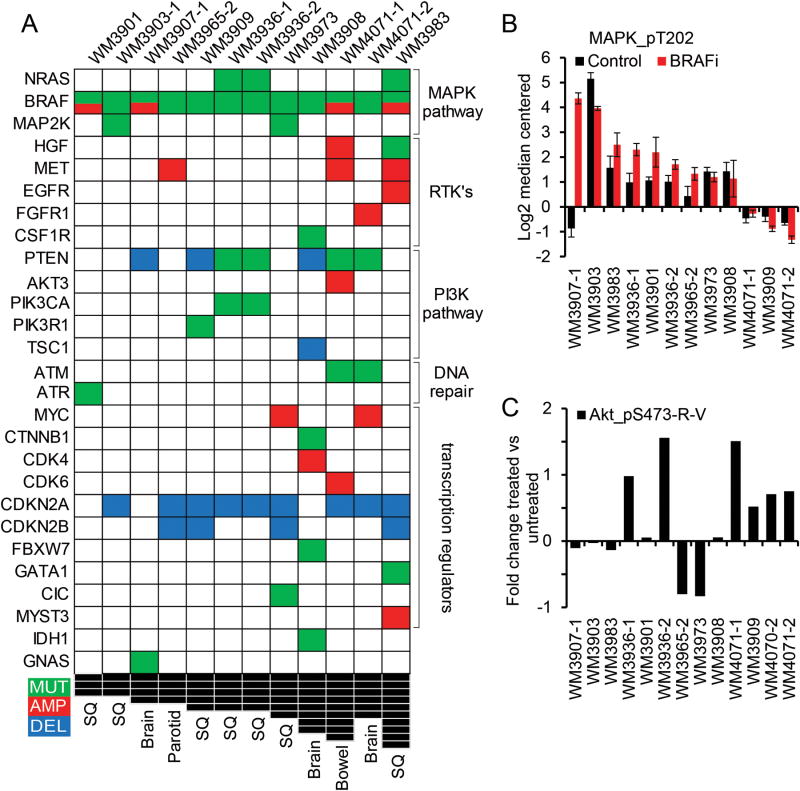
To characterize the resistance mechanisms in these models and assess how well they would recapitulate the known biology of resistance in patients, targeted next generation sequencing was performed on all PDX expanded under BRAF inhibition with a median exon coverage of 713 using the Foundation One panel (Foundation Medicine, Cambridge, MA). A median of 11.5 somatic short variants of known, likely and unknown significance were identified, with one PDX containing 111 variants in the 343 exons and introns assessed and complete results are provided in (Supplementary Figure S3). The BRAF V600E variant was confirmed in all samples. Importantly, at least 2 and up to 9 known deleterious concomitant alterations (mutations, amplifications, deletions) were found in each of the 12 PDX samples (Fig 2A), including genes in the MAPK and PI3K pathways, the receptor tyrosine kinase family, transcription regulators, and DNA repair genes. The most common alteration was loss of CDKN2A in 9/12 samples (23). Manygenetic aberrations found through this approach were previously associated with resistance to BRAF inhibitors. For instance, 3/12 PDX had activating NRAS mutations (24), 2/12 had activating MAP2K1 mutations (Q56P and K57E, functional analysis in (25) and (6) respectively), 4/12 had BRAF amplification (8), and7/12 samples had deletion or mutation of PTEN (8). Moreover, in several cases multiple candidate resistance mechanisms co-occurred (e.g. PIK3CA and NRAS). Lastly, some potentially actionable alterations detected were not previously described in the context of BRAF inhibitor resistance, such as MET amplification in 3 PDX models (WM3965-2 with a calculated copy number (CN) of 16, WM3983 with a CN of 9, and WM4071-1 with a CN of 93).
Figure 2. targeted sequencing to identify targetable alterations.
A DNA alterations identified in 12 PDX samples. Only known somatic short-variants, deletions and amplifications in found in at least one PDX are shown; the full data set of alterations in343 exons and introns can be found in supplementary figure S3 as an excel file. PDX sorted by number of concomitant alterations are in columns, genes sorted by biological pathway are in rows. Mutations are in green, amplifications in red, deletions in blue, and black squares indicate the number of concomitant alterations. B. Levels of ERK/MAPK protein phosphorylation, measured by RPPA, as a surrogate for MAPK pathway activation; BRAFi mice were under continuous BRAF inhibitor therapy at time of harvest, mean of 3 biological replicates is shown, error bars are SD. C. Fold change in AKT phosphorylation between untreated and BRAF inhibitor treated tumors as a surrogate for PI3K pathway re-activation under therapy, mean of 3 biological replicates is shown.
Matched samples were collected from several patients: WM4007 is a pre-treatment lesion to WM3901 and does not have amplified BRAF; WM3936-1 and -2 are both from the same relapsed lesion at different time points and after aggressive growth under therapy, but are both remarkably similar; finally WM4071-1 and -2 are from therapy resistant bowel and brain metastases respectively and although the 2 lesions have distinct mutation profiles pERK and pAKT and other protein levels were concordant in both PDX.
BRAF short splicing variants have been reported in BRAF inhibitor progressed patient samples at a frequency of 13-32% (6, 26, 27). However, all of our PDX models were determined to be negative for BRAF splice variants by protein and RNA analysis (Supplementary Figure S4).
To complement genomic profiling with an assessment of pathway activation status, reverse phase protein arrays (RPPA) were run for all PDX. To differentiate between genomic/epigenomic changes versus signaling feedback loops due to continued BRAF inhibition, an analysis of differential protein signaling between all untreated PDX by unsupervised hierarchical clustering was performed (Supplementary Figure S2A). Principal component analysis (PCA) was performed on three groups identified in the clustering, but failed to distinguish among the groups due to the lack of similarly expressed proteins (Supplementary Figure S2B). Further, attempting to identify signaling feedback loops we analyzed protein fold changes between treated and untreated tumors using unsupervised hierarchical clustering (Supplementary Figure S2C). Again, PCA did not succeed in defining commonly changed pathway. Instead it highlighted the heterogeneity of resistance mechanisms within our relatively small tumor subset (Supplementary Figure S2D). However, MAPK pathway re-activation was identified as a putative mechanism of resistance in the majority of PDX (Fig 2B). Fold change in pAKT levels between BRAF inhibitor treated vs. untreated PDX tumors indicated the PI3K pathway as a possible compensatory mechanism in 5 PDX models (Fig 2C). Although we did not see a negative correlation between pERK and pAKT, the increase of pAKT while on drug indicates that continued pathway inhibition in the resistant setting might lead to upregulation of PI3K signaling through crosstalk between these two pathways (28).
Rational dual MAPK and PI3K pathway inhibition inhibits tumor growth in vivo
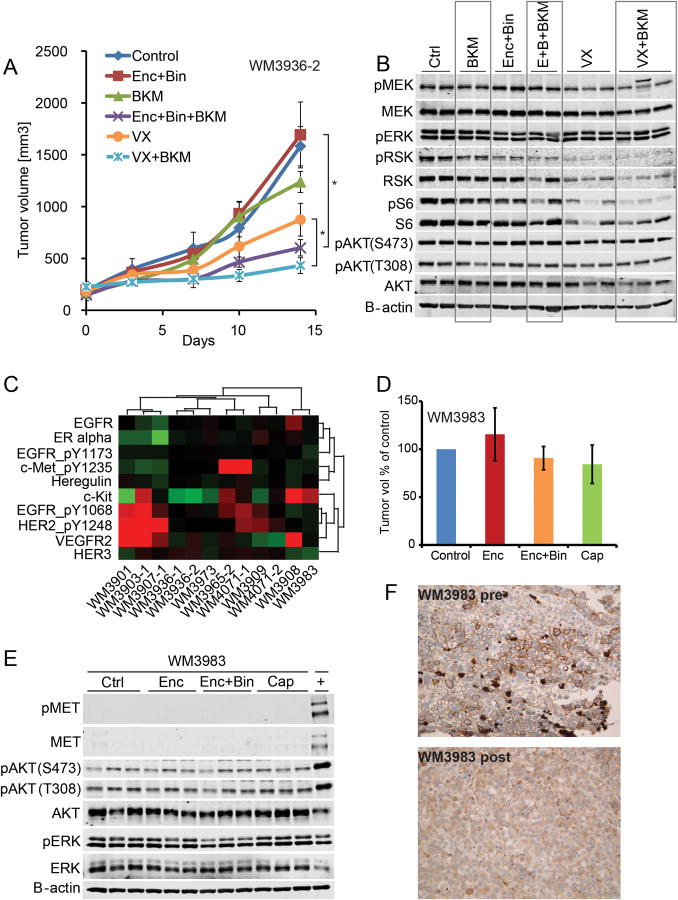
To test the hypothesis of dual core pathway inhibition based on genomic and proteomic data we selected a MAPK and PI3K hyper-activated model for a multi arm PDX in vivo study. The patient whose tumor tissue was used in this study had received dabrafenib in a clinical trial with an excellent clinical response, but developed a new subcutaneous thigh lesion after 9 months of therapy which was then biopsied (WM3936-1). The patient was transitioned to commercial vemurafenib but aggressive growth of that same lesion was observed under therapy so that this progressing thigh lesion was surgically excised after 3 months on vemurafenib (WM3936-2). We found that both PDX had similar mutation profiles and had acquired NRAS and PIK3CA mutations. Available next generation sequencing data of a pre-therapy lesion biopsy indicated NRAS wild type and PIK3CA wild type status at least to the depth of sequencing performed. WM3936-1 and -2 were both derived from the same patient lesion progressing on dabrafenib and subsequently vemurafenib and both harboredNRASQ61K heterozygous, PTENC105Y homozygous, and PIK3CAH1047Yheterozygous mutations as potential resistance mechanisms that would be expected to lead to re-activation of the MAPK and compensatory activation of the PI3K pathway as confirmed in the RPPA data. Neither the PIK3CA or NRAS mutations were detected in a pre-therapy patient lesion; PTEN status could not be assessed. Based on genomic and RPPA (2A,C) data, we designed a rational second line combination therapy to target all candidate resistance mechanisms centered on the pan-PI3K inhibitor BKM120 (29) in combination with either a BRAF/MEK inhibitor combination using encorafenib and binimetinib currently in clinical trials (NCT01543698 and ASCO 2015 abstract 9007) or the ERK inhibitor VX-11e (30). For this 6-arm in vivo study, the WM3936-2 PDX model was expanded until tumor grafts could be implanted simultaneously into a cohort of 60 NSG mice. Once tumors were established, animals were dosed for 2 weeks. As expected, the tumors were resistant to BRAF/MEK inhibitor combination therapy as well as to PI3K inhibition alone. The ERK inhibitor inhibited tumor growth as a single agent compared to control (p<0.0001), but targeting both MAPK and PI3K signaling using either of 2 strategies: triple combination encorafenib/binimetinib/BKM120 (p<0.0001) or double combination VX-11e/BKM120 (p<0.0001) resulted in significantly improved tumor growth control (Fig 3A). The difference between triple and double therapy was not significant. This result confirmed the utility of rationally designed MAPK/PI3K inhibitor combination therapy based on genomic and proteomic data. Tumor tissue harvested at the end of study was assessed for pathway inhibition, BKM120 as a single agent did not decrease pAKT signaling and this difficulty in assessing PI3K inhibition at the level of AKT has been previously described. The combination of encorafenib/binimetinib resulted in a modest inhibition of pRSK as a downstream target of the MAPK pathway and pS6, downstream of MAPK and PI3K pathways. Similarly, ERK inhibition by VX-11eresulted in robust inhibition of pRSK reflected in tumor growth inhibition. However, only the encorafenib/binimetinib/BKM120triple combination led to a complete inhibition of both pRSK and pS6 (Fig 3B).
Figure 3. dual pathway inhibition controls tumor growth.
A tumor growth curves of WM3936-2 PDX. Animals were treated with vehicle control, encorafenib 20mg/kg QD+binimetinib 3mg/kg QD (Enc+Bin), BKM120 30mg/kg QD, the triple combination encorafenib+binimetinib+BKM120, VX-11e 50mg/kg BID (VX), or VX-11e+BKM120. Dosing was started with established tumors, all compounds were administered orally, n=10/group, error bars are SEM, * indicates p<0.0001. B. immunoblot of tumors harvested at end of study, 4 hours post last dose. The membrane was probed with indicated antibodies. B-Actin was included to ensure equal loading. C. levels of RTK proteins assessed by RPPA, red higher than median, green lower than median, unsupervised hierarchical clustering, data is mean of 3 biological replicates. D. relative tumor growth (final volume/days to max volume) of WM3983 PDX relative to vehicle control. In two separate experiments, animals were treated with 1) vehicle control, encorafenib 20mg/kg QD, encorafenib+binimetinib 3mg/kg QD and 2) vehicle control, capmatinib 25mg/kg QD (Cap). In both experiments, dosing was started with established tumors. All compounds were administered orally for 14 days, n=10/group, error bars are SEM. E. immunoblot of tumor grafts harvested after 3 days of dosing 4 hours post last dose. The membrane was probed with indicated antibodies. B-Actin was included to ensure equal loading.+ denotes the WM3965 tumor graft tissue included as a positive control with elevated levels of MET and pMET. F. IHC staining for MET of patient's melanoma lesion. The pre BRAF inhibitor biopsy shows a strong membrane stain for MET, but only in a subpopulation. The post progression biopsy is negative for MET, the positive cells are macrophages (this lesion was used to establish the PDX).
MET amplification alone is not sufficient to predict effective therapy
We then analyzed the activation status of receptor tyrosine kinases (RTK) present on the RPPA (Fig 3C) since these have been reported as potential resistance mechanisms in melanoma (31). Clusters of PDX with upregulation in the EGFR/HER2/HER3 family of RTKs, c-Kit upregulation, as well as MET were identified. We selected the proto oncogene MET (32, 33) as another promising target for second line therapies. The 25% (3/12) incidence of MET amplifications in this set of BRAF inhibitor progressed PDX was significantly higher than in the melanoma cancer genome atlas (TCGA) at 8/129 BRAF hotspot mutated, 0/17 BRAF non hotspot, and 3/149 BRAF wild type yielding a total of 11/299 or 3.7%. Thus, MET amplification and BRAF hotspot mutation had a significant tendency towards co-occurrence (p-value 0.035). In the TCGA database, out of 9 MET amplified melanomas with available RPPA data only 2 showed more than 2-fold increase in pMET. Indeed, in one of the three MET amplified PDX, an isolated progression of a scalp lesion, pMET was not increased compared to the median. This was possibly due to being part of a much broader amplicon including EGFR and possibly the entire chromosomal arm 7, and despite being amplified 9-fold. In a study of 1115 patients, MET amplifications were detected in 2.5% of solid tumors (melanoma 2/61) and these patients presented with more metastatic sites then non MET amplified tumors (26). This led us to hypothesize that MET amplifications represent either preexisting or acquired mechanisms of resistance and would predict response to the MET antagonist capmatinib (34). Indeed, MET pY1235 (pMET) levels significantly higher than the median were confirmed in 2/3 PDX models with MET amplification (Fig 3C). High levels of pMET were seen in WM3965 and WM4071-1 PDX tumors, with the latter derived from a bowel metastasis. A brain metastasis from the same patient and also collected post progression (WM4071-2) did not have amplified MET or increased pMET, although this could relate to difference in sequencing depth and an ability to make amplification calls.
To test whether genomic data alone would be sufficient to design a rational second line therapy in a MET amplified setting, we expanded the WM3983 PDX in vivo and confirmed it to be completely resistant to encorafenib (even exhibiting increased proliferation as compared to vehicle control) and encorafenib/binimetinib combination therapy (Fig 3D). Importantly, while WM3983 demonstrated amplified MET, it did not have pMET signaling (Fig 3C). Thus, the MET inhibitor capmatinib had no anti-tumor effect in this model in vivo (Fig 3D), concordant with un-detectable levels of pMET in control tumors harvested at the end of study (Fig 3E). Lack of pERK and pMEK inhibition in encorafenib/binimetinib treated vs untreated animals (Fig 3E) supported a MAPK reactivation mechanism of resistance in line with the NRAS mutant, BRAF amplified genotype of this tumor. To rule out the possibility that the PDX had lost its pMET phenotype due to changes in the murine environment, we analyzed patient samples from before and after BRAF inhibitor progression. Although MET positive subpopulations of tumor cells were found in the pre-therapy lesion, these had disappeared in the progression biopsy used to generate the PDX (Fig 3F).
Integrating genomic and proteomic data for the design of second line combination therapies
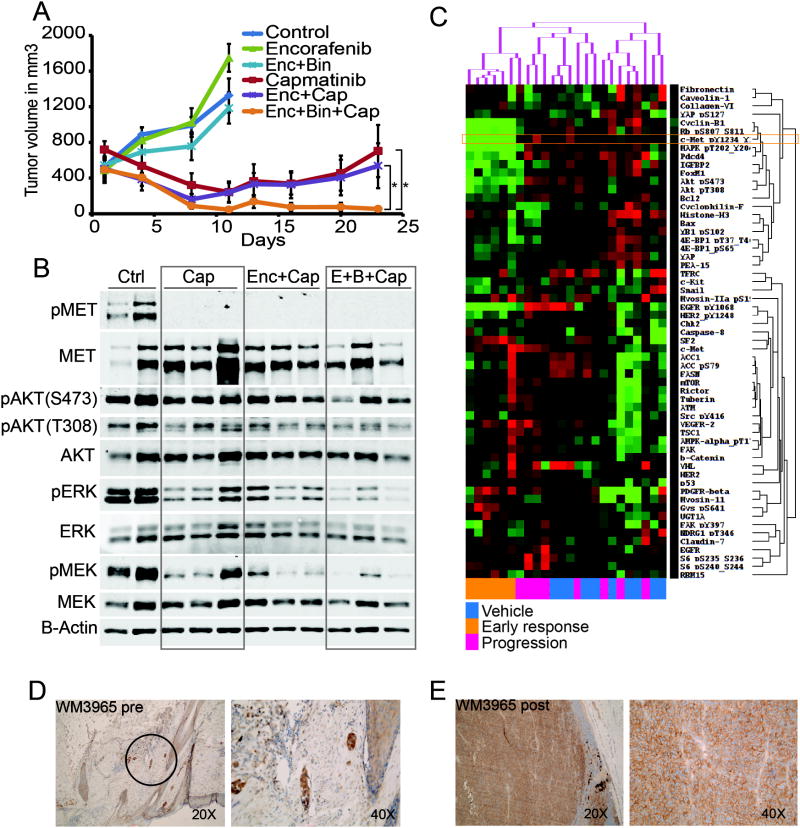
Based on these findings, we hypothesized that integration of genomic and protein data may provide greater information content than either alone and selected the MET amplified and high pMET signaling PDX model WM3965 for a multi-arm combination study centered on capmatinib (Fig 4A). The patient whose tumor was used to generate the PDX had received vemurafenib in a neoadjuvant setting, but after only 3 months developed early PD in the right parotid gland, which was surgically excised and used to generate the PDX. The 6 arm design included a vehicle control group showing rapid tumor growth, encorafenib single agent (accelerated tumor growth, p=0.020), and encorafenib/binimetinib combination arms (no anti-tumor effect) confirming the aggressive and MAPK pathway inhibitor resistant phenotype of this PDX model. Remarkably, the tumors in the other 3dosing groups (capmatinib single agent, capmatinib/encorafenib, and capmatinib/encorafenib/binimetinib) all rapidly regressed after as short as 3 days of dosing. This trend continued until more than 2 weeks of daily dosing, at which point a separation of the growth curves became apparent: whereas the tumor grafts on capmatinib single agent and capmatinib/encorafenib developed therapy resistance, albeit with variable tumor growth kinetics, the capmatinib/encorafenib/binimetinib (triple combination) treated tumors showed complete tumor regression in 10/10 animals after 21 days of dosing with no evidence of therapy resistance (Fig 4A).
Figure 4. integrating genomic and protein signaling results in an effective triple therapy pre-clinical in vivo trial.
A tumor growth curves of WM3965 PDX (MET amplified, high pMET);dosing was started with well-established tumors expanded on BRAF inhibitor diet followed by a washout period before start of dosing. Animals received either vehicle control, encorafenib 20mg/kg QD, binimetinib 3mg/kg QD, capmatinib 25mg/kg QD as single agents or combinations as indicated. All compounds were administered orally, n=10/group, error bars are SEM, * indicates p<0.05. B. immunoblot of tumors harvested after 3 days of dosing and 4 hours post last dose. The membrane was probed with indicated antibodies and B-Actin was included to ensure equal loading. C. RPPA analysis of tumors harvested either at the end of the efficacy experiments (A) or 3 days of dosing (B). Mice without palpable tumors were not included (all triple combo animals). Color coding on the lower x-axis denotes the following groups: blue are control animals (vehicle), orange are the 6animals treated with capmatinib orencorafenib/capmatinib for 3 days (early responders), and pink were treated with capmatinib or encorafenib/capmatinib in the efficacy cohorts and progressed after initial response (progression). Proteins with similar expression along all samples were excluded. The proteins that vary across the samples over a cutoff of 0.4 standard deviations are shown on the y axis. Green is down, red is up regulated. Unsupervised hierarchical clustering of the log2 median centered data is shown on the top x axis. D. MET IHC staining (brown) of FFPE patient tissue. The pre BRAF inhibitor sample is the safety margin around the primary lesion and shows residual melanomanests in the dermal layer (black circle). E. a lymph node metastasis of the same patient after progression (this biopsy was used to establish the PDX).
High levels of pMET protein were confirmed in vehicle treated tumors by Western blot, and high pERK and pAKT levels indicated active signaling through both MAPK and PI3K pathways respectively (Fig 4B). Further, the highly proliferative phenotype could be demonstrated by strong Ki67 staining (Supplementary Figure S6A). In contrast, animals dosed for 3 days with capmatinib showed complete abrogation of pMET (Fig 4B). Importantly, capmatinib single agent did not lead to meaningful decreases in pAKT or pERK signaling whereas the triple combination resulted in almost complete inhibition of pAKT and pERK. This observation correlated with increased apoptosis as measured by cleaved caspase 3 staining at this early time point (Supplementary Figure S6B).
Tumor tissue of vehicle treated, capmatinib +/- encorafenib responding and progressing animals was submitted for RPPA analyses. Intriguingly, three distinct clusters could be observed: one containing all early responders and the other two randomly distributed untreated and progressed tumors (Fig 4C). The capmatinib responding tumor cluster was predominately defined by pMET, pEGFR, and pHER2 down-regulation in association with down-regulation of their downstream effectors pMAPK, pRB, Cyclin D1, pAKT, p4E-BP1, IGFBP2, and FOXM1. Interestingly, glycogen synthase (GYS) phosphorylation was inversely up-regulated indicating a decrease in glycogen production ability in these tumors. The observation that the control and progression samples were interspersed is consistent with a loss of MET inhibitory effect and reversal of the signaling profile back to the untreated state. Still, two distinct population were apparent in this PDX model as defined by protein expression profile, confirming the heterogeneity found in PDX tumors. We then performed PCA comparing the 2 clusters and found evidence for differences in metabolism, PI3K signaling, and RTKs (Figure S7) although these did not correspond with time to progression. Since resistance occurred at roughly the same time and relatively quickly after initial regression, this may suggest a common (adaptive) resistance mechanism.
Finally, patient biopsies from the therapy naïve primary lesion (Fig 4D) and a post-progression metastasis used to generate the PDX (Fig 4E) both stained highly positive for MET, indicating that the amplification of MET might have been pre-existing in the primary melanoma and thus acted as an intrinsic mechanism of resistance leading to early relapse with only 3 months progression free survival.
Discussion
Patient derived xenografts provide sustainable models for personalized therapy. The key advantage of these models is their surprising biological and genetic stability when implanted into mice, as reflected in our current study. This allowed for a comprehensive analysis of drivers of resistance to targeted therapy, and the design of effective second line combination therapies tailored to each model. Although patient specific real-time “co-clinical” trials are feasible, obstacles such as timing and regulatory issues may hinder progress. Instead, the development of biomarkers using this approach might offer a higher benefit ratio. A single patient based approach is further complicated by the fact that PDX models do currently not allow for a comprehensive assessment of immune therapies, which are rapidly becoming first line therapy for melanoma patients. While efforts are ongoing to move this technology into “humanized” mice, which have a reconstituted human immune system, this will prove both challenging and extremely costly. Our studies are therefore restricted to direct targeting of signaling pathways in melanoma cells, and possibly the murine stroma. These PDX models with acquired resistance to targeted therapy provide an effective way to expand tissue for multiple methods (sequencing, RPPA, etc.) that do not face the same challenges as growing these cells on plastic (loss of architecture, single cell clonality) and as we show in this study, provide alternative therapeutic targets in relapsed patients. Overall, PDX can play a major role in exploring novel targets and combination therapies based on the increasingly detailed picture of genomic and proteomic aberrations in cancer cells.
In this study, we utilized tumor samples from patients who reflected the wide range of responses seen in the clinic (35). The relative uniformity in tumor grafting in vivo and invariable resistance to dosing with a BRAF inhibitor, indicated that once resistance had occurred all tumors were capable of tumor initiation in vivo irrespective of time to progression in patients. Next generation sequencing of patients lesions using targeted platforms is transforming personalized cancer therapy by uncovering actionable genomic alterations in a majority of solid tumors such as lung cancer (36). We used the same approach to uncover possible mechanisms of intrinsic and acquired resistance. Many of these alterations such as NRAS (24) and MAP2K1 (25, 37) mutations as well as PTEN deletion and BRAF amplification (8) were previously described to confer resistance to BRAF inhibitors. MAPK pathway hyper-activating alterations (BRAF or RTK amplification, NRAS and MAP2K1 mutations) were found in 11/12 samples (most often mutually exclusive) and RPPA analyses confirmed active MAPK signaling under drug pressure in the majority of PDX concordant with recently published results (6, 38). Interestingly, all three brain metastasis derived PDX had aberrant PTEN, whereas only one of the extra-cerebral metastases had a PTEN deletion, which had been previously described as a poor prognostic marker (39). Taking into account the intra-patient heterogeneity and possible clonal selection in the generation of a PDX model, it is likely that not the whole spectrum of tumor cell population in a patient will be represented in a mouse avatar. Thus, mapping of patients for driver mutations and their possible convergence on the same biological pathways (40), can be studied by generating multiple PDX from the same patient.
The identification of multiple concomitant alterations per PDX further emphasized the challenges of personalized therapy selection in the clinic and was consistent with published reports (27). To determine whether multiple genomic aberrations integrated into a limited number of pathways, we extended our analysis to signaling on the phospho protein level using the RPPA platform. RPPA relies on validated antibodies and well characterized targets, allowing us to confirm sustained signaling through the MAPK pathway in the majority of PDX and increased activation of the PI3K pathway in a smaller subset. We did not focus our study on the discovery of novel targets since studies of patient samples directly are much more suited to this approach. Rather, the strength of our PDX platform lies in translating findings to in vivo target validation.
Indeed, using either a BRAF/MEK inhibitor or ERK inhibitor combination strategy with the pan PI3K inhibitor BKM120 proved effective in abrogating tumor growth in a MAPK and PI3K pathway activated mouse avatar. This confirmed previous reports in cell lines with acquired NRAS mutations resistant to dabrafenib (41) and in those with increased RTK signaling leading to increased PI3K signaling (42). Therefore, we could establish that genomic profiling and assessment of associated signaling pathway activity is a viable strategy to design rational second line combination therapies in vivo. Also, we can postulate that the corresponding patient likely would not have benefited from dabrafenib/trametinib combination therapy, but that the inhibition of both MAPK and PI3K pathways would have been critical in achieving a response. This combination efficacy may be the first clear example of co-occurrence of redundant mechanisms of resistance.
On the other hand, a MET amplified BRAF and combined BRAF/MEK inhibitor resistant model did not respond to MET inhibition in vivo and we therefore concluded that amplification of MET is not sufficient to define it as a driver of resistance and that a second readout, such as pMET protein levels would be necessary. This was in line with a previously published study of a large cohort of over 1000 patients where MET amplification did not correlate with response to a MET inhibitor and where MET protein levels were not assessed (43). Targeting MET has proved effective in MET-amplified gastric cancer using the inhibitor volitinib (44) and MET has been described as a novel target for adjuvant therapy for melanoma (45). In our study, the triple combination of MET, BRAF, and MEK inhibition was exceptionally effective in vivo, with profound MAPK pathway inhibition. This observation might be explained by HGF/MET mediated RAS activation (46) leading to BRAF dimerization and thus resistance to vemurafenib (47). We therefore propose that increased MET protein phosphorylation with or without MET amplification should be assessed as a biomarker of response to MET inhibitor combination therapies. This will be of highest priority in MAPK pathway inhibitor resistant patients since increases in MET RNA levels have been described at an increased frequency in this patient cohort (48). However, since targeted genomic sequencing is currently the gold standard of personalized therapies, a preselection of patients for protein signaling analysis currently not in wide-spread use for clinical treatment selection, will be advantageous.
Although we did not extend our study to patient treatment after determining efficacious second line therapies in the PDX models, a clinical trial with parallel assignment into 5 treatment arms based on sequencing data is currently ongoing in relapsed melanoma patients (ClinicalTrials.gov: NCT02159066). The results from the PDX pre-clinical studies clearly argue that genomic and proteomic approaches should be integrated to increase the success rates of personalized cancer therapies, as this approach allowed us to outline and confirm personalized medicine strategies. These models can be used to refine precision medicine approaches and to develop biomarkers of response for future clinical trials and avoid treatment failures for patients. Future studies using humanized mouse models with reconstituted T cell function will be of major importance to integrate the findings described here into an immunotherapy landscape of melanoma.
Supplementary Material
Statement of translational relevance.
Basket trials to assign patients into treatment arms according to targetable alterations are an important development in personalized cancer medicine. However, selecting the appropriate combinations for each patient can be challenging. Especially in patients with acquired resistance to kinase inhibitors, intra-patient heterogeneity and concurrent mutations within the same lesion occur frequently. In this study, we used PDX from targeted therapy progressed patients to test personalized combinations based not only on genomic data, but validated by proteomic analysis. By using a multi-arm pre-clinical trial design, we were able to identify efficacious precision medicine approaches and highlight the importance of assessing pathway activation status at the protein level.
Acknowledgments
We thank the Animal and Imaging Core Facilities at the Wistar Institute. We thank the Tumor Tissue and Biospecimen Bank (TTAB) at the University of Pennsylvania Abramson Cancer Center. We thank Gideon Bollag at Plexxikon (Berkeley, CA) for providing PLX4720.
Grant Support: Support for Shared Resources utilized in this study was provided by Cancer Center Support Grant (CCSG) P30CA010815 to the Wistar Institute. This work was supported by NIH grants PO1 CA114046, P01 CA025874, P30 CA010815, R01 CA047159, and a research grant by Novartis to MH, CCSG grant P30CA016672 to GBM, RO1 CA174746-01 to ATW, and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Potential conflicts of interest: Part of the studies was supported by a research grant from Novartis.
References
- 1.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved Survival with Vemurafenib in Melanoma with BRAF V600E Mutation. New England Journal of Medicine. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med. 2015;372:30–9. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 3.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. New England Journal of Medicine. 2010;363:711–23. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, et al. Nivolumab plus Ipilimumab in Advanced Melanoma. New England Journal of Medicine. 2013;369:122–33. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, et al. Safety and Tumor Responses with Lambrolizumab (Anti–PD-1) in Melanoma. New England Journal of Medicine. 2013;369:134–44. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rizos H, Menzies AM, Pupo GM, Carlino MS, Fung C, Hyman J, et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: spectrum and clinical impact. Clin Cancer Res. 2014;20:1965–77. doi: 10.1158/1078-0432.CCR-13-3122. [DOI] [PubMed] [Google Scholar]
- 7.Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer discovery. 2013;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer discovery. 2013;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Connell MP, Marchbank K, Webster MR, Valiga AA, Kaur A, Vultur A, et al. Hypoxia Induces Phenotypic Plasticity and Therapy Resistance in Melanoma via the Tyrosine Kinase Receptors ROR1 and ROR2. Cancer Discovery. 2013;3:1378–93. doi: 10.1158/2159-8290.CD-13-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwong LN, Davies MA. Targeted therapy for melanoma: rational combinatorial approaches. Oncogene. 2014;33:1–9. doi: 10.1038/onc.2013.34. [DOI] [PubMed] [Google Scholar]
- 12.Merlino G, Flaherty K, Acquavella N, Day CPP, Aplin A, Holmen S, et al. Meeting report: The future of preclinical mouse models in melanoma treatment is now. Pigment cell & melanoma research. 2013:26. doi: 10.1111/pcmr.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hidalgo M, Amant F, Biankin AV, Budinská E, Byrne AT, Caldas C, et al. Patient-Derived Xenograft Models: An Emerging Platform for Translational Cancer Research. Cancer discovery. 2014;4:998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJC. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–8. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stebbing J, Paz K, Schwartz GK, Wexler LH, Maki R, Pollock RE, et al. Patient-derived xenografts for individualized care in advanced sarcoma. Cancer. 2014;120:2006–15. doi: 10.1002/cncr.28696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Einarsdottir BO, Bagge RO, Bhadury J, Jespersen H, Mattsson J, Nilsson LM, et al. Melanoma patient-derived xenografts accurately model the disease and develop fast enough to guide treatment decisions. Oncotarget. 2014;5:9609–18. doi: 10.18632/oncotarget.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quintana E, Piskounova E, Shackleton M, Weinberg D, Eskiocak U, Fullen DR, et al. Human melanoma metastasis in NSG mice correlates with clinical outcome in patients. Sci Transl Med. 2012;4:159ra49. doi: 10.1126/scitranslmed.3004599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–31. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5:2512–21. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 20.Akbani R, Ng PK, Werner HM, Shahmoradgoli M, Zhang F, Ju Z, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat Commun. 2014;5:3887. doi: 10.1038/ncomms4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grippo JF, Zhang W, Heinzmann D, Yang KH, Wong J, Joe AK, et al. A phase I, randomized, open-label study of the multiple-dose pharmacokinetics of vemurafenib in patients with BRAF V600E mutation-positive metastatic melanoma. Cancer Chemotherapy and Pharmacology. 2014;73:103–11. doi: 10.1007/s00280-013-2324-5. [DOI] [PubMed] [Google Scholar]
- 22.Jang S, Atkins MB. Which drug, and when, for patients with BRAF-mutant melanoma? The Lancet Oncology. 2013;14:e60–e9. doi: 10.1016/S1470-2045(12)70539-9. [DOI] [PubMed] [Google Scholar]
- 23.Bastian BC. The molecular pathology of melanoma: an integrated taxonomy of melanocytic neoplasia. Annual review of pathology. 2014;9:239–71. doi: 10.1146/annurev-pathol-012513-104658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20411–6. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–90. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, et al. Acquired Resistance and Clonal Evolution in Melanoma during BRAF Inhibitor Therapy. Cancer discovery. 4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends in biochemical sciences. 2011;36:320–8. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burger MT, Pecchi S, Wagman A, Ni ZJ, Knapp M, Hendrickson T, et al. Identification of NVP-BKM120 as a Potent, Selective, Orally Bioavailable Class I PI3 Kinase Inhibitor for Treating Cancer. ACS medicinal chemistry letters. 2011;2:774–9. doi: 10.1021/ml200156t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aronov AM, Tang Q, Martinez-Botella G, Bemis GW, Cao J, Chen G, et al. Structure-guided design of potent and selective pyrimidylpyrrole inhibitors of extracellular signal-regulated kinase (ERK) using conformational control. J Med Chem. 2009;52:6362–8. doi: 10.1021/jm900630q. [DOI] [PubMed] [Google Scholar]
- 31.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–9. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gentile A, Trusolino L, Comoglio P. The Met tyrosine kinase receptor in development and cancer. Cancer Metastasis Rev. 2008;27:85–94. doi: 10.1007/s10555-007-9107-6. [DOI] [PubMed] [Google Scholar]
- 33.Boccaccio C, Comoglio PM. MET, a driver of invasive growth and cancer clonal evolution under therapeutic pressure. Current opinion in cell biology. 2014;31C:98–105. doi: 10.1016/j.ceb.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Wang Q, Yang G, Marando C, Koblish HK, Hall LM, et al. A novel kinase inhibitor, INCB28060, blocks c-MET-dependent signaling, neoplastic activities, and cross-talk with EGFR and HER-3. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:7127–38. doi: 10.1158/1078-0432.CCR-11-1157. [DOI] [PubMed] [Google Scholar]
- 35.McArthur GA, Chapman PB, Robert C, Larkin J, Haanen JB, Dummer R, et al. Safety and efficacy of vemurafenib in BRAF(V600E) and BRAF(V600K) mutation-positive melanoma (BRIM-3): extended follow-up of a phase 3, randomised, open-label study. The Lancet Oncology. 2014;15:323–32. doi: 10.1016/S1470-2045(14)70012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drilon A, Wang L, Arcila ME, Balasubramanian S, Greenbowe JR, Ross JS, et al. Broad, hybrid capture-based next-generation sequencing identifies actionable genomic alterations in “driver-negative”. lung adenocarcinomas Clinical Cancer Research. 2015 doi: 10.1158/1078-0432.CCR-14-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villanueva J, Infante JR, Krepler C, Reyes-Uribe P, Samanta M, Chen HY, et al. Concurrent MEK2 mutation and BRAF amplification confer resistance to BRAF and MEK inhibitors in melanoma. Cell Rep. 2013;4:1090–9. doi: 10.1016/j.celrep.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwong LN, Boland GM, Frederick DT, Helms TL, Akid AT, Miller JP, et al. Co-clinical assessment identifies patterns of BRAF inhibitor resistance in melanoma. The Journal of clinical investigation. 2015;125:1459–70. doi: 10.1172/JCI78954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bucheit AD, Chen G, Siroy A, Tetzlaff M, Broaddus R, Milton D, et al. Complete loss of PTEN protein expression correlates with shorter time to brain metastasis and survival in stage IIIB/C melanoma patients with BRAFV600 mutations. Clin Cancer Res. 2014;20:5527–36. doi: 10.1158/1078-0432.CCR-14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nature genetics. 2014;46:225–33. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greger JG, Eastman SD, Zhang V, Bleam MR, Hughes AM, Smitheman KN, et al. Combinations of BRAF, MEK, and PI3K/mTOR Inhibitors Overcome Acquired Resistance to the BRAF Inhibitor GSK2118436 Dabrafenib, Mediated by NRAS or MEK Mutations. Molecular Cancer Therapeutics. 2012;11:909–20. doi: 10.1158/1535-7163.MCT-11-0989. [DOI] [PubMed] [Google Scholar]
- 42.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer cell. 18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fontes Jardim DL, Tang C, De Melo-Gagliato D, Falchook GS, Hess KR, Janku F, et al. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014. Analysis of 1,115 patients tested for MET amplification and therapy response in the MD Anderson Phase I clinic. [DOI] [PubMed] [Google Scholar]
- 44.Gavine PR, Ren Y, Han L, Lv J, Fan S, Zhang W, et al. Volitinib, a potent and highly selective c-Met inhibitor, effectively blocks c-Met signaling and growth in c-MET amplified gastric cancer patient-derived tumor xenograft models. Mol Oncol. 2015;9:323–33. doi: 10.1016/j.molonc.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Etnyre D, Stone AL, Fong JT, Jacobs RJ, Uppada SB, Botting GM, et al. Targeting c-Met in melanoma: mechanism of resistance and efficacy of novel combinatorial inhibitor therapy. Cancer biology & therapy. 2014;15:1129–41. doi: 10.4161/cbt.29451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Graziani A, Gramaglia D, dalla Zonca P, Comoglio PM. Hepatocyte growth factor/scatter factor stimulates the Ras-guanine nucleotide exchanger. J Biol Chem. 1993;268:9165–8. [PubMed] [Google Scholar]
- 47.Poulikakos PI, Rosen N. Mutant BRAF melanomas--dependence and resistance. Cancer Cell. 2011;19:11–5. doi: 10.1016/j.ccr.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 48.Hugo W, Shi H, Sun L, Piva M, Song C, Kong X, et al. Non-genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell. 2015;162:1271–85. doi: 10.1016/j.cell.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.